Abstract
Recently, we mapped the sites of transcription across ∼30% of the human genome and elucidated the structures of several hundred novel transcripts. In this report, we describe a novel combination of techniques including the rapid amplification of cDNA ends (RACE) and tiling array technologies that was used to further characterize transcripts in the human transcriptome. This technical approach allows for several important pieces of information to be gathered about each array-detected transcribed region, including strand of origin, start and termination positions, and the exonic structures of spliced and unspliced coding and noncoding RNAs. In this report, the structures of transcripts from 14 transcribed loci, representing both known genes and unannotated transcripts taken from the several hundred randomly selected unannotated transcripts described in our previous work are represented as examples of the complex organization of the human transcriptome. As a consequence of this complexity, it is not unusual that a single base pair can be part of an intricate network of multiple isoforms of overlapping sense and antisense transcripts, the majority of which are unannotated. Some of these transcripts follow the canonical splicing rules, whereas others combine the exons of different genes or represent other types of noncanonical transcripts. These results have important implications concerning the correlation of genotypes to phenotypes, the regulation of complex interlaced transcriptional patterns, and the definition of a gene.
A significant challenge to a functional understanding of the human genome is obtaining a detailed knowledge of its transcriptional output. Ideally, a catalog of transcriptional activities in a genome should include all isoforms of coding and noncoding transcripts that are present in all tissue and cell types. Two large-scale efforts, sponsored independently by the National Cancer Institute (Strausberg et al. 1999, 2000, 2002) and RIKEN (Okazaki et al. 2002; Ota et al. 2004), have significantly contributed to the current understanding of the complexity of the human transcriptome. Several recent reports have also provided substantial evidence that the transcriptional output of the human genome is far more complex than can be explained by current collections of partial or full-length cDNAs (Chen et al. 2002; Kapranov et al. 2002; Okazaki et al. 2002; Saha et al. 2002; Rinn et al. 2003; Kampa et al. 2004; Ota et al. 2004; Cheng et al. 2005). The majority of recently detected regions of transcriptional activity in the human genome lies outside of the annotated areas and may, therefore, represent novel transcriptional units or hitherto undiscovered isoforms of known genes.
Recently, we described the sites of transcription at a 5-bp resolution for 10 human chromosomes (30% of the nonrepeat portion of the human genome) (Cheng et al. 2005). As part of this study a total of 768 randomly selected unannotated regions of transcription were studied using a combination of RACE and high-density arrays to validate the presence of transcription occurring at the selected sites and to better understand the structures of the unannotated transcripts. A total of 634 of the 768 loci (82.6%) yielded a set of 5′- and/or 3′-RACE products, and ∼61% of surveyed loci show evidence of overlapping transcription on the positive and negative strands of the genome. RT-PCR reactions were conducted on 250 (57%) of the genomic loci that produced 5′- and 3′-RACE products from at least one genomic strand. In this report, we explore in depth the complex pattern and transcript structures observed in this genome survey. Using the combination of RACE and high-density arrays, transcript structures corresponding to unannotated array-detected regions mapping within as well as outside of the bounds of well-characterized coding genes were studied in depth. Examples of complex overlapping sense/antisense transcription within the bounds of known genes emerging from these studies are presented and discussed. The complexity of the organization and structure of the RNAs detected are consistent with the existence of a complex transcriptome whose organization and transcript structure have potentially important implications about the regulation of transcription and the possible interpretation of the naturally occurring genetic variation in humans.
Methods
Cell lines and nucleic acid material
Human cell lines Jurkat (ATCC no. TIB-152), HepG2 (ATCC no. HTB-8065), FHs 738Lu (ATCC no. HTB-157), and U87MG (ATCC no. HTB-14) were grown to confluency in either RPMI (Jurkat) or DMEM (HepG2, FHs 738Lu, or U87 MG) supplemented with 10% FBS. Total cytosolic RNA and its poly(A)+ fraction were isolated sequentially using RNeasy and Oligotex kits (Qiagen) following the manufacturer's instructions. Cytosolic poly(A)+ RNA treated with RNAse-free DNAse I (Roche) as described previously (Cawley et al. 2004) was used for all RACE and RT-PCR assays described in this report. RACE and RT-PCR assays were conducted either on pools of all four cell lines or on individual cell lines, as indicated (Supplemental Table S1). Common RACE primers (3′-CDS, UPS, UPL, and NUP) were synthesized by Qiagen and HPLC-purified. Gene-specific RACE and RT-PCR primers were synthesized by Integrated DNA Technologies either individually or in a 96-well format using the standard desalting step. All cDNA synthesis steps, as well as RACE and RT-PCR amplifications, were done in PE 9600 PCR thermocyclers. Unless strand information was available, that is, in the case of exons of known genes, 3′- and 5′-RACE assays were performed on both genomic strands. In other words, for each novel array-detected region, two separate (forward and reverse) 3′-RACE as well as two separate (forward and reverse) 5′-RACE assays were conducted. To ensure that RACE/array profiles were not contaminated by residual genomic DNA in the cases where novel transcripts showed no evidence of splicing, control RACE/array profiles were performed in which the reverse transcription step was omitted.
High-density arrays
The following tiling arrays were used in this study: DGCR array with probe spacing every 1 bp (Fig. 2 below; Supplemental Fig. S1A; Kapranov et al. 2002); ENCODE arrays with spacing every 22 bp (Fig. 6 below); or tiling arrays spanning 10 human chromosomes with probe spacing every 5 bp (Figs. 3, 4, 5, 7 below; Supplemental Figs. S1B-D, S2, and S3).
Figure 2.
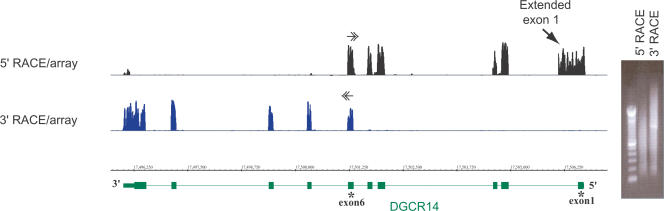
RACE/array profiling of the DGCR14 gene, located on the bottom strand of Chromosome 22. 5′- and 3′-RACE profiling with RACE primers selected at exon-exon junctions of exons 6/7 and 5/6, correspondingly. In this and the following figures, positions and directions (5′-3′) of the RACE primers are indicated by horizontal double arrows, and annotations are represented by UCSC Known Genes or RefSeqs (http://www.genome.ucsc.edu) (green). In addition, RACE/array data are always represented as graphs of signal intensities for every probe on an array. The gel image of electrophoretic profiles of the RACE reactions prior to array-hybridizations are also shown. Profiles of known exons (bottom) can be clearly discerned from the RACE/array maps.
Figure 6.
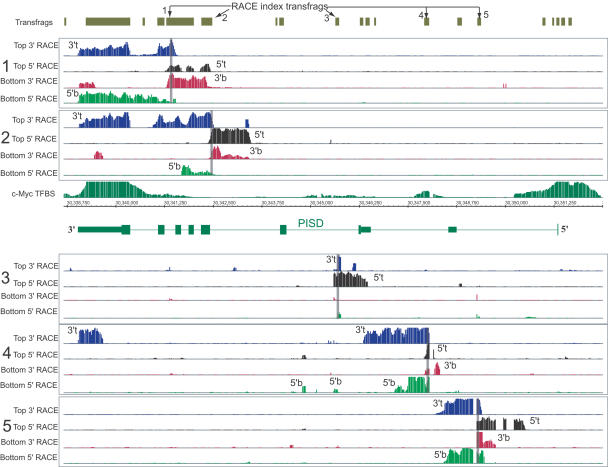
Overlapping sense/antisense transcripts in the PISD locus. RACE/array profiling of five novel transfrags found within an isoform of the PISD gene (accession no. BC001482), encoding phosphatidylserine decarboxylase, reveals a complex pattern of sense/antisense transcription. For each index transfrag (panels 1-5 and indicated by arrows), the presence of a transcript on either the bottom-sense strand or top-antisense strand, with respect to PISD, was interrogated using two 5′-RACE/array and two 3′-RACE/array assays with RACE primers designed to query for transcription on either strand. The position of the index transfrag is also indicated by the opaque vertical lines in the corresponding group of four RACE maps (5′/3′-RACE on the top and 5′/3′-RACE on the bottom strand). Approximate positions of the 5′- and 3′-ends of the transcripts on the top or bottom strand are indicated as “5′t,” “3′t,” “5′b,” and “3′b.” ChIP-CHIP profiles with anti-cMyc anti-body compared to input control DNA are also shown (Cawley et al. 2004). Two prominent sites at both 5′- and 3′-ends of the PISD gene, together with sites of lower affinity, can be seen.
Figure 3.
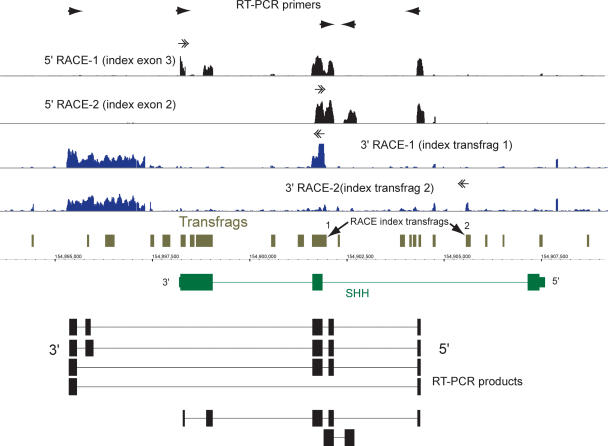
Several novel exons can be found in the gene SHH, which encodes a human homolog of Sonic hedgehog. The 5′-RACE/array profiling was performed with primers designed in exons 2 and 3 of the annotated form. The 3′-RACE/array profiling was performed with primers positioned in two unannotated transfrags in intron 1 (indicated by the arrows). A significant amount of signal can be seen in intron 1 of the gene, representing alternative exons of this gene. The annotated form of exon 3 appears to be represented as two different exons. An alternative 3′-exon was detected downstream of the annotated gene bounds. The maps of cloned RT-PCR products representing different alternative isoforms of transcripts containing novel exons are shown at the bottom.
Figure 4.
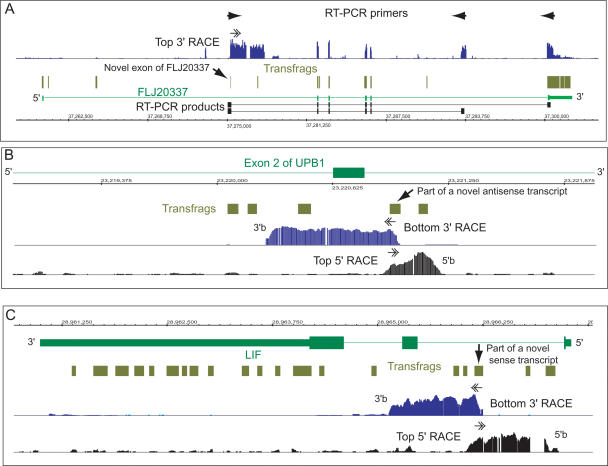
Different classes of novel intronic transfrags revealed by RACE/array. (A) A novel transfrag (shown by an arrow) within RIKEN cDNA FLJ20337 (top strand, Chromosome 6) represents a novel exon of this gene, as evidenced by the 3′-RACE/array profile and sequences of two different RT-PCR products. (B) Several novel intronic transfrags represent a bottom-strand transcript antisense to the UPB1 gene (top strand, Chromosome 22), as evidenced by 5′- and 3′-RACE/array analyses. (C) Intronic transfrags within the LIF gene (bottom strand, Chromosome 22) represent internal sense transcripts as evidenced by 5′- and 3′-RACE/array analysis. Approximate positions of the 5′- and 3′-ends of the transcripts on the bottom strand in panels B and C are indicated as “5′b” and “3′b.”
Figure 5.
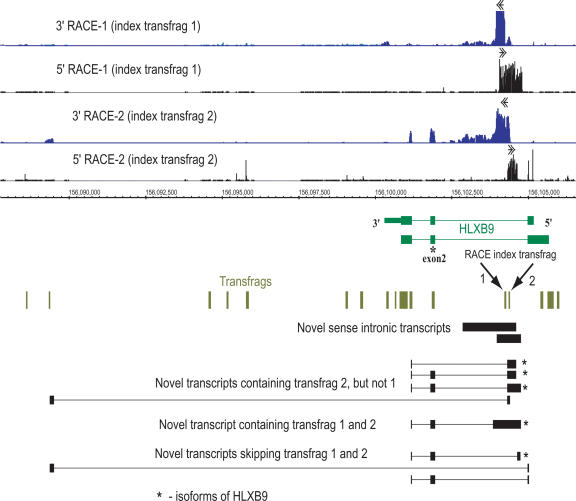
Overlapping sense transcripts in the HLXB9 locus. RACE/array analysis of two non-exonic transfrags (transfrags 1 and 2, depicted by arrows) of the HLXB9 gene, encoding a homeobox protein, is shown. Both 5′- and 3′-RACE/array profiles are shown for each of the two transfrags as 5′/3′-RACE-1 and 5′/3′-RACE-2, correspondingly. Both transfrags are connected together in a novel sense transcript located within intron 1 of HLXB9 on the bottom strand of Chromosome 7. Transfrag 2, however, is also represented as an alternative exon in several novel HXLB9 transcripts. This transfrag is also connected to a distant downstream region located ∼16 kb away. The maps of cloned RT-PCR products representing different alternative isoforms of transcripts containing novel exons are shown at the bottom.
Figure 7.
RACE/array transcript profiling in a novel locus on Chromosome 6, Chr6-74. The position of the index transfrag for RACE/array profiling is shown by an arrow on the track of the array-detected transfrags in this region. The presence of a transcript on either the top or bottom strand was interrogated using two 5′-RACE/array and two 3′-RACE/array assays with RACE primers designed to query for transcription on either strand. For the gene on the top strand, two 5′-RACE primers, separated by 428 bp, were used and are represented by 5′-RACE-1 and 5′-RACE-2 tracks. Transfrags not connected by RACE/array indicate the presence of additional transcripts in this locus.
3′-RACE/array
In the RACE reactions, thermostable reverse transcriptases (RTs) such as ThermoScript (Invitrogen) and C. therm polymerases (Roche) were able to identify the structure of novel RNA transcripts in a more consistent fashion than enzymes that elongate at 42°C (data not shown). This could be explained by the extensive secondary RNA structures of some transcripts that might interfere with cDNA synthesis. The advantages of using alternative RTs have been noted previously (Hawkins et al. 2003). Therefore, the cDNA templates for both 5′- and 3′-RACE assays were routinely prepared using two or more different RTs and then pooled prior to amplification with gene-specific primers.
3′-RACE reactions were performed on a common pool of oligo(dT) cDNA templates using a modified version of the SMART II protocol (BD Biosciences). First-strand cDNA was synthesized using two separate reactions with two different RTs, Superscript II (Invitrogen) and Thermoscript (Invitrogen). In each case, 5 μg of RNA was combined with 50 pmol of 3′-CDS primer [5′-AAGCAGTGGTATCAACGCAGAGTAC(T)30VN-3′], heated to 70°C for 2 min and cooled to 4°C for 2 min, at which point the respective reaction buffers, DTT, dNTPs, and RTs were added to the respective concentrations of 1×, 10 mM, 0.5 mM, and 75 U/reaction (ThermoScript) or 1000 U/reaction (Superscript II) in the final volume of 50 μL. After addition of these reagents, the reactions were incubated for an additional 3 min at 4°C and then rapidly ramped to 42°C (SuperScript II) or slowly ramped (30 min duration) to 60°C (ThermoScript). The cDNA synthesis was allowed to proceed for 90 min, after which SuperScript II was inactivated by incubation at 75°C for 15 min, while ThermoScript reactions were purified directly without enzyme inactivation.
The cDNA synthesis reactions were purified using a Qiagen PCR purification kit. SuperScript II and ThermoScript cDNA synthesis reactions were pooled and used directly for RACE PCR amplification reactions. Each reaction was done on cDNA corresponding to 100 ng of the starting poly(A)+ RNA. Two rounds of nested PCR were used, resulting in a total of four PCR reactions/region. PCR amplification was conducted using the Advantage II PCR enzyme system (BD Biosciences) under the following conditions. The first round of amplification was done in 1× vendor's reaction buffer, 0.2 mM dNTPs, 1 μL Advantage II polymerase mix, 0.3 μM of transfrag-specific primer, 0.04 μM UPL primer (5′-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′), and 0.2 μM UPS primer (5′-CTAATACGACTCACTATAGGGC-3′). The second round of amplification was conducted on 1 μL of a 1:100 dilution of the products from the first round of PCR under the same conditions, with the exception that the UPS/UPL pair of primers was substituted with 0.2 μM NUP primer (5′-AAGCAGTGGTATCAACGCAGAGT-3′). All amplifications were done in 50 μL under the following conditions: 40 cycles of denaturation at 94°C for 5 sec, annealing at 64°C for 10 sec, and extension at 72°C for 10 min. Amplification reactions with each transfrag-specific primer were done separately. PCR amplicons were purified using a PCR purification kit (Qiagen). Amplification products were pooled, fragmented with DNase I, end-labeled with biotin, and hybridized to whole-genome tiled arrays as described previously (Kapranov et al. 2002).
5′-RACE/array
The cDNA templates for 5′-RACE were synthesized using gene-specific primers. Given the relatively small average size (<100 bp) of array-detected transcribed fragments (transfrags), two such primers, forward and reverse, were selected from within 50 bp of the ends of the transfrags. The cDNA synthesis reactions were performed with individual gene-specific primers or pools of up to 12 gene-specific primers in the same reaction volume. The procedure described below is for a pool of eight gene-specific primers, but it can be used for 1-12 gene-specific primers under the same reaction conditions. cDNA synthesis was performed in a 96-well format with a pool of eight primers in three separate reactions using SuperScript II (Invitrogen), ThermoScript (Invitrogen), and Transcriptor (Roche) reverse transcriptases. For each reaction, 1 μg of DNAse I treated cytosolic poly(A)+ RNA was mixed with 15 pmol of each of the eight primers, heated to 70°C for 2 min, cooled to 4°C for 2 min, at which point a corresponding vendor's reaction buffer, DTT (SuperScript II and ThermoScript only), dNTPs, and corresponding reverse transcriptase (200 U of Superscript II [Invitrogen], 15 U of Thermoscript [Invitrogen], or 10 U of Transcriptor [Roche]) were added to the final concentrations of 1×, 10 mM, and 0.5 mM (1 mM for Transcriptor RT) in the final reaction volume of 20 μL. The reactions were incubated at 4°C for an additional 3 min, after which they were rapidly ramped to 42°C (SuperScript II) or 55°C (Transcriptor) or slowly ramped (30 min) to 60°C (ThermoScript). cDNA synthesis was allowed to proceed for 90 min, after which reactions were heat-inactivated (SuperScript II reactions for 15 min at 75°C; Transcriptor reactions for 5 min at 85°C), pooled, and purified using the QIAquick 96 well system (Qiagen).
The purified first-strand cDNA template was heated to 94°C for 2 min and chilled on ice, at which point the tailing reaction buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl at pH 8.3), 0.2 mM dATP, and 200 U of recombinant terminal transferase (Roche) were added. The tailing reaction was carried out for 30 min at 37°C in a total volume of 25-75 μL. The reaction was terminated by incubation at 70°C for 10 min. An aliquot of the tailing reaction, corresponding to 100 ng of starting poly(A)+ material, was used for 5′-RACE PCR without any further purification. 5′-RACE amplification was done with the same primers and amplification conditions as for the 3′-RACE, with the exception that in the first round of PCR, a 3′-CDS oligonucleotide was used as the common 5′-primer at a concentration of 0.2 μM, and in the second round, the common 5′-primer was represented by UPL (0.04 μM) and UPS (0.2 μM). PCR reactions were then purified using the QIAquick 96 well system (Qiagen), pooled, fragmented with DNase I, end-labeled with biotin, and hybridized to the arrays, as in the 3′-RACE procedure.
Cloning and sequencing of cDNAs corresponding to RACE/array products
RT-PCR primers were designed based on the results of the RACE/array analysis. RT-PCR amplification was done using the same amount of oligo(dT)-primed template as for the 3′-RACE, with the exception that amplification was done with two gene-specific primers. Two rounds of nested RT-PCR with 30-40 cycles each were commonly used with the Advantage II enzyme system and the same PCR conditions as for the 5′/3′-RACE (see above). The RT-PCR products were gel-purified and cloned into pGEM-T Easy vectors (Promega) and sequenced. All sequence coordinates mentioned in the text are based on the April 2003 release NCBIv33. Sequences of RT-PCR products reported in this manuscript were deposited in GenBank under accession nos. AY927416-AY927467.
Analysis of RACE/array results with genomic annotations
After hybridization of RACE reactions to tiling arrays, RACE/array maps were generated using methodologies previously described (Kampa et al. 2004). Positive probes on RACE/array maps were grouped together into RACE transfrags using appropriate positive probe thresholds, maxgap and minrun parameters (Kampa et al. 2004). RACE/array transfrag maps were related to the following annotations from the UCSC Genome Browser database (Kent et al. 2002; Karolchik et al. 2003): Class 1, “Known,” which is a combination of RefSeq and UCSC Known Genes exons; Class 2, “mRNA,” which is mRNAs from GenBank that do not overlap Class 1; and Class 3, “EST,” which represents all publicly available ESTs that are not represented in Classes 1 or 2. Visualization of data collected by RACE/array was carried out using a freely available integrated genome browser (IGB; http://www.affymetrix.com/support/developer/downloads/TilingArrayTools/index.affx). The RACE/array data reported in this manuscript can be downloaded at http://transcriptome.affymetrix.com/publication/race/. All RACE/array maps and annotations are based on the April 2003 release NCBIv33.
Results
Application of RACE/array to unravel the complexities of individual loci
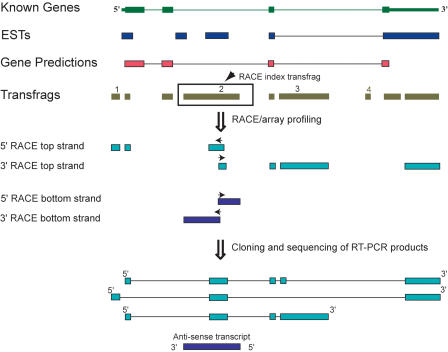
A RACE/array methodology was developed to assist in filling the gap between identifying transfrags in the genome using array-based technology and elucidating structures of the corresponding transcripts by using RACE. Five crucial pieces of information are derived from using a RACE/array approach, namely, (1) the strand of origin of the transcript, (2) array-detected exons that are connected to the index exon used as a template for the RACE reaction, (3) maximal number of exons that comprise a full-length transcript, (4) estimated maximal lengths and genomic positions of each of the exons, and (5) the estimated maximal lengths of the transcript and the extent of the genome that is covered by the RACE-associated exons. As shown below, RACE/array profile often represents a summary profile of several transcripts sharing an index transfrag. Below, we discuss how these properties of RACE/array can be used to characterize simultaneously the transcripts emanating from multiple genomic loci, either defined by the bounds of an annotated gene or by transfrags lying outside known annotations.
A hypothetical example outlining the utility of RACE/array in untangling the transcriptional complexity of a genomic locus is shown in Figure 1. In this figure and in all subsequent figures, the data derived from array profiling of cellular RNA are represented by a map of array-detected transfrags, typically shown in relation to the annotations available for a given genomic locus. The transfrag data shown on the subsequent figures are derived from cytosolic poly(A)+ RNA from the HepG2 cell line (Cheng et al. 2005). Since data derived from tiling arrays represent a sum of all transcripts made within the bounds of the locus, neighboring transfrags can be part of the same or different transcripts (Fig. 1). Deciphering which transfrags are connected together in mature transcripts often requires additional experimental information. Thus, to explore the transcriptional output of any given locus, some or all transfrags will be selected as index regions for RACE/array analysis. In the example shown in Figure 1, RACE/array transcript profiling from unannotated array-detected transfrags (transfrag 2) reveals the presence of one or more transcripts derived from both strands of the genome. The precise transcript structures are further elucidated using RT-PCR and cloning/sequencing efforts, which are guided by the available RACE/array map data. Some of the transcripts represented by the transfrags are alternative isoforms of the known gene, while others represent antisense transcripts. The hypothetical example shown in Figure 1 also illustrates an important point, namely, that RACE/array analysis starting with a single index transfrag often does not connect all of the transfrags in a locus. In Figure 1, RACE/array profiling connects novel transfrags 1, 2, and 3 together, while transfrag 4 is predicted to be part of a different, as-yet-unannotated transcript. This outcome was frequently observed and is illustrated by all our subsequent results.
Figure 1.
A schematic representation of the transcriptome data and RACE/array workflow. An example of a locus defined by bounds of a known gene is shown together with additional existing genome annotations, such as ESTs and GenScan predictions. Exons are represented by boxes and introns by lines. Throughout all of the figures, transcriptome data derived from array profiling of cellular RNA are represented by transcribed fragments (transfrags; olive green).
Given that all cDNA species in a RACE reaction are tailed with the same sequences on either 5′- or 3′-ends, specific amplification of the intended target can only be attempted by using one gene-specific primer. This can result in a complex outcome as often manifested by a very complicated electrophoretic pattern or smear of RACE-generated PCR products, making isolation of a specific cDNA a lengthy and complicated cloning and sequencing process. This problem becomes even more significant in the case of low-abundance transcripts, which is not easily circumvented by using nested primers (data not shown).
While the nonspecific primer extension and subsequent amplification reactions can significantly plague the direct cloning of RACE products, this is usually not a significant problem if the RACE reaction is analyzed using the tiling-arrays-based hybridization. This is illustrated in Figure 2 for the DGCR14 gene, where the primers flanking exon 6 were used in 5′- and 3′-RACE reactions. In this case, both the 3′- and 5′-RACE reactions produce a mixture of amplified products, with perhaps a single band in the 3′-RACE (Fig. 2). However, hybridization of the RACE products to a tiling array reveals the clear exon-intron structure of the locus (Fig. 2).
Detection of unannotated isoforms of well-characterized genes
Approximately 45% of all unannotated transfrags could be detected within boundaries of known genes, suggesting that the presence of unannotated isoforms of known genes might be a common phenomenon (Kampa et al. 2004; Cheng et al. 2005). In order to test this assumption, seven known protein-coding genes (DGCR14, CTCLC1, EP300, GTSE1, SHH, SEC14L2, and EPI64) were analyzed by the RACE/array approach, by using one or two annotated exons as the index points for 5′- and 3′-RACE (Figs. 2, 3; Supplemental Fig. S1A-D). With the exception of SHH, the known exon-intron structures were recapitulated for the remaining six genes (Fig. 2; Supplemental Fig. S1A-D). However, the RACE/array hybridization patterns also indicated the presence of one or more of the following characteristics: (1) novel exons, (2) extended isoforms of annotated exons, or (3) novel transcript isoforms extending beyond the annotated gene bounds. Overall, six genes (DGCR14, CTCLC1, EP300, GTSE1, SEC14L2, and SHH) showed examples of characteristics described in either points 1 and/or 2. In addition, genes GTSE1, SEC14L2, and EPI64 also showed evidence of additional transcripts extending beyond the annotated gene boundaries. These data are consistent with the high degree of alternative splicing that was observed in other studies (Rinn et al. 2003; Kampa et al. 2004).
The percentage of unannotated transcription as compared to the total transcription detected by RACE/array varied from 11.2% to 48.7% for the seven genes tested (average of 25%) (Supplemental Table S1). Overall, a total of ∼38.9 kb of DNA represented in cytosolic polyadenylated transcripts was detected by RACE/array for all seven genes combined, of which ∼9.8 kb (25%) did not overlap any annotated exons, including ESTs. These regions of transcription could either represent exons of unannotated transcripts, which overlap the coding genes, or novel exonic isoforms of the genes.
A large extent of unannotated transcriptionally active sequence coverage was quite surprising, given that very few RACE primers were used in these assays (only one or two exons from each gene were used as the index points for the RACE analyses). Different primers used for different index exons within the same gene produced overlapping, yet slightly different RACE/array patterns. This result suggests that each gene is likely to be represented by a mixture of transcripts (Figs. 2 and 3; see also Supplemental Fig. S1). Therefore, a large number of additional isoforms of a gene is likely to be uncovered by a systematic application of the RACE/array approach, using primers for every exon of a given gene.
This is exemplified by the Sonic hedgehog gene (SHH) (Marigo et al. 1995), one of the three human homologs of the Drosophila Hedgehog protein, which represents a striking example of a complex transcriptional activity. Array-based mapping data revealed the presence of multiple unannotated transfrags in this locus, suggesting the presence of several unannotated transcripts (Fig. 3). To investigate this assumption further, 5′- and 3′-RACE/array experiments were initiated from the previously annotated exons as well as from intronic transfrags derived from the array-based mapping. The 5′-RACE/array profile with two primers positioned in either exons 2 or 3, identified several novel exons of SHH, compared to the annotated form of this gene (accession no. L38518), as well as additional isoforms of all three known exons (5′-RACE-1 and 5′-RACE-2) (Fig. 3). On the other hand, 3′-RACE/array analysis, initiated from two unannotated intronic transfrags, revealed a novel 3′-UTR of this gene (3′-RACE-1 and 3′-RACE-2) (Fig. 3). The six isoforms shown in Figure 3 represent the longest cloned and sequenced RT-PCR products that correspond to the 5′- and 3′-RACE/array maps. Most of these novel isoforms contain exon 2, which includes approximately half of the highly conserved N-terminal HH signaling domain of the Hedgehog family of proteins. This domain is reported to be released via auto-proteolysis of the full-length Hedgehog protein by the protease domain encoded in the C-terminal part of the protein, and then modified by addition of lipophilic moieties (Bijlsma et al. 2004). However, the novel SHH isoforms presented here appear to lack most or the entire C-terminal protease domain encoded by exon 3. Thus, the novel SHH isoforms combine the exon 2 portion of the N-terminal domain, with different amino acid sequences that do not share similarities to the HH domain or any other known proteins. The predicted proteins encoded by these novel cDNAs range from 123 to at least 171 amino acids in length (as compared with the 462 amino acids of the annotated form of SHH), and hence might represent additional forms of SHH protein with potentially unknown biological functions. However, whether any of the novel isoforms of the known genes described above, including SHH, represent functionally important entities in a cell requires additional experimentation.
Detection and characterization of unannotated transcripts
As mentioned above, intronic transfrags represent a significant portion of all unannotated transfrags. When used as index points for RACE/array analyses, such transfrags were typically found to represent a variety of different and often overlapping transcripts that can be generally characterized as (1) unannotated exons of annotated genes or length variants of annotated exons or (2) unidentified transcriptional structures overlapping, on the same or opposite strand, the annotated gene. Four sets of examples are presented below to help illustrate the organization and structures of such complex transcripts.
Unannotated sense and antisense transcripts
An unannotated transfrag in the FLJ20337 locus represents a novel exon of this gene (Fig. 4A). An example of a transcript antisense to exon 2 of the UPB1 gene on Chromosome 22, encoding an enzyme β-ureidopropionase, is shown in Figure 4B and an unannotated sense transcript in the LIF gene, encoding leukemia inhibitory factor, is shown in Figure 4C. Thus, even when transcription is detected only on one strand, a transfrag could be part of multiple distinct transcripts.
Mixtures of overlapping sense transcripts located at the same genomic loci
RACE/array analysis starting with a single index transfrag often does not connect all of the transfrags in a locus (Figs. 4A-C). This is consistent with the presence of additional transcripts, which do not contain the index region. Therefore, a comprehensive characterization of any locus requires selecting all transfrags for RACE/array analysis. This point is illustrated below in the examples of two loci, HLXB9 and PISD, where all unannotated transfrags were used as index transfrags for RACE/array analysis.
The HLXB9 locus, encoding a homeobox transcription factor (Ross et al. 1998), represents a seemingly simple case, where two intronic transfrags located at genomic positions 156104223-156104282 and 156104370-156104419 of Chromosome 7 do not overlap with the annotated exons of the gene in the HepG2 cell line (Fig. 5). Although appearing straightforward, the two transfrags represent a glimpse of a complex web of transcripts emanating from this locus. While closely spaced in the genome (87 bp apart), they are parts of the same, as well as different, transcripts (Fig. 5). 5′- and 3′-RACE-1 based on transfrag 1 revealed the presence of an unannotated group of sense transcripts confined entirely within the first intron of the HLXB9 gene and overlapping transfrag 2 (Fig. 5). However, while the 5′- and 3′-RACE-2 based on transfrag 2 detected the same group of sense transcripts as 5′- and 3′-RACE-1, it could also connect transfrag 2 to all three exons of HLXB9. This indicates that transfrag 2 also represents an alternative exon of the HLXB9 (Fig. 5). In fact, cloning and sequencing of some RT-PCR products (Fig. 5) confirmed the presence of cDNA species where an unannotated exon of HLXB9 contained transfrag 1, but not transfrag 2 (Fig. 5; Supplemental Fig. S2).
HLXB9 transcript isoforms containing this novel exon also include an alternative, truncated form of 3′-UTR (Fig. 5). An un-anticipated transcript structure was also revealed in the 3′-RACE-2 reaction, where the unannotated exon represented by transfrag 2 is connected to another unannotated transfrag ∼16 kb downstream and thus located outside of the HLXB9 gene boundary in a structure that is otherwise unrelated to the known form of the HLXB9 gene (Fig. 5).
RACE performed with one gene-specific primer is likely to identify the most abundant isoforms present in a sample, while PCR with two gene-specific primers would also identify minor isoforms as long as these transcripts contain sites where both gene-specific primers could anneal. In addition to the transcripts whose structures were directly suggested by RACE reactions, we extended our RT-PCR analysis to design RT-PCR primers based on either 3′-RACE of transfrag 1 and 5′-RACE of transfrag 2 and vice versa. As a result, we could identify additional RT-PCR products where now both index transfrags were present in unannotated exons of HLXB9 gene (Fig. 5).
It is important to point out that sometimes RT-PCR products with primers designed on the very 5′- and 3′-ends of the RACE maps do not contain regions that overlap with the original index transfrag. This is illustrated in the bottom group of RT-PCR products in Figure 5. This most likely reflects the presence of multiple transcripts in the same cell line. Depending on their relative abundance, sequence composition, and length, various isoforms are preferentially detected either in RACE/array or RT-PCR experiments.
Sequence analysis of the novel isoforms of HLXB9 retaining the annotated exon 2 (Fig. 5) revealed two potential major open reading frames: one encoding a novel protein of 119 amino acids with no significant similarity to any known protein and another encoding a novel protein retaining the HLXB9 homeodomain and thus potentially functioning as a transcription factor (data not shown).
Mixtures of overlapping sense and antisense genic transcripts
The gene encoding enzyme phosphatidylserine decarboxylase (PISD) represents a much more complicated case that illustrates a significant number of sense-antisense transcripts being produced from a single locus (Fig. 6). The gene itself is annotated with four known isoforms, of which we interrogated one (accession no. BC001482). Of the 12 non-exonic transfrags located within introns of this particular isoform, six do not align with any known annotations including ESTs (Fig. 6). RACE/array profiling of five of the six such transfrags revealed that they are part of at least nine overlapping sense/antisense transcripts (five sense, four antisense). No signal was detected in the minus RT step controls (data not shown).
Interestingly, previous analysis of the regulatory regions of this gene using ChIP-CHIP (Cawley et al. 2004) identified two prominent c-Myc transcription-factor-binding sites at both the 5′- and 3′-ends of PISD (Fig. 6). The occurrence of a c-Myc binding site at the 3′-end is consistent with the presence of a regulatory region, possibly directing the production of the antisense transcripts that overlap the 3′-UTR of the gene. In addition, two more c-Myc-binding sites of a lower frequency of occupancy were detected within the gene boundaries consistent with the observed internal transcriptional activity (Fig. 6). While this locus appears to be a striking example of sense-antisense transcription, it may represent a fairly common phenomenon. Overall, based on the analysis of a representative subset of 768 randomly selected regions within the 10 analyzed human chromosomes, we found that 60% of all novel transfrags represent transcripts from opposite strands in the genome and that 50% of intronic transfrags represent antisense transcripts (Cheng et al. 2005). In these analyses, we also observed a significant base pair overlap between the transcripts found on both strands.
Mixtures of intergenic transcription
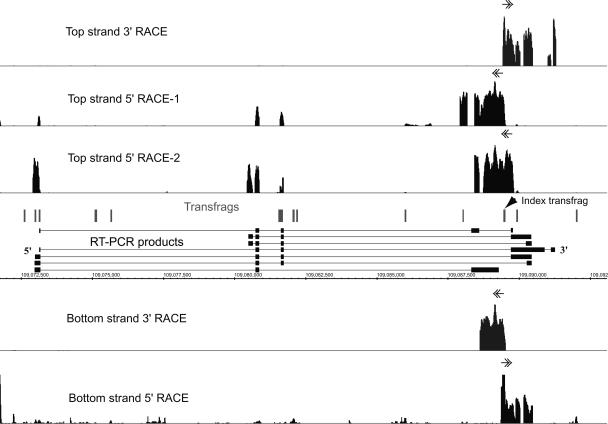
More then half of the array-detected transcription in human cells is found in the intergenic regions (Kampa et al. 2004; Cheng et al. 2005). Transcriptional output of such intergenic regions can also be complex, as illustrated by a multitude of transcripts detected for a novel locus Chr6-74 on Chromosome 6 (Fig. 7). Characterization of this region started with RACE/array analysis of an unannotated transfrag found in the HepG2 cell line and located on Chromosome 6 (position 109089460-109089531). The RACE/array maps derived from this index transfrag revealed the presence of at least one unannotated spliced transcript group on the top strand whose 5′- and 3′-boundaries are at coordinates 109072940-109091320 (Top strand 3′-RACE, Top strand 5′-RACE-1) (Fig. 7). However, similar to the case of the HLXB9 gene, an overlapping and different transcript could be observed using a different set of 5′-RACE primers located within the 3′-RACE extension, but separated from the original 5′-RACE primers by 428 bp (Top strand 5′-RACE-2) (Fig. 7). Since the second transcript is composed of a different set of exons of this gene, it is likely that these exons are not connected to the novel index transfrag at 109089460-109089531. Overall, cloning and sequencing of the RT-PCR cloned products derived from the RACE maps revealed at least seven isoforms of this novel gene (Fig. 7). In addition to the top-strand transcript, an overlapping unspliced bottom-strand transcript was detected from base pairs 109088679 to 109090492 (Bottom strand 3′/5′-RACE) (Fig. 7).
Identification and characterization of noncanonical transcripts
In addition to identifying unannotated transcripts, RACE/array also provided evidence for existence of various types of transcripts that appear not to follow canonical splicing rules. Below, we discuss examples of two such classes of RNA molecules: (1) transcripts containing exons of different genes and (2) spliced antisense transcripts with exon structures closely mirroring exon-intron structures of the sense transcripts.
As mentioned above, RACE/array profiling of three annotated genes (Supplemental Figs. S3A and S1C,D) revealed the presence of transcripts extending beyond the annotated gene bounds. As shown in Supplemental Figure S3A, RACE/array profiling using exon 3 of EPI64 as the index region revealed the presence of a group of alternatively spliced transcripts where exons 3-8 and, in some transcripts, exon 9 (accession nos. AF331308 and AK074656), appear to be connected to exons of another downstream gene of unknown function (accession nos. BC021927 and AK055587). Similar types of transcription events are also observed for SEC14L2 and GTSE1 genes (Supplemental Fig. S1C,D). In all such cases, the presence of such “chimeric” transcripts was further characterized by cloning and sequencing of RT-PCR products. It is possible that such transcripts were generated via transcriptional read-through followed by canonical cis-splicing. Whether these specific transcripts are translated or used for possible regulatory functions, this finding illustrates a potential mechanism that can increase the complexity of the human proteome.
Another type of noncanonically processed transcripts was observed as sense-antisense transcript pairs with very similar exon-intron junctions. An example is shown in Supplemental Figure S3B, where a RACE/array analysis initiated from an unannotated transfrag adjacent to exon 2 of gene C20orf31, encoding putative α-mannosidase, identified an antisense top-strand transcript with a very similar RACE/array pattern to the spliced annotated form C20orf31 (Supplemental Fig. S3B). The RACE/array profile of the bottom-strand transcript identified yet another transcript on the same strand as gene C20orf31, but different from the sense spliced form of this gene. This gene is unique in the genome, as indicated by the BLAT analysis of its cDNA. Thus, it is unlikely that the antisense transcript comes from a processed pseudogene elsewhere in the genome.
Discussion
The data presented here continue to support the concept that the transcriptional complexity of the human genome is significantly underestimated. This appears to be the case even with respect to both well-characterized regions, such as annotated protein-coding genes as well as unannotated transcribed regions. We identified additional isoforms for all seven well-characterized protein-coding genes. In addition, our results paint a picture of a highly overlapping, complex, and dynamic nature of the human transcriptome, where one base pair can be part of many transcripts emanating from both strands of the genome. The data further suggest that base pairs normally thought to contribute to transcripts from different genes can be joined together in a single RNA molecule.
The biological importance of the unannotated transcripts described in this report and in Cheng et al. (2005) is as yet unknown. It is possible that some or most of these unannotated transcripts are part of the biological noise that is likely to be the consequence of complex cellular expression programs and as such may be functionally unimportant. Alternatively, these noncoding and noncanonical transcripts might not themselves be functional, but instead reflect transcriptional processes that function to increase accessibility of various regions of each chromosome to the regulatory machinery, thereby facilitating transcription of the coding genes. It is also possible that the many complex transcripts observed are by-products of the ongoing splicing processing within cells. This line of reasoning is especially attractive given several factors: (1) the low abundance and cell-type specificity of many of these unannotated transcripts, (2) the noncanonical structures observed for some of the forms of these transcripts, (3) lack of prior experimental evidence that points to the biological need for such a collection of unannotated transcripts, and (4) lack of evolutionary conservation for a majority of the unannotated regions, as reported previously (Cawley et al. 2004).
While straightforward and appealing, such reasoning essentially favors a less complex model of the genome as embodied by the current set of genomic annotations and relegates a large body of complicated array-based data presented in this work to the category of nonfunctional biological noise. Faced with this possibility, it might be useful to point out the difference between biologically essential versus influential processes. The complex transcripts that we report here may underlie processes that have more subtle effects at a cellular level (e.g., cell cycle or growth rate changes) that could serve to modulate a cell's response to its environment. The transcriptional elements that are part of these controls might not be easy to identify since classic mutational analyses of such transcripts are likely to yield subtle phenotypes. Thus, such transcriptional elements might possess properties unlike those observed in well-characterized coding transcripts. It is conceivable that such subtle phenotypes could be modulated by cell-type-specific and/or low abundant transcripts. This might also be a reason why such transcriptional products as reported here have not been observed previously in the large data sets that underlie collections of our current annotations. Although Nobrega et al. (2004) did not observe phenotypic effect of the deletion of two large regions (∼1.5 and 0.8 Mb) in the mouse genome, subtle biological effect cannot be excluded.
It is thus important to explore the possible functional roles that some of these unannotated transcripts might fulfill. One attractive idea that may prove fruitful to investigate is to better understand the expression and role of such transcripts of unknown function (TUFs) during early developmental stages.
Acknowledgments
The authors thank Dione Bailey for helpful discussions; Bradley Bernstein, John Manak, Krzysztof Szczyglowski, and Katie Kong for helpful comments and edits on the manuscript; Jean Stevens for administrative support; and the entire Affymetrix Transcriptome Group for their encouragement and support. This work has been funded in part with Federal Funds from the National Cancer Institute, National Institutes of Health, under Contract No. N01-CO-12400, and by Affymetrix, Inc.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3455305.
Footnotes
[Supplemental material is available online at www.genome.org. Sequences of RT-PCR products reported in this manuscript were deposited in GenBank under accession nos. AY927416-AY927467.]
References
- Bijlsma, M.F., Spek, C.A., and Peppelenbosch, M.P. 2004. Hedgehog: An unusual signal transducer. Bioessays 26: 387-394. [DOI] [PubMed] [Google Scholar]
- Cawley, S., Bekiranov, S., Ng, H.H., Kapranov, P., Sekinger, E.A., Kampa, D., Piccolboni, A., Sementchenko, V., Cheng, J., Williams, A.J., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499-509. [DOI] [PubMed] [Google Scholar]
- Chen, J., Sun, M., Lee, S., Zhou, G., Rowley, J.D., and Wang, S.M. 2002. Identifying novel transcripts and novel genes in the human genome by using novel SAGE tags. Proc. Natl. Acad. Sci. 99: 12257-12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., Long, J., Stern, D., Tammana, H., Helt, G., et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science http://www.sciencemag.org/cgi/content/abstract/1108625v2. [DOI] [PubMed]
- Hawkins, P.R., Jin, P., and Fu, G.K. 2003. Full-length cDNA synthesis for long-distance RT-PCR of large mRNA transcripts. Biotechniques 34: 768-770, 772-773. [DOI] [PubMed] [Google Scholar]
- Kampa, D., Cheng, J., Kapranov, P., Yamanaka, M., Brubaker, S., Cawley, S., Drenkow, J., Piccolboni, A., Bekiranov, S., Helt, G., et al. 2004. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 14: 331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916-919. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo, V., Roberts, D.J., Lee, S.M., Tsukurov, O., Levi, T., Gastier, J.M., Epstein, D.J., Gilbert, D.J., Copeland, N.G., Seidman, C.E., et al. 1995. Cloning, expression, and chromosomal location of SHH and IHH: Two human homologues of the Drosophila segment polarity gene hedgehog. Genomics 28: 44-51. [DOI] [PubMed] [Google Scholar]
- Nobrega, M.A., Zhu, Y., Plajzer-Frick, I., Afzal, V., and Rubin, E.M. 2004. Megabase deletions of gene deserts result in viable mice. Nature 431: 988-993. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., Wakamatsu, A., Hayashi, K., Sato, H., Nagai, K., et al. 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36: 40-45. [DOI] [PubMed] [Google Scholar]
- Rinn, J.L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N.M., Hartman, S., Harrison, P.M., Nelson, F.K., Miller, P., Gerstein, M., et al. 2003. The transcriptional activity of human Chromosome 22. Genes & Dev. 17: 529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, A.J., Ruiz-Perez, V., Wang, Y., Hagan, D.M., Scherer, S., Lynch, S.A., Lindsay, S., Custard, E., Belloni, E., Wilson, D.I., et al. 1998. A homeobox gene, HLXB9, is the major locus for dominantly inherited sacral agenesis. Nat. Genet. 20: 358-361. [DOI] [PubMed] [Google Scholar]
- Saha, S., Sparks, A.B., Rago, C., Akmaev, V., Wang, C.J., Vogelstein, B., Kinzler, K.W., and Velculescu, V.E. 2002. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20: 508-512. [DOI] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Klausner, R.D., and Collins, F.S. 1999. The mammalian gene collection. Science 286: 455-457. [DOI] [PubMed] [Google Scholar]
- Strausberg, R.L., Buetow, K.H., Emmert-Buck, M.R., and Klausner, R.D. 2000. The cancer genome anatomy project: Building an annotated gene index. Trends Genet. 16: 103-106. [DOI] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Grouse, L.H., Derge, J.G., Klausner, R.D., Collins, F.S., Wagner, L., Shenmen, C.M., Schuler, G.D., Altschul, S.F., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 99: 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://transcriptome.affymetrix.com/publication/race/; RACE/array data of this paper.
- http://www.affymetrix.com/support/developer/downloads/TilingArrayTools/index.affx; Integrated Genome Browser.
- http://www.genome.ucsc.edu; UCSC Genome Browser.