Abstract
We have developed a mutation-scanning approach suitable for whole population screening for unknown mutations. The method, meltMADGE, combines thermal ramp electrophoresis with MADGE to achieve suitable cost efficiency and throughput. The sensitivity was tested in blind trials using 54 amplicons representing the BRCA1 coding region and a panel of 94 unrelated family breast cancer risk consultands previously screened in a clinical diagnostic laboratory. All 10 common polymorphisms, 15/15 previously identified disease-causing mutations, and three previously untested single base changes were identified. Assays of LDLR exons 3 and 8 were validated in 460 familial hypercholesteremics and detected 8/9 known variants. We then applied the exon 3 assay in several DNA banks representing ∼8000 subjects with known cholesterol values and applied both assays in one DNA bank (n = 3600). In exon 3 we identified one previously reported moderate mutation, P84S (n = 1), also associated with moderate hypercholesteremia in this subject; an unreported silent variant, N76N (n = 1); and known severe hypercholesteremia splice mutation 313+1G→A (n = 2). Around exon 8 we identified a paucimorphism (n = 35) at the splice site 1061–8T→C (known to be in complete linkage disequilibrium with T705I) and unreported sequence variants 1186+11G→A (n = 1) and D335N G→A (n = 1). The cholesterol value for D335N was on the 96.2 percentile and for T705I, 2/35 carriers were above the 99th percentile. Thus, variants with predicted severe, moderate, and no effect were identified at the population level. In contrast with case collections, CpG mutations predominated. MeltMADGE will enable definition of the full population spectrum of rare, paucimorphic, severe, moderate (forme fruste), and silent mutations and effects.
The prevailing hypothesis for the molecular basis of common diseases is that it involves the combinatorial action of common polymorphic alleles of minor effect (common disease/common variant, CD/CV hypothesis). A contrasting approach has been the study of very highly selected cases and families by linkage and mutation detection techniques to identify rare mutations of large effect on a gene, often private to a single family (rare disease/rare variant, RD/RV hypothesis). However, intermediate possibilities exist. Sequence changes at an intermediate frequency (paucimorphisms) may exist and may have a moderate effect (Day et al. 2004). Additionally, rare sequence variants of moderate effect may be cumulatively common in the population. Theoretical and observational literature relevant to these possibilities has been recently reviewed (Day et al. 2004). Several different loci may predispose to the same disease, although only one paucimorphic allele of one particular gene will be found in any one individual. It is also possible that large numbers of “private” mutations of moderate effect could cumulatively account for a significant fraction of disease in a population. Exploring these hypotheses will require mutation detection applied both at the level of large numbers of relatively unselected cases and at the population level.
In this study, we developed and applied meltMADGE (see Supplemental Fig. 1 and Methods) for population studies. MeltMADGE combines the properties of MADGE (Day and Humphries 1994; Gaunt et al. 2003) with a reconfiguration of denaturing gradient gel electrophoresis (DGGE) (Fischer and Lerman 1979), using a thermal ramp in time rather than a linear gradient in space, to increase the sample parallelism and reduce the costs of mutation scanning by one to two orders of magnitude. Here we describe the development of the method, its validation in the detection of unknown single base and small insertion/deletion variants in BRCA1 and LDLR, and the study of regions of the LDLR gene in relation to cholesterol levels in population samples representing ∼8000 subjects. We chose to use mutations in BRCA1 identified and validated in a clinical diagnostics laboratory for initial development and validation of the sensitivity of the meltMADGE method. However, for both ethical and interpretative reasons, we chose LDLR to undertake proof-of-principle population studies. Interpretation should be simpler, although not simple, for well-characterized quantitative traits than for case events and for a gene with a very well-characterized protein product, such as LDLR. Importantly, our cohort consents covering cholesterol data are such that no individual at high clinical risk (and deductively a proband potentially for a family) would be identified without a mechanism for clinical feedback.
The low-density lipoprotein receptor (LDLR) removes LDL-cholesterol particles from the circulation (Brown and Goldstein 1986). Mutations in LDLR (Sudhof et al. 1985) lead to an accumulation of LDL-cholesterol in plasma, resulting in the classical familial hypercholesteremia (FH) phenotype. The frequency of heterozygous FH is estimated to be ∼1/500 in the general population (Goldstein et al. 1995). Most such mutations (Hobbs et al. 1992) fully inactivate alleles (e.g., deletions and stop codons), although some amino acid changes (e.g., W66G) (Moorjani et al. 1993) may only cause partial haploinsufficiency. We previously estimated that among FH cases (heterozygotes and homozygotes) there is a fivefold underrepresentation of amino acid changes, compared with codon mutations to stop codons (Day et al. 1997). This analysis was based on comparison of expected versus observed distribution of single base changes across the LDLR coding sequence. Thus, ∼1/100 people might possess an amino acid substitution not causing a severe classical FH heterozygote phenotype. However, almost all mutation scanning has been undertaken only in cases selected for severe hypercholesteremia. Only population-based studies could fully define the wider spectrum of mutational effects in LDLR covering severe, plus possible moderate, silent, or protective effects and the full spectrum of polymorphisms, paucimorphisms (arbitrarily, alleles 0.0005 < q < 0.05) (Day et al. 2004), and “private” sequence variation that may exist. Worldwide, more than 850 sequence variants and mutations have been described in the LDLR gene (http://www.ucl.ac.uk/fh). Although there are several methods available for mutation scanning, their throughput and cost make them unsuitable for population studies (Cotton 1998). Therefore, the extent of such variation in natural populations and its impact on common traits have not been fully evaluated. For this study, we selected LDLR exon 3, representing part of the ligand-binding domain and with a high density of mutations identified in FH cases, and exon 8, representing a region with a lower density of mutation in FH cases.
Results
In effect, the system we have developed achieves a reconfiguration of DGGE, such that using a thermal ramp instead of a spatial gradient, 10–11 small gels, each directly compatible with a 96-well microplate, can be electrophoresed in parallel in a 1–2-h run in a 2-L tank. This achieves a large throughput increment at low costs using simple equipment.
Initial assessment of features of meltMADGE assays
We examined the relationship between the predicted Tm (Lerman and Silverstein 1987) and the suitable temperature ramp range for meltMADGE assays (data not shown). In general, ramp ranges from (predicted Tm - 4°C) to (predicted Tm or predicted Tm + 1) were found to be suitable for heteroduplex (but not necessarily homoduplex) resolution. For heterozygote recognition, heteroduplex resolution was always sufficient for identification of mutation-positive samples. We also investigated amplicon length and found that amplicons in the range 180–350 bp were suitable, although longer amplicons (e.g., >400 bp) may compromise resolution. Resolution at different positions in the gel track arrays of MADGE (which could influence heteroduplex resolution either through thermal inhomogeneities in our prototype apparatus or through anodal to cathodal ionic inhomogeneities in the gel) was checked by loading a heterozygous sample in every well: track to track variation was found to be minor and neither could we detect thermal inhomogeneity using a high-precision platinum resistivity thermometer (data not shown). The overall process displayed in Supplemental Figure 1 functioned efficiently and reliably.
Development phase: Detection of common polymorphisms and some known mutations in BRCA1
We screened 20 anonymous normal samples for polymorphisms in all 24 exons of the BRCA1 gene, expecting to detect most polymorphisms with minor allele frequency >0.07. The Breast Cancer Information Core (BIC) database lists, in the sequences we scanned, 10 such common polymorphisms that are believed to be functionally neutral. The 3426-bp-long exon 11 was scanned using 17 overlapping amplimers. Alternative amplimers were tested for ability to detect the same polymorphism(s), to examine the tolerance of meltMADGE to alternative amplimer designs. Ten polymorphisms (1186A→G, 2196G→A, 2201C→T, 2430T→C, 2731C→T, 3232A→G, 3238G→A, 3667A→G, 4427T→C, 4956A→G) were detected in our assays, in exon 11B, 11G, 11H, 11J, 11L, 11M, 11O, exon 13, and exon 16, respectively. Figure 1 shows a range of examples of different amplicons, different polymorphisms, and different mutations. Samples with known mutations (1218insA, 2441T→A, 3881delGA, 4176G→T, and 4158A→G) were also included (respectively positive in assays 11G, 11I, 11P, and 11Q) during the initial assay development, and each showed a split band pattern in contrast with the wild-type band.
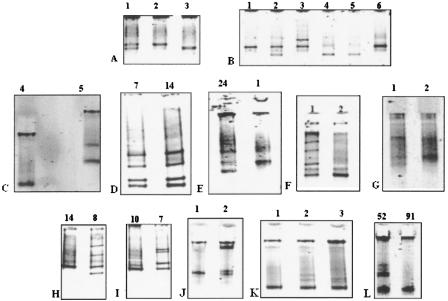
Figure 1.
BRCA1 meltMADGE variant patterns. The track width is 2 mm: total migration distances range from ∼5 to 12 mm. All samples have been re-run in the thermal ramp system for illustration purposes in H-PAGE format (Day and Humphries 1994) except for panel C, which is in MADGE format. Tracks are labeled by subject number. (A) Amplimer BRCA1x8 displays the three genotypes of the common SNP, IVS7–34C→T. The homoduplexes, distinguished by one hydrogen-bond difference (G:C duplex vs. A:T duplex) are well resolved. (B) Patterns of BRCA1x11M containing 3232A→G (G allele frequency 0.33) and 3238G→A (A allele frequency 0.02) polymorphisms. Sample 2 is heterozygous for 3232A→G, and sample 3 is heterozygous for 3238G→A. (C) BRCA1x11B: track 5 displays 1218insA and track 4 displays wild type. (D) Four band heterozygote patterns of the common polymorphism, 2731C→T, in two heterozygous samples, in amplicon 11J. (E) Amplicon 11J: 2804delAA frameshift mutation is seen in sample 24 compared with wild type (sample 1). (F) BRCA1x11G: track 2 represents the wild-type homoduplex, and track 1 displays the heterozygote formed pattern for SNP 2201C→T. (G) BRCA1x11O: 3825delAA heterozygous mutation (track 1) compared with wild type (track 2). The two homoduplexes resolved, but the two heteroduplexes appear to coelectrophorese. (H) BRCA1x11H: SNP 2401 C→T heterozygote (track 8) and CC homozygote (track 14). (I) BRCA1x11I: SNP 2401C→T heterozygote (track 7) and CC homozygote (track 10). (J) BRCA1x11P: track 1 represents wild type. In track 2, the heteroduplexes for heterozygous mutation 3875del4 (GTCT) were retarded at an early stage near the well, whereas the homoduplex mobilities differentiated at a later stage in the thermal ramp. (K) BRCA1x11O: track 1 represents wild type. Track 2 represents a heterozygote for SNP 3667A<G. Track 3 represents a heterozygote for mutation 3694insT. Although the thermal ramp used did not resolve the homoduplexes, two heteroduplexes are resolved both in track 2 and track 3, the heteroduplex bands in track 3 clearly differing from these in track 2. (L) BRCA1x20: track 91 represents wild type, whereas track 52 represents a subject heterozygous for 5396+47ins12; in track 52 the two heteroduplexes appear to coelectrophorese, as do the two homoduplexes.
BRCA1 meltMADGE assays of a panel of 94 unrelated familial breast cancer risk consultands
Samples had been previously screened for mutations in the BRCA1 gene using SSCP/HA and PTT in the Wessex Regional Genetics Laboratory, (Salisbury, UK). In all, 94 anonymized DNA samples were rescanned using meltMADGE by author M.A.A., blind to sample identity, followed by sequencing of mutation positives identified. Of 15 mutations previously identified, 13 were identified, one mutation in exon 2, 11 mutations in exon 11, and one mutation in exon 20. Two mutations (2773delTC, 2804delA) in amplicon 11J in exon 11 were not recognized because of PCR failures but were identified clearly in subsequent runs. In Figure 2, the gel image for the same panel, for BRCA1x11B, is shown. In this, 10 subjects were identified with a polymorphism (1187A→G) and three different mutations, each with a different band pattern and mutation, were also identified (1138delG, 1218insA, 1131A→T). Table 1 shows the comparison of different mutation-scanning methods for this sample set. Additionally, three single base changes each present in one subject, which would not have been identified during diagnostic screening, were found. These were 2413A→G (E765G), 1967T→G (S616S), and 5143C→T (T1675I). The former has previously been reported by others to be probably clinically deleterious (Fleming et al. 2003), but the role of the other two is unknown and further investigation was beyond the scope of the present work. No false positives were picked during these analyses, and independent workers identified the same set of true positives.
Figure 2.
MeltMADGE analysis (undertaken blind to sample information) of amplicon 11B in a panel of 94 cases. Three different mutations, circled numbers 71, 83, and 87 (1138delG, 1218insA, 1131A→T, respectively), and one polymorphism (1187A→G), samples 5, 8, 23, 29, 47, 51, 60, 61, 72, 76, and 92, were identified. Adjacent tracks outside the 8 × 12 96-well array were loaded with known mutation amplicons (tracks A, B, and C) and wild-type amplicon (D).
Table 1.
