Abstract
In yeast, the TREX complex contains the THO transcription elongation complex, which functions in direct cotranscriptional recruitment of the mRNA export proteins Sub2 and Yra1 to nascent transcripts. Here we report the identification of the human THO complex and show that it associates with spliced mRNA, but not with unspliced pre-mRNA in vitro. Transcription is not required for this recruitment. We also show that the human THO complex colocalizes with splicing factors in nuclear speckle domains in vivo. Considering that splicing occurs cotranscriptionally in humans, our data indicate that recruitment of the human TREX complex to spliced mRNA is not directly coupled to transcription, but is instead coupled to transcription indirectly through splicing.
Keywords: Spliceosome, THO complex, TREX complex, mRNA export, splicing, transcription
During expression of protein-coding genes, pre-mRNAs are transcribed in the nucleus and undergo several RNA-processing steps, including capping, splicing, and polyadenylation. The mature mRNA is then exported from the nucleus to the cytoplasm for translation. Many of the steps in gene expression are coupled to each other via an extensive network of physical and functional interactions among the cellular machines that carry out each step of gene expression (for reviews, see Hirose and Manley 2000; Bentley 2002; Maniatis and Reed 2002; Reed 2003). This extensive coupling is likely to function as a quality control mechanism by ensuring that each step occurs efficiently only if the proper contacts are made to the other steps in the pathway.
In yeast, the multi-subunit TREX complex plays a role in coupling transcription to mRNA export (Fischer et al. 2002; Jimeno et al. 2002; Huertas and Aguilera 2003; Rondon et al. 2003a; Abruzzi et al. 2004). This complex contains the mRNA export factors Sub2 and Yra1 as well as the THO complex, which functions in transcription elongation (Fischer et al. 2002; Jimeno et al. 2002). The THO complex contains four subunits, Tho2, Hpr1, Mft1, and Thp2 (Piruat and Aguilera 1998). In addition, Tex1, a protein of unknown function, is present in the TREX complex (Fischer et al. 2002). In contrast to Sub2 and Yra1, Tex1 and the THO components are not essential for viability (Chavez et al. 2000). However, deletion mutants of THO components show defects in transcription elongation and have a hyper-recombination phenotype (Chavez et al. 2000; Rondon et al. 2003a). The elongation defects and hyper-recombination appear to be due to the presence of RNA–DNA hybrids that form between the nascent RNA and the DNA template (Huertas and Aguilera 2003).
Hyper-recombination and transcription defects are also observed with mutations in Sub2 and Yra1, as well as with mutations in other proteins involved in mRNA export including Sac3, Thp1, Mex67, Mtr2, and Nab2 (Fischer et al. 2002; Jimeno et al. 2002; Gallardo et al. 2003). Reciprocally, mRNA export defects are observed with null mutants of THO components (Jimeno et al. 2002; Libri et al. 2002; Strasser et al. 2002). The THO complex associates with actively transcribed genes (Fischer et al. 2002; Hammell et al. 2002; Abruzzi et al. 2004; Kim et al. 2004) and functions in the cotranscriptional loading of Sub2 and Yra1 onto the nascent transcripts, both containing and lacking introns (Zenklusen et al. 2002; Abruzzi et al. 2004). The Drosophila counterpart of the THO complex was also recently identified and contains homologs of Tho2 and Hpr1 as well as three proteins, THOC5, THOC6, and THOC7, which do not have apparent yeast homologs (Rehwinkel et al. 2004). The Drosophila Tex1 protein is present in substoichiometric amounts in the dTHO complex. RNA interference showed that the dTHO complex functions in mRNA export, but genome-wide studies led to the conclusion that the vast majority of mRNAs are transcribed and exported independently of the dTHO complex (Rehwinkel et al. 2004) (see Discussion). An important unanswered question is whether the dTHO complex is cotranscriptionally recruited to active genes.
In human, UAP56 and Aly colocalize with splicing factors in nuclear speckle domains (for reviews, see Izaurralde 2002; Reed and Hurt 2002) and confocal microscopy studies in mammalian cells indicate that UAP56 and Aly are recruited to sites of active transcription in a splicing-dependent manner (Custodio et al. 2004). In previous work, we identified human Tho2 and Hpr1 by virtue of their association with GST-UAP56 (Strasser et al. 2002). We have now characterized the human TREX complex in detail, showing that it contains Aly, UAP56, and the human counterpart of the THO complex. Surprisingly, we find that the human THO complex specifically associates with spliced mRNA and not with unspliced pre-mRNA. Together, our data indicate that recruitment of the human TREX complex to spliced mRNA occurs by a splicing-coupled mechanism rather than by the direct transcription-coupled mechanism that occurs in yeast.
Results and Discussion
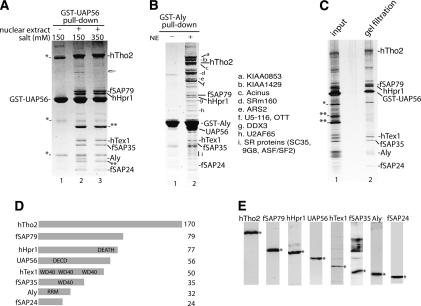
To characterize the human TREX complex, we used optimized conditions for a GST-UAP56 pull-down from RNase-treated HeLa nuclear extract, and a discrete set of bands of approximately equal stoichiometry was detected (Fig. 1A, lane 2). Mass spectrometry identified hTho2, hHpr1, hTex1, and Aly, as observed previously (Fischer et al. 2002). Three other proteins, designated fSAP79, fSAP35, and fSAP24, are also specifically associated with UAP56 (Fig. 1A, lane 2). When the UAP56 pull-down is washed in high salt (350 mM), only Aly dissociates, indicating that this protein is less tightly bound (Fig. 1A, lane 3). When the pull-down is carried out with GST-Aly (in low salt), hTho2, fSAP79, hHpr1, UAP56, hTex1, fSAP35, and fSAP24 are detected (Fig. 1B). We conclude that the human TREX complex contains hTho2, fSAP79, hHpr1, UAP56, hTex1, fSAP35, Aly, and fSAP24. The GST-Aly pull-down also contains several other proteins, labeled a–i, most of which were previously detected in the spliceosome, consistent with the observation that Aly binds to mRNA during splicing (Neubauer et al. 1998; Jurica et al. 2002; Makarov et al. 2002; Rappsilber et al. 2002; Zhou et al. 2002).
Figure 1.
Characterization of the human TREX complex. (A) GST-UAP56 pull-down in the absence (lane 1) or presence (lanes 2,3) of RNase-treated HeLa nuclear extract. The GST-UAP56 pull-down was washed with 150 mM salt (lane 2) or 350 mM salt (lane 3). The indicated bands were identified by mass spectrometry. (*) GST-UAP56 dimer and breakdown products; (**) GST-binding protein in HeLa nuclear extract; the fish icon indicates ARS2, a protein associated with the hTREX complex at low salt and substoichiometrically. (B) GST-Aly pull-down in the absence (lane 1) or presence (lane 2) of RNase-treated HeLa nuclear extracts. a–i are Aly-associated proteins not present in the hTREX complex. The identity of these proteins is shown to the right of B. Abundant proteins such as ribosomal proteins are not indicated. (C) The GST-UAP56 pull-down was eluted with glutathione, run on a Sephacryl S-500 gel filtration column, and fractions run on an SDS gel followed by silver staining. (Lane 1) Input. (Lane 2) Gel filtration fraction. (D) Schematics of hTREX complex proteins. (DEATH) Death domain; (DECD) DECD RNA helicase motif; (WD40) WD 40 repeat; (RRM) RNA recognition motif. (E) Western analysis of nuclear extract with the indicated antibody. The specific protein is designated with an asterisk.
To determine whether all of the hTREX components exist together in one complex, we carried out a GST-UAP56 pull-down and then eluted using glutathione. The eluate was run on a gel filtration column and aliquots of the fractions were examined on a silver-stained SDS gel. Significantly, the eight hTREX proteins are present in the same fraction and are purified away from abundant contaminants (Fig. 1C). The hTREX complex appears to be stable as there was one main peak, and most of the gel filtration fractions contained all of the hTREX proteins (data not shown).
The domain organizations of the hTREX proteins are shown in Figure 1D. To investigate these proteins further, we obtained cDNAs encoding hTho2, fSAP79, hHpr1, hTex1, fSAP35, and fSAP24, and raised antibodies to all of them. We had previously raised antibodies to Aly (Zhou et al. 2000) and UAP56 (Luo et al. 2001). Western analysis with each of these antibodies detects a band of the appropriate size in HeLa cell nuclear extracts (Fig. 1E).
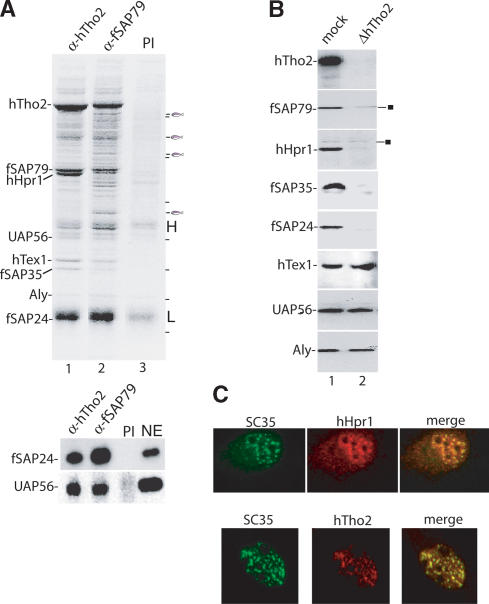
To further characterize the hTREX complex, we carried out immunoprecipitations from RNase-treated HeLa extracts followed by mass spectrometry of specific bands detected on Coomassie-stained gels. Using the hTho2 antibody, hTho2 as well as fSAP79, hHpr1, UAP56, hTex1, fSAP35, Aly, and fSAP24 are detected in the immunoprecipitate (Fig. 2A, lane 1; the antibody light chain is the dark band above fSAP24). The same set of hTHO proteins was detected using the fSAP79 antibody for immunoprecipitation (Fig. 2A, lane 2; the lower panel shows Western blots probed with fSAP24 and UAP56 antibodies). These proteins are not detected using preimmune serum or nonrelated antibodies (Fig. 2A, lane 3; data not shown). We conclude that hTREX exists as a preformed complex in HeLa nuclear extract.
Figure 2.
Identification of the human THO complex and its colocalization with splicing factors in nuclear speckles. (A) Immunoprecipitations from RNase-treated HeLa nuclear extracts using anti-hTho2 antibody (lane 1), anti-fSAP79 antibody (lane 2), and preimmune serum (lane 3). (H) Heavy chain; (L) light chain. Proteins were identified by mass spectrometry. Bands indicated by the fish icon are, from top to bottom, RAD 50 homolog, SAP 145, CDC27 homolog, and NMT 55. A dash indicates molecular weight markers, from top to bottom (in kilodaltons): 181, 115, 82, 63, 48, 37, and 26. (Lower panel) Western analysis of immunoprecipitated samples. (NE) Nuclear extract. (B) Western analysis of mock- and hTho2-immunodepleted nuclear extracts. The square indicates nonspecific bands. (C) Immunofluorescence of HeLa cells. Merge shows coimmunofluorescence with the SC35 antibody.
Identification of the human THO complex
To determine whether a salt-stable human THO complex exists, we carried out Western analysis of nuclear extracts immunodepleted with the hTho2 antibody in high salt (375 mM). hTho2, as well as hHpr1 and fSAP79, fSAP35, and fSAP24, are completely depleted by this antibody, whereas UAP56, Aly, and hTex1 are not (Fig. 2B). These data, together with the gel filtration analysis (Fig. 1C), indicate that all of the fSAP79, fSAP35, fSAP24, and hHpr1 in the nuclear extract exist together with hTho2 in a single salt-stable complex. The hTHO complex resembles that of Drosophila, which contains counterparts of Tho2 and Hpr1 as well as of fSAP79, fSAP35, and fSAP24 (designated THOC5–7) (Rehwinkel et al. 2004). There are no apparent Mft1 or Thp2 homologs in human or Drosophila and conversely no fSAP79, fSAP35, and fSAP24 homologs in yeast.
Aly and UAP56 are localized in the nucleus with a pattern similar to that of most splicing factors. Specifically, these proteins have both a diffuse nucleoplasmic staining and also colocalize with SC35, an SR protein-splicing factor that is tightly localized to nuclear speckle domains (Spector 1993; Zhou et al. 2000; Gatfield et al. 2001). Significantly, both hTho2 and hHpr1 show a similar staining pattern to Aly, UAP56, and most splicing factors, with a diffuse nucleoplasmic staining and colocalizing with SC35 (Fig. 2C). We conclude that the human TREX complex colocalizes with the splicing factors in the nucleus.
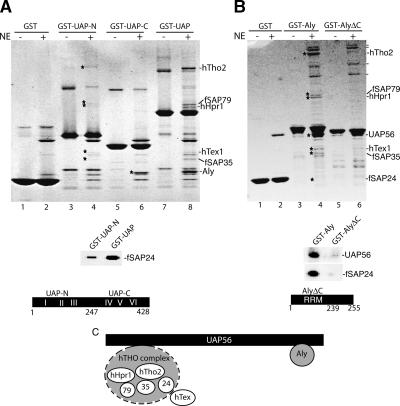
To gain insight into how the human THO complex associates with UAP56 and Aly, we used mutant forms of these proteins for GST pull-downs from RNase-treated nuclear extract (Fig. 3). This analysis revealed that UAP56-N does not interact with Aly but does associate at a low level with hTho2, fSAP79, hHpr1, hTex1, fSAP35, and fSAP24 (Fig. 3A, lane 4; these bands were detected faintly on the Coomassie-stained gel but were readily identified by mass spectrometry and fSAP24 was also identified by a Western blot, lower panel). Aly interacts with UAP56-C as efficiently as with full-length UAP56, indicating that the C terminus is sufficient for Aly binding (Fig. 3A, lane 6).
Figure 3.
Aly and the hTHO complex bind to distinct regions of UAP56. (A,B) GST pull-downs with the indicated proteins in the absence (–) or presence (+) of RNase-treated HeLa nuclear extracts. hTHO components are indicated with stars. On the right of the gel shown in B, a dash indicates Aly-associated proteins identified in Figure 1B. (Lower panels) Western analysis with the indicated antibodies from the GST-UAP-N, GST-UAP, GST-Aly, and GST-AlyΔC eluates used in A and B. The structures of UAP-N (amino acids 1–247), UAP-C (amino acids 247–428), and AlyΔC (amino acids 1–239) are indicated. (C) A model for interaction of the hTHO complex and Aly with UAP56. The hTHO complex is shown with a dashed line because it binds weakly to UAP-N, indicating that some other region of UAP56 is required for efficient binding.
Both UAP56 and the hTHO complex interact with GST-Aly, but not with GST-AlyΔC (Fig. 3B, and see Western blot, lower panel). Thus, the C terminus of Aly is required for binding of both UAP56 and the hTHO complex. Previous studies with recombinant proteins showed that the C terminus of Aly is required for direct interactions with UAP56 (Luo et al. 2001). A preliminary model for the interactions in the hTREX complex is shown in Figure 3C. The C terminus of UAP56 is sufficient for Aly binding. In contrast, the N terminus of UAP56 binds only a low level of the hTHO complex (designated by dashed line in Fig. 3C), indicating that other regions of UAP56 are required for maximal binding.
Transcription-independent recruitment of the human TREX complex to spliced mRNA in vitro
To investigate whether the human THO complex, like its yeast counterpart (Chavez et al. 2001; Jimeno et al. 2002; Rondon et al. 2003b), functions in transcription, we used the hTho2-innumodepleted nuclear extract to assay initiation and elongation with linear or supercoiled plasmids containing the CMV promoter fused to templates ranging in length from 200 to 8000 bp. No effect on transcription was observed with the depleted extract compared to either mock or Aly-immunodepleted extracts (data not shown). A similar assay was successfully used to detect a role for the THO complex in yeast extracts (Rondon et al. 2003b). Thus, it is possible that the human THO complex has a different function than the yeast THO complex or that our assays are not appropriate for detecting a role for the human complex in transcription.
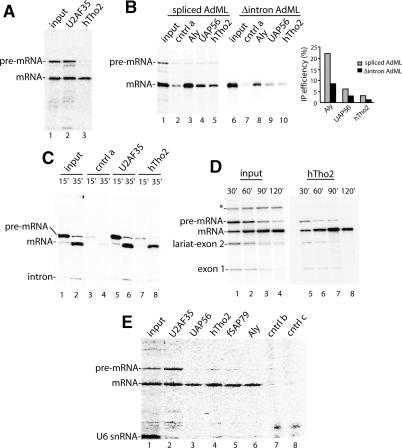
Nuclear extracts depleted of Aly, UAP56, or the hTHO complex were also used to assay for splicing in vitro. No effect on spliceosome assembly, splicing, or RNA stability was observed (data not shown). All of the TREX components were nevertheless previously detected by mass spectrometry in purified spliceosomes (Neubauer et al. 1998; Jurica et al. 2002; Makarov et al. 2002; Rappsilber et al. 2002; Zhou et al. 2002). Thus, we next asked when during the splicing pathway the TREX complex associates with the spliceosome. To do this, we carried out immunoprecipitations of splicing reactions containing 32P-labeled AdML (Fig. 4A) or Ftz (data not shown) pre-mRNA. Both pre-mRNA and mRNA are immunoprecipitated by the positive control antibody to the splicing factor U2AF35 (Fig. 4A, lane 2). Unexpectedly, antibodies to hTho2, though its yeast counterpart functions in transcription, efficiently immunoprecipitate the spliced mRNA but not unspliced pre-mRNA (Fig. 4A, lane 3).
Figure 4.
The hTREX complex is recruited to spliced mRNAs by a transcription-independent mechanism. (A) T7 AdML pre-mRNA was incubated under splicing conditions for 90 min, and aliquots (5 μL) of the reaction were used for immunoprecipitations. (B) T7-AdML pre-mRNA (lanes 1–5) or corresponding cDNA transcript (lanes 6–10) was incubated under splicing conditions, followed by immunoprecipitations. The percentage immunoprecipitated relative to input was quantitated by PhosphorImager. One-fourth of the input was loaded on the gel. (C–E) Ftz pre-mRNA was synthesized and spliced in an RNAP II transcription/splicing system in HeLa nuclear extract for the times indicated (input). An aliquot (5 μL) from each time point was used for immunoprecipitations. One-fourth (C) or one-tenth (D,E) of the input was loaded on the gel. (*) Nonspecific band. U6 snRNA is labeled during incubation of the RNAP II transcription/splicing system. Control antibodies (cntrl a–c) were rabbit antibodies to unrelated proteins.
To determine whether the hTREX complex is also recruited to AdML cDNA transcripts (Δintron AdML), we carried out immunoprecipitations side by side with spliced mRNA using antibodies to hTho2, Aly, and UAP56. As shown in Figure 4B, the Δintron AdML is immunoprecipitated less efficiently than spliced mRNA. Abundant RNA-binding proteins, including hnRNP proteins, splicing factors, and export factors, bind to cDNA transcripts and random RNAs depending on the sequences of the RNAs (data not shown). Thus, although this type of analysis is consistent with previous studies in vivo and in vitro indicating that recruitment of UAP56 and Aly to mRNA is splicing-dependent (Custodio et al. 2004; Zhou et al. 2000), further studies are required to support this conclusion for the hTHO complex. In any case, the observation that the hTREX antibodies immunoprecipitate the spliced mRNA and the cDNA transcript (albeit less efficiently) but not the unspliced pre-mRNA indicates that the hTREX complex is specifically excluded from the unspliced pre-mRNA.
To determine whether the hTREX complex might require RNAP II transcription to load onto the unspliced pre-mRNA, we carried out immunoprecipitations using an RNAP II-coupled transcription/splicing system in HeLa nuclear extract (R. Das and R. Reed, in prep.). In this system, a DNA template containing the CMV promoter and encoding Ftz pre-mRNA was transcribed for 15 min to generate unspliced pre-mRNA or 35 min to generate both unspliced and spliced products (Fig. 4C, lanes 1,2). As shown in Figure 4C, the U2AF35 antibody immunoprecipitated both the unspliced pre-mRNA and spliced mRNA (lanes 5,6). In contrast, the hTho2 antibody efficiently immunoprecipitated only the spliced mRNA and not the unspliced pre-mRNA (Fig. 4C, lanes 7,8). When a longer transcription/splicing time course was carried out, hTho2 remained associated with the spliced mRNA (Fig. 4D, lanes 1–8). Thus, even when the pre-mRNA was generated by transcription, hTho2 was not associated with it until after splicing.
Significantly, antibodies to other hTREX components (UAP56, fSAP79, and Aly) also efficiently immunoprecipitate the spliced mRNA and not the unspliced pre-mRNA (Fig. 4E). It is unlikely that our failure to detect significant levels of immunoprecipitation of the unspliced pre-mRNA is due to a buried epitope as antibodies to so many different hTREX components gave the same result. We conclude that the hTHO complex, as well as UAP56 and Aly, are recruited to spliced mRNA, and this recruitment does not occur cotranscriptionally in our in vitro system.
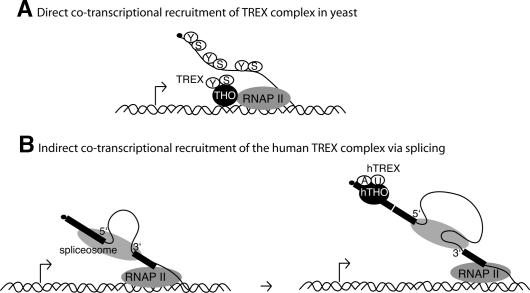
In this study, we have characterized the human counterpart of the yeast TREX complex, which has been extensively studied as a factor involved in the direct cotranscriptional recruitment of the yeast mRNA export machinery (Lei et al. 2001; Lei and Silver 2002; Libri et al. 2002; Strasser et al. 2002; Zenklusen et al. 2002; Abruzzi et al. 2004; Kim et al. 2004). Surprisingly, our in vitro studies indicate that the human TREX complex, while largely conserved in composition, is recruited to spliced mRNAs at a late step in the splicing reaction rather than by direct cotranscriptional recruitment as in yeast. Assuming that this splicing-coupled mechanism also occurs in vivo in human cells, it is possible that there are distinct mechanisms for TREX complex recruitment: splicing-coupled and direct transcription-coupled. These mechanisms may differ in yeast versus human because most yeast genes lack introns, whereas the converse is the case in metazoans. In metazoans, splicing is thought to occur cotranscriptionally (for review, see Bentley 2002). Thus, as shown in the model (Fig. 5), recruitment of the hTREX complex to pre-mRNAs containing multiple introns will obviously be cotranscriptional, but this recruitment will only occur as a consequence of splicing. Significantly, confocal studies in mammalian cells support the conclusion that the export machinery is cotranscriptionally recruited in a splicing-dependent manner (Custodio et al. 2004).
Figure 5.
Models for TREX complex recruitment. (A) Direct cotranscriptional recruitment of Yra1 (Y) and Sub2 (S) via the THO complex (THO) to nascent yeast transcript. (B) Indirect cotranscriptional recruitment of the hTREX complex to spliced mRNA UAP56 (U), Aly (A).
Our data show that the hTREX complex is assembled on every mRNA examined (this study; data not shown). Similarly, in yeast chromatin immunoprecipitations assays, the TREX complex has been detected on every gene examined (Zenklusen et al. 2002; Abruzzi et al. 2004; Kim et al. 2004; our unpublished observations). Considering the general association of the TREX complex with a wide variety of mRNAs, together with the striking conservation of this complex, it seems plausible that export via the TREX complex is a general physiological pathway for most mRNAs. The failure to detect a general mRNA export role for these factors when they are depleted by RNAi may be because a nonphysiological pathway substitutes when TREX is absent.
Materials and methods
GST pull-downs
GST-binding proteins were precleared from 200 μL of RNase-treated nuclear extract by mixing it with 4 volumes of PBS/0.1% Triton X-100/0.2 mM PMSF (buffer A), protease inhibitor EDTA-free (Roche), 8 μg of GST protein, and 20 μL of glutathione Sepharose 4B (Amersham). The reaction mixture was rotated at 4°C for 2 h. After a brief spin, the supernatant was transferred to a new tube and mixed with 20 μL of glutathione Sepharose 4B at 4°C for 2 h. The supernatant was then mixed with 8 μg of GST-UAP56 or GST-Aly and 20 μL of glutathione beads at 4°C overnight. The beads were washed four times with buffer A, then eluted with SDS loading buffer containing 0.5 mM DTT. After boiling, samples were loaded on a 4%–12% gradient gel (Invitrogen). Gel filtration column isolation of the hTREX complex was performed by carrying out a GST-UAP56 pull-down, eluting the bound material with glutathione, and loading the eluate on a Sephacryl S500 column (equilibrated in PBS/0.1% Triton X-100/0.2 mM PMSF). Aliquots of the column fractions were run on an SDS gel followed by silver staining.
Plasmids and cDNAs
The hTho2 clone was described (Strasser et al. 2002). Full-length hHpr1, hTex1, fSAP79, fSAP35, and fSAP24 clones were obtained from I.M.A.G.E., and their sequences were confirmed by DNA sequencing. The accession numbers for full-length hHpr1, hTex1, fSAP79, fSAP35, and fSAP24 clones are BC010381, BC006849, BC003615, BC003118, and AK027098, respectively. Each cDNA was subcloned into a pGEX vector (Amersham) to express GST fusion proteins.
Antibodies
To obtain recombinant proteins for raising antibodies, full-length hTex1 and fSAP35, hTho2 C terminus or hHpr1 C terminus (amino acids 465–657) were fused to GST and transformed into BL21. Full-length fSAP24 was fused to MBP. Full-length his-tagged fSAP79 cDNA was produced in baculovirus. Antibodies to fSAP35 and fSAP24 were raised in rat and all others in rabbit. Immunofluorescence was carried using the indicated primary antibodies and conjugated secondary antibodies (Harlow 1988; Zhou et al. 2000).
Immunoprecipitations
Antibodies were covalently cross-linked to protein A Sepharose in dimethylpimelimidate (Sigma) at a 2:1 ratio of beads to antibody (Harlow 1988). For coimmunoprecipitations, 200 μL of nuclear extract was mixed with 5 volumes of buffer A, protease inhibitor EDTA-free (Roche), and 20 μL of antibody-cross-linked beads. The mixture was rotated overnight. The beads were washed four times with buffer A, and mixed with protein gel loading buffer without DTT. After incubating at room temperature for 5 min, the eluate was recovered to a new tube. DTT was added to the eluate to a final concentration of 0.5 mM. After boiling, the samples were loaded onto 4%–12% gradient gels (Invitrogen).
Immunodepletion
Antibody-cross-linked beads were prepared as described above, except that a 1:4 ratio of beads to antibody was used. Nuclear extract (400 μL) in high salt (350 mM KCl) was mixed with 100 μL of antibody-cross-linked beads and rotated at 4°C for 2 h. After a brief spin, the supernatant was recovered, and a fresh 100-μL aliquot of antibody-cross-linked beads was added. The supernatant was recovered and mixed with another fresh 100-μL aliquot of antibody-cross-linked beads. The depleted nuclear extracts were stored at –80°C.
RNAP II transcription/splicing and RNA immunoprecipitation
A coupled RNAP II transcription/splicing system will be described elsewhere (R. Das and R. Reed, in prep.). Briefly, a PCR product (200 ng) containing the CMV promoter fused to an AdML or Ftz splicing template containing one intron and two exons was incubated in a 25-μL reaction mixture containing 1 μL of 32P-UTP (800 Ci/mmol), 15 μL of HeLa nuclear extract, 0.5 mM ATP, 3.5 mM MgCl2, and 20 mM creatine phosphate. Reactions were incubated at 30°C for the times indicated. For immunoprecipitation, antibodies were coupled at a 1:2 ratio of protein A Sepharose beads to antibody. Immunoprecipitations were carried out using 10 μL of antibody-coupled beads, 5 μL of RNAP II transcription/splicing or T7 splicing reaction, and 100 μL of high-salt buffer (250 mM KCl, 20 mM HEPES at pH 7.9, 0.1% Triton, 0.25 mM EDTA) and rotated overnight at 4°C. The immunoprecipitates were washed with high-salt buffer, total RNAs were ethanol-precipitated, and analyzed on a 6.5% denaturing polyacrylamide gel.
Acknowledgments
We are grateful to Dr. K. Nagata for the gift of GST-UAP56-N and GST-UAP56-C plasmids. We thank Sabine Merker, Petra Ihrig (Dr. J. Lechner, BZH Mass Spectrometry), and Jennifer Shuler (Nikon Imaging Center, Harvard Medical School) for technical assistance. HeLa cells were obtained from the National Cell Culture Center (Minneapolis, MN).
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1302205.
References
- Abruzzi K.C., Lacadie, S., and Rosbash, M. 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 23: 2620–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. 2002. The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14: 336–342. [DOI] [PubMed] [Google Scholar]
- Chavez S., Beilharz, T., Rondon, A.G., Erdjument-Bromage, H., Tempst, P., Svejstrup, J.Q., Lithgow, T., and Aguilera, A. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19: 5824–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S., Garcia-Rubio, M., Prado, F., and Aguilera, A. 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 7054–7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N., Carvalho, C., Condado, I., Antoniou, M., Blencowe, B.J., and Carmo-Fonseca, M. 2004. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA 10: 622–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Strasser, K., Racz, A., Rodriguez-Navarro, S., Oppizzi, M., Ihrig, P., Lechner, J., and Hurt, E. 2002. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21: 5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M., Luna, R., Erdjument-Bromage, H., Tempst, P., and Aguilera, A. 2003. Nab2p and the Thp1p–Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 278: 24225–24232. [DOI] [PubMed] [Google Scholar]
- Gatfield D., Le Hir, H., Schmitt, C., Braun, I.C., Kocher, T., Wilm, M., and Izaurralde, E. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11: 1716–1721. [DOI] [PubMed] [Google Scholar]
- Hammell C.M., Gross, S., Zenklusen, D., Heath, C.V., Stutz, F., Moore, C., and Cole, C.N. 2002. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 22: 6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow L. 1988. Antibodies: A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Hirose Y. and Manley, J.L. 2000. RNA polymerase II and the integration of nuclear events. Genes & Dev. 14: 1415–1429. [PubMed] [Google Scholar]
- Huertas P. and Aguilera, A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12: 711–721. [DOI] [PubMed] [Google Scholar]
- Izaurralde E. 2002. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur. J. Cell Biol. 81: 577–584. [DOI] [PubMed] [Google Scholar]
- Jimeno S., Rondon, A.G., Luna, R., and Aguilera, A. 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 21: 3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M.S., Licklider, L.J., Gygi, S.R., Grigorieff, N., and Moore, M.J. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Ahn, S.H., Krogan, N.J., Greenblatt, J.F., and Buratowski, S. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23: 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P. and Silver, P.A. 2002. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes & Dev. 16: 2761–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P., Krebber, H., and Silver, P.A. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 15: 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen, T.H. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22: 8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.J., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M., and Reed, R. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647. [DOI] [PubMed] [Google Scholar]
- Makarov E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., and Luhrmann, R. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Maniatis T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Neubauer G., King, A., Rappsilber, J., Calvio, C., Watson, M., Ajuh, P., Sleeman, J., Lamond, A., and Mann, M. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20: 46–50. [DOI] [PubMed] [Google Scholar]
- Piruat J.I. and Aguilera, A. 1998. A novel yeast gene, THO2, is involved in RNA Pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 17: 4859–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ryder, U., Lamond, A.I., and Mann, M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15: 326–331. [DOI] [PubMed] [Google Scholar]
- Reed R. and Hurt, E. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108: 523–531. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Herold, A., Gari, K., Kocher, T., Rode, M., Ciccarelli, F.L., Wilm, M., and Izaurralde, E. 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 11: 558–566. [DOI] [PubMed] [Google Scholar]
- Rondon A.G., Garcia-Rubio, M., Gonzalez-Barrera, S., and Aguilera, A. 2003a. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 22: 612–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon A.G., Jimeno, S., Garcia-Rubio, M., and Aguilera, A. 2003b. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 278: 39037–39043. [DOI] [PubMed] [Google Scholar]
- Spector D.L. 1993. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9: 265–315. [DOI] [PubMed] [Google Scholar]
- Strasser K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 28: 28. [DOI] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra, P., Wyss, J.C., and Stutz, F. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22: 8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Luo, M.J., Straesser, K., Katahira, J., Hurt, E., and Reed, R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Licklider, L.J., Gygi, S.P., and Reed, R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. [DOI] [PubMed] [Google Scholar]