Abstract
Pcf11 is one of numerous proteins involved in pre-mRNA 3′-end processing and transcription termination. Using elongation complexes (ECs) formed from purified yeast RNA polymerase II (Pol II), we show that a 140-amino acid polypeptide from yeast Pcf11 is capable of dismantling the EC in vitro. This action depends on the C-terminal domain (CTD) of the largest subunit of Pol II and the CTD-interaction domain (CID) of Pcf11. Our experiments reveal a novel termination mechanism whereby Pcf11 bridges the CTD to the nascent transcript and causes dissociation of both Pol II and the nascent transcript from the DNA in the absence of nucleotide hydrolysis. We posit that conformational changes in the CTD are transduced through Pcf11 to the nascent transcript to cause termination.
Keywords: Polyadenylation, termination, RNA polymerase II, Pcf11, CTD
Transcription termination in eukaryotes is essential for recycling polymerase II (Pol II) and for preventing Pol II from perturbing promoters of genes located downstream from a transcription unit (Proudfoot et al. 2002; Proudfoot 2004). The mechanism of Pol II termination is poorly understood. Termination depends on the polyadenylation site in the nascent transcript. Numerous proteins involved in cleavage and polyadenylation also appear to participate in termination since mutations in several of these proteins affect both RNA processing and termination (Birse et al. 1998; Sadowski et al. 2003). Termination also depends on the C-terminal domain (CTD) of the largest subunit of Pol II (McCracken et al. 1997; Park et al. 2004). The CTD is composed of repeating heptapeptide motifs with the consensus YSPTSPS, and undergoes cycles of phosphorylation and dephosphorylation during the transcription process (Buratowski 2003). It functions as both a binding site and an allosteric regulator of the RNA processing machinery and serves to couple RNA processing and transcription elongation (Bentley 2002).
In addition to the termination that occurs at the end of a transcription unit, premature termination within the body of genes can affect the processivity of Pol II in ways that regulate gene expression. One of the clearest examples of this is HIV. In the absence of the virally encoded protein Tat, transcription from the LTR results in accumulation of short transcripts in the cytoplasm due to premature termination (Feinberg et al. 1991; Kessler and Mathews 1992). HIV Tat modifies the elongation complex (EC) by recruiting the kinase, P-TEFb (Price 2000). P-TEFb phosphorylates the CTD of Pol II, and phosphorylation of the CTD increases the processivity of Pol II at least in part by counteracting the inhibitory action of two elongation factors, NELF and DSIF (Wada et al. 1998; Yamaguchi et al. 1999, 2002; Renner et al. 2001). Several studies have provided evidence that cellular activators stimulate gene expression by increasing the processivity of Pol II, although it is not clear if this occurs by preventing premature termination or by reducing the frequency of pauses and arrests (Krumm et al. 1993; Yankulov et al. 1994; Bentley 1995; Blau et al. 1996; Lis 1998; Lis et al. 2000; Barboric et al. 2001).
Isolated Pol II ECs are remarkably stable, resisting disruption by high salt, sarkosyl, heparin, and even proteinase K (Ackerman et al. 1983; Brown et al. 1996; Gnatt et al. 1997; Zhang et al. 2004). This raises the question of how the EC is dismantled from the DNA. Moreover, how does the CTD impinge on the processivity since the CTD projects from the body of the Pol II molecule in an unstructured conformation and is attached by a flexible linker (Meinhart and Cramer 2004; Noble et al. 2005)? The negative elongation factors NELF and DSIF slow the rate of elongation but do not appear to dissociate the EC (Wada et al. 1998; Yamaguchi et al. 1998, 1999). Nucleosomes also inhibit elongation, but they alone do not appear to induce termination (Izban and Luse 1991; Kireeva et al. 2005). A protein called TTF2 (also known as factor 2) disrupts both Pol II ECs in an ATP-dependent fashion, but this protein appears to function during mitosis to clear ECs from the DNA during chromosome condensation (Jiang et al. 2004). Moreover, since TTF2 also acts upon Pol I, its function does not appear to be through the CTD (Hara et al. 1999).