Diagnostic laboratory information for BRCA1-positive samples included in panel of 94 samples assayed blind by meltMADGE
| Diagnostic laboratory information
|
||||
|---|---|---|---|---|
| Sample ID | Amplicon ID | Mutation | Method | MeltMADGE detection |
| 5 | Exon 2 | 185delAG | HA | + |
| 71 | 11B | 1138delG | HA | + |
| 83 | 11B | 1218insA | DS | + |
| 87 | 11B | 1131A→T | PTT | + |
| 52 | 11D | 1445T→A | DS | + |
| 70 | 11G | 2187delA | PTT | + |
| 7 | 11I | 2594delC | SSCP | + |
| 88 | 11J | 2773delTC | PTT | See Discussion |
| 90 | 11J | 2804delAA | HA | See Discussion |
| 10 | 11N | 3519G→T | PTT | + |
| 60 | 11O | 3695insT | HA | + |
| 95 | 11O | 3826delAA | PTT | + |
| 72 | 11P | 3882delAA | HA | + |
| 81 | 11Q | 4184del4 | SSCP/HA | + |
| 30,34 | Exon 20 | 5382insC | HA | + |
Validation of meltMADGE method on LDLR amplicons chosen for population study
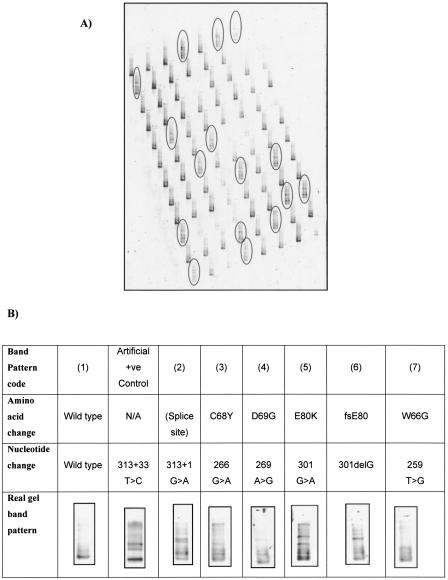
To examine the sensitivity of meltMADGE, 460 DNA samples from the Simon Broom Familial Hypercholesterolemia (SBFH) register (Betteridge et al. 1999; Neil et al. 2004), previously screened for mutations of LDLR using the SSCP technique (Whittall et al. 1995), were used. MeltMADGE mutation scanning was undertaken by author K.K.A. in the laboratory of author I.N.M.D., both blind to the SSCP data of authors R.A.W. and S.E.H. until the meltMADGE scanning had been completed. Six different mutant band patterns were identified (Fig. 3), representing eight different mutations (D69G, C68Y, 313+1, E80K, C83F, W66G, fsE80, and new fsV45) involving 71 out of 460 SBFH cases scanned (Table 2). The band pattern for fsV45 was similar to that for W66G, and the band pattern for C83F was similar to that for E80K, prospectively classified as the same although retrospectively showing differences. An artificial mutant (see Fig. 3B and Methods) also gave a split band pattern compared with the single band pattern of wild type. MeltMADGE identified two mutations, fsV45 and C83F, which had not been identified by SSCP in this case set—both were confirmed by direct sequencing. All sequence variation detected by SSCP was detected by meltMADGE. FsV45 (deletion GT at 196–197) appears to represent a novel mutation not previously described in familial hypercholesteremia. In most mutation band patterns, three or four bands were observed, representing two heteroduplexes nearest the wells and two closely spaced (or coelectrophoresing) homoduplexes. However, indels tended to produce a two-band pattern, assumed to represent two coelectrophoresing heteroduplexes and two coelectrophoresing homoduplexes.
Figure 3.
Example of meltMADGE scanning in familial hypercholesteremia case collection. (A) Single gel image (one of six gels run in the same tank at the same time) meltMADGE scan of LDLR exon 3 for 92 (out of 460) SBFH cases; conditions were linear ramp 59°–64°C for 3 h at 50 V/2 A. The 16 tracks with mutant band patterns are ringed. Other tracks show identical wild-type pattern. (B) Examples of band patterns for wild type, artificial +ve control (COANN), and FH mutations in LDLR exon 3.
Table 2.
Summary of meltMADGE findings for LDLR exon 3 in 460 unrelated familial hypercholesterolemia previously scanned by SSCP
| Mutation | Original detect method | No. of subjects | Detection by meltMADGE | Classified band pattern |
|---|---|---|---|---|
| delGT196_197 (fsV45)a | Not detected by SSCP | 1 | Yes | 7 |
| W66G | Detected | 5 | Yes | 7 |
| C68Y | Detected | 3 | Yes | 3 |
| D69G | Detected | 2 | Yes | 4 |
| fsE80 | Detected | 3 | Yes | 6 |
| E80K | Detected | 27 | Yes | 5 |
| C83Fa | Not detected by SSCP | 1 | Yes | 5 |
| 313+1G→A | Detected | 29 | Yes | 2 |
Detected by meltMADGE, followed up by direct sequencing
LDLR exon 3 scanning in cohorts
Approximately 8000 subjects, including SBFH, were scanned for exon 3 of LDLR (Table 3). Three variant band patterns were observed and relevant amplicons were subjected to direct sequencing.
Table 3.
Summary of population findings by meltMADGE and followup sequencing in LDLR exon 3 and 8
| LDLR | Cohort ID | No. of subjects | Nucleotide change | Amino acid change | Reference | Remarks |
|---|---|---|---|---|---|---|
| Exon 3 | BWHHS | 3600 | 291C→T | N76N (silent) | Not previously described | One subject. |
| At CpG site. | ||||||
| Hertfordshire | 2500 | 313+1G→A | Splice site | Hobbs et al. 1992 | Two subjects. | |
| At CpG site. | ||||||
| Known severe FH mutation. | ||||||
| Subject with highest cholesterol level within cohort. | ||||||
| SAS | 1500 | 313C→T | P84S | Vuorio et al. 1997 | One subject. | |
| At CpG site. | ||||||
| P84S is a known mild LDLR mutation. | ||||||
| Exon 8 | BWHHS | 3600 | 1061-8T→C | Intron 7–8 | Jensen et al. 1996 | 35 subjects. |
| 2/35 subjects above 99th percentile for cholesterol level. | ||||||
| 1078G→A | D335N | Not previously described | One subject. | |||
| At CpG site. | ||||||
| Subject above 95th percentile for cholesterol level. | ||||||
| 1186+11G→A | Intron 8+11 | Not previously described | One subject. | |||
| At CpG site. |
313+1G→A
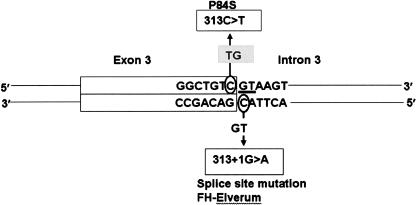
The splice site mutation (313+1G→A) was identified in two subjects (2/7600), both in the Hertfordshire population cohort. The plasma total cholesterol value was 12.6 mmol/L total and LDL-cholesterol 10.1 mmol/L representing the highest value in this cohort. This G→A mutation is at a CpG site bridging the exon 3/intron 3 boundary and represents an antisense strand CpG→TpG mutation (Fig. 4).
Figure 4.
Schematic of exon 3/intron 3 boundary showing severe and moderate mutations, presumed to represent deamination on opposite strands at the same CpG site. Obligate splice-site bases (GT) are underlined. 313C→T changes the first base of codon84 (CCC, proline) to T (TCC, serine).
313C→T [P84S]
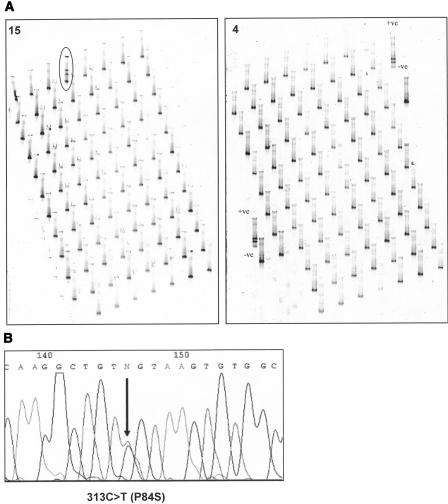
The mutation 313C→T was identified in one subject (1/7600), who was in the SAS cohort. This female subject aged 66 yr, was not taking cholesterol-lowering medication, had a body mass index of 18.9, and displayed a cholesterol level of 7.2 mmol/L. In Figure 5 we show an example of this cohort scan, which embodied 17 arrays scanned in two tank runs (all shown in Supplemental Fig. 2), which identified 16 arrays displaying only the wild-type pattern (such as array 4) and one array (array 15) containing 1/96 tracks with a split band pattern (Fig. 5A); a closeup of this 2.6-cm track (compared with a wild-type track) and subsequent sequencing is shown in Figure 5B. The other 6560 subjects' data (a further 72 arrays) representing the Hertfordshire, BWHHS, and SBFH collections are not shown. This C→T mutation is at a CpG site bridging the exon 3/intron 3 boundary and represents a sense strand CpG→TpG mutation (Fig. 4).
Figure 5.
Overview of a mutation scan by meltMADGE of one cohort (LDLR exon 3, SAS). (A) Annotated enlarged gel images for array 4 and array 15 (artificial +ve control; and control WT labeled). (B) Sense strand sequencing of subject E1 in array 15. Heterozygous mutation 313C→T (P84S) is marked by an arrow.
291C→T [N76N]
One subject (1/7600) was shown to possess 291C→T (N76N), a previously unknown, apparently neutral sequence change at a CpG site. This subject had a plasma cholesterol level of 5.6 mmol/L (18.1 percentile) and LDL-cholesterol of 3.2 mmol/L, which are on middle percentiles.
LDLR exon 8 region: Private variants and paucimorphism
An assay of exon 8 (validated on SBFH in a process the same as for exon 3) was applied to the BWHHS cohort. No variants in exon 8 were identified in the SBFH sample. An infrequent polymorphism (Ala370Thr) is not detected by this assay (see Discussion). Two new variants (each observed in one individual) and one paucimorphism (observed in 35 individuals) were identified. All were characterized by direct sequencing (Table 3).
1186+11G→A (IVS8+11)
One subject (1/3600) displayed 1186+11G→A (intron8+11), a previously unreported CpG site mutation. The subject was female, aged 64 yr, with plasma cholesterol of 5.7 mmol/L (20.0 percentile) and LDL-cholesterol of 3.2 mmol/L.
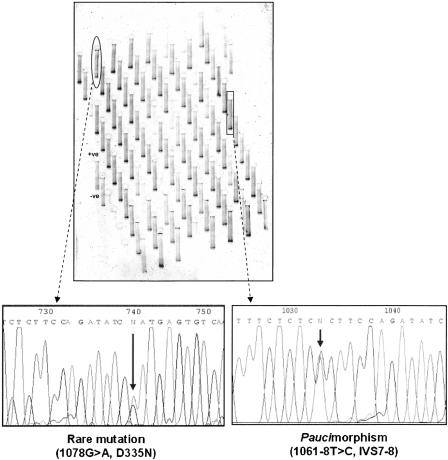
1078G→A [D335N]
One previously unreported CpG mutation (1078G→A, D335N), was found in exon 8 of LDLR (Fig. 6) in a single subject (1/3600) with total cholesterol on the 96.2 percentile (8.8 mmol/L) and also with high LDL-cholesterol (6.4 mmol/L).
Figure 6.
One gel image (out of 40) for meltMADGE scan of LDLR exon 8 (BWHHS cohort). Two variant band patterns were identified (ringed). Direct sequencing, as shown, identified these subjects to be heterozygous, respectively, for 1078G→A [D335N] and 1061-8T→C [IVS7–8].
1061–8T→C (IVS7–8) [T705I]
Thirty-five subjects (35/3600) were detected who were heterozygous for a paucimorphism, at position 1061–8T→C in intron 7 (Fig. 6), which has been reported previously (Jensen et al. 1996). This base change is in the polypyrimidine tract of the intron 7 splice acceptor site. There was no age difference between carriers and noncarriers (0.15 yr; 95% C.I. -1.68, 1.98 yr; p = 0.87). The age-adjusted mean cholesterol level in carriers was not significantly higher than for noncarriers (0.29 mmol/L; 95% C.I. -0.13, 0.70 mmol/L; p = 0.18). In post hoc examination of distribution of carriers' cholesterol levels, two subjects (2/35) displayed cholesterol values above the 99th percentile (99.1 and 99.4 percentiles, respectively) for the cohort (test for two proportions: fWT = 0.01, NWT = 3565, fT705I = 0.057, NT705I = 35, z = -2.7, p = 0.007). Direct sequencing for possible co-occurrence of mutation T705I (Hobbs et al. 1992) in exon 15 showed all 35 subjects to be positive, consistent with very strong linkage disequilibrium between 1061T→C and T705I.
Discussion
The ability to undertake mutation scanning at the level of the “whole” population was enabled by the development and application of meltMADGE methodology. The approach is sensitive to most single base changes and relies on minimal capital, consumable, or personnel expenditure (Supplemental Table 2; Sevilla et al. 2003). An unbiased insight into the prevalence and effects of rarer sequence variation is derived from unselected subjects, contrasting with previous studies focused on mutation scanning in highly selected clinical cases.
Validation of the technique
MeltMADGE assays are capable of detecting most mutations present in an amplicon. A priori, the similarity of the approach to DGGE would predict sensitivity and performance characteristics similar to DGGE. Criteria for successful development of DGGE assays have been described previously (Wu et al. 1998). Most exons are of suitable size and melting characteristics, and it should be possible to adopt established DGGE primer pairs and clamps, requiring only checks of suitable thermal ramp start and finish temperatures to achieve a validated assay. Track location within gels and gel location amongst the 10 gels in a tank did not affect detection of heteroduplexes. In the absence of availability of known natural sequence variants, the generation of an artificial positive control was found to be useful during large-scale running of assays in which most samples are expected to be negative for sequence variants.
For BRCA1, using a set of 54 assays representing the entire coding region, all 10 common polymorphisms, 15/15 mutations previously found by a clinical diagnostics laboratory, and three single base mutations that would not have been identified during diagnostic screening were identified by meltMADGE.
For LDLR exon 3, eight mutations were identified in 71 subjects from a collection of 460 FH cases. SSCP had detected six of these mutations. These mutations involved both transitions, transversions, and small insertion–deletions and were distributed throughout the sequence of exon 3 and its intronic boundaries; thus it is reasonable to assume that most mutations present in any case study or cohort study would be detected. The only exception in these studies was for an infrequent polymorphism (StuI RFLP, Ala370Thr) near the 3′-end of the LDLR exon 8 amplicon used. While this was convenient for our mutation-scanning studies, it indicates that not every heteroduplex will resolve in a meltMADGE assay. We confirmed this polymorphism to be present by restriction digest as expected with a rarer allele frequency of 0.05–0.1. Moving the GC-rich clamp to the 5′-end of the amplicon did not alter this situation. There is a small pocket of significant GC richness centered around this StuI site. Pockets of atypical GC content (high or low) have been noted to be problematic in other techniques such as dHPLC, and homogeneous melting domains (rather than biphasic or graded melting domains) are well known to be favorable for DGGE. For diagnostic applications and where a large number of positive controls exist, revised primer locations can be explored. False-positive tracks have not proven to be a problem. Heteroduplex yield is substantial, and theoretically 25% of total reannealed amplicon should be found in each of the two heteroduplex bands. Although this amount is not always found, true heteroduplexes are prominent compared with the occasional minor background band observed in meltMADGE assays. While the background of the large family of PCR error heteroduplexes is compressed in the short electrophoresis track lengths of MADGE, this faint background smear (if observed) does not obscure the prominent heteroduplexes. In population studies, there are thousands of tracks defining the wild-type track. Reruns for occasional tracks giving uncertainty can be loaded along with usual negative and artificial positive controls in the two control rows of six wells located outside the main 8 × 12-well array in MADGE gels. Lastly, as for many mutation-scanning techniques, presence of SNPs in a region can confound scanning for rare mutations since SNP heteroduplex and rare mutation heteroduplex patterns may look similar. Direct SNP assay or coannealing then meltMADGE of SNP heterozygote and suspected rare mutation heterozygote amplicons, which would form double heteroduplexes (Nissen et al. 1998), represent two possible ways to address this.
Specific comments on sequence variants identified from cohort studies
Mutation 313+1G→A is a classical FH-mutation known as FH-Elverum in Norway (Leren et al. 1994), and its effects on splicing have been characterized in detail (Sun et al. 1995). We observed this mutation only in the Hertfordshire cohort, not in cohorts from other regions. As we have recently observed for chromosome-Y haplogroups (Chen et al. 2004), there might be a distribution reflecting the region of Danelaw, representing Viking influx. However, the mutation has been widely observed in other countries (http://www.ucl.ac.uk/fh). That the mutation conferred the highest cholesterol level in the entire cohort is not surprising.
Mutation 313C→T [P84S] has only been described previously in a mutation scan of 18 Finnish subjects selected for moderate hypercholesteremia (Vuorio et al. 1997). The subject identified in our study also displays moderate hypercholesteremia. P84S may, like 313+1G→A, be prevalent throughout Europe, but its total effect in the population might not be recognized from FH case collections. Familial segregation may be less consistent for moderate phenotypes, since other factors will play a relatively greater role, and intra-individual phenotypic variability may be greater also. This is well exemplified by the APOB mutation R3500Q (familial defective apolipoprotein B, FDB) which also causes a moderate hypercholesteremia (Miserez and Keller 1995). These points will make it more difficult to fully evaluate the effects of potential moderate effect mutations.
313+1G→A and P84S represent mutations at the same CpG site bridging exon 3 and intron 3. Assuming that both of these mutations represent typical CpG mutation by deamidation of methylated C base to T (Cooper and Krawczak 1990), then 313+1G→A represents antisense strand deamination and P84S represents sense strand deamination (Fig. 4). Since our studies are of essentially unselected subjects, our findings of both strand mutations suggest that this particular CpG site might be an extreme mutational hotspot, perhaps influenced by flanking bases (Ollila et al. 1996; Krawczak et al. 1998). It is notable that five out of six mutations identified in unselected population in this study were at CpG sites (313G→A; P84S; N76N; 1186+11G→A; and D335N). In mutation studies of highly selected cases, overrepresentation of CpG mutation is well known, but nevertheless very many mutations and FH mutations are not at CpG sites. Recent estimates based on extensive case data (Kondrashov 2003) have suggested that CpG increases base substitution rate by an order of magnitude. However, since CpG sites are depleted and infrequent in the mammalian genome (Cooper and Krawczak 1989), such a mutation rate could not account for their predominance over any other type of base change, as found in our population study. Case selection may exert strong ascertainment bias toward other non-CpG mutations, or toward particular CpG mutations—either way, highly distorted estimates of mutational rates may be obtained. Therefore, at the population level, a greater degree of disease burden might be attributable to CpG mutation, than has hitherto been suspected.
Mutation 1078G→A [D335N] has not been observed previously. In FH case collections, D335Y (codon GAT to TAT) and D335H (codon GAT to CAT) have both been observed previously at the same CpG site, respectively, leading to A or G tranversions of the presumed methylated C on the antisense strand. D335N represents the typical C-to-T presumed deamination event of CpG mutation, leading to G-to-A transition on the sense strand. The other typical mutation, assuming the full methylation of this CpG site (Reik et al. 2001), would be silent, namely, I334I. Other mutational mechanisms than deamination may determine transversion mutations (Cooper and Krawczak 1990; Yoon et al. 2001; Zhao and Boerwinkle 2002). The observation to date only of transversion at case level and only of transition at population level underscore potential major differences in estimation of mutational events based mainly on selected disease cases.
The cholesterol level for this subject (not on cholesterol-lowering medication) was above the 95th percentile for the BWHHS cohort. This by definition is a significant hypercholesteremia. Family studies would be useful, but the written consent for this cohort (as for many cohort studies) is solely for anonymised study. D335N lies in an epidermal growth factor homology domain, in a Ca2+-binding loop important in pH-dependent ligand release and receptor recycling following internalization. A cluster of amino acids is involved in calcium binding, including D333, E336, and N349, in a loop stabilized by a disulfide bridge between C337 and C447. Multiple sequence alignment of LDLR protein sequence from different species shows that codon 335 is either aspartic acid (D) or asparagine (N). Tolerability of differences between species may not, however, be a good indicator of tolerability of effects within a species (Vitkup et al. 2003), and more subtle effects will be harder to predict. D335N may thus be a key determinant in causing this subject's plasma cholesterol level to be above the 95th percentile, but may not have been observed in severe FH case collections (cholesterol values often far above the 99th percentile such as 313+1G→A described above) because its effects are milder than D335Y and D335H, which are predicted on the basis both of sequence conservation and amino acid change to be more disruptive.
1061–8T→C is a paucimorphism, with allele frequency ∼0.5%. Previous literature (Jensen et al. 1996; Heath et al. 2000; Mozas et al. 2000) had indicated significant linkage disequilibrium with mutation T705I in exon 15. Our direct tests for T705I in subjects positive for 1061–8T→C were all positive, whereas other subjects tested were negative. T705I was initially reported among FH case collections and was designated FH-Paris-9. In this study, the mean cholesterol level for 35 subjects positive for the haplotype bearing both 1061–8T→C and T705I, was 0.3 mmol/L higher than that of the whole cohort, a finding of no clinical or statistical significance. However, we did observe in post hoc examination of distribution of cholesterol levels that 2/35 carriers were above the 99th percentile for cholesterol for this female cohort (p = 0.007). A previous study of T705I in men (Heath et al. 2000) did not identify any mean difference, and reanalysis by centiles showed no significant distributional skewing into upper centiles, although 1/30 carriers was above 99th percentile (3.6%) compared with 29/2243 (1.3%) below 99th perentile. We also noted that the 1061–8T→C variant was approximately twice as common in the FH case collection (9/460) as in the BWHHS cohort (35/3600), but not statistically significant (χ2 = 3.69, p = 0.055). T705I resides in a set of serine and threonine residues in exon 15 that undergo O-linked glycosylation, which seems to protect the cell surface receptor from proteases, thus stabilizing it (Kozarsky et al. 1988). 1061–8T→C, although in the polypyrimidine tract of a splice acceptor site, is not predicted to have a significant effect on splicing. It is possible that either variant (e.g., in the presence of mutation in the other LDLR allele or other disease) might exert conditional effects.
In conclusion, we have developed a technique, meltMADGE, for cost-efficient and high-throughput mutation scanning. We have evaluated its sensitivity to base changes in a wide variety of sequence contexts in BRCA1 and LDLR. In population studies of LDLR, both severe, moderate, and silent variants were identified, at the population level. In conrast with case collections, CpG mutations predominated. MeltMADGE, on account of its high throughput and cost efficiency, will contribute to research of population-based `reference ranges' for rarer sequence variation; characterization of `paucimorphisms'; research of `formes frustes' milder mutaions; and identification of severe mutations at the population level.
Methods
DNA samples for BRCA1 mutation-scanning trials of meltMADGE
For this, 94 anonymized DNA samples from consultands at high risk of breast/ovarian cancer predisposition gene mutations and previously scanned by the Wessex Regional Genetics Laboratory, UK, were assorted into one microplate. Mutation identities for known positive samples (tested by standard single-strand conformation polymorphism [SSCP], heteroduplex analysis [HA], and protein truncation test [PTT] methods) were known only to author J.S. until all analysis had been completed by M.A.A. supervised by D.M.E. and I.N.M.D.
Primer design and PCR for BRCA1 meltMADGE assays and sequencing
Amplicons were designed to give a single flat melting domain, using an MS Office program, Tixis (E. Spanakis and I.N.M. Day, unpubl.) based on melt87 (Lerman and Silverstein 1987) and using GC-rich clamps as described previously for DGGE (Sheffield et al. 1989). PCR primers, optimal Mg and temperature conditions are listed in Supplemental Table 1; 20-μL reactions were as in O'Dell et al. (2000). DNA sequencing followed the manufacturers' instructions using an ABI PRISM 377 DNA sequencer (Perkin Elmer) and BigDye Terminator Cycle Sequencing Ready Reaction Kit (ABI) and used the same primers as for PCR.
Cholesterol characterized case and cohort collections for LDLR mutation scanning
Genomic DNA was extracted from nucleated white blood cells in whole blood as previously described (Miller et al. 1988). The Simon Broome Familial Hypercholesterolemia (SBFH) case collection was from 11 hospital outpatient lipid clinics in the United Kingdom (Neil et al. 2004), all with a diagnosis of definite FH. There were 460 mutation-characterized samples available for meltMADGE assay validation. The Hertfordshire cohorts included 2500 subjects (1390 men and 1110 women) with measured plasma cholesterol (and for some, estimated LDL-cholesterol) (Barker et al. 1992). The Southampton Atherosclerosis Study (SAS) cohort represents 1500 consecutive Caucasian patients undergoing diagnostic coronary angiography in the Wessex Cardiothoracic Unit, Southampton General Hospital, United Kingdom (Ye et al. 2003), with measured plasma cholesterol levels. The British Women's Heart and Health Study (BWHHS) cohort included women aged 60–79 yr recruited from 23 centers in England, Wales, and Scotland, with ∼150–200 from each town (Ebrahim et al. 2004), and plasma cholesterol levels available on all. In all, 3600 DNA samples were available for mutation analysis. In all collections, cholesterol assays were subject to national quality control. Estimated LDL-cholesterol values were by the Friedwald formula and were available in the Hertfordshire and BWHHS cohorts.
PCR of LDLR exons 3 and 8
PCR of LDLR gene exons was performed essentially as described previously (Gaunt et al. 2001). Primers were from MWG-Biotech (http://www.mwgdna.com): LDLR exon 3, 5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGTCGGCCTCAGTGGGTCTTTC-3′ (sense) and 5′-ACTCCCCAGGACTCAGATAGGC-3′ (antisense); exon 8, 5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGTCCCCACCAAGCCTCTTTCTCTC-3′ (sense) and 5′-CCACCACTGCTGCCTGTAAG-3′ (antisense).
MeltMADGE
MADGE formers and glass plates were as previously described (Day and Humphries 1994; Gaunt et al. 2003). Large batches of gels were poured in a purpose-built pouring box (Supplemental Fig. 1). Each 50 mL of gel mixture contained 10 mL of 30% acrylamide-bisacrylamide (19:1), 5 mL of 10× TAE buffer, 6 M urea, and 35 mL of warmed (30°–40°C) dH2O to dissolve the urea. After cooling to room temperature, 100 μL of 20% APS and 100 μL of TEMED were added. Gels were set for at least 40 min, before prising open-faced gels (anchored on one glass plate) from formers. Approximately 2 μL of PCR product were loaded from microplates by passive transfer using a 96-slot pin replicator (Supplemental Fig. 1). Each gel was covered by sliding a clean (untreated) glass plate over it; 2–3 mL of buffer (or water) were dropped onto the edges of the gel in order to facilitate sliding of the cover and to eliminate formation of air bubbles in wells. The assembly was secured with two stationery rubber bands (Supplemental Fig. 1). The long edge of the gel was sealed with silicon rubber tubing stretched and inserted between the glass plates in order to prevent electrophoretic edge artifacts (Supplemental Fig. 1).
Prototype melt-MADGE electrophoresis tanks were 23 cm long (anode to cathode), 11 cm wide, and 15 cm high (Supplemental Fig. 1Ea) and contained two platinum electrodes, a motorized propeller stirrer, a glass serpent, and a removable gel rack (Supplemental Fig. 1). The tank was made of 0.5-cm-thick polypropylene. The electrodes were connected through the cover of the tank to a commercial 200 V, 2 A power supply (Supplemental Fig. 1Eb); spatial thermal homogeneity was achieved by vigorous stirring. The glass serpent was connected to a programmable heating–cooling circulator (Supplemental Fig. 1Ed), and a digital thermometer (Supplemental Fig. 1Ec) regularly calibrated and certified to UK national standards was used to monitor the temperature. Electrophoresis was for 2–3 h at 50 V and 2 A with a linear ramp temperature from 59°C to 64°C (LDLR exon 3) or from 60°Cto65°C(LDLR exon 8), regularly calibrated to national standards.
Gels were stacked separated by spacers for staining in 100 mL of 1× TAE buffer with 10 μL of Vistra Green (Molecular Probes) on a shaker at minimum speed for 15 min, and visualized using a Fluorimager 595 (Molecular Dynamics, Amersham Pharmacia Biotech) as described (Gaunt et al. 2003). Ethidium bromide and a UV transilluminator can also be used.
Generation of artificial positive controls for LDLR amplicons
An artificial positive control was generated by synthesis of amplicon using a primer (5′-ACTCCCCAGGACTCAGACAGGC-3′ for exon 3 and 5′-CCACCACTGCTGCCTGCAAG-3′ for exon 8) with a one-base chemical mutation (at position -4 from the 3′-end) and coannealing with a similar quantity of “wild-type” (WT) amplicon (synthesized using primers perfectly matched to the genomic template). An equal volume of the mutant PCR product (designated “MUT”) was mixed with WT amplicon, and the mixture of PCR products (designated “MIX”) was coannealed (designated “COANN” or “+ve control”) to generate heteroduplexes. This heteroduplex generator step was also carried out for all test samples in the cohort studies. The steps were 95°C for 3 min and then 40°C for 5 min.
Direct sequencing of LDLR amplicons
Big Dye Terminator cycle sequencing was applied to meltMADGE amplicons displaying variant patterns, and products resolved by ABI PRISM 377 DNA sequencer (Applied Biosystems; www.appliedbiosystem.com). Residual PCR product from meltMADGE assays was used as template, sequencing primers were 5′-GCCTCAGTGGGTCTTTCCTT-3′ sense and 5′-CCAGGACTCAGATAGGCTCAA-3′ antisense for exon 3, and 5′-TCCCCACCAAGCCTCTTTCTCTC-3′ sense and 5′-CCACCACTGCTGCCTGTAAG-3′ antisense for exon 8. Sequencing of exon 15 was undertaken for samples positive for 1061–8T→C in exon 8 since it was suspected (Jensen et al. 1996; Heath et al. 2000; Mozas et al. 2000) that mutation T705I and i7–8 would co-occur. Residual genomic DNA in the respective meltMADGE exon 8 PCR products was used as template and primers both for exon 15 PCR and subsequent sequencing were 5′-AGGCGCACACCTATGAGAAG-3′ (sense) and 5′-GTGAGGACGACACCTGGACT-3′ (antisense).
Acknowledgments
K.K.A. was the recipient of a PhD Scholarship from King Saud University, Saudi Arabia. M.A.A. was the recipient of a PhD scholarship from the Ministry of Higher Education, Saudi Arabia. The project was also supported by the Breast Cancer Campaign, UK and the UK Department of Health (National Genetics Reference Laboratory). E.S. was a Wessex Medical Trust Senior Fellow. L.H. was the recipient of a British Heart Foundation PhD Studentship. I.N.M.D. was a Lister Institute Professor. Support for meltMADGE development was from the UK Medical Research Council and from the Department of Health–UK National Genetics Reference Laboratory (Wessex) (I.N.M.D./X.C.). The cohort collections and DNA banks were supported by the British Heart Foundation and the UK Medical Research Council. S.E.H. and R.A.W. are supported by the British Heart Foundation (PG 2000/15) and the Simon Broome DNA collection by RG3008. D.A.L. is funded by a (UK) Department of Health Career Scientist Award. BWHHS is funded by the (UK) Department of Health. The views expressed in this paper are those of the authors and not necessarily those of any funding body. Santiago Rodriguez is thanked for helpful comments on the manuscript.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3313405.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Barker, D.J., Meade, T.W., Fall, C.H., Lee, A., Osmond, C., Phipps, K., and Stirling, Y. 1992. Relation of fetal and infant growth to plasma fibrinogen and factor VII concentrations in adult life. BMJ 304: 148-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betteridge, D.J., Broome, K., Durrington, P.N., Hawkins, M.M., Humphries, S.E., Mann, J.I., Miller, J.P., Neil, H.A.W., Thompson, G.R., and Thorogood, M. 1999. Mortality in treated heterozygous familial hypercholestrolaemia: Implications for clinical management. Atherosclerosis 142: 105112 (Abstr.). [Google Scholar]
- Brown, M.S. and Goldstein, J.L. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science 232: 34-47. [DOI] [PubMed] [Google Scholar]
- Chen, X.-H., Rodriguez, S., Hawe, E., Talmud, P.J., Miller, G.J., Underhill, P., Humphries, S.E., and Day, I.N. 2004. Evidence of admixture from haplotyping in an epidemiological study of UK Caucasian males: Implications for association analyses. Hum. Hered. 57: 142-155. [DOI] [PubMed] [Google Scholar]
- Cooper, D.N. and Krawczak, M. 1989. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum. Genet. 83: 181-188. [DOI] [PubMed] [Google Scholar]
- ———. 1990. The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum. Genet. 85: 55-74. [DOI] [PubMed] [Google Scholar]
- Cotton, R.G. 1998. Mutation detection and mutation databases. Clin. Chem. Lab Med. 36: 519-522. [DOI] [PubMed] [Google Scholar]
- Day, I.N. and Humphries, S.E. 1994. Electrophoresis for genotyping: Microtiter array diagonal gel electrophoresis on horizontal polyacrylamide gels, hydrolink, or agarose. Anal. Biochem. 222: 389-395. [DOI] [PubMed] [Google Scholar]
- Day, I.N., Whittall, R.A., O'Dell, S.D., Haddad, L., Bolla, M.K., Gudnason, V., and Humphries, S.E. 1997. Spectrum of LDL receptor gene mutations in heterozygous familial hypercholesterolemia. Hum. Mutat. 10: 116-127. [DOI] [PubMed] [Google Scholar]
- Day, I.N., Alharbi, K.K., Smith, M., Aldahmesh, M.A., Chen, X.-H., Lotery, A.J., Pante-de-sousa, G., Hou, G., Ye, S., Eccles, D., et al. 2004. Paucimorphic alleles versus polymorphic alleles and rare mutations in disease causation: Theory, observation and detection. Curr. Genomics 5: 431-438. [Google Scholar]
- Ebrahim, S., Montaner, D., and Lawlor, D.A. 2004. Clustering of risk factors and social class in childhood and adulthood in British women's heart and health study: Cross sectional analysis. BMJ 328: 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, S.G. and Lerman, L.S. 1979. Length-independent separation of DNA restriction fragments in two-dimensional gel electrophoresis. Cell 16: 191-200. [DOI] [PubMed] [Google Scholar]
- Fleming, M.A., Potter, J.D., Ramirez, C.J., Ostrander, G.K., and Ostrander, E.A. 2003. Understanding missense mutations in the BRCA1 gene: An evolutionary approach. Proc. Natl. Acad. Sci. 100: 1151-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt, T.R., Cooper, J.A., Miller, G.J., Day, I.N., and O'Dell, S.D. 2001. Positive associations between single nucleotide polymorphisms in the IGF2 gene region and body mass index in adult males. Hum. Mol. Genet. 10: 1491-1501. [DOI] [PubMed] [Google Scholar]
- Gaunt, T.R., Hinks, L.J., Rassoulian, H., and Day, I.N. 2003. Manual 768 or 384 well microplate gel `dry' electrophoresis for PCR checking and SNP genotyping. Nucleic Acids Res. 31: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, J.L., Hobbs, H.H., and Brown, M.S. 1995. The metabolic bases of inherited disease. In The metabolic bases of inherited disease (eds. C.R. Scriver et al.), pp. 1981-2030. McGraw Hill, New York.
- Heath, K.E., Whittal, R.A., Miller, G.J., and Humphries, S. 2000. I705 variant in the low density lipoprotein receptor gene has no effect on plasma cholesterol levels. J. Med. Genet. 37: 713-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs, H.H., Brown, M.S., and Goldstein, J.L. 1992. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum. Mutat. 1: 445-466. [DOI] [PubMed] [Google Scholar]
- Jensen, H.K., Jensen, L.G., Hansen, P.S., Faergeman, O., and Gregersen, N. 1996. High sensitivity of the single-strand conformation polymorphism method for detecting sequence variations in the low-density lipoprotein receptor gene validated by DNA sequencing. Clin. Chem. 42: 1140-1146. [PubMed] [Google Scholar]
- Kondrashov, A.S. 2003. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum. Mutat. 21: 12-27. [DOI] [PubMed] [Google Scholar]
- Kozarsky, K., Kingsley, D., and Krieger, M. 1988. Use of a mutant cell line to study the kinetics and function of O-linked glycosylation of low density lipoprotein receptors. Proc. Natl. Acad. Sci. 85: 4335-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak, M., Ball, E.V., and Cooper, D.N. 1998. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet. 63: 474-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leren, T.P., Solberg, K., Rodningen, O.K., Tonstad, S., and Ose, L. 1994. Two founder mutations in the LDL receptor gene in Norwegian familial hypercholesterolemia subjects. Atherosclerosis 111: 175-182. [DOI] [PubMed] [Google Scholar]
- Lerman, L.S. and Silverstein, K. 1987. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 155: 482-501. [DOI] [PubMed] [Google Scholar]
- Miller, S.A., Dykes, D.D., and Polesky, H.F. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miserez, A.R. and Keller, U. 1995. Differences in the phenotypic characteristics of subjects with familial defective apolipoprotein B-100 and familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 15: 1719-1729. [DOI] [PubMed] [Google Scholar]
- Moorjani, S., Roy, M., Torres, A., Betard, C., Gagne, C., Lambert, M., Brun, D., Davignon, J., and Lupien, P. 1993. Mutations of low-density-lipoprotein-receptor gene, variation in plasma cholesterol, and expression of coronary heart disease in homozygous familial hypercholesterolaemia. Lancet 341: 1303-1306. [DOI] [PubMed] [Google Scholar]
- Mozas, P., Cenarro, A., Civeira, F., Castillo, S., Ros, E., and Pocovi, M. 2000. Mutation analysis in 36 unrelated Spanish subjects with familial hypercholesterolemia: Identification of 3 novel mutations in the LDL receptor gene. Hum. Mutat. 15: 483-484. [DOI] [PubMed] [Google Scholar]
- Neil, H.A., Seagroatt, V., Betteridge, D.J., Cooper, M.P., Durrington, P.N., Miller, J.P., Seed, M., Naoumova, R.P., Thompson, G.R., Huxley, R., et al. 2004. Established and emerging coronary risk factors in patients with heterozygous familial hypercholesterolaemia. Heart 90: 1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, H., Day, L.B., Horder, M., Humphries, S.E., and Day, I.N. 1998. Denaturing gradient gel electrophoretic analysis of codons 3456–3553 of the apolipoprotein-B gene in 106 type 11a hyperlipoproteinaemic individuals. Ann. Clin. Biochem. 35: 137-139. [DOI] [PubMed] [Google Scholar]
- O'Dell, S.D., Chen, X., and Day, I.N. 2000. Higher resolution microplate array diagonal gel electrophoresis: Application to a multiallelic minisatellite. Hum. Mutat. 15: 565-576. [DOI] [PubMed] [Google Scholar]
- Ollila, J., Lappalainen, I., and Vihinen, M. 1996. Sequence specificity in CpG mutation hotspots. FEBS Lett. 396: 119-122. [DOI] [PubMed] [Google Scholar]
- Reik, W., Dean, W., and Walter, J. 2001. Epigenetic reprogramming in mammalian development. Science 293: 1089-1093. [DOI] [PubMed] [Google Scholar]
- Sevilla, C., Julian-Reynier, C., Eisinger, F., Stoppa-Lyonnet, D., Bressac-de Paillerets, B., Sobol, H., and Moatti, J.P. 2003. Impact of gene patents on the cost-effective delivery of care: The case of BRCA1 genetic testing. Int. J. Technol. Assess. Health Care 19: 287-300. [DOI] [PubMed] [Google Scholar]
- Sheffield, V.C., Cox, D.R., Lerman, L.S., and Myers, R.M. 1989. Attachment of a 40-base-pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. 86: 232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof, T.C., Goldstein, J.L., Brown, M.S., and Russell, D.W. 1985. The LDL receptor gene: A mosaic of exons shared with different proteins. Science 228: 815-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X.M., Patel, D.D., Bhatnagar, D., Knight, B.L., and Soutar, A.K. 1995. Characterization of a splice-site mutation in the gene for the LDL receptor associated with an unpredictably severe clinical phenotype in English patients with heterozygous FH. Arterioscler. Thromb. Vasc. Biol. 15: 219-227. [DOI] [PubMed] [Google Scholar]
- Vitkup, D., Sander, C., and Church, G.M. 2003. The amino-acid mutational spectrum of human genetic disease. Genome Biol. 4: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorio, A.F., Turtola, H., and Kontula, K. 1997. A novel point mutation (Pro84→Ser) of the low density lipoprotein receptor gene in a family with moderate hypercholesterolemia. Clin. Genet. 51: 191-195. [DOI] [PubMed] [Google Scholar]
- Whittall, R., Gudnason, V., Weavind, G.P., Day, L.B., Humphries, S.E., and Day, I.N. 1995. Utilities for high throughput use of the single strand conformational polymorphism method: Screening of 791 patients with familial hypercholesterolaemia for mutations in exon 3 of the low density lipoprotein receptor gene. J. Med. Genet. 32: 509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Hayes, V.M., Osinga, J., Mulder, I.M., Looman, M.W., Buys, C.H., and Hofstra, R.M. 1998. Improvement of fragment and primer selection for mutation detection by denaturing gradient gel electrophoresis. Nucleic Acids Res. 26: 5432-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, S., Dunleavey, L., Bannister, W., Day, L.B., Tapper, W., Collins, A.R., Day, I.N., and Simpson, I. 2003. Independent effects of the -219 G>T and ε2/ε3/ε4 polymorphisms in the apolipoprotein E gene on coronary artery disease: The Southampton atherosclerosis study. Eur. J. Hum. Genet. 11: 437-443. [DOI] [PubMed] [Google Scholar]
- Yoon, J.H., Smith, L.E., Feng, Z., Tang, M., Lee, C.S., and Pfeifer, G.P. 2001. Methylated CpG dinucleotides are the preferential targets for G-to-T transversion mutations induced by benzo[a]pyrene diol epoxide in mammalian cells: Similarities with the p53 mutation spectrum in smoking-associated lung cancers. Cancer Res. 61: 7110-7117. [PubMed] [Google Scholar]
- Zhao, Z. and Boerwinkle, E. 2002. Neighboring-nucleotide effects on single nucleotide polymorphisms: A study of 2.6 million polymorphisms across the human genome. Genome Res. 12: 1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.appliedbiosystem.com; Applied Biosystems.
- http://www.mwgdna.com; MWG-Biotech.
- http://www.ucl.ac.uk/fh; familial hypercholesteremia LDLR mutation data.