Abstract
Mutations in distant regulatory elements can have a negative impact on human development and health, yet because of the difficulty of detecting these critical sequences, we predominantly focus on coding sequences for diagnostic purposes. We have undertaken a comparative sequence-based approach to characterize a large noncoding region deleted in patients affected by Van Buchem (VB) disease, a severe sclerosing bone dysplasia. Using BAC recombination and transgenesis, we characterized the expression of human sclerostin (SOST) from normal (SOSTwt) or Van Buchem (SOSTvbΔ) alleles. Only the SOSTwt allele faithfully expressed high levels of human SOST in the adult bone and had an impact on bone metabolism, consistent with the model that the VB noncoding deletion removes a SOST-specific regulatory element. By exploiting cross-species sequence comparisons with in vitro and in vivo enhancer assays, we were able to identify a candidate enhancer element that drives human SOST expression in osteoblast-like cell lines in vitro and in the skeletal anlage of the embryonic day 14.5 (E14.5) mouse embryo, and discovered a novel function for sclerostin during limb development. Our approach represents a framework for characterizing distant regulatory elements associated with abnormal human phenotypes.
Deleterious mutations in distant regulatory elements postulated to have a dramatic impact on human development and health have been minimally explored. This problem is in large part due to the fact that there are no simple ways to discern regulatory elements from nonfunctional sequences or to ascertain whether mutant phenotypes are caused by regulatory mutations. Among Mendelian disorders associated with noncoding mutations, only a few cases are described that clearly link alterations in distant cis-acting regulatory regions to the cause of the disease (Ionasescu et al. 1996; Wang et al. 2000; Enattah et al. 2002; Lettice et al. 2003; Tsui et al. 2003), and these documented cases predominantly correspond to large chromosomal aberrations (Curtin et al. 1985; Curtin and Kan 1988; Cimbora et al. 2000; Kleinjan et al. 2001; Chuzhanova et al. 2003). Structural variation in the human genome described as large-scale polymorphisms has been recently shown to be more common than previously anticipated (Sebat et al. 2004); therefore, the extent to which large noncoding duplications and deletions have an impact on human biology remains a largely unanswered question. In this study, we demonstrate that a very important skeletal dysplasia, Van Buchem (VB) disease, associated with a large noncoding deletion is caused by the removal of a bone-specific distant enhancer element.
Van Buchem disease (MIM 239100) is a homozygous recessive disorder (Van Hul et al. 1998; Balemans et al. 2002; Staehling-Hampton et al. 2002) that maps to Chromosome 17p21 and results in progressive increase in bone density (Wergedal et al. 2003). The accumulation of bone mass gives rise to facial distortions, enlargement of the mandible and head, entrapment of the cranial nerves, increase in bone strength, and excessive weight (Van Hul et al. 1998; Balemans et al. 2002; Staehling-Hampton et al. 2002). Sclerosteosis (MIM 269500) is a cranio-tubular hyperosteosis that is phenotypically indistinguishable from Van Buchem (VB) disease except that it is more severe and occasionally displays syndactyly of the digits (Beighton et al. 1977; Balemans et al. 1999; Brunkow et al. 2001; Hamersma et al. 2003), a trait absent in VB patients.
An exciting development has been the recent discovery of a negative regulator of bone formation, sclerostin (SOST) (Balemans et al. 2001; Brunkow et al. 2001), whose expression is affected in both sclerosteosis and Van Buchem disease. Whereas sclerosteosis patients carry homozygous null SOST mutations, VB patients lack any SOST coding mutations (Staehling-Hampton et al. 2002; Winkler et al. 2003; Van Bezooijen et al. 2004). They do, however, carry a homozygous 52-kb noncoding deletion (vbΔ) ∼35 kb downstream of the SOST transcript and ∼10 kb upstream of the downstream gene, MEOX1, on human Chromosome 17p21 (Fig. 1A; Balemans et al. 2002; Staehling-Hampton et al. 2002). The shared clinical similarities between VB and sclerosteosis along with their strong genetic linkage to the SOST locus on Chromosome 17q12 suggests that they are allelic, and that the deletion in VB patients removes an enhancer element essential for directing the expression of human SOST in the adult skeleton. To gain insight into the mechanism by which this newly discovered gene has an impact on bone patterning and remodeling in Van Buchem disease, as well as to characterize the transcriptional regulation of sclerostin, we have characterized human BAC SOST transgenic mice carrying either a normal (SOSTwt) or an allele with the VB-associated deletion (SOSTvbΔ). Only the SOSTwt allele faithfully expresses human SOST in the adult bone and has an impact on bone metabolism, consistent with the model that the VB noncoding deletion removes a SOST-specific regulatory element. By exploiting cross-species sequence comparisons with in vitro and in vivo enhancer assays, we have identified a potential enhancer element that drives human SOST expression in the skeletal anlage, and discovered a novel function for sclerostin during limb development, demonstrating that this very important skeletal dysplasia, Van Buchem disease, is caused by the removal of bone-specific distant enhancer elements and is allelic to sclerosteosis.
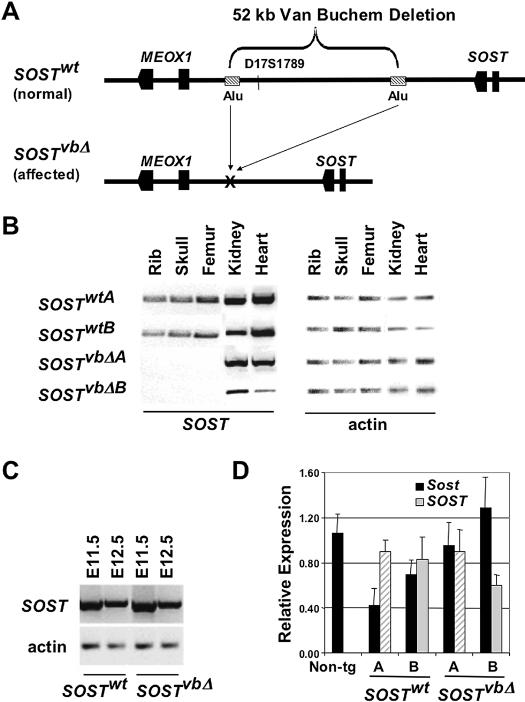
Figure 1.
Generation and characterization of Van Buchem transgenic mouse models. (A) A 158-kb human BAC (SOSTwt) spanning SOST and MEOX1 was engineered using in vitro BAC recombination in Escherichia coli (Lee et al. 2001) by deleting the 52-kb noncoding region missing in VB patients (SOSTvbΔ). Human SOST expression was analyzed by rtPCR in adult tissues (B), embryos (C), and measured by quantitative rtPCR in E10.5 embryos (D) from two independent lines of each SOSTwt and SOSTvbΔ transgene.
Results
Molecular and phenotypic characterization of Van Buchem transgenic mouse models
An ∼158-kb human BAC (RP11–209M4; SOSTwt) encompassing the 3′-end of the DUSP3 gene, SOST, MEOX1, and the ∼90-kb noncoding intergenic interval separating sost from the neighboring gene, MEOX1, was engineered using homologous recombination in bacteria (Lee et al. 2001) to delete the 52-kb region missing in VB patients and to create a construct that mimics the VB allele, SOSTvbΔ (Fig. 1A). Three independent founder lines of each transgenic construct were generated using standard transgenic procedures (Nobrega et al. 2003). Similar to the endogenous mouse Sost expression and the reported human expression (Balemans et al. 2001; Brunkow et al. 2001), all lines of SOSTwt transgenic animals reliably expressed human SOST in the mineralized bone of neonatal and adult mice (skull, rib, and femur), while all of the SOSTvbΔ lines had dramatically reduced levels of human SOST mRNA expression, as determined by rtPCR and qPCR. All lines (SOSTwt and SOSTvbΔ) also consistently expressed human SOST in the adult kidney and heart. Here, we present data on the top expressing lines from each transgenic construct, which are referred to in the manuscript as the A and B versions of each transgene (Fig. 1B). Expression in the lung, brain, and spleen varied among lines, while no BAC transgenic line expressed in the liver contrary to the reported human tissue-specific expression (Brunkow et al. 2001). These data demonstrate that in vivo, the VB allele confers dramatically reduced SOST expression in the adult bone and suggests that the vbΔ contains essential bone-specific enhancer elements.
Sclerostin is an osteocyte-expressed negative regulator of bone formation that is structurally most closely related to the DAN/Cerberus family of BMP antagonists (Winkler et al. 2003; Van Bezooijen et al. 2004). In the mouse, several members of this family including noggin and gremlin are expressed embryonically in the developing limb (Brunet et al. 1998; Khokha et al. 2003); therefore, we examined human SOST expression in the early mouse embryo. rtPCR analysis of RNA isolated from whole embryos showed high levels of human SOST transgenic expression in all lines from both SOSTwt and SOSTvbΔ transgenic animals (Fig. 1C). SOST expression precedes endochondral ossification, and was detected as early as embryonic day 9.5 (E9.5). Since the VB deletion did not have an impact on human SOST embryonic expression, we used E10.5 embryonic RNA to quantify the level of transgene expression in different SOSTwt and SOSTvbΔ transgenic founder lines (Fig. 1D). Comparable expression levels were also confirmed in the kidneys of SOSTwt and SOSTvbΔ animals. These data strengthen the evidence that the lack of human SOST bone expression in SOSTvbΔ animals is dependent on the 52-kb noncoding deletion, rather than reflecting an artifact due to transgene copy number or site-of-integration position effect.
The availability of BAC transgenic animals carrying wild-type and VB alleles also allowed us to address whether the other gene flanking the vbΔ region, the transcription factor MEOX1, is affected by removing the 52-kb noncoding region. MEOX1 has been previously shown to be involved in skeletal myogenesis (Mankoo et al. 2003; Petropoulos et al. 2004); therefore, it has been uncertain whether it also plays a role in the phenotypic outcomes of Van Buchem Disease, particularly since previous experiments using human patient samples prevented researchers from directly examining the effect of the noncoding deletion on gene expression (Staehling-Hampton et al. 2002). We have examined MEOX1 expression in all available transgenic lines from both SOSTwt and SOSTvbΔ constructs, and unfortunately were not able to detect any significant human MEOX1 expression in adult tissues (data not shown). Based on these results, we have concluded that the 209M4 BAC does not possess sufficient MEOX1-specific regulatory elements to recapitulate wild-type human MEOX1 expression in the SOSTwt transgenics; therefore, our experiments cannot evaluate the impact the VB deletion has on MEOX1 expression.
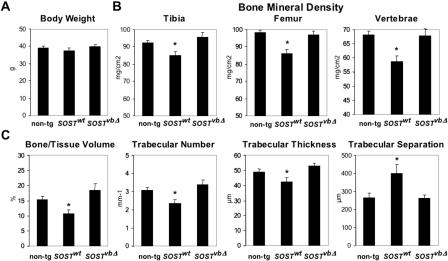
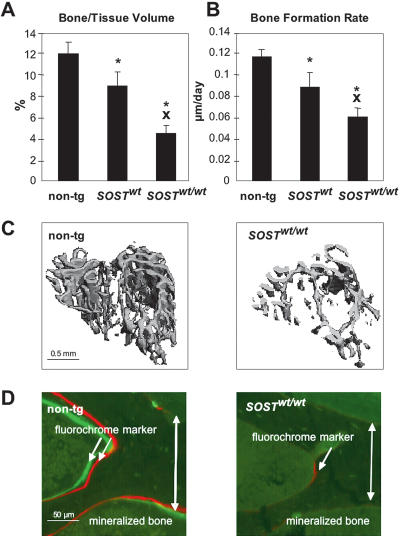
Since lack of sclerostin causes increased bone density (Brunkow et al. 2001), we investigated whether elevated levels of human sclerostin have opposite effects on bone mass. Consistent phenotypic data have been obtained for lines A of SOSTwt and SOSTvbΔ transgenic constructs (Fig. 2; Supplemental material). Animals from these lines expressed equivalent amounts of embryonic human SOST (Fig. 1D, gray shaded bars). For the rest of the manuscript we refer to these mice as SOSTwt and SOSTvbΔ. SOSTwt transgenics grew to skeletal maturity with normal body size and weight (Fig. 2A); however, the animals displayed decreased bone mineral density in the appendicular and axial skeleton, as evaluated by dual energy X-ray absorptiometry (DEXA) analysis (Fig. 2B). Micro-Computed-Tomography (microCT) analysis of three-dimensional cancellous bone structures revealed that the mice have decreased bone volume, trabecular number, thickness, and increased trabecular separation (Fig. 2C). In contrast, the bone parameters of SOSTvbΔ transgenics were indistinguishable from nontransgenic littermate controls. The observed osteopenia was gene dose-dependent. SOSTwt transgenic mice bred to homozygosity revealed a further dramatic decrease in tibial cancellous bone volume (Fig. 3A,C). Histomorphometric analysis revealed that these animals display further decreased bone formation rates at skeletal maturity reflected in decreased fluorochrome marker uptake into mineralizing bone both in cancellous (Fig. 3B,D) and cortical bone (tibia: non-tg = 0.319 ± 0.016 μm/d vs. SOSTwt/wt = 0.110 ± 0.027 μm/d; p < 0.001) in both the appendicular (Fig. 3B,C) and the axial skeleton. Neither the number of terminally differentiated bone-forming cells, the osteocytes, nor the number of bone-resorbing cells, the osteoclasts, were significantly affected by the transgene expression (data not shown).
Figure 2.
SOST transgenic expression has a negative impact on bone parameters. (A) Body weight measurements of 5-mo-old male mice (non-tg = 13; SOSTwt = 15; SOSTvbΔ = 14; animals were pooled from two lines of SOSTwt and two lines of SOSTvbΔ; Supplemental material). (B) Bone mineral density in the tibia, femur, and lumbar spine as evaluated by DEXA. (C) Bone volume, trabecular number, thickness, and separation as evaluated in the cancellous bone compartment of the proximal tibia metaphysis by microCT. Mean ± SEM; (*) p < 0.05 vs. non-tg.
Figure 3.
Human SOST dose effect on bone metabolism in the proximal tibia metaphysis of 5-mo-old male mice (non-tg = 5; SOSTwt = 7; SOSTwt/wt = 4). (A) Bone volume and (B) bone formation rates as determined by microCT scans and histomorphometric analysis, respectively. Mean ± SEM; (*) p < 0.05 vs. non-tg; (X) p < 0.05 vs. SOSTwt. (C) Cancellous bone compartment of nontransgenic and SOSTwt/wt mice. (D) Fluorochrome marker uptake at site of active mineralization of bone matrix laid down by osteoblasts in wild-type and transgenic mice at the interface between endocortex and cancellous bone.
In contrast to SOSTwt transgenics, SOSTvbΔ animals did not display an osteopenic bone phenotype in either the appendicular or the axial mature skeleton, even in the homozygous configuration (Fig. 2B,C; Supplemental material). These data demonstrate that modulation of SOST expression has a dramatic impact on bone formation in the adult mammalian skeleton. Most importantly, these phenotypic data suggest that overexpressing human SOST under the control of its own proximal promoter elements in concert with the downstream VB region negatively modulates adult bone mass. In contrast, bone mass is unaffected in transgenic animals that lack the 52-kb VB region, in a construct that mimics the allele carried by VB patients, consistent with the model that Van Buchem disease is caused by removing a bone-specific regulatory element.
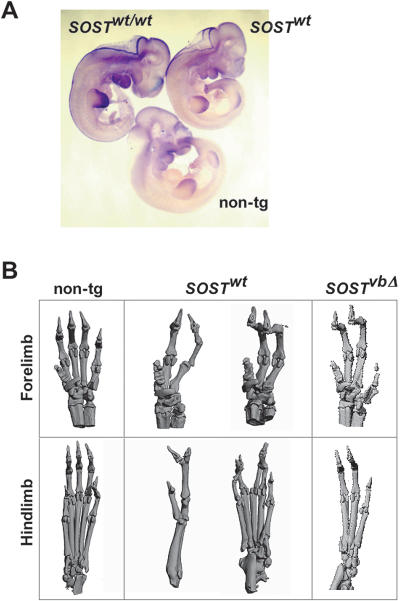
Interestingly, and consistent with the observed embryonic expression, elevated levels of human SOST result in abnormal digit development in both SOSTwt and SOSTvbΔ BAC transgenics bred to homozygosity. The forelimbs and hindlimbs of these animals display a wide range of fused and missing digits as visualized by autoradiography (data not shown) and microCT (Fig. 4B). rtPCR data correlate SOST expression with the severity of digit abnormalities (data not shown). Mouse whole mount in situ hybridization revealed SOST to be expressed as early as E9.5, predominantly in the mesenchymal tissue of the developing limb bud (Fig. 4A). These findings imply that SOST embryonic expression is controlled by a transcriptional regulatory element different from the one driving the adult bone expression, consistent with the observation that both sclerosteosis and VB patients suffer from abnormal bone mass accumulation, while only sclerosteosis patients exhibit syndactyly of the digits (Staehling-Hampton et al. 2002).
Figure 4.
Embryonic sost expression and limb deformity in SOSTwt and SOSTvbΔ transgenic mice. (A) High levels of embryonic SOST expression were predominantly detected in the developing limb bud of E10.5 defective mice, as visualized by whole-mount in situ hybridization using a SOST probe that detects human and mouse transcripts. (B) microCT scans of defective SOSTwt and SOSTvbΔ adult limbs overexpressing human SOST.
Comparative sequence analysis and enhancer assays
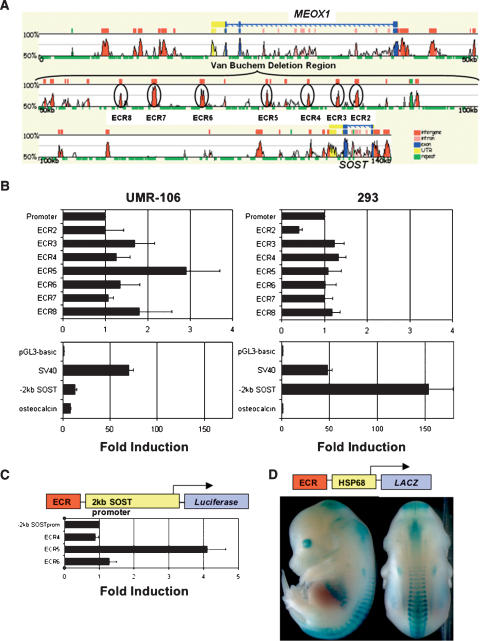
Given the striking bone phenotypes observed in both VB and sclerosteosis patients, we next focused on the identification of noncoding sequences required for SOST bone-specific expression through a combination of comparative sequence analysis and transient transfection assays. We aligned an ∼140-kb human SOST region (RP11–209M4; AQ420215, AQ420216) (http://zpicture.dcode.org/; Ovcharenko et al. 2004) to the corresponding mouse sequences from Chromosome 11 (Mouse chr11: 101,489,231–101,688,385; Oct.03 Freeze) (Fig. 5A). A stringent requirement of at least 80% identity (% ID) over a 200-bp window (≥80% ID; ≥200 bp) identified seven evolutionarily conserved regions (ECR2–8) within the vbΔ genomic interval, which were prioritized for in vitro enhancer analysis. ECR2–8 were tested for their ability to stimulate a heterologous promoter (SV40) in osteoblastic (UMR-106) and kidney (293) derived cell lines. One element, ECR5, was able to stimulate transcription in UMR106 cells (Fig. 5B), but not in the kidney cell line, suggesting that ECR5 enhancer function is specific to the osteoblastic lineage. We also tested the transcriptional activity of the human SOST proximal promoter region (2-kb region upstream of the 5′-UTR) in the two cell types and compared it to the SV40 and the osteoblast-specific osteocalcin promoter (Og2). The SV40 promoter showed comparable activity in both cell lines and, as expected, Og2 was only active in the UMR-106 cells. The human SOST promoter stimulated transcription in the osteoblastic cells similarly, albeit with slightly higher activity than the Og2 promoter, while it demonstrated stronger activity in kidney cells (Fig. 5B). These data suggest that SOST kidney expression may be due to proximal promoter sequences, whereas strong expression in osteoblast cells requires the activity of the ECR5 element. Consistent with the results obtained from transfecting SV40 promoter constructs, only ECR5 was capable of activating the human SOST promoter (4X) in UMR106 cells (Fig. 5C), while all other ECR-constructs had background level expression. Thus, a small sequence element within the vbΔ region (ECR5) was identified that confers in vitro osteoblast-specific enhancer activity onto both the human SOST and the SV40 heterologous promoter.
Figure 5.
Enhancer activity of evolutionarily conserved noncoding sequences from the Van Buchem deletion region. (A) Human/mouse genomic alignment generated using the zPicture alignment engine (http://zpicture.dcode.org/). Exons are in blue, untranslated regions in yellow, repetitive elements in green, and noncoding sequences in red (intragenic) or pink (intronic). Seven highly conserved elements (≥200 bp; ≥80% ID; ECR2–8) within VBΔ were tested in rat osteosarcoma (UMR-106) and kidney cells (293) for the ability to enhance luciferase expression from the SV40 promoter (B) or human SOST promoter (C). ECR5 activates the human SOST promoter in rat osteosarcoma cells (C), and drives the hsp68 promoter in the skeleton of E14.5 mouse embryos (D).
To test ECR5's ability to drive expression in the skeletal structures of the mouse embryo, we expressed an ECR5-hsp68-LacZ construct in transgenic mice (Fig. 5D; Nobrega et al. 2003). Transient transgenic animals were created using standard techniques (Mortlock et al. 2003) and F0 pups were stained for β-galactosidase (LacZ) expression at E14.5 (Nobrega et al. 2003). Transgenic embryos expressed LacZ in cartilage of the ribs, vertebrae, and skull plates (Fig. 5D), and the expression was identical in all positive transgenic embryos obtained from two independent injections (N = 2). In parallel we also injected ECR4-hsp68-LacZ and ECR6-hsp68-LacZ constructs and assayed LacZ expression at E12.5 and E14.5. None of the ECR4 (N = 6) and ECR6 (N = 2) positive embryos expressed LacZ at these time points. These data highly suggest that the 250-bp ECR5 element contained within the 52 kb deleted in VB patients functions to drive SOST bone-specific expression in vivo.
Discussion
Sclerosing bone dysplasias are rare genetic disorders in which excessive bone formation occurs because of defects in bone remodeling (Van Hul et al. 2001). Identifying the responsible genes, their regulation, and mechanisms of action will provide useful insights into bone physiology and potentially benefit the treatment of these disorders, as well as facilitate the development of therapies for replenishing bone loss in osteoporosis. In this study we have demonstrated that the 52-kb noncoding deletion present in Van Buchem patients removes a distant SOST-specific regulatory element, and therefore Van Buchem disease is hypomorphic to sclerosteosis. Currently, we don't have a clear view of how the lack of sclerostin promotes osteogenesis; therefore, elucidating its transcriptional regulation is key to understanding the interconnection between its expression pattern in osteogenic cells and its mode of action either as a BMP-antagonist (Winkler et al. 2003) or WNT-antagonist (Li et al. 2005). The elaborate expression pattern we detect along with the multitude of putative enhancer elements that have the potential to have a positive or negative impact on SOST in a spatial and temporal precise manner attest to this molecule's complexity and functional versatility. Consistent with this view, our analysis provides robust in vivo evidence for the role of sclerostin during bone formation, modulation of adult bone mass, and for a novel function during limb development.
In general, the osteopenic phenotype we observed is consistent with reports describing transgenic mice overexpressing BMP-antagonists from cDNA constructs driven by the osteocalcin (Og2) promoter (Devlin et al. 2003; Winkler et al. 2003). Unlike Og2>SOST transgenic animals (Winkler et al. 2003), we never observed architectural disorganization of the lumbar vertebrae. We believe the osteopenic phenotypic variations between the cDNA and BAC SOST transgenic mice are most likely attributed to the transcriptional control of human SOST in each transgenic construct. SOSTwt BAC transgenics more faithfully mirror the proper regulatory control exerted on the SOST gene in the endogenous context of the human genome, while the Og2>SOST transgenic expression is ectopic and highlights the transcriptional specificity of the osteocalcin promoter.
Since sclerosteosis is caused by SOST-null mutations (Balemans et al. 2001; Brunkow et al. 2001), our results indicate that VB disease and sclerosteosis are allelic, and VB patients are hypomorphic for the SOST gene and lack SOST expression in the adult bone. Our data suggest that SOST embryonic expression is unaltered in VB patients, who also never display syndactyly of the digits, indicating that both reduced and elevated levels of human sclerostin have a negative impact on limb development and digit formation, a novel function attributed to this molecule.
Our findings provide evidence that noncoding regions in the VB deletion control sclerostin expression levels and modulate BMD in mice; therefore, an important question is whether variation in BMD in the general population could also be directly impacted by sequence variants in key noncoding regions of the VB deletion. A recent new study investigated the association between common polymorphisms in the SOST gene region with BMD in elderly whites (Uitterlinden et al. 2004). From a set of eight polymorphisms, one 3-bp deletion (SRP3) from the SOST promoter region was associated with decreased BMD in women, and a polymorphic variant (SRP9) from the VB deletion region was associated with increased BMD in men. Whereas this SRP9 does not map on any human–mouse conserved region in the VB deletion, an important question for future studies is whether this SNP is in linkage disequilibrium with ECR5 or if additional functional SNPs could be identified in this or other SOST-specific enhancer elements.
The genetic factors that contribute to susceptibility to bone loss are extremely heterogeneous; therefore, murine models that affect bone development and growth can provide invaluable insights into the molecular mechanisms of progressive bone loss in humans. Human genetic diseases of the skeleton such as sclerosteosis and Van Buchem disease provide a starting point for understanding the modulation of anabolic bone formation, and ultimately have the potential to identify key molecular components that can be used as new therapeutic agents to treat individuals suffering from bone loss disorders. Our study also provides strong support for the utilization of comparative sequence analysis to dramatically filter through nonfunctional regions in the human genome and enhance the discovery of noncoding disease-causing mutations both in discrete enhancer elements or in large noncoding deletions. This study represents a clear and unambiguous case in which altering noncoding genomic content has a deleterious impact on gene expression, demonstrating that mutations in distant regulatory elements are able to cause congenital abnormalities analogous to coding mutations.
Methods
Generating transgenic mice
An FRT-kan-FRT cassette was excised from a pICGN21 vector (KpnI; SacI) and inserted into pUC18 to create pUC18.kan.FRT. Homologous arms were PCR-amplified from 209M4 BAC DNA and cloned into a pUC18.kan.FRT vector using EcoRI/SacI sites for the left arm (VBDelH1: fwd, 5′-TTGGTACCGGATTGAAGTGATCCCCAGCTGGA-3′; rvd, 5′-TTGAGCTCCAATCTCCTGACCTTGTGATCCGC-3′), and the SmaI site for the right arm (VbDelH2: fwd, 5′-TTCCCGGGCGCTTGAACCCAGTAGGTGGAGG-3′; rvd, 5′-TTCCCGGGTACCAAGGGATGGACAGAAGACAGGCAG-3′) to create the recombination vector pUC18.kan.FRT.VBDel. Then 200–300 ng of KpnI-digested VBDelH1-FRT-kan-FRT-VbdelH2 fragment was electroporated into EL250–209M4 cells. Recombinant BACs were identified by PCR and pulse-field gel analysis, were isolated at a final concentration of 1 ng/mL, and microinjected into fertilized FVB mouse eggs using standard procedures. Transgenic mice were genotyped using PCR analysis of DNA prepared from tail DNA of founder animals using the following primer pair: 5′-ATGTCCACCTTGCTGGACTC-3′ and 5′-GTCTGTGGGCTGGTTTGCAT-3′. Transgenic mice were maintained on an FVB background.
RT-PCR, quantitative RT-PCR, and in situ hybridization
Total RNA was isolated with Trizol reagent (Invitrogen) and reverse-transcribed into cDNA (Superscript II; GIBCO) using standard methods. cDNA was amplified using a GC-Melt PCR kit (Clontech) (65°C annealing/3 min extension/35 cycles) using human (fwd, 5′-AGAGCCTGTGCTACTGGAAGGTGG-3′, rvd, 5′-TAGGCGTTCTCCAGCTCGGCC-3′) and mouse (fwd, 5′-GACTGGAGCCTGTGCTACCGA-3′, rvd, 5′-CTTGAGCTCCGACTGGTTGTGGAA-3′) SOST primer sets. Mouse β-actin (fwd, 5′-CCTCTATGCCAACACAGTGC-3′, rvd, 5′-CTGGAAGGTGGACAGTGAG G-3′) was used as control (58°C annealing/30 sec extension/25 cycles). Quantitative rtPCR expression analysis was performed using an ABI Prism 7900HT sequence detection system, TaqMan Universal PCR Master mix, human 18S rRNA pre-developed TaqMan assay reagent for normalization, and TaqMan Assay-on-Demand products for mouse, rat, and human SOST, all from Applied Biosystems. We considered noon on the day that we found a vaginal plug to be E0.5. We carried out RNA localization by whole-mount in situ hybridization according to established protocols. RNA antisense probes were labeled with digoxigenin and were synthesized with T7 RNA polymerase as previously described.
Dual energy X-ray absorptiometry (DEXA) analysis
Tibial, femoral, and lumbar vertebral bone mineral density (in milligrams per square centimeter) was measured using a regular Hologic QDR-1000 instrument (Hologic). A collimator with 0.9-cm-diameter aperture and an ultrahigh resolution mode (line spacing, 0.0254 cm; resolution, 0.0127 cm) was used. The excised long bones were placed in 70% alcohol onto a resin platform provided by the company for soft tissue calibration. Daily scanning of a phantom image controlled the stability of the measurements. Instrument precision and reproducibility had been previously evaluated by calculating the coefficient of variation of repeated DEXA and had been found to be below 2%. Coefficients of variation were 0.5% to 2% for all evaluated parameters. A set of 5-mo-old male mice was analyzed (non-tg = 13 littermates of all analyzed lines; SOSTwt = 15 heterozygous mice from two SOSTwt BAC lines; SOSTvbΔ = 14 offspring of heterozygous matings from two SOSTvbΔ lines).
Micro-Computed-Tomography (microCT) analysis
Cancellous bone structure was evaluated in the proximal tibia metaphysis using a Scanco vivaCT20 (Scanco Medical AG). The nonisometric voxels had a dimension of 12.5 μm × 12.5 μm × 12.5 μm. From the cross-sectional images the cancellous bone compartment was delineated from cortical bone by tracing its contour at every tenth section. In all the other slices, boundaries were interpolated based on the tracing to define the volume of interest. In all, 660 slices covering a total length of 0.8 mm within the area of the secondary spongiosa (1.3 mm from the proximal end) were evaluated. A threshold value of 175 was used for the three-dimensional evaluation of trabecular number, thickness, and separation. Both sets of male 5-mo-old mice on which DEXA and histomorphometric analysis have been performed were analyzed. A voxel size of 25 μm × 25 μm × 25 μm was chosen for visualization of the digits of the forelimbs and hindlimbs.
Histomorphometric analysis
After dissection, the tibia and lumbar vertebrae were placed for 24 h in Karnovsky's fix, dehydrated in ethanol at 4°C, and embedded in methylmethacrylate. A set of 4-μm and 8-μm-thick nonconsecutive microtome sections were cut in the frontal mid-body plane for evaluation of fluorochrome-label-based dynamic and cellular parameters of bone turnover. The 4-μm-thick sections were stained with TRAP and Giemsa stain. The sections were examined using a Leica DM microscope (Leica) fitted with a camera (SONY DXC-950P) and adapted Quantimet 600 software (Leica). Two sections/animal were sampled for all sets of parameters. Microscopic images of specimens were evaluated semiautomatically digitally (400× magnification). All parameters were measured and calculated according to Paritt et al. (1987). Fluorochrome label bone formation dynamics were evaluated on unstained 8-μm-thick sections. Bone perimeter, single- and double-labeled bone perimeter, and interlabel width were measured. Mineralized perimeter (%), mineral apposition rates (micrometers/day; corrected for section obliquity in the cancellous bone compartment), and daily bone formation rates (daily bone formation rate/bone perimeter; micrometer/day) were calculated. Osteoclast numbers (osteoclast number/bone perimeter; per millimeter) and perimeter values (osteoclast perimeter/bone perimeter; percent) were determined on the TRAP-stained slides, and osteocyte number (osteocyte number/bone perimeter; per millimeter) on the Giemsa-stained slides. All parameters were evaluated in the spongiosa and at the endocortex. A set of 5-mo-old male mice from one SOSTwt line was analyzed (non-tg = 5, SOSTwt = 7, SOSTwt/wt = 4).
In vitro enhancer assays
ECRs were PCR-amplified with 5′-NheI-linkers, TOPO-cloned into a pCR2.1 vector (Invitrogen), then shuttled into NheI/XhoI sites of a pGL3 promoter (Promega) or HindIII/PstI of hsp68-LacZ (B. Black). The following primers were used to amplify human DNA (62°C annealing/30 sec extension/35 cycles): ECR2 (545 bp), 5′-AGCAACGCAGGGCAGGAGCCAAGA-3′, 5′-TAGCTGGCCTCTCCTGGGCGTCTT-3′; ECR3 (410 bp), 5′-GGGGGCTGTATGGAAAGGAGACAT-3′, 5′-CTTGAGCAGTAGGGCCAAGCCCT-3′; ECR4 (296 bp), 5′-TGACAAACAGGAAGGTGGCAGGGC-3′, 5′-CCCCCAACATTCCTGTCCCCTTG-3′; ECR5 (259 bp), 5′-TCCTTGCCACGGGCCACCAGCTTT-3′, 5′-CCCCCTCATGGCTGGTCTCATTTG-3′; ECR6 (666 bp), 5′-CCCTGAGAAACATGCCTCTGTCCC-3′, 5′-CTTAGCAATCTGGGTGACCCTGGG-3′; ECR7 (568 bp), 5′-AAACTGCCAAGCCCCAGCTGGCTA-3′, 5′-GCCCAGGGCTCAGAAATGTGTGGA-3′; ECR8 (352 bp), 5′-TTCCTACCAAGGTGGCTGCCACC-3′, 5′-CCTTCAGAGAAGCAAATGGCTGGGG-3′; –2 kb promoter, 5′-CAGCAGAAGATGTCACAGCAGG-3′, 5′-GAGCTGCATGGTACCAGCCAGA-3′. The human SOST promoter sequence (2 kb upstream of the 5′-UTR) was PCR-amplified with SmaI linkers and transferred into the SmaI site of pGL3basic (Promega). A luciferase reporter plasmid containing the mouse osteocalcin (Og2) promoter sequence from –1323 to +10 in pGL3basic was kindly obtained from B. Fournier (Novartis Basel, Switzerland). Reporter plasmids containing ECR-4, ECR-5, or ECR-6 upstream of the human SOST promoter were generated by inserting the ECR elements into the NheI site. Plasmid DNA was isolated using standard endotoxin-free methods (QIAGEN). FuGene (Roche) and a CMV-bgal reporter plasmid (Clontech) as internal control were used for transient transfections of rat UMR-106 and human 293 cells. Cells were incubated for 24 h at 37°C, and luciferase and galactosidase expression were measured using standard assay kits (Promega).
Transient transgenic analysis
For transient transgenic analysis, 500 mg of DNA was linearized with NotI, followed by CsCl gradient purification, and 2–5 ng was used for pronuclear injections of FVB embryos. E12.5–E14.5 embryos were dissected in ice-cold PBS, and were fixed in 4% paraformaldehyde at 4°C for 1–2 h, and stained for LacZ as described. Transgenic embryos were detected by PCR from tail DNA (fwd, 5′-TTTCCATGTTGCCACTCGC-3′; rvd, 5′-AACGGCTTGCCGTTCAGCA-3′;55°C annealing/30 sec extension/25 cycles).
Acknowledgments
The authors thank V. Afzal, M. Bruederlin, E. Kuhn, H. Jeker, M. Merdes, A. Studer, and J. Wirsching for excellent technical help and B. Fournier for sharing the mouse osteocalcin reporter plasmid. Many thanks to M. Brunkow and D. Winkler for providing us with the original 209M4 human BAC. The authors are deeply indebted to Lisa Stubbs, Richard Harland, Mustafa Khokha, and Marcelo Nobrega for invaluable advice and comments on the manuscript. G.G.L. was supported by the Department of Energy Alexander Hollaender Fellowship and by the NIH (HD47853-01). J.C. and N.M.C. were supported by NIH (HD47853-01). This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Berkeley National Laboratory, Contract No. AC0376SF00098 and Lawrence Livermore National Laboratory Contract No. W-7405-Eng-48.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3437105. Article published online before print in June 2005. Freely available online through the Genome Research Immediate Open Access option.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: B. Black, B. Fournier, M. Brunkow, and D. Winkler.]
References
- Balemans, W., Van Den Ende, J., Freire Paes-Alves, A., Dikkers, F.G., Willems, P.J., Vanhoenacker, F., de Almeida-Melo, N., Alves, C.F., Stratakis, C.A., Hill, S.C., et al. 1999. Localization of the gene for sclerosteosis to the van Buchem disease-gene region on chromosome 17q12–q21. Am. J. Hum. Genet. 64: 1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balemans, W., Ebeling, M., Patel, N., Van Hul, E., Olson, P., Dioszegi, M., Lacza, C., Wuyts, W., Van Den Ende, J., Willems, P., et al. 2001. Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Hum. Mol. Genet. 10: 537-543. [DOI] [PubMed] [Google Scholar]
- Balemans, W., Patel, N., Ebeling, M., Van Hul, E., Wuyts, W., Lacza, C., Dioszegi, M., Dikkers, F.G., Hildering, P., Willems, P.J., et al. 2002. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J. Med. Genet. 39: 91-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton, P., Davidson, J., Durr, L., and Hamersma, H. 1977. Sclerosteosis—An autosomal recessive disorder. Clin. Genet. 11: 1-7. [DOI] [PubMed] [Google Scholar]
- Brunet, L.J., McMahon, J.A., McMahon, A.P., and Harland, R.M. 1998. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280: 1455-1457. [DOI] [PubMed] [Google Scholar]
- Brunkow, M.E., Gardner, J.C., Van Ness, J., Paeper, B.W., Kovacevich, B.R., Proll, S., Skonier, J.E., Zhao, L., Sabo, P.J., Fu, Y., et al. 2001. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am. J. Hum. Genet. 68: 577-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuzhanova, N., Abeysinghe, S.S., Krawczak, M., and Cooper, D.N. 2003. Translocation and gross deletion breakpoints in human inherited disease and cancer II: Potential involvement of repetitive sequence elements in secondary structure formation between DNA ends. Hum. Mutat. 22: 245-251. [DOI] [PubMed] [Google Scholar]
- Cimbora, D.M., Schubeler, D., Reik, A., Hamilton, J., Francastel, C., Epner, E.M., and Groudine, M. 2000. Long-distance control of origin choice and replication timing in the human β-globin locus are independent of the locus control region. Mol. Cell. Biol. 20: 5581-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin, P.T. and Kan, Y.W. 1988. The inactive β globin gene on a γ δ β thalassemia chromosome has a normal structure and functions normally in vitro. Blood 71: 766-770. [PubMed] [Google Scholar]
- Curtin, P., Pirastu, M., Kan, Y.W., Gobert-Jones, J.A., Stephens, A.D., and Lehmann, H. 1985. A distant gene deletion affects β-globin gene function in an atypical γ δ β-thalassemia. J. Clin. Invest. 76: 1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, R.D., Du, Z., Pereira, R.C., Kimble, R.B., Economides, A.N., Jorgetti, V., and Canalis, E. 2003. Skeletal overexpression of noggin results in osteopenia and reduced bone formation. Endocrinology 144: 1972-1978. [DOI] [PubMed] [Google Scholar]
- Enattah, N.S., Sahi, T., Savilahti, E., Terwilliger, J.D., Peltonen, L., and Jarvela, I. 2002. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 30: 233-237. [DOI] [PubMed] [Google Scholar]
- Hamersma, H., Gardner, J., and Beighton, P. 2003. The natural history of sclerosteosis. Clin. Genet. 63: 192-197. [DOI] [PubMed] [Google Scholar]
- Ionasescu, V.V., Searby, C., Ionasescu, R., Neuhaus, I.M., and Werner, R. 1996. Mutations of the noncoding region of the connexin32 gene in X-linked dominant Charcot-Marie-Tooth neuropathy. Neurology 47: 541-544. [DOI] [PubMed] [Google Scholar]
- Khokha, M.K., Hsu, D., Brunet, L.J., Dionne, M.S., and Harland, R.M. 2003. Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat. Genet. 34: 303-307. [DOI] [PubMed] [Google Scholar]
- Kleinjan, D.A., Seawright, A., Schedl, A., Quinlan, R.A., Danes, S., and van Heyningen, V. 2001. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum. Mol. Genet. 10: 2049-2059. [DOI] [PubMed] [Google Scholar]
- Lee, E.C., Yu, D., Martinez de Velasco, J., Tessarollo, L., Swing, D.A., Court, D.L., Jenkins, N.A., and Copeland, N.G. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73: 56-65. [DOI] [PubMed] [Google Scholar]
- Lettice, L.A., Heaney, S.J., Purdie, L.A., Li, L., de Beer, P., Oostra, B.A., Goode, D., Elgar, G., Hill, R.E., and de Graaff, E. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12: 1725-1735. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Kang, H., Liu, W., Liu, P., Zhang, J., Harris, S.E., and Wu, D. 2005. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J. Biol. Chem. 280: 19883-19887. [DOI] [PubMed] [Google Scholar]
- Mankoo, B.S., Skuntz, S., Harrigan, I., Grigorieva, E., Candia, A., Wright, C.V., Arnheiter, H., and Pachnis, V. 2003. The concerted action of Meox homeobox genes is required upstream of genetic pathways essential for the formation, patterning and differentiation of somites. Development 130: 4655-4664. [DOI] [PubMed] [Google Scholar]
- Mortlock, D.P., Guenther, C., and Kingsley, D.M. 2003. A general approach for identifying distant regulatory elements applied to the Gdf6 gene. Genome Res. 13: 2069-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega, M.A., Ovcharenko, I., Afzal, V., and Rubin, E.M. 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413. [DOI] [PubMed] [Google Scholar]
- Ovcharenko, I., Loots, G.G., Hardison, R.C., Miller, W., and Stubbs, L. 2004. zPicture: Dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 14: 472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt, A.M., Drezner, M.K., Glorieux, F.H., Kanis, J.A., Malluche, H., Meunier, P.J., Ott, S.M., and Recker, R.R. 1987. Bone histomorphometry: Standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2: 595-610. [DOI] [PubMed] [Google Scholar]
- Petropoulos, H., Gianakopoulos, P.J., Ridgeway, A.G., and Skerjanc, I.S. 2004. Disruption of Meox or Gli activity ablates skeletal myogenesis in P19 cells. J. Biol. Chem. 279: 23874-23881. [DOI] [PubMed] [Google Scholar]
- Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525-528. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton, K., Proll, S., Paeper, B.W., Zhao, L., Charmley, P., Brown, A., Gardner, J.C., Galas, D., Schatzman, R.C., Beighton, P., et al. 2002. A 52-kb deletion in the SOST-MEOX1 intergenic region on 17q12–q21 is associated with van Buchem disease in the Dutch population. Am. J. Med. Genet. 110: 144-152. [DOI] [PubMed] [Google Scholar]
- Tsui, F.W., Tsui, H.W., Cheng, E.Y., Stone, M., Payne, U., Reveille, J.D., Shulman, M.J., Paterson, A.D., and Inman, R.D. 2003. Novel genetic markers in the 5′-flanking region of ANKH are associated with ankylosing spondylitis. Arthritis Rheum. 48: 791-797. [DOI] [PubMed] [Google Scholar]
- Uitterlinden, A.G., Arp, P.P., Paeper, B.W., Charmley, P., Proll, S., Rivadeneira, F., Fang, Y., van Meurs, J.B., Britschgi, T.B., Latham, J.A., et al. 2004. Polymorphisms in the sclerosteosis/van Buchem disease gene (SOST) region are associated with bone-mineral density in elderly whites. Am. J. Hum. Genet. 75: 1032-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bezooijen, R.L., Roelen, B.A., Visser, A., Van Der Wee-Pals, L., De Wilt, E., Karperien, M., Hamersma, H., Papapoulos, S.E., Ten Dijke, P., and Lowik, C.W. 2004. Sclerostin is an osteocyte-expressed negative regulator of bone formation, but not a classical BMP antagonist. J. Exp. Med. 199: 805-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hul, W., Balemans, W., Van Hul, E., Dikkers, F.G., Obee, H., Stokroos, R.J., Hildering, P., Vanhoenacker, F., Van Camp, G., and Willems, P.J. 1998. Van Buchem disease (hyperostosis corticalis generalisata) maps to chromosome 17q12–q21. Am. J. Hum. Genet. 62: 391-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hul, W., Vanhoenacker, F., Balemans, W., Janssens, K., and De Schepper, A.M. 2001. Molecular and radiological diagnosis of sclerosing bone dysplasias. Eur. J. Radiol. 40: 198-207. [DOI] [PubMed] [Google Scholar]
- Wang, H.L., Wu, T., Chang, W.T., Li, A.H., Chen, M.S., Wu, C.Y., and Fang, W. 2000. Point mutation associated with X-linked dominant Charcot-Marie-Tooth disease impairs the P2 promoter activity of human connexin-32 gene. Brain Res. Mol. Brain Res. 78: 146-153. [DOI] [PubMed] [Google Scholar]
- Wergedal, J.E., Veskovic, K., Hellan, M., Nyght, C., Balemans, W., Libanati, C., Vanhoenacker, F.M., Tan, J., Baylink, D.J., and Van Hul, W. 2003. Patients with Van Buchem disease, an osteosclerotic genetic disease, have elevated bone formation markers, higher bone density, and greater derived polar moment of inertia than normal. J. Clin. Endocrinol. Metab. 88: 5778-5783. [DOI] [PubMed] [Google Scholar]
- Winkler, D.G., Sutherland, M.K., Geoghegan, J.C., Yu, C., Hayes, T., Skonier, J.E., Shpektor, D., Jonas, M., Kovacevich, B.R., Staehling-Hampton, K., et al. 2003. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 22: 6267-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]