Abstract
At present, hundreds of microbial genomes have been sequenced, and hundreds more are currently in the pipeline. The Virtual Institute for Microbial Stress and Survival has developed a publicly available suite of Web-based comparative genomic tools (http://www.microbesonline.org) designed to facilitate multispecies comparison among prokaryotes. Highlights of the MicrobesOnline Web site include operon and regulon predictions, a multispecies genome browser, a multispecies Gene Ontology browser, a comparative KEGG metabolic pathway viewer, a Bioinformatics Workbench for in-depth sequence analysis, and Gene Carts that allow users to save genes of interest for further study while they browse. In addition, we provide an interface for genome annotation, which like all of the tools reported here, is freely available to the scientific community.
Functional genomic studies have generated a large and growing database of experimental results for eukaryotic model organisms, such as yeast. For most prokaryotic model organisms, fewer direct experimental data are available, yet this apparent deficit is compensated by the wealth of genomic sequence available. Although a wide variety of tools are available to study various aspects of genomic sequence, many are not designed to facilitate direct comparison of multiple genomes and thus fully exploit this source of information. Furthermore, many tools are implemented in different Web sites, and use different gene nomenclature, which makes it inconvenient to perform a deep analysis on a single gene and then return to the original Web site to identify more genes to analyze. To perform basic analyses such as multiple sequence alignment, users often need to download gene sequences, manipulate them into a particular file format (e.g., FASTA), and cut and paste them into another Web site. To continue further analysis, such as constructing a phylogenetic tree, users may be required to paste the resulting alignment into a third Web site.
Several other Web sites and software tools have been described that assist in the annotation and exploration of comparative genomic data. The Prolinks and STRING databases offer convenient tools for browsing predicted functional associations among proteins, while GenDB and other systems allow for detailed annotation of individual genomes (Snel et al. 2000; Meyer et al. 2003; Bowers et al. 2004; von Mering et al. 2005). The ERGO system combines some of these features, but a full version of the system is not publicly available (Overbeek et al. 2003). The TIGR Comprehensive Microbial Resource (CMR) also offers some useful tools such as genome alignment and precomputed BLAST and TIGRfam results (Peterson et al. 2001).
We have compiled a list of genome analysis tools that we believe to be among the most useful of those currently available, and have combined all of these tools on a single Web site (http://www.microbesonline.org). To make it as simple as possible for users to select genes of interest while they are browsing, we have implemented a “Gene Cart” feature analogous to the “Shopping Cart” common to many commercial Web sites. Genes can be added to the cart from most of the pages on the MicrobesOnline Web site, and then saved for further study, downloaded to a local computer, or analyzed using the Bioinformatics Workbench. Users can create and save any number of Gene Carts for further study. The Workbench currently includes basic sequence analysis tools for multiple sequence alignment and for building phylogenetic trees.
In addition to collecting these tools in one location, the MicrobesOnline Web site offers several new features as well as extensions of existing tools to facilitate comparative genomics. In particular, metabolic pathway maps allow for the comparison of two different genomes to highlight differences in their expected physiological capabilities. The Gene Ontology browser we developed, called VertiGO, differs from similar tools in that it allows any number of the genomes to be displayed at the same time. A multispecies genome browser capable of zoom and scroll actions allows users to simultaneously align any number of the 200+ genomes currently hosted at MicrobesOnline, quickly access additional info on displayed genes, and save displayed genes to a user's Gene Cart.
In addition to these comparative genomic features, MicrobesOnline hosts additional genome annotations produced by the VIMSS group including predicted pseudogenes, complete operon predictions, as well as “regulon” predictions based on both comparative genomic and (for some genomes) experimental gene expression data that may be useful for identifying groups of genes with related functions. Finally, the MicrobesOnline Web site provides an opportunity for microbiologists to annotate individual genes using a simple Web-based interface, and to share those annotations with the microbial research community at large.
Results and Discussion
Finding genes of interest
Most browsing sessions begin with a search for a gene of interest. The Keyword Search window provides a simple yet powerful interface for identifying genes with a particular function. Users can search one or more specific genomes, high-level taxonomic groups (e.g., Firmicutes), or the entire database. Because genome annotations are often incomplete, and because no single protein family database is best for all situations, the Keyword Search feature returns hits to a wide number of annotation types: (1) genes with a matching name or synonym (e.g., from a search for “trpA”); (2) genes with a matching gene description (e.g., from a search for “tryptophan”); (3) genes mapping to a matching description in the COG database; (4) genes with a matching Interpro domain description; and (5) genes assigned to a matching GO category. This comprehensive search feature allows users to combine searches across individual databases such as Pfam (Bateman et al. 2004), TIGRfam (Haft et al. 2003), or COG (Tatusov et al. 1997) to identify genes that might have a particular function. It should be noted, however, that the primary databases might have more up-to-date information than the combined database at MicrobesOnline. An advanced search feature is also provided, which allows further text/Boolean searches for genes by KEGG metabolic pathways (Kanehisa 2002), enzyme commission (EC) numbers, Gene Ontology (GO) identifiers (Camon et al. 2003), or COGs (Tatusov et al. 1997).
Often it is desirable to locate genes that have sequence identity to an existing gene for which there is little available annotation information. For these cases, users can search available genomes using a Web-based BLAST interface. Results are displayed in the traditional BLAST output format, but hits to genes in the MicrobesOnline database are hyperlinked to the “Locus Info” pages with more detailed descriptions of each gene.
Multispecies Genome Browser
The MicrobesOnline Comparative Genome Browser allows the analysis of genes based on their physical position on the chromosome, and highlights conservation of gene order across distantly related species, which generally indicates functionally related genes in conserved operon structures (Dandekar et al. 1998; Overbeek et al. 1999; Lathe III et al. 2000), as well as large-scale synteny between closely related genomes. From the Comparative Genome Browser (Fig. 1) users can select any number of genomes and align them using an “anchor” gene. Users can add or delete selected genomes from the view, zoom in or out, and scan upstream or downstream. All genes within a view are color-coded according to predicted orthology relationships. Each gene on the browser is a hyperlink that can perform one of three actions: (1) load the Locus Info Page for that gene (described in more detail below); (2) realign genomes by selecting a new gene as the anchor; or (3) add that gene to the user's Gene Cart (described below). Figure 1 shows an example of how the browser was used to investigate the evolutionary history of an operon identified in Escherichia coli.
Figure 1.
Comparative Genome Browser. In a subgroup of the γ-Proteobacteria, two ancient operons for adjacent steps in the TCA cycle, sdhCDAB and sucABCD, have been merged into one larger operon. This view from the Comparative Genome Browser shows the sdhA locus across an archaeon (Halobacterium sp.), a Gram-positive bacterium (Mycobacterium tuberculosis), and a diverse range of proteobacteria. Note that the larger operon is present in only a single clade, yet it is highly conserved within this group, suggesting that it may reflect a recent innovation within the γ-Proteobacteria.
VertiGO: Gene Ontology in multiple genomes
The Gene Ontology (GO) is a controlled vocabulary that describes biological roles associated with gene products in a species-independent manner (Camon et al. 2003). This standardized biological vocabulary makes it possible to compare functional annotations across species. Although users of the site can make their own annotations of gene function using the GO terms, users will find pre-existing genome-wide GO assignments for each of the genomes hosted at MicrobesOnline that are based on homology to the InterPro database (Mulder et al. 2005) and based on any EC numbers assigned to that gene. These automated assignments can help to provide an overview of functional capabilities even for newly sequenced or otherwise sparsely annotated genomes.
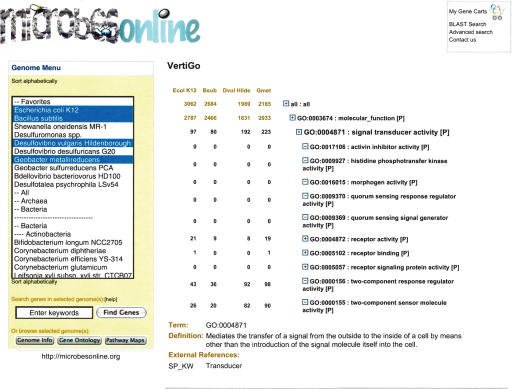
VertiGO, our multispecies GO browser, is similar in design to the AmiGo browser (http://www.godatabase.org/dev/), which displays GO terms downloaded from the Gene Ontology consortium in a hierarchical style. Unlike AmiGo, however, VertiGO is designed to compare multiple genomes at once. Users navigate the GO hierarchy by clicking to expand the three top-level terms initially displayed, or by clicking on a GO term from the Locus Info page. The number of unique genes that are linked to a GO term is displayed for each genome. Many GO terms are not exclusive since a single gene can be involved in multiple biological processes and/or molecular functions. As a result, the number of unique genes in a parent GO term is not always the total count of genes in its child terms. For each GO term with <200 genes, a link is provided to a detailed list of individual genes along with short descriptions. VertiGO allows users to quickly get an overview of the functional profile of a genome, and identify differences among a set of genomes. Figure 2 shows an example of how the VertiGO browser can be used to identify physiological differences between different species or strains. In the figure, the unusually high number of signal transduction proteins found in metal-reducing δ-proteobacteria is highlighted when compared to the model organisms E. coli and Bacillus subtilis.
Figure 2.
The VertiGO Browser. The VertiGO Browser uses the Gene Ontology (Camon et al. 2003) to display and highlight differences in the physiological capabilities among a set of bacteria. In the figure, the unusually large number of signal transduction proteins in the metal-reducing bacteria, D. vulgaris and Geobacter metallireducens, is evident when compared to other model organisms. Users can view any number of genomes in this way by selecting them from the menu at left, and navigate the GO hierarchy by clicking the boxes to the left of each term. In addition, users can retrieve all genes that match a particular GO annotation by clicking the number corresponding to the appropriate term and species (for terms with <200 genes).
Metabolic Pathway Browser
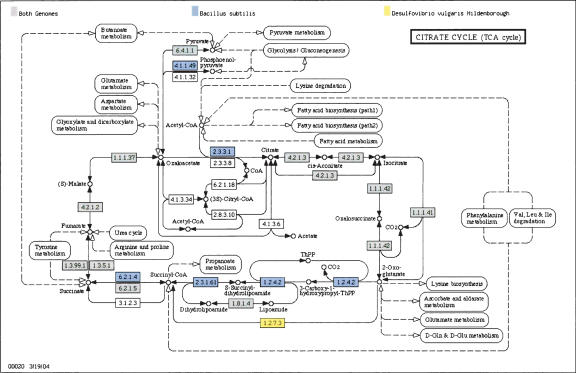
The metabolic capability of a microbial cell is one of the most important phenotypic characteristics for differentiating among species. KEGG maps offer a convenient graphical representation of most metabolic pathways (Kanehisa et al. 2004). The Metabolic Pathway Browser allows users to view the KEGG maps with reactions predicted to be present highlighted. A reaction is present if the enzyme that catalyzes the reaction is found in the genome either by protein sequence homology to a known enzyme, annotations from the KEGG database, or directly from user annotations. The browser can also superimpose metabolic maps from two different genomes and highlight the metabolic differences between them. Figure 3 shows an example comparing the citric acid cycle in B. subtilis and Desulfovibrio vulgaris. The former has a complete pathway, while the metal-reducing anaerobe D. vulgaris is shown to have an incomplete pathway. From the metabolic maps, links to the Locus Info pages for each enzyme are provided to explore individual genes in greater depth.
Figure 3.
Comparative Metabolic Maps. Differences in the tricarboxylic acid (TCA) cycle are shown for the Gram-positive bacterium B. subtilis and the Gram-negative Proteobacterium D. vulgaris. Reactions highlighted with filled boxes indicate that corresponding enzymes were identified in both species (gray boxes), only in B. subtilis (blue boxes), or only in D. vulgaris (yellow boxes). As expected, B. subtilis is found to contain a complete complement of enzymes for the TCA cycle. In contrast, the strictly anaerobic bacterium D. vulgaris is shown to lack a complete cycle, and likely uses different enzymes for conversion between 2-oxoglutarate and succinate.
Locus Info Pages
Detailed information on each gene is displayed on the Locus Info Page, which includes six sections: (1) Gene Info and annotation history, (2) Operon and Regulon Browsers, (3) protein domain alignments, (4) homologs, (5) access to sequences, and (6) an annotation editor. We describe several of these features in more detail below.
Once users have selected a gene of interest, the Microbes-Online Web site provides several ways to learn more about the functional interactions associated with that gene. Information on cotranscription with other genes in an operon, correlation of gene expression profiles, and positional clustering of orthologs in distantly related organisms are displayed graphically in the Operon and Regulon Browsers (the Regulon Browser is shown in Fig. 4).
Figure 4.
Regulon Browser. The Regulon Browser page for flgH of Shewanella oneidensis MR-1 is shown. Genes predicted to be possible transcriptional regulators are highlighted with yellow boxes; other genes are blue. Genes in the same predicted operon are connected, and operons in the same predicted regulon appear inside the same gray box (e.g., the two operons in the top right regulon). Red lines connecting regulons indicate strong similarity in expression patterns, while blue lines indicate chromosomal proximity of the orthologs, one or more component genes from each regulon in several distantly related species. Most of the genes highlighted in this view are flagellar-related, and the fliA gene highlighted in yellow is the alternative σ factor responsible for expression of flagellar genes.
The Operon Browser provides a graphical representation of the predicted operon structure at a given gene locus. As described in more detail in Methods, the operon predictions are derived from a statistical model trained independently for each genome, and have been validated using microarray data in a diverse set of prokaryotes (Price et al. 2005). When available, experimentally identified transcripts are also shown.
The Regulon Browser, shown in Figure 4, presents a high-level summary of the predicted transcriptional regulation of a group of genes related to the query gene (shown on left high-lighted with a blue outline). First, individual genes are grouped into their predicted operons (joined boxes). These operons are further clustered into predicted “regulons” (within the same large gray boxes) if their component genes tend to be in conserved operons in other species even though they are transcribed separately in the target genome (more details in Methods). We have observed that such genes tend to have highly correlated gene expression patterns in microarray experiments (E.J. Alm, R.P. Koche, and A.P. Arkin, unpubl.), and therefore refer to these larger groups as predicted regulons. Finally, these regulons are linked together if they tend to be coexpressed in gene expression experiments (red lines), or if they have a subset of genes that cluster together in unrelated species (blue lines). Genes annotated as possible transcriptional regulators according to their GO classification are colored in yellow.
Functional links to proteins with annotated functions can be used to infer a possible function for uncharacterized genes, or to provide additional support for existing annotations. For each of the genes in the Regulon Browser, GO terms are identified that are enriched 10 times or more over that expected by chance. These terms are listed below the Regulon Browser together with information on which genes were used to infer each functional annotation (data not shown).
Creating annotations
As well as hosting annotations made with external tools, the MicrobesOnline Web site includes a simple interface to add new gene annotations or edit existing annotations. Annotations are stored along with user identity and date of entry. Annotations can change specific attributes of a gene including: the gene name (e.g., trpA), short description (e.g., tryptophan synthase, α subunit), EC numbers, and GO terms. Any number of EC or GO terms may be assigned to or removed from a given gene from the annotation page. A free-text field is provided to address user-formatted attributes in addition to those currently tracked by the system, such as experimental results regarding the gene or links to relevant literature. The use of free-text entries is also encouraged as a way to explain the evidence used to make each annotation.
To add annotations to the database, users must register, an automatic process providing a contact e-mail address so that users can be informed if any changes are made to their annotations. User annotations are visible immediately via the Annotation History table on the Locus Info page, and every 24 h, annotations are parsed and entered into the database, at which point changes to EC and GO assignments are reflected in the metabolic maps and the VertiGO browser.
Since annotations are immediately released into the public domain, the MicrobesOnline Web portal provides a public repository for gene annotations, and community knowledge. To ensure access for the microbiological research community, annotations are available upon request and can be redistributed free of charge or other restrictions.
Gene Carts and the Bioinformatics Workbench
A unique feature of the MicrobesOnline Web site is that each page that displays genes, including the Genome Browser, allows users to identify genes of interest and store them in a Gene Cart for later analysis using the Bioinformatics Workbench. Gene Carts are modeled on the Shopping Cart features common on many commercial Web sites, and they eliminate the need for users to interrupt their browsing session to collect gene sequences for future analysis. Instead, users click to add genes to their Cart from any page within the MicrobesOnline Web site, and continue browsing on the same page. Users can save Gene Carts permanently and create any number of new Carts.
Genes added to a user's Cart can be downloaded to a local computer, or further analyzed using tools provided in the Bioinformatics Workbench. Currently, basic analysis tools for protein or nucleotide sequences are provided including CLUSTALW (Thompson et al. 1994) for multiple sequence alignment and TREE-PUZZLE (Schmidt et al. 2002) for building phylogenetic trees. For users with Java-compatible browsers, a Jalview Web-applet is provided for visualization of sequence alignments (Clamp et al. 2004) in addition to the standard CLUSTALW output. Figure 5 shows an example using the Gene Cart and Bioinformatics Workbench to infer phylogenetic relationships among dissimilatory sulfite reductase genes. This particular example highlights how copies from sulfate-reducing bacteria (D. vulgaris, Desulfovibrio desulfuricans, Desulfotalea psychrophila, and the archeon Archaeglobus fulgidis) form well-supported clades for both genes despite the phylogenetic diversity of the organisms themselves. The phylogenetic tree-building interface supports a choice of substitution matrices, automatic alignment “trimming,” and includes a custom interface for viewing trees.
Figure 5.
Bioinformatics Workbench. The Bioinformatics Workbench allows users to perform basic sequence analysis on the genes and features of interest they have collected in their Gene Carts while browsing the MicrobesOnline Web site. (A) Gene Cart including α and β subunits of dissimilatory sulfite reductase (dsrAB), and a related hydrogenase as an outgroup. (B,C) A multiple sequence alignment and phylogenetic tree generated for the genes in the Gene Cart using the built-in tools. Colors in the tree indicate taxonomy, and numbers show quartet-puzzling scores from TREE-PUZZLE (Schmidt et al. 2002).
To interface with applications not currently implemented on the Web site, users can download their genes in several formats (short description, protein FASTA, and nucleotide FASTA) for use in other software applications.
Conclusions
We present a set of tools that can facilitate the interpretation of the wealth of publicly available microbial sequence data. The power of the MicrobesOnline tools comes from focusing efforts away from single genome analysis toward comparative genomics. In addition, every tool has been designed to allow users to save genes or features of interest to a central workbench area, where users can either conduct bioinformatics analysis within the MicrobesOnline system, or download sequences for use locally. Finally, the MicrobesOnline Web site offers an opportunity for microbiologists to pool their genome annotation efforts by offering a freely accessible central repository for manually curated annotations.
Methods
Constructing the database
To provide a comprehensive view of genome structure for as many species as possible, we imported a complete set of microbial genomes from NCBI (http://www.ncbi.nih.gov/) as well as several draft-quality genomes of particular interest to our group (200+ genomes at the time this manuscript was written). For genomes with gene models deposited at NCBI, we used those models; otherwise protein-coding genes were identified using CRITICA (Badger and Olsen 1999) supplemented with nonoverlapping high-scoring hits from Glimmer (Delcher et al. 1999). Additional RNAs were identified using tRNAscan-SE (Lowe and Eddy 1997) and BLASTn (Altschul et al. 1990), and potential pseudogenes were identified as described below.
For each protein-coding gene, we use a comprehensive set of sequence databases to identify conserved domain structure and to provide additional sources of annotations such as Enzyme Commission (EC) numbers, GO terms (Camon et al. 2003), and membership in COGs (Clusters of Orthologous Groups of proteins) (Tatusov et al. 1997; Marchler-Bauer et al. 2002, 2003). Furthermore, all protein gene models are compared to each other using BLASTp, and the results are stored in the MicrobesOnline database. After applying sequence analysis methods to individual genes, we looked for possible associations between genes using our operon and regulon prediction algorithms (Price et al. 2005). Details on these analyses are provided below.
Protein domains
The Domain Alignments section displays predicted domains and motifs within each protein. The coverage of the sequence by each domain is displayed graphically. The domains and motifs identified are from the publicly available set of databases included in the InterPro (Mulder et al. 2005) database compilation, and currently include: PROSITE (Hulo et al. 2004), UniProt (Bairoch et al. 2005), PRODOM (Servant et al. 2002), Pfam (Bateman et al. 2004), PRINTS (Attwood et al. 2003), SMART (Letunic et al. 2004), PIR SuperFamily (Wu et al. 2003), SUPERFAMILY (Gough et al. 2001), and TIGRFAM (Haft et al. 2003). From the Domain Alignments page, links are included to both the InterPro page for that domain, and to the external databases responsible for the domain definition.
Homologs
The Homologs section displays all BLASTp hits to other genes in the MicrobesOnline database as well as hits to the KEGG (Kanehisa 2002) and SWISS-PROT (Boeckmann et al. 2003) databases, and RPS-BLAST hits to the COG families in the CDD database (Marchler-Bauer et al. 2002, 2003). As in the Domain Alignments section, a graphical view of the sequence coverage is provided. In addition, a short description of the hit, the species of origin, a link to the Locus pages for that hit, and a link to add that hit to the user's Gene Cart are provided. The display can also be limited to a subset of genomes, to predicted orthologs, or to paralogs in the same genome.
GO assignments
GO terms are assigned to genes based on the InterPro domains found within each gene using the external reference file provided by Gene Ontology Consortium, or by manual annotation. Genes with an EC number (except those with one or more dashes) assigned are also mapped to GO terms using this reference file. For InterPro annotated genes, we filtered out GO terms that have a parent-child relationship—only the most specific GO terms are assigned to a gene.
Orthologs
Genes from two organisms are labeled as orthologs if they are bidirectional best BLASTp hits and the sequence alignment coverage is at least 75% of the length of both genes.
COG assignment
We use RPS-BLAST to search against the NCBI COGs included in the CDD database (Marchler-Bauer et al. 2003) and assign COG numbers to the best hit with an E-value < 1e-5 and alignment coverage >60% of the COG.
EC numbers
We used annotations directly from the KEGG database if available. For genomes that are not yet included in the KEGG database, we assigned EC numbers using several lines of evidence. First, if a genome is already annotated in the KEGG database, we use those assignments and do not attempt to assign additional EC numbers. If not, we take EC assignments annotated for TIGRFam equivalogs, which are identified as part of the InterPro pipeline. In addition, we include EC numbers for genes that are orthologs to manually curated E. coli enzymes. Finally, we assign an EC number if that number occurs for >40% of all BLASTp hits to the KEGG database with an E-value ≤ 1e-5, identity >35%, and an alignment length covering >75% of the sequence.
Pseudogene predictions
We did not attempt to distinguish between pseudogenes and unannotated ORFs. To identify pseudogenes of protein-coding genes, we took every intergenic region and used BLASTn to compare it to annotated ORFs. We used tRNAscan-SE (Lowe and Eddy 1997) to find tRNA pseudogenes. We considered only matches >150 bp long, as shorter regions often appear to be remnants of recombination. We also excluded regions that appeared to be truncated from adjacent genes, where the candidate pseudogene and a gene adjacent to it were homologous to the same ORF and matched distinct regions in the correct orientation. This approach identified 6942 pseudogenes in 125 different genomes, including 135 in E. coli and 182 in Bacillus anthracis compared to just two in B. subtilis. On the Web site, each pseudogene is shown with its unique identifier and description of the matching gene. Of the predicted pseudogenes, 1459 (21%) were homologous to the adjacent gene (but not truncations), suggesting that they might have arisen by partial duplication of the gene rather than by decay of a functioning ORF. These cases are labeled in their description, which is accessible from the Locus Info page.
Operon predictions
Two genes on the same strand of DNA that have a very short intergenic distance or are found to be adjacent in multiple unrelated genomes are likely to be in the same operon. In E. coli, the intergenic distance between two adjacent genes in the same operon is usually within ∼50 bp. However, the E. coli distance model is not necessarily suitable for all prokaryotic genomes; therefore, we build genome-specific distance models using an unsupervised machine learning algorithm (Price et al. 2005). Our method performs comparably to other prediction methods in E. coli and B. subtilis, with accuracy >80% on candidate gene pairs (adjacent genes on the same strand). Our analysis of microarray data from E. coli, B. subtilis, Helicobacter pylori, Chlamydia trachomatis, Synechocystis sp. PCC 6803, and the archaeon Halobacterium sp. NRC-1 suggests that our predictions are broadly effective across the prokaryotes. Predictions and a complete description of the method are available from the MicrobesOnline Web site (http://www.microbesonline.org/operons).
Regulon links
Genes whose orthologs tend to be colocalized on the chromosome of multiple microbial genomes are predicted to be functionally related even when they are not nearby in the genome of interest (Dandekar et al. 1998; Overbeek et al. 1999; Lathe III et al. 2000). Most likely these represent genes that tend to be in conserved operons in other species. We have observed that gene pairs identified in this way display strong correlation in gene expression patterns (E.J. Alm, R.P. Koche, A.P. Arkin, unpubl.), and therefore refer to these groups as predicted regulons. Based on these results, we group operons into predicted regulons if each pair of operons in a regulon shares a pair of genes linked by this clustering in unrelated genomes.
Once operons are clustered into regulons, functional links between regulons are computed based on expression profiles and positional clustering as described above. If an operon has genes that are positionally clustered to some, but not all, of the operons in a given regulon, it will not be joined by the “complete linkage” clustering described above. In those cases, the regulons containing both operons will be joined by a blue line. Red links indicate regulons with similar expression profiles (measured using the Pearson correlation coefficient) for genomes with a large enough amount of expression data stored in the MicrobesOnline database (currently E. coli, B. subtilis, and Shewanella oneidensis). Correlations are computed using the average expression profile across all genes in each regulon, as this was shown to improve the functional homogeneity of resulting clusters, most likely by reducing experimental noise (E.J. Alm, R.P. Koche, A.P. Arkin, unpubl.). The threshold for considering two regulons to be correlated is picked manually for each genome to minimize false-positive connections, and is updated regularly as new expression data become available.
Updating genomes
Periodically, new annotations or sequences become available for previously published genomes. In the former case, we simply import the new annotations, and replace gene models with newer variants if the two models have the same stop codon. In the latter case, when there are changes to the nucleotide sequence, we use the nucmer program from the MUMMER software suite (Kurtz et al. 2004) to align the nucleotide sequences before trying to identify equivalent gene models. Equivalent gene models in both cases are assigned the same locusId with different version numbers.
Acknowledgments
The Virtual Institute for Microbial Stress and Survival (VIMSS) and the MicrobesOnline comparative genomics Web site are sponsored by the US Department of Energy Genomics GTL grant (DE-AC03-76SF00098). A.P.A. also acknowledges the support of the Howard Hughes Medical Institute.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3844805. Freely available online through the Genome Research Immediate Open Access option.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Attwood, T.K., Bradley, P., Flower, D.R., Gaulton, A., Maudling, N., Mitchell, A.L., Moulton, G., Nordle, A., Paine, K., Taylor, P., et al. 2003. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 31: 400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger, J.H. and Olsen, G.J. 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16: 512-524. [DOI] [PubMed] [Google Scholar]
- Bairoch, A., Apweiler, R., Wu, C.H., Barker, W.C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. 2005. The Universal Protein Resource (UniProt). Nucleic Acids Res. 33: D154-D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M.C., Estreicher, A., Gasteiger, E., Martin, M.J., Michoud, K., O'Donovan, C., Phan, I., et al. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31: 365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, P.M., Pellegrini, M., Thompson, M.J., Fierro, J., Yeates, T.O., and Eisenberg, D. 2004. Prolinks: A database of protein functional linkages derived from coevolution. Genome Biol. 5: R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon, E., Magrane, M., Barrell, D., Binns, D., Fleischmann, W., Kersey, P., Mulder, N., Oinn, T., Maslen, J., Cox, A., et al. 2003. The Gene Ontology Annotation (GOA) project: Implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 13: 662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp, M., Cuff, J., Searle, S.M., and Barton, G.J. 2004. The Jalview Java alignment editor. Bioinformatics 20: 426-427. [DOI] [PubMed] [Google Scholar]
- Dandekar, T., Snel, B., Huynen, M., and Bork, P. 1998. Conservation of gene order: A fingerprint of proteins that physically interact. Trends Biochem. Sci. 23: 324-328. [DOI] [PubMed] [Google Scholar]
- Delcher, A.L., Harmon, D., Kasif, S., White, O., and Salzberg, S.L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27: 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, J., Karplus, K., Hughey, R., and Chothia, C. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313: 903-919. [DOI] [PubMed] [Google Scholar]
- Haft, D.H., Selengut, J.D., and White, O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31: 371-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo, N., Sigrist, C.J., Le Saux, V., Langendijk-Genevaux, P.S., Bordoli, L., Gattiker, A., De Castro, E., Bucher, P., and Bairoch, A. 2004. Recent improvements to the PROSITE database. Nucleic Acids Res. 32: D134-D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. 2002. The KEGG database. Novartis Found. Symp. 247: 91-101; discussion 101-103, 119-128, 244-252. [PubMed] [Google Scholar]
- Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32: D277-D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz, S., Phillippy, A., Delcher, A.L., Smoot, M., Shumway, M., Antonescu, C., and Salzberg, S.L. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathe III, W.C., Snel, B., and Bork, P. 2000. Gene context conservation of a higher order than operons. Trends Biochem. Sci. 25: 474-479. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Bork, P. 2004. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32: D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer, A., Panchenko, A.R., Shoemaker, B.A., Thiessen, P.A., Geer, L.Y., and Bryant, S.H. 2002. CDD: A database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30: 281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer, A., Anderson, J.B., DeWeese-Scott, C., Fedorova, N.D., Geer, L.Y., He, S., Hurwitz, D.I., Jackson, J.D., Jacobs, A.R., Lanczycki, C.J., et al. 2003. CDD: A curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31: 383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, F., Goesmann, A., McHardy, A.C., Bartels, D., Bekel, T., Clausen, J., Kalinowski, J., Linke, B., Rupp, O., Giegerich, R., et al. 2003. GenDB—An open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31: 2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder, N.J., Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Binns, D., Bradley, P., Bork, P., Bucher, P., Cerutti, L., et al. 2005. InterPro, progress and status in 2005. Nucleic Acids Res. 33: D201-D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek, R., Fonstein, M., D'Souza, M., Pusch, G.D., and Maltsev, N. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. 96: 2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek, R., Larsen, N., Walunas, T., D'Souza, M., Pusch, G., Selkov Jr., E., Liolios, K., Joukov, V., Kaznadzey, D., Anderson, I., et al. 2003. The ERGO genome analysis and discovery system. Nucleic Acids Res. 31: 164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, J.D., Umayam, L.A., Dickinson, T., Hickey, E.K., and White, O. 2001. The Comprehensive Microbial Resource. Nucleic Acids Res. 29: 123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M.N., Huang, K.H., Alm, E.J., and Arkin, A.P. 2005. A novel method for accurate operon prediction in all sequenced prokaryotes. Nucleic Acids Res. 33: 880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, H.A., Strimmer, K., Vingron, M., and von Haeseler, A. 2002. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502-504. [DOI] [PubMed] [Google Scholar]
- Servant, F., Bru, C., Carrere, S., Courcelle, E., Gouzy, J., Peyruc, D., and Kahn, D. 2002. ProDom: Automated clustering of homologous domains. Brief Bioinform. 3: 246-251. [DOI] [PubMed] [Google Scholar]
- Snel, B., Lehmann, G., Bork, P., and Huynen, M.A. 2000. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28: 3442-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278: 631-637. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering, C., Jensen, L.J., Snel, B., Hooper, S.D., Krupp, M., Foglierini, M., Jouffre, N., Huynen, M.A., and Bork, P. 2005. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33: D433-D437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.H., Yeh, L.S., Huang, H., Arminski, L., Castro-Alvear, J., Chen, Y., Hu, Z., Kourtesis, P., Ledley, R.S., Suzek, B.E., et al. 2003. The Protein Information Resource. Nucleic Acids Res. 31: 345-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.godatabase.org/dev/; AmiGo browser.
- http://www.microbesonline.org; MicrobesOnline.
- http://www.ncbi.nih.gov/; NCBI.