Previously, we discovered that an antibody interacting with the CTD of Drosophila Pol II caused Pol II to release the nascent transcript (Zhang et al. 2004). A search for a Drosophila protein that had similar activity resulted in the identification of a protein with sequence similarity to yeast Pcf11 (Z. Zhang and D.S. Gilmour, in prep.). Mutations in yeast Pcf11 cause Pol II to read through a transcription terminator in vivo (Sadowski et al. 2003). Moreover, Pcf11 associates with the CTD, and a defect in termination correlates with mutations in the CTD-interacting domain (CID) of Pcf11 (Sadowski et al. 2003). Since genetic data implicated Pcf11 in Pol II termination in yeast, we investigated what effect yeast Pcf11 has on an EC formed from purified yeast Pol II. We identify a novel CTD-dependent mechanism for dismantling the EC and propose that Pcf11 as the engine that drives some termination reactions.
Results
Formation of stalled ECs from purified yeast Pol II and a tailed template
We adopted a procedure first used to study murine and yeast Pol I termination to generate a stalled EC that could be used as a model for Pol II termination (Kuhn et al. 1990; Lang et al. 1994; Zhang et al. 2004). Figure 1A illustrates the stalled complex. Transcription by purified yeast Pol II was initiated at the end of a tailed template with the dinucleotide UpG. Transcription of the G-less cassette in the presence of radiolabeled UTP but the absence of GTP resulted in a stalled EC having a radiolabeled nascent transcript. Analysis of the transcript produced under a variety of conditions verifies that the EC formed as anticipated. The transcript was sensitive to digestion by RNase A but not RNase H (Fig. 1B, lanes 2,3), indicating that the nascent transcript had not formed a heteroduplex along its entire length as is sometimes the case when purified Pol II transcribes tailed templates (Dedrick and Chamberlin 1985). Transcription required the dinucleotide UpG (Fig. 1B, lane 5), indicating that initiation occurred at the underlined AC in Figure 1A. Results in Figure 1C show that the stalled ECs were transcriptionally active, since they resumed elongation upon addition of GTP.
Figure 1.
Generation of a stalled EC from purified yeast Pol II and a tailed template. (A) Schematic of the stalled EC formed from highly purified yeast Pol II and a special template. In the presence of the UpG dinucleotide, purified Pol II initiates transcription at the underlined AC and elongates to the end of a 68-nt-long G-less cassette (for further description, see Zhang et al. 2004). The bottom strand has a biotin tag at its 5′-end for immobilizing the DNA template. (B) Evaluation of transcripts produced under various conditions. All reactions contained 500 μM ATP, 500 μM UTP, and 25 μM UTP supplemented with 2 μCi of α-labeled 32P UTP. For samples in lanes 2 and 3, RNase treatments were performed on stalled ECs. (C) The stalled ECs resume elongation when supplied GTP. (Lane 1) Transcripts recovered from complexes stalled by the absence of GTP. The weak band marked by an asterisk appears to be due to contaminating GTP, since it can be eliminated by including 3′-methoxy-GTP in the reaction (Zhang et al. 2004). (Lane 2) Run-off transcripts produced when stalled ECs were supplied GTP. Radioactive transcripts were detected in denaturing gels with a PhosphorImager.
yPcf11 dismantles the EC
ECs containing radiolabeled RNA migrated as one prominent complex following electrophoresis on a native gel (Fig. 2A, lane 1). Treatment of the EC with full-length yPcf11 or the CID of Pcf11 prior to electrophoresis significantly diminished the amount of complex detected in the gel (Fig. 2A, lanes 2,3). To determine if the decrease in complex detected on the native gel corresponded to a complete dismantling of the EC, we formed ECs on an immobilized DNA template and determined if treatment of these ECs affected the association of Pol II and the nascent transcript. RNA and Pol II remained bound (B) to the immobilized template when treated with BSA (Fig. 2B [lanes 1,2], C [lanes 9,10]). In contrast, treatment of the EC with the CID caused transcript and Pol II to be released (R) from the immobilized template (Fig. 2B [lanes 5,6], C [lanes 1,2]). Dismantling of the EC occurred in the absence of nucleotides as these were washed away during preparation of the immobilized complexes.
Figure 2.
The CID of Pcf11 dismantles an EC. (A) Native gel analysis of ECs treated with intact Pcf11, the CID, or mutant derivatives. The amino acid changes associated with the M1, M2, and M3 mutations are provided above lanes 4–6. (Lane 1) ECs generated from purified Pol II and treated with BSA. (Lanes 2–6) ECs treated for 30 min with Pcf11, CID, or its derivatives prior to electrophoresis on the native gel. The complexes were detected by the presence of the radiolabeled nascent transcript. (B) Measurement of transcripts released from immobilized ECs following treatment with derivatives of Pcf11. The ECs were formed on DNA templates that had been immobilized on magnetic beads via an avidin-biotin linkage (Zhang et al. 2004). The immobilized ECs were incubated for 30 min with various derivatives of Pcf11 and then separated into bound (B) and released (R) fractions. The radiolabeled transcripts in each fraction were isolated and analyzed on a denaturing polyacrylamide gel. (C) Western blot analysis of the Pol II recovered from the bound and released fractions following treatment of immobilized ECs with derivatives of the CID. Pol II was detected in the bound and released fractions using the Pol II antibody, ARNA-3 (Research Diagnostics, Inc.).
Identification of mutations in the CID that impair the dismantling activity
A three-amino acid mutation at positions 68–70 in the CID of yeast Pcf11 had previously been found to impair termination in vivo (Sadowski et al. 2003). To determine if the dismantling activity observed in vitro correlated with termination in vivo, a CID carrying the corresponding mutation was analyzed. The three-amino acid mutation, CID-M3, completely abrogated the dismantling activity as measured by native gel electrophoresis (Fig. 2A, lane 6), release of nascent transcript (Fig. 2B, lanes 11,12), and release of Pol II (Fig. 2C, lanes 7,8). The crystal structure of the yeast CID predicts that amino acids 68 and 69 make contact with the CTD while amino acid 70 projects into the hydrophobic core of the protein (Meinhart and Cramer 2004). Mutating the two amino acids that contact the CTD resulted in partial loss of dismantling activity (Fig. 2A [lane 5], B [lanes 9,10], C [lanes 5,6]). A CID mutant with a single change at position 66 was also tested because this mutation in the CID, along with several other mutations elsewhere in the protein, impaired termination in vivo (Sadowski et al. 2003). The single-amino acid mutation at position 66 in the CID also impaired the dismantling activity (Fig. 2A [lane 4], B [lanes 7,8], C [lanes 3,4]). These results are consistent with the notion that the dismantling activity of the CID of Pcf11 detected in vitro is involved in Pol II termination in vivo.
The dismantling reaction requires the CTD of Pol II
The CTD of the largest Pol II subunit has been implicated in transcription termination in mammalian cells (McCracken et al. 1997). To test if the dismantling activity of the CID involved the CTD, we compared the sensitivity of Pol IIA and Pol IIB ECs to the action of the CID. Pol IIA has an intact CTD and was the version of Pol II used in the experiments presented in Figures 1 and 2. Pol IIB lacks the CTD and can be produced by treating Pol IIA with chymotrypsin, which selectively removes the CTD (Dezelee et al. 1976; Zehring et al. 1988). Removal of the CTD can be detected as a shift in the mobility of the largest subunit on SDS-PAGE (Fig. 3B). Treatment of Pol II with chymotrypsin prior to formation of the ECs resulted in complexes with significantly faster mobility than Pol IIA ECs on the native gel (Fig. 3A, cf. lanes 1 and 4), similar to what was previously seen for Drosophila Pol IIA and IIB ECs (Zhang et al. 2004). PMSF was used to inhibit chymotrypsin after trimming off the CTD. When PMSF was added before the chymotrypsin, Pol II retained the CTD as evidenced by the similarity in the mobility of the ECs to those generated by untreated Pol II (Fig. 3, lanes 1,2). This Pol IIA EC remained sensitive to dismantling by the CID (Fig. 3A, lane 3). In contrast, the Pol IIB EC was completely resistant to the CID (Fig. 3A, lane 5). These data indicate that the CID requires the presence of the CTD on Pol II to dismantle the EC.
Figure 3.
The effects of the CID on Pol IIA and Pol IIB ECs. (A) Detection of Pol IIA and Pol IIB ECs before and after treatment with CID. Prior to forming ECs, a portion of Pol IIA was converted to Pol IIB by treatment with chymotrypsin. Another portion of the Pol IIA was treated first with PMSF and then chymotrypsin, so no proteolysis occurred. (Lane 1) Pol IIA EC treated with buffer. (Lanes 2,3) Pol IIA EC formed from Pol IIA that had been incubated first with PMSF followed by chymotrypsin. The EC in lane 3 received additional treatment with the CID prior to electrophoresis. (Lanes 4,5) Pol IIB EC formed from Pol IIB generated by treatment first with chymotrypsin followed by PMSF. The EC in lane 5 received additional treatment with CID prior to electrophoresis. (B) Western blot analysis detecting the shift in mobility of the largest Pol II subunit following removal of the CTD.
Pcf11 binds both the CTD and RNA, and the dismantling reaction requires both interactions
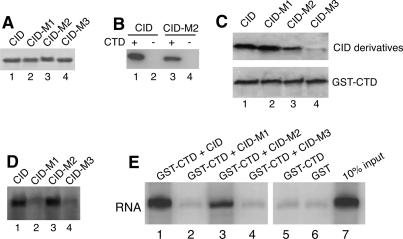
Because the dismantling reaction required the CTD, we tested the ability of each CID derivative to bind an immobilized version of the CTD. Figure 4A shows SDS-PAGE analysis of each CID derivative expressed in and purified from Escherichia coli. CID and CID-M2 bound to immobilized GST-CTD but not GST (Fig. 4B, cf. lanes 1,3 and lanes 2,4). More CID than CID-M2 associated with the CTD (Fig. 4B, cf. lanes 1 and 3). Further analysis showed that the CID and the M1 mutant bound equally well to the CTD (Fig. 4C, lanes 1,2); M2 exhibited weaker binding and M3 exhibited the weakest binding (Fig. 3C, lanes 3,4). Thus, the strong effect of the M3 mutation and the partial effect of the M2 mutation on the dismantling activity (Fig. 2) might be attributed to defects in CTD binding.
Figure 4.
Evidence that Pcf11 dismantles the EC by bridging the CTD to the nascent transcript. (A) Coomassie blue-stained gel showing purified preparations of derivatives of the CID. (B) Equal amounts of the CID (lanes 1,2) and CID-M2 derivative (lanes 3,4) were incubated with immobilized GST-CTD (lanes 1,3) or GST (lanes 2,4). After washing to remove unbound protein, the bound material was analyzed by SDS-PAGE and Western blotting with antibody against the 6-histidine tag associated with CID and its derivative. (C) CTD pull-down analysis for binding between CID derivatives and the CTD. GST-CTD fusion protein with a 6-histidine tag at the C terminus was immobilized on glutathione Sepharose and separated into four aliquots. Each aliquot of immobilized CTD was incubated with a different derivative of the CID. The immobilized material was extensively washed and then analyzed by Western blotting with antibody against the 6-histidine tag residing on both the CID and GST-CTD. (D) Protein–RNA cross-linking assay for CID–RNA binding. One microgram of each CID derivative was incubated with radiolabeled RNA and cross-linked with UV light for 2 min. The RNA was degraded with RNase A and proteins retaining small radioactive tags were detected after SDS-PAGE using a PhosphorImager. (E) CID forms a bridge between RNA and an immobilized form of the CTD. Glutathione Sepharose beads were first loaded with GST-CTD (lanes 1–5), or GST-CTD (lane 6). Following several washes, the beads were incubated with CID and its derivatives as indicated above the lanes. Beads were again washed and then incubated with radiolabeled RNA followed by washing to remove unbound RNA. The radiolabeled RNA was isolated from the beads and analyzed on a denaturing polyacrylamide gel. Lane 7 shows 10% of the total amount of RNA added to each of the samples.
Rho-dependent termination in bacteria provides a precedent for the potential contribution a protein–RNA interaction might make toward termination (Richardson 2002). To determine if the CID binds RNA, a UV cross-linking analysis was done. We incubated each derivative of the CID with a radiolabeled RNA fragment, and UV-irradiated the mixtures. RNA was degraded with RNase, and the relative levels of radiolabeled RNA cross-linked to each CID were determined following SDS-PAGE. The CID and the M2 mutant cross-linked to RNA equally well, while the M1 and M3 mutants cross-linked significantly more poorly (Fig. 4D). These results suggest that the CID and the M2 mutant bind RNA with higher affinity than the M1 and M3 mutants. Thus, the effect of the M1 mutation on dismantling activity (Fig. 2) could be attributed to a defect in RNA binding.
The CID can form a bridge between the CTD and RNA
Since the CID associates with RNA and the CTD, we tested if the CID could form a bridge between the CTD and RNA. Immobilized GST-CTD or GST exhibits very little binding to RNA (Fig. 4E, lanes 5,6). In contrast, addition of CID causes ∼10% of the RNA to partition with the GST-CTD (Fig. 4E, lane 1). The M3 mutation that lacks both CTD- and RNA binding activity and the M1 mutation that lacks RNA binding activity fail to bring RNA into the bound fraction (Fig. 4E, lanes 2,4). The M2 mutation, which retains RNA binding activity but has reduced CTD binding activity, brings approximately threefold less RNA into the bound fraction than the CID (Fig. 4E, lane 3). Collectively, our results indicate that the CID can form a bridge between RNA and the CTD.
Hybridization of an oligonucleotide to the nascent transcript blocks the dismantling reaction
The CTD is thought to extend in an unstructured conformation and be attached to the rest of the Pol II molecule by a flexible linker (Meinhart and Cramer 2004; Noble et al. 2005). How Pcf11 might dismantle the elongation posed a conundrum if its only point of contact with the EC was the CTD. Our finding that Pcf11 can bridge the CTD to RNA offers a possible mechanism for the dismantling activity of Pcf11. Figure 5A presents a model. Conformational changes in the CTD might be transmitted via Pcf11 to the nascent transcript. Force exerted on the nascent transcript, in turn, could cause the EC to dissociate from the DNA. This model is analogous to the mechanism by which Rho is thought to terminate transcription, although there is the important distinction that Pcf11 does not require ATP hydrolysis, whereas Rho does (Richardson 2002).
Figure 5.
DNA oligonucleotide blocking test of a Pcf11-dependent dismantling mechanism. (A) A possible mechanism by which Pcf11 disrupts the EC. By forming a bridge between the CTD and the nascent transcript, movement of the CTD might be transduced via the Pcf11 to the nascent transcript, and the resulting force on the nascent transcript could disrupt the EC. See text for further discussion. (B) Illustration of the DNA oligonucleotide blocking experiment. DNA oligonucleotides (Oligo) 1, 2, and 3 are complementary to parts of the nascent transcript corresponding to regions 1, 2, and 3, respectively (sequences are provided in Materials and Methods). (C) ECs remaining after pretreatment with DNA oligonucleotides followed by CID treatment. ECs were formed and then incubated for 10 min with buffer (lane 1), all three DNA oligonucleotides (lane 2), or one of the three individual oligonucleotides (lanes 3–5). Next, buffer (lanes 1,2) or 3 μg of CID was added to the EC, and the mixtures were incubated an additional 30 min before analyzing the ECs on a native gel.
To test our model, we determined if DNA oligonucleotides complementary to different parts of the nascent transcript affected the capacity of the CID to dismantle the EC. A DNA oligonucleotide hybridized to the transcript might interfere with the dismantling reaction by blocking association between the CID and the nascent transcript. Three oligonucleotides were made that were complementary to three regions in the nascent transcript as illustrated in Figure 5B (see Material and Methods for specific sequences). ECs were first incubated with oligonucleotides, and then incubated with CID. The resulting ECs were analyzed on a native gel. The addition of oligonucleotides alone had no effect on the EC (Fig. 5C, lane 2). Oligonucleotide 3 failed to block the dismantling activity of the CID (Fig. 5C, lane 3). This was expected since oligonucleotide 3 was unlikely to have access to its complementary region buried inside the EC. In contrast, oligonucleotide 2 provided complete protection against the CID (Fig. 5C, lane 4). This region is complementary to the 27-nucleotide (nt) region of the nascent transcript that is immediately adjacent to the point where RNA exits Pol II (Fig. 5B). The results are consistent with the hypothesis that the CID must interact with the nascent transcript in this region to dismantle the EC. An oligonucleotide hybridizing to the last 25 nt of the nascent transcript weakly blocked CID activity. The results suggest that the dismantling activity involves contact between the CID and the region of the nascent transcript in close proximity to Pol II. Bridging the CTD to a region close to the exit point of the RNA might be important so that force generated by the CTD is not dissipated by a flexible region of RNA.
Discussion
We have discovered that the protein Pcf11 dismantles a Pol II EC by a mechanism that involves the CTD of Pol II. We observe that Pcf11 associates with the CTD, in accordance with previous reports (Barilla et al. 2001; Licatalosi et al. 2002; Sadowski et al. 2003). We also find that Pcf11 binds RNA, thus allowing Pcf11 to form a bridge between the CTD and the nascent transcript. We speculate that this bridge transmits force generated by conformational changes occurring at the CTD to the nascent transcript. The force exerted on the nascent transcript could then disrupt the 8-nt heteroduplex formed between the 3′-end of the nascent transcript and the DNA template within the EC, thus resulting in total dissolution of the EC (Kireeva et al. 2000; Komissarova et al. 2002). This mechanism is analogous to the mechanism by which Rho is thought to disrupt ECs in bacteria (Richardson 2002), but with an important distinction: Pcf11 dismantles the EC in the absence of nucleotide hydrolysis. The Pcf11-mediated reaction is not unique to the yeast proteins used in this study: The same reaction occurs when Drosophila Pcf11 is incubated with ECs formed from purified Drosophila Pol II (Z. Zhang and D.S. Gilmour, in prep.).
The discovery of this Pcf11-mediated reaction offers a new perspective on how the CTD of Pol II might regulate transcription elongation. The CTD is required for terminating transcription at the ends of genes (McCracken et al. 1997). In yeast, Pcf11 is part of a complex called CF1 that is involved in cleaving and polyadenylating pre-mRNA (Gross and Moore 2001a). In vivo cross-linking analyses show that Pcf11 is concentrated in the vicinity of the polyadenylation signal relative to the rest of the transcription unit (Kim et al. 2004a). CF1 recognizes part of the polyadenylation signal in the nascent transcript (Gross and Moore 2001b), thus providing a way to recruit Pcf11 to the EC at a point that would be appropriate for termination. We propose that Pcf11 is the engine that disrupts the EC. In accord with this, we have observed that a three-amino acid mutation in Pcf11 previously shown to impair termination in vivo (Sadowski et al. 2003), also abrogates Pcf11's dismantling activity. Characterization of multiple alleles of Pcf11 has provided evidence that Pcf11 contains functionally distinct domains (Sadowski et al. 2003). Mutations in the CID impair termination, while mutations elsewhere in the protein impair the cleavage and polyadenylation reactions. Importantly, some alleles that impair the RNA processing reactions do not impair termination. This suggests that cleavage of the nascent transcript during the polyadenylation reaction is not essential for termination.
Recruitment of Pcf11 by the polyadenylation signal provides a mechanism by which polyadenylation and termination can be coupled. Termination occurs downstream from the polyadenylation signal, possibly in a stochastic manner (Orozco et al. 2002; Kim and Martinson 2003). We propose that Pcf11 is transferred from the polyadenylation signal in the nascent transcript to the EC, where it becomes poised to trigger transcript release. Evidence suggests that termination is preceded by pausing (Orozco et al. 2002; Park et al. 2004), and our analysis of the dismantling reaction with Drosophila Pol II and Pcf11 indicates that Pcf11 only acts on paused Pol II (Z. Zhang and D.S. Gilmour, in prep.). This fits our model for the dismantling reaction. When Pol II is undergoing active elongation, RNA reeling out of the Pol II will inhibit the transfer of force from the CTD to the heteroduplex buried inside the EC.
An alternative mechanism for coupling polyadenylation and termination involves an RNA exonuclease that initiates degradation at the uncapped 5′-end of the nascent transcript generated when the polyadenylation machinery cleaves the transcript (Kim et al. 2004b; Luo and Bentley 2004; West et al. 2004). An important role for this reaction in yeast is indicated by the widespread impairment of termination observed in a Rat1 mutant strain (Kim et al. 2004b). Rat1 is a 5′-to-3′ exonuclease that is thought to chase down the EC as it degrades the uncapped nascent transcript. However, it seems unlikely that Rat1 alone can cause termination, since ECs remain intact even after nascent transcripts have been extensively degraded with various ribonucleases (Gu et al. 1996). We propose that Rat1 facilitates the action of Pcf11. By shortening the transcript, Rat1 could reduce the distance between Pcf11's point of contact on the nascent transcript and the RNA exit channel. A short distance between these two points would be necessary if force generated by conformational changes in the CTD is to impact on the nascent transcript within the EC. The Rat1 homolog Xrn1 might function to facilitate Pcf11 in human cells as well (Teixeira et al. 2004; West et al. 2004).
The CTD also controls the elongation properties of Pol II as it clears the promoter and traverses the body of the gene. Efficient elongation correlates with hyperphosphorylation of the CTD (Price 2000). In vivo cross-linking analysis shows that Pcf11 is present throughout transcription units in yeast, albeit at a level lower than found in the vicinity of the polyadenylation signal (Kim et al. 2004a). Given its distribution on the gene, Pcf11 could be poised to induce premature termination under circumstances that cause Pol II to pause. Hence, Pcf11 might be a new target that is counteracted by the action of activators and elongation factors involved in promoting transcriptional elongation in vivo.
Materials and methods
Purification of yeast Pol II
Yeast Pol II was purified from a protease-deficient haploid strain (CB010) using the TAP tagging strategy (Rigaut et al. 1999). A TAP-tag was incorporated into the C terminus of the Pol II subunit Rpb4 in the manner described previously (Bushnell and Kornberg 2003). Briefly, following cell lysis with bead-beating, lysate was clarified by ultracentrifugation and subjected to the TAP purification procedure. High concentrations of salt (500 mM KCl in a Tris-HCl buffer) were then used throughout the tandem affinity columns (IgG-Calmodulin) to wash away contaminants as well as several transcription factors that usually associate with the polymerase. The eluent from the TAP purification was further cleaned using heparin resin (Amersham-Pharmacia), followed by DEAE HPLC as the final purification step (Edwards et al. 1990). A cocktail of protease inhibitors was kept in solutions throughout the purification.
Purification of Pcf11 and CