Abstract
Objective
We sought to measure the deformation of tibiofemoral cartilage immediately following a 3-mile treadmill run, as well as the recovery of cartilage thickness the following day. To enable these measurements, we developed and validated deep learning models to automate tibiofemoral cartilage and bone segmentation from double-echo steady-state magnetic resonance imaging (MRI) scans.
Design
Eight asymptomatic male participants arrived at 7 a.m., rested supine for 45 min, underwent pre-exercise MRI, ran 3 miles on a treadmill, and finally underwent post-exercise MRI. To assess whether cartilage recovered to its baseline thickness, participants returned the following morning at 7 a.m., rested supine for 45 min, and underwent a final MRI session. These images were used to generate 3D models of the tibia, femur, and cartilage surfaces at each time point. Site-specific tibial and femoral cartilage thicknesses were measured from each 3D model. To aid in these measurements, deep learning segmentation models were developed.
Results
All trained deep learning models demonstrated repeatability within 0.03 mm or approximately 1 % of cartilage thickness. The 3-mile run induced mean compressive strains of 5.4 % (95 % CI = 4.1 to 6.7) and 2.3 % (95 % CI = 0.6 to 4.0) for the tibial and femoral cartilage, respectively. Furthermore, both tibial and femoral cartilage thicknesses returned to within 1 % of baseline thickness the following day.
Conclusions
The 3-mile treadmill run induced a significant decrease in both tibial and femoral cartilage thickness; however, this was largely ameliorated the following morning.
Keywords: Auto-segmentation, Cartilage deformation, Cartilage thickness, Magnetic resonance imaging, UNet
1. Introduction
Advances in medical imaging techniques have enabled in vivo measurement of tissue biomechanical function and properties [[1], [2], [3]]. These insights are particularly critical to developing a greater understanding of joint health [4]. For example, there is significant interest in determining how activities such as running impact the function of articular cartilage [5,6]. To probe this question, magnetic resonance imaging (MRI)-based studies have investigated how acute bouts of running induce cartilage strain (i.e. change in cartilage thickness normalized to baseline thickness) within the tibiofemoral joint [[7], [8], [9]]. However, there is a paucity of data describing whether cartilage thickness recovers after running at time scales relevant to habitual loading, such as daily exercise [7]. Investigating cartilage thickness over time may provide insights into the influence of running on cartilage health.
In vivo measurement of articular cartilage thickness and strain often requires MRI-derived 3D models of the cartilage and bone. While some prior studies have utilized manual segmentation to create these models, this process can be time consuming [10,11]. In recent years, deep learning has demonstrated promise in automating some previously manual medical imaging segmentation tasks. One particularly successful deep learning segmentation architecture is the UNet, which several groups have successfully implemented to automate the segmentation of bone, cartilage, ligament, and muscle from MRI scans [[12], [13], [14], [15], [16], [17], [18], [19]]. Inspired by these prior works, we sought to develop deep learning models to automate the segmentation of the tibiofemoral bone and cartilage for our dataset of double-echo steady-state (DESS) MRI scans [7], in order to aid in the efficient measurement of cartilage thickness and strain.
Thus, the objectives of this work were to measure changes in cartilage thickness before and after a 3-mile treadmill run and to assess whether cartilage thickness recovered the following day. This was enabled via the development of deep learning models to automate segmentation of the tibiofemoral bone and cartilage from DESS knee MRI scans. Using our trained deep learning models to expedite segmentation, we hypothesized that both tibial and femoral cartilage would experience significant decreases in cartilage thickness following the 3-mile run and that these deformations would largely recover the next day.
2. Methods
2.1. Study design
Following institutional review board approval, eight asymptomatic males (age: 27–40 years; BMI: 18–25 kg/m2) were recruited to participate in this study [7,20]. The study design consisted of an MRI-based protocol that began with participants arriving at approximately 7 a.m. and resting supine for 45 min to allow for consistent cartilage unloading before undergoing a pre-exercise DESS MRI scan [7,20]. All DESS MRI scans were collected on the same 3.0 T scanner (TrioTim; Siemens Healthcare; Malvern, Pennsylvania) with identical imaging parameters (field of view = 16 cm × 16 cm; image resolution = 0.3 × 0.3 × 1.0 mm; flip angle = 25°; repetition time = 17 ms; echo time = 6 ms). Following the first scan, participants performed a 3-mile treadmill run, followed immediately by a post-exercise DESS MRI scan. Participants returned the following morning at approximately 7 a.m., rested supine for 45 min, and underwent a recovery DESS MRI scan. This scan occurred an average of 22 h and 46 min (standard deviation = 5 min) after the end of the treadmill run the previous morning.
2.2. Segmentation model development & testing
In order to analyze the aforementioned data, we first sought to develop deep learning models to aid in the segmentation of the tibiofemoral bone and cartilage. All data utilized for model development were obtained from 6 previously published IRB-approved studies [[21], [22], [23], [24], [25], [26]]. Data were aggregated from 2 of these studies to train the tibia and femur bone models, and from all 6 of these studies to train the tibial and femoral cartilage models. Table 1 presents the participant demographics for each aggregated dataset. The primary differences between the bone and cartilage datasets were that the cartilage datasets included data from more participants and participants with a more diverse injury status. Participants were not included if they exhibited high-grade chondral defects or full thickness cartilage loss. The inclusion of additional participants for the cartilage models was driven by the need to have more training data due to the increased variability of cartilage morphology compared to bone.
Table 1.
Population variables used for model development.
For each dataset involved in model development, all participants completed two DESS knee MRI scans, all of which were acquired using the same MRI scanner and imaging parameters described in the study design. MRI scans were previously segmented in prior works using commercially available solid modeling software Rhinoceros 4.0 (Robert McNeel and Associates; Seattle, Washington) by 6 researchers (E.C.O., M.R.W., C.S.P., B.S.C., N.K.L., E.G.S.) who were blinded to the order of scan acquisition when applicable. Each researcher received the same formalized task-specific training using a standardized training dataset, following standard guidelines, and was required to pass quality control testing administered by an expert radiologist prior to performing the segmentation task. The reviewing radiologist was fellowship-trained, currently with over 35 years of musculoskeletal (MSK) imaging experience and particular expertise in MRI and MSK biomechanics (C.E.S.). Subsequently, the same radiologist reviewed all segmentations of the study with the researcher. Furthermore, in each study, select data was resegmented to demonstrate reproducibility. Using custom MATLAB (Mathworks; Natick, Massachusetts) software, segmentations were converted into binary masks and automatically cropped from 512 × 512 pixels to 256 × 256 pixels surrounding the tissue of interest to aid in computational efficiency. All data were then randomly split into sets for training (cartilage: 100 scans/12,000 images; bone: 32 scans/3840 images), validation (cartilage: 22 scans/2640 images; bone: 6 scans/720 images), and testing (cartilage: 22 scans/2640 images; bone: 4 scans/480 images).
Both 2D- and 3D-UNet models were trained for each segmentation task in order to determine the optimal model architecture. All models were coded in Python 3.9 with Tensorflow 2.5 and Keras 2.5. Model training was performed using the Duke Compute Cluster, which features a variety of available GPUs. To optimize hyperparameter selection, a gridsearch for each model was performed over the following parameters: batch size, filter size, kernel size, and learning rate. Additionally, the 3D-UNet models included patch volume and stride length as parameters in the gridsearch with the following options implemented using the python module patchify: [volume: 256 × 256 × 112 pixels, stride: not applicable], [volume: 256 × 256 × 32 pixels, stride: 16], [volume: 256 × 256 × 16 pixels, stride: 8], [volume: 112 × 112 × 112 pixels, stride: 72], and [volume: 64 × 64 × 64 pixels, stride: 48]. Model depth was held constant at 5, and 2D and 3D models were trained for 500 and 2000 epochs, respectively, using early stopping with a patience set to 100 epochs. The Dice Similarity Coefficient (DSC) loss function was used for all models, which is equal to 1 – DSC and is a measure of the overlap between the manually segmented and model predicted masks [27]. Optimal models for each of the four tissues were determined by the model that achieved the highest validation set DSC during training. Subsequently, each of the four optimal models was applied to their corresponding testing set, and a new DSC was calculated.
Following model development and testing, the next objective was to evaluate the repeatability of using the trained bone segmentation models to create 3D models of the tibia and femur. To accomplish this, we utilized a separate dataset not involved in model development, consisting of seven new participants (sex: 3F/4M, age: 22–39 years, BMI: 19–28 kg/m2) with no history of knee injury or surgery. For each participant, two MRI scans were acquired using the same imaging parameters as the data utilized for model development. Assuming that the tibia and femur bones should not deform significantly between the repeated scans, the objective was to use the trained bone segmentation models to create 3D models of the tibia and femur and measure bone surface distances between the repeated scans. These distances represent differences in how the trained segmentation models predicted the scans and can be used as a measure of scan-to-scan repeatability.
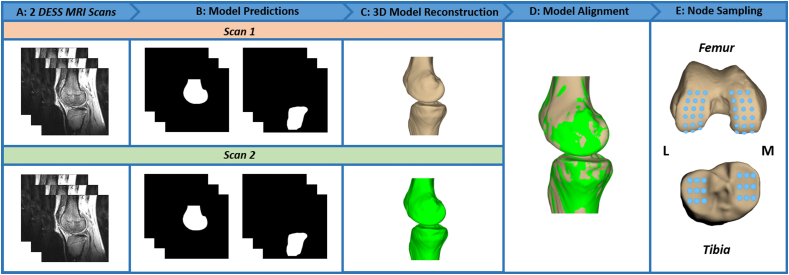
The pipeline for this process began by inputting the two DESS MRI scans of the same participant into the trained segmentation models, which generated masks for each of the bones (Fig. 1). Outlier pixels disconnected from the main bone regions of interest were manually removed and mask contours were extracted and converted into point clouds for all images stacked together. Point clouds were then reconstructed into 3D surface mesh models using Geomagic Studio 11 (3D Systems; Research Triangle Park, North Carolina), and the 3D models of each bone were aligned with one another [7]. Finally, surface distances were calculated in the x- (anterior-posterior), y- (superior-inferior), and z- (medial-lateral) directions at different regions of interest (within a 2.5 mm radius) sampled evenly across the femoral condyles and tibial plateaus. A total of 36 regions of interest were measured across the femoral condyles and 18 regions of interest were measured across the tibial plateaus [28]. Mean distances in the x-, y-, and z-directions for each participant were used to calculate a group mean and 95 % confidence interval for each measurement direction.
Fig. 1.
Bone Surface DistanceRepeatability Analysis Pipeline. (A) To assess bone surface repeatability, two DESS MRI scans from a single participant are inputted into the trained tibia and femur segmentation models. (B) The trained models predict the tibia and femur masks. (C) The outer contours from the predicted masks are extracted and reconstructed into 3D models using Geomagic Studio 11 (3D Systems; Research Triangle Park, North Carolina). (D) The tibia and femur models from Scan 1 and Scan 2 are aligned using an iterative closest point technique [6]. (E) Bone surface distances in the x-, y-, and z-directions are calculated between the two scans across all regions of interest [28]. M = medial and L = lateral.
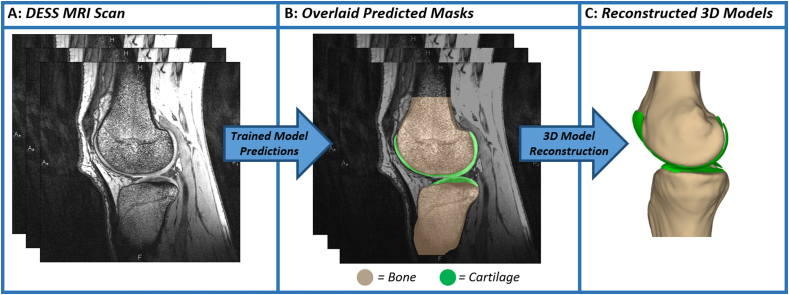
The next task was to assess the repeatability of cartilage thickness measurements by evaluating cartilage thickness from scans acquired on different days. Using the same dataset as was previously described in the study design, we used the pre-exercise and recovery scans, as these scans both represent unloaded time points when the cartilage is at baseline thickness. It should be noted that day-to-day fluctuations in cartilage thickness could impact measurement repeatability. In order to enable repeated site-specific cartilage thickness measurements, 3D surface mesh models of the associated tibia and femur from each scan were created and aligned together along with the corresponding cartilage surfaces (Fig. 2). Cartilage thickness was then measured by sampling and averaging 18 locations across the tibial plateaus and 36 locations across the femoral condyles (Fig. 1). At each location, all cartilage vertices within a 2.5 mm radius were included. Cartilage thickness at each vertex was defined as the distance to the nearest point on the bone surface. Finally, the thicknesses at all vertices were averaged together to produce a thickness for each location across the tibial plateaus and femoral condyles [28]. Prior to measuring cartilage thickness, outlier pixels disconnected from the main cartilage regions were manually removed and bone models were thoroughly reviewed. Cartilage thickness measurement reliability was evaluated by calculating a two-way, mixed effects, multiple-measurement, absolute agreement intraclass correlation coefficient for each cartilage surface. Additionally, we determined the coefficient of variation and standard deviation between the thickness measurements taken on different days.
Fig. 2.
Bone and Cartilage 3D Model Reconstruction Pipeline. (A) Each DESS MRI scan is inputted into the 4 trained segmentation models, and binary masks are outputted for each of the 4 tissues. (B) Visualization of the predicted bone and cartilage masks overlaid on the original DESS MRI scan. (C) Mask contours are extracted, converted into point clouds, and reconstructed into 3D surface mesh models using Geomagic Studio 11 (3D Systems; Research Triangle Park, North Carolina). Tan color indicates the tibia and femur, while green indicates the tibial and femoral cartilage.
2.3. Running-induced cartilage strain & recovery
Following testing of the bone and cartilage segmentation models, the trained models were used to perform the primary objective of this work, which was to measure tibial and femoral cartilage strain and recovery following a 3-mile run. In this analysis, all scans were manually cropped around the regions of interest, predicted by the trained segmentation models, reviewed, and compiled to generate 3D models of the bone and cartilage from which we could measure cartilage thickness. Obtaining thickness measurements at all three time points allowed for the comparison of cartilage thickness between the pre-exercise and recovery scans, as well as the calculation of cartilage strain post-exercise (Equation (1)).
| Eq 1 |
Two repeated measures ANOVAs were performed to determine the influence of time point (pre-exercise vs. post-exercise vs. recovery) on tibial and femoral cartilage thicknesses. Subsequent Fisher’s Least Significant Difference post-hoc tests were performed to detect differences between means as appropriate. Differences were considered statistically significant with p < 0.05.
3. Results
3.1. Model development & testing
In terms of testing set DSCs, the 2D-UNet models outperformed the 3D-UNet models across all four tissues (Table 2). While the difference between the 2D- and 3D-UNet models was relatively small (Δ DSC <0.01), the difference was most pronounced for the cartilage models (Δ DSC for tibial cartilage = 0.006 and Δ DSC for femoral cartilage = 0.008). After optimal models were determined, they were subsequently applied to the testing set for each tissue (Table 2). Both bone models achieved testing set DSCs greater than 0.980, and both cartilage models achieved testing set DSCs greater than 0.900.
Table 2.
Dice Similarity Coefficient for bone and cartilage by dataset.
| Tissue | Validation Set |
Testing Set | |
|---|---|---|---|
| 2D-UNet | 3D-UNet | ||
| Femoral bone | 0.989 | 0.987 | 0.990 |
| Tibial bone | 0.984 | 0.984 | 0.988 |
| Femoral cartilage | 0.921 | 0.913 | 0.913 |
| Tibial cartilage | 0.909 | 0.903 | 0.901 |
Testing set Dice Similarity Coefficients were calculated using the 2D-UNet.
Evaluation of scan-to-scan repeatability demonstrated that bone surface distance group means were less than 0.03 mm for both the tibia and femur across all three directions (Table 3). Day-to-day measurements of cartilage thickness similarly revealed excellent reliability and precision, with ICCs of 0.984 and 0.987, and CVs of 1 % (standard deviation 0.03 mm) and 1 % (standard deviation 0.02 mm) for the tibial and femoral cartilage, respectively. Further, using our trained deep learning models, the total time required to segment the bone and cartilage in the tibiofemoral joint decreased from an order of hours or days when performed manually to an order of minutes when performed using deep learning.
Table 3.
Mean (±95 % CI) scan-to-scan bone surface distances (mm).
| Direction | Tibia | Femur |
|---|---|---|
| X | 0.00 (0.04) | 0.01 (0.03) |
| Y | 0.02 (0.04) | 0.02 (0.03) |
| Z | 0.00 (0.03) | 0.01 (0.02) |
3.2. Running-induced cartilage strain & recovery
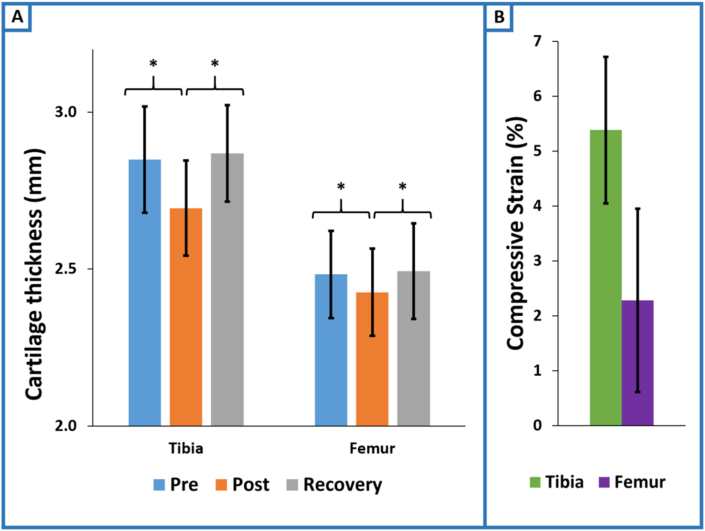
In general, the 3-mile treadmill run induced compressive cartilage strains, and these strains were largely attenuated the morning following exercise (Fig. 3). An overall effect of time on cartilage thickness was observed for both the tibial (p < 0.0001) and femoral (p < 0.01) cartilage (Fig. 4). Tibial cartilage thickness changed by −0.15 (95 % CI = −0.20 to −0.11; p < 0.0001) mm pre-to post-exercise and recovered to 0.02 (95 % CI = −0.02 to 0.06; p = 0.37) mm the following day relative to baseline. Similarly, femoral cartilage thickness changed by −0.06 (95 % CI = −0.10 to −0.02; p < 0.01) mm pre-to post-exercise and recovered to 0.01 (95 % CI = −0.02 to 0.04; p = 0.60) mm the following day relative to baseline. Three miles of treadmill running resulted in compressive strains of 5.4 % (95 % CI = 4.1 to 6.7) and 2.3 % (95 % CI = 0.6 to 4.0) for the tibial and femoral cartilage, respectively (Fig. 4).
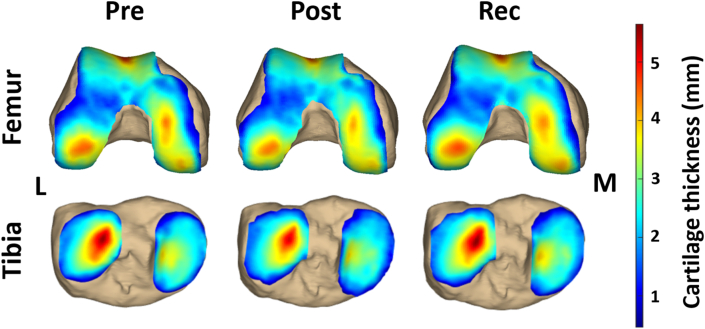
Fig. 3.
Cartilage thickness maps for one participant pre-exercise, post-exercise, and after 24-h of recovery. Red represents thicker regions of cartilage, while blue represents thinner regions. Post-exercise cartilage thickness decreased compared to the pre-exercise and recovery time points, which were relatively similar. M = medial and L = lateral.
Fig. 4.
Running-induced changes to cartilage thickness and strain. (A) Cartilage thickness decreased significantly pre-to post-exercise for both the tibial (p < 0.0001) and femoral (p < 0.01) cartilage. Subsequently, cartilage thickness increased back towards baseline thickness at the recovery time point for both the tibial (p < 0.0001) and femoral (p < 0.01) cartilage. Significant differences between baseline and recovery were not detected for either the tibial (p = 0.37) or femoral (p = 0.60) cartilage. (B) Running induced post-exercise compressive strains of 5.4 % (95 % CI = 4.1 to 6.7) and 2.3 % (95 % CI = 0.6 to 4.0) for the tibial and femoral cartilage, respectively.
4. Discussion
This work sought to measure the deformation of tibiofemoral cartilage thickness immediately following a 3-mile treadmill run, as well as the recovery of cartilage thickness the following morning. In these analyses, both tibial and femoral cartilage thickness significantly decreased following the 3-mile run and recovered to within 1 % of pre-exercise thickness after one day of recovery. Additionally, deep learning models were trained to automate segmentation of the tibiofemoral bone and cartilage in order to enable the efficient measurement of cartilage thickness. All models were reliable and demonstrated precision similar to that of manual segmentation [28].
In terms of deep learning model development, we successfully trained models to segment the tibiofemoral bone and cartilage from DESS knee MRI scans. For each task, we trained both 2D- and 3D-UNet models to ensure that the optimal model architecture was chosen for each specific tissue. In terms of DSCs, the 2D models moderately outperformed the 3D models for each of the four tasks. While the primary benefit of the 3D-UNet architecture is the inclusion of inter-slice context, training of the 3D-UNet models was inherently constrained due to the computationally expensive nature of the 3D network and the relatively large image volumes. These factors led to a tradeoff between image volume size and maximum hyperparameter complexity due to memory constraints of the GPUs available for model training. Ultimately, the added inter-slice context of the 3D-UNet models may have not outweighed training restrictions, leading to the 2D models performing better. In terms of the optimal model testing set DSCs for each of the four tasks, our models compared favorably with the previously reported best scores [6,13,15,16,19,[29], [30], [31]]. It should be noted, however, that it is difficult to make direct comparisons between testing set DSCs across studies, as groups typically use different testing sets, which may impact the ultimate score. Despite this, the testing set DSCs from the present study reflect significant agreement between manually and automatically derived segmentations. One factor that likely contributed to our success in training models specific for our use-case was the consistency of data utilized for model development. All scans were collected on the same 3.0 T MRI scanner with identical imaging parameters [[21], [22], [23], [24], [25], [26]], and all segmentations were reviewed by the same MSK radiologist with more than 35 years of experience. A limitation of our model development techniques was that training a specific model for each segmentation task now requires application of 4 separate models to predict the tissues of interest, whereas a multiclass model could have predicted each tissue with a single model. However, training a separate model for each segmentation task enabled task-specific hyperparameter optimization. This also enabled cropping of the image volume to each tissue, which decreased computational cost and ultimately enabled more hyperparameter combinations to be tested during model training. While the objective of this work was to train models to segment the tibiofemoral bone and cartilage from DESS knee MRI scans with specific imaging parameters, a limitation is therefore that the application of these models is thus specific in nature. Specifically, it is not well known how they would perform with imaging data that differs from that which the models were trained on.
Following model development, we evaluated the repeatability of using the trained segmentation models to create 3D models of the tibia and femur, as well as measure cartilage thickness. Beginning with the tibia and femur, we utilized two scans per participant that were obtained approximately 30 min apart after the participant was repositioned and measured scan-to-scan group mean distances less than 0.03 mm for the x-, y-, and z-directions. While some distances represent a small positive bias, the 95 % confidence intervals of all distances overlap with zero. Further, while these distances should largely represent differences in scan-to-scan segmentation, they may also encompass some error introduced from 3D model alignment. However, most importantly, the bone surface distances measured here all fall within the previously reported manual measurement resolution for this technique [28], meaning that the repeatability of using the trained bone models is comparable to that of trained manual segmenters plus review from a fellowship-trained radiologist. The repeatability of creating bone 3D models is important, as it has been suggested that bone shape may change over time with injury, aging, and OA status [[32], [33], [34], [35], [36]]. Next, we evaluated the repeatability of using the bone and cartilage segmentation models to measure cartilage thickness. A limitation of this work is that we did not perform manual segmentations to compare manual and automated strain measurements. However, in order to assess measurement reliability, we measured cartilage thickness in the same cohort of participants across two different days. Assessing measurement reliability across different days in this way has an advantage over comparison to manual segmentations as it removes intra- and inter-segmenter variability that may bias the perceived validity of the model. The ICCs for this analysis were greater than 0.980 for both the tibial and femoral cartilage, indicating excellent measurement reliability [37]. Further, the day-to-day measurement of cartilage thickness resulted in CVs of 1 % for both the tibial and femoral cartilage. The present results indicate that our resolution for measuring cartilage thickness is similar to the previously established resolution of this technique using manual segmentation (∼1 %) [28].
The primary objective of this work was to use our trained models to measure tibial and femoral cartilage strain and recovery in response to a 3-mile treadmill run. Prior work has investigated running-induced changes to cartilage thickness and volume with varying results [7,[38], [39], [40], [41], [42]]. While several studies have demonstrated changes in cartilage thickness and volume in some compartments after running [38,[40], [41], [42]], others have failed to detect significant changes in tibiofemoral cartilage thickness [39]. In the present study, using site-specific measurements of cartilage thickness, we detected a significant decrease in both the tibial (5.4 %) and femoral (2.3 %) cartilage (Fig. 4). The tibial and femoral cartilage strain magnitudes in this study align with prior work that reported higher strains in the tibial cartilage compared to the femoral cartilage in response to walking and hopping [21,22,43]. Potential explanations for why femoral cartilage experiences less compressive strain include differences in the mechanical properties and loading between the femur and tibia [22,44]. Specifically, prior work has reported that femoral cartilage is stiffer and less permeable compared to tibial cartilage [44,45]. With regard to loading differences, as the knee flexes during dynamic activities, the regions of the femoral cartilage in contact change more than those of the tibial cartilage [22,43]. These differences may cause the loading to be more dispersed, reducing strain on the femoral cartilage [22,43].
Prior work has also utilized quantitative MRI techniques, such as T1rho and T2 relaxation mapping to evaluate changes in cartilage composition following loading [20,[38], [39], [40],46]. For example, a recent study from our lab found that running results in decreased tibial, femoral, and patellar cartilage T1rho relaxation times, indicative of an increase in the concentration of proteoglycans due to expulsion of water [20]. Similarly, decreased T2 relaxation times have been measured in the superficial regions of tibiofemoral cartilage immediately following running, likely resulting from a reduction in water content [46]. Thus, our present findings support the hypothesis that running results in immediate post-exercise changes in both tibial and femoral cartilage thickness due to water loss from the cartilage.
In addition to probing the immediate influence of a 3-mile run, tibiofemoral cartilage thickness was also measured the following morning to elucidate the ability of cartilage to recover at a time scale relevant to habitual exercise. At the recovery time point, both the tibial and femoral cartilage thickness had significantly increased from the post-exercise thickness, and recovered to within 1 % of baseline. With regard to prolonged effects after exercise, several prior studies have probed cartilage thickness and composition between 2 and 12 h following a marathon run [[47], [48], [49]]. However, in these studies, cartilage thickness and composition were not measured immediately following exercise, making it difficult to draw direct comparisons to the present study. Prior work from our lab evaluated tibial, femoral, and patellar cartilage T1rho relaxation times before, immediately following, and approximately 24 h following a 3 or 10 mile run [20]. T1rho relaxation times were significantly decreased immediately post-exercise, and returned to pre-exercise levels after approximately 24 h of recovery [20]. Our present findings, in conjunction with these prior results, suggest that tibiofemoral cartilage thickness and composition have the ability to recover from a 3-mile run within 24 h. However, a limitation of this work is that the participants were relatively young and healthy. Future work is needed to investigate how repetitive bouts of running may alter cartilage strain and recovery as it relates to long-term joint health, in addition to evaluating the influence of age, sex, BMI, and knee health status.
In conclusion, we measured tibiofemoral cartilage strain and recovery following a 3-mile treadmill run, enabled via the development of deep learning models to automate segmentation of the tibiofemoral cartilage and bone. In this analysis, we demonstrated that a 3-mile run induced both tibial and femoral cartilage compressive strains, and after approximately one day, the cartilage had largely returned to baseline thickness. This suggests that a single 3-mile run may be a safe magnitude of loading for tibial and femoral cartilage in a relatively young and healthy cohort of participants. Future work will seek to expand upon these findings in a more diverse participant population and under varying run distances and intensities.
Author contributions
PXB: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing – Original draft preparation, Writing – Reviewing and editing; SYK: Methodology, Software, Data curation, Writing – Reviewing and editing; BSB: Validation, Formal analysis, Investigation, Writing – Reviewing and editing; ADR: Conceptualization, Methodology, Investigation, Data curation, Writing – Reviewing and editing; ATC: Investigation, Data curation, Writing – Reviewing and editing; LNH: Conceptualization, Methodology, Investigation, Data curation, Writing – Reviewing and editing; ECO: Investigation, Data curation, Writing – Reviewing and editing; MRW: Investigation, Data curation, Writing – Reviewing and editing; CSP: Investigation, Data curation, Writing – Reviewing and editing; BSC: Investigation, Data curation, Writing – Reviewing and editing; NKL: Investigation, Data curation, Writing – Reviewing and editing; EGS: Investigation, Data curation, Writing – Reviewing and editing; BPM: Conceptualization, Methodology, Writing – Reviewing and editing; CES: Conceptualization, Methodology, Writing – Reviewing and editing; LED: Conceptualization, Methodology, Resources, Supervision, Project administration, Funding acquisition, Investigation, Writing – Reviewing and editing.
Role of the funding source
This work was supported by NIH grants R01AR074800, R01AR065527, R01AR082665, and R01AR079184.
Conflict of interest
This work was prepared while Dr. Amber Collins was employed at Duke University School of Medicine. The opinions expressed in this article are the author’s own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government. The remaining authors have no conflicts of interest to disclose.
Acknowledgements
Writing support was provided by Donald T. Kirkendall.
Handling Editor: Professor H Madry
References
- 1.Chan D.D., Cai L., Butz K.D., Trippel S.B., Nauman E.A., Neu C.P. In vivo articular cartilage deformation: noninvasive quantification of intratissue strain during joint contact in the human knee. Scientific Reports. 2016;6 doi: 10.1038/srep19220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X., Benjamin Ma C., Link T.M., Castillo D.D., Blumenkrantz G., Lozano J., et al. In vivo T1ρ and T2 mapping of articular cartilage in osteoarthritis of the knee using 3T MRI. Osteoarthritis Cartilage. 2007;15:789–797. doi: 10.1016/j.joca.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckstein F., Wluka A.E., Wirth W., Cicuttini F. 30 Years of MRI-based cartilage & bone morphometry in knee osteoarthritis: from correlation to clinical trials. Osteoarthritis Cartilage. 2024;32:439–451. doi: 10.1016/j.joca.2024.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Guilak F. Biomechanical factors in osteoarthritis. Best Practice & Research Clinical Rheumatology. 2011;25:815–823. doi: 10.1016/j.berh.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coburn S.L., Crossley K.M., Kemp J.L., Warden S.J., West T.J., Bruder A.M., et al. Is running good or bad for your knees? A systematic review and meta-analysis of cartilage morphology and composition changes in the tibiofemoral and patellofemoral joints. Osteoarthritis Cartilage. 2023;31:144–157. doi: 10.1016/j.joca.2022.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Khan S., Azam B., Yao Y., Chen W. Deep collaborative network with alpha matte for precise knee tissue segmentation from MRI. Comput Methods Programs Biomed. 2022;222 doi: 10.1016/j.cmpb.2022.106963. [DOI] [PubMed] [Google Scholar]
- 7.Heckelman L.N., Riofrio A.D., Vinson E.N., Collins A.T., Gwynn O.R., Utturkar G.M., et al. Dose and recovery response of patellofemoral cartilage deformations to running. Orthopaedic Journal of Sports Medicine. 2020;8 doi: 10.1177/2325967120967512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenneman Wilson E.C., Gatti A.A., Maly M.R. A new technique to evaluate the impact of running on knee cartilage deformation by region. Magnetic Resonance Materials in Physics, Biology and Medicine. 2021;34:593–603. doi: 10.1007/s10334-020-00896-8. [DOI] [PubMed] [Google Scholar]
- 9.Eckstein F., Lemberger B., Gratzke C., Hudelmaier M., Glaser C., Englmeier K.H., et al. In vivo cartilage deformation after different types of activity and its dependence on physical training status. Annals of the Rheumatic Diseases. 2005;64:291–295. doi: 10.1136/ard.2004.022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coppock J.A., Zimmer N.E., Spritzer C.E., Goode A.P., DeFrate L.E. Automated segmentation and prediction of intervertebral disc morphology and uniaxial deformations from MRI. Osteoarthritis and Cartilage Open. 2023;5 doi: 10.1016/j.ocarto.2023.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heckelman L.N., Soher B.J., Spritzer C.E., Lewis B.D., DeFrate L.E. Design and validation of a semi-automatic bone segmentation algorithm from MRI to improve research efficiency. Scientific Reports. 2022;12:7825. doi: 10.1038/s41598-022-11785-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ronneberger O., Fischer P., Brox T. In: Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Navab N., Hornegger J., Wells W.M., Frangi A.F., editors. Springer International Publishing; Cham: 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 13.Kim-Wang S.Y., Bradley P.X., Cutcliffe H.C., Collins A.T., Crook B.S., Paranjape C.S., et al. Auto-segmentation of the tibia and femur from knee MR images via deep learning and its application to cartilage strain and recovery. Journal of Biomechanics. 2023;149 doi: 10.1016/j.jbiomech.2023.111473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flannery S.W., Kiapour A.M., Edgar D.J., Murray M.M., Fleming B.C. Automated magnetic resonance image segmentation of the anterior cruciate ligament. Journal of Orthopaedic Research. 2021;39:831–840. doi: 10.1002/jor.24926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaj S., Yang M., Nakamura K., Li X. Automated cartilage and meniscus segmentation of knee MRI with conditional generative adversarial networks. Magnetic Resonance in Medicine. 2019;84:437–449. doi: 10.1002/mrm.28111. [DOI] [PubMed] [Google Scholar]
- 16.Norman B., Pedoia V., Majumdar S. Use of 2D U-net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology. 2018;288:177–185. doi: 10.1148/radiol.2018172322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulseng C.P.S., Nainamalai V., Grøvik E., Geitung J.T., Årøen A., Gjesdal K.I. Automatic segmentation of human knee anatomy by a convolutional neural network applying a 3D MRI protocol. BMC Musculoskelet Disorder. 2023;24:41. doi: 10.1186/s12891-023-06153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai A.D., Caliva F., Iriondo C., Mortazi A., Jambawalikar S., Bagci U., et al. The international workshop on osteoarthritis imaging knee MRI segmentation challenge: a multi-institute evaluation and analysis framework on a standardized dataset. Radiology: Artificial Intelligence. 2021;3 doi: 10.1148/ryai.2021200078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatti A.A., Maly M.R. Automatic knee cartilage and bone segmentation using multi-stage convolutional neural networks: data from the osteoarthritis initiative. Magma. 2021;34:859–875. doi: 10.1007/s10334-021-00934-z. [DOI] [PubMed] [Google Scholar]
- 20.Heckelman L.N., Smith W.A.R., Riofrio A.D., Vinson E.N., Collins A.T., Gwynn O.R., et al. Quantifying the biochemical state of knee cartilage in response to running using T1rho magnetic resonance imaging. Scientific Reports. 2020;10:1870. doi: 10.1038/s41598-020-58573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crook B.S., Collins A.T., Lad N.K., Spritzer C.E., Wittstein J.R., DeFrate L.E. Effect of walking on in vivo tibiofemoral cartilage strain in ACL-deficient versus intact knees. Journal of Biomechanics. 2021;116 doi: 10.1016/j.jbiomech.2020.110210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lad N.K., Liu B., Ganapathy P.K., Utturkar G.M., Sutter E.G., Moorman C.T., et al. Effect of normal gait on in vivo tibiofemoral cartilage strains. Journal of Biomechanics. 2016;49:2870–2876. doi: 10.1016/j.jbiomech.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okafor E.C., Utturkar G.M., Widmyer M.R., Abebe E.S., Collins A.T., Taylor D.C., et al. The effects of femoral graft placement on cartilage thickness after anterior cruciate ligament reconstruction. Journal of Biomechanics. 2014;47:96–101. doi: 10.1016/j.jbiomech.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paranjape C.S., Cutcliffe H.C., Grambow S.C., Utturkar G.M., Collins A.T., Garrett W.E., et al. A new stress test for knee joint cartilage. Scientific Reports. 2019;9:2283. doi: 10.1038/s41598-018-38104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutter E.G., Liu B., Utturkar G.M., Widmyer M.R., Spritzer C.E., Cutcliffe H.C., et al. Effects of anterior cruciate ligament deficiency on tibiofemoral cartilage thickness and strains in response to hopping. The American Journal of Sports Medicine. 2019;47:96–103. doi: 10.1177/0363546518802225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Widmyer M.R., Utturkar G.M., Leddy H.A., Coleman J.L., Spritzer C.E., Moorman I.I.I.C.T., et al. High body mass index is associated with increased diurnal strains in the articular cartilage of the knee. Arthritis & Rheumatism. 2013;65:2615–2622. doi: 10.1002/art.38062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou K.H., Warfield S.K., Bharatha A., Tempany C.M., Kaus M.R., Haker S.J., et al. Statistical validation of image segmentation quality based on a spatial overlap index. Academic Radiology. 2004;11:178–189. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coleman J.L., Widmyer M.R., Leddy H.A., Utturkar G.M., Spritzer C.E., Moorman C.T., 3rd, et al. Diurnal variations in articular cartilage thickness and strain in the human knee. Journal of Biomechanics. 2013;46:541–547. doi: 10.1016/j.jbiomech.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambellan F., Tack A., Ehlke M., Zachow S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: data from the Osteoarthritis Initiative. Medical Image Analysis. 2019;52:109–118. doi: 10.1016/j.media.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Latif M.H.A., Faye I. Automated tibiofemoral joint segmentation based on deeply supervised 2D-3D ensemble U-Net: data from the Osteoarthritis Initiative. Artificial Intelligence in Medicine. 2021;122 doi: 10.1016/j.artmed.2021.102213. [DOI] [PubMed] [Google Scholar]
- 31.Panfilov E., Tiulpin A., Klein S., Nieminen M.T., Saarakkala S.S. IEEE/CVF International Conference on Computer Vision Workshop (ICCVW) 2019. Improving robustness of deep learning based knee MRI segmentation: mixup and adversarial domain adaptation; pp. 450–459. 2019. [Google Scholar]
- 32.Pedoia V., Lansdown D.A., Zaid M., McCulloch C.E., Souza R., Ma C.B., et al. Three-dimensional MRI-based statistical shape model and application to a cohort of knees with acute ACL injury. Osteoarthritis Cartilage. 2015;23:1695–1703. doi: 10.1016/j.joca.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haverkamp D.J., Schiphof D., Bierma-Zeinstra S.M., Weinans H., Waarsing J.H. Variation in joint shape of osteoarthritic knees. Arthritis & Rheumatism. 2011;63:3401–3407. doi: 10.1002/art.30575. [DOI] [PubMed] [Google Scholar]
- 34.Ding C., Cicuttini F., Scott F., Cooley H., Jones G. Association between age and knee structural change: a cross sectional MRI based study. Annals of the Rheumatic Diseases. 2005;64:549–555. doi: 10.1136/ard.2004.023069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowes M.A., Maciewicz R.A., Waterton J.C., Hunter D.J., Conaghan P.G. Bone area provides a responsive outcome measure for bone changes in short-term knee osteoarthritis studies. Journal of Rheumatology. 2016;43:2179–2182. doi: 10.3899/jrheum.151118. [DOI] [PubMed] [Google Scholar]
- 36.Bredbenner T.L., Eliason T.D., Potter R.S., Mason R.L., Havill L.M., Nicolella D.P. Statistical shape modeling describes variation in tibia and femur surface geometry between Control and Incidence groups from the osteoarthritis initiative database. Journal of Biomechanics. 2010;43:1780–1786. doi: 10.1016/j.jbiomech.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koo T.K., Li M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. Chiropractic Journal of Medicine. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gatti A.A., Noseworthy M.D., Stratford P.W., Brenneman E.C., Totterman S., Tamez-Peña J., et al. Acute changes in knee cartilage transverse relaxation time after running and bicycling. Journal of Biomechanics. 2017;53:171–177. doi: 10.1016/j.jbiomech.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 39.Subburaj K., Kumar D., Souza R.B., Alizai H., Li X., Link T.M., et al. The acute effect of running on knee articular cartilage and meniscus magnetic resonance relaxation times in young healthy adults. The American Journal of Sports Medicine. 2012;40:2134–2141. doi: 10.1177/0363546512449816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenneman Wilson E.C., Gatti A.A., Keir P.J., Maly M.R. Daily cumulative load and body mass index alter knee cartilage response to running in women. Gait Posture. 2021;88:192–197. doi: 10.1016/j.gaitpost.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 41.Van Ginckel A., Verdonk P., Victor J., Witvrouw E. Cartilage status in relation to return to sports after anterior cruciate ligament reconstruction. The American Journal of Sports Medicine. 2013;41:550–559. doi: 10.1177/0363546512473568. [DOI] [PubMed] [Google Scholar]
- 42.Boocock M., McNair P., Cicuttini F., Stuart A., Sinclair T. The short-term effects of running on the deformation of knee articular cartilage and its relationship to biomechanical loads at the knee. Osteoarthritis and Cartilage. 2009;17:883–890. doi: 10.1016/j.joca.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Sutter E.G., Widmyer M.R., Utturkar G.M., Spritzer C.E., Garrett W.E., Jr., DeFrate L.E. In vivo measurement of localized tibiofemoral cartilage strains in response to dynamic activity. The American Journal of Sports Medicine. 2015;43:370–376. doi: 10.1177/0363546514559821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treppo S., Koepp H., Quan E.C., Cole A.A., Kuettner K.E., Grodzinsky A.J. Comparison of biomechanical and biochemical properties of cartilage from human knee and ankle pairs. Journal of Orthopaedic Research. 2000;18:739–748. doi: 10.1002/jor.1100180510. [DOI] [PubMed] [Google Scholar]
- 45.Wong B.L., Sah R.L. Mechanical asymmetry during articulation of tibial and femoral cartilages: local and overall compressive and shear deformation and properties. Journal of Biomechanics. 2010;43:1689–1695. doi: 10.1016/j.jbiomech.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crowder H.A., Mazzoli V., Black M.S., Watkins L.E., Kogan F., Hargreaves B.A., et al. Characterizing the transient response of knee cartilage to running: decreases in cartilage T(2) of female recreational runners. Journal of Orthopaedic Research. 2021;39:2340–2352. doi: 10.1002/jor.24994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang M., Li Y., Feng R., Wang Z., Wang W., Zheng N., et al. Change in susceptibility values in knee cartilage after marathon running measured using quantitative susceptibility mapping. Journal of Magnetic Resonance Imaging. 2021;54:1585–1593. doi: 10.1002/jmri.27745. [DOI] [PubMed] [Google Scholar]
- 48.Zhang P., Yu B., Zhang R., Chen X., Shao S., Zeng Y., et al. Longitudinal study of the morphological and T2∗ changes of knee cartilages of marathon runners using prototype software for automatic cartilage segmentation. British Journal of Radiology. 2021;94 doi: 10.1259/bjr.20200833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z., Ai S., Tian F., Liow M.H.L., Wang S., Zhao J., et al. Higher body mass index is associated with biochemical changes in knee articular cartilage after marathon running: a quantitative T2-relaxation MRI study. Orthopaedics Journal of Sports Medicine. 2020;8 doi: 10.1177/2325967120943874. [DOI] [PMC free article] [PubMed] [Google Scholar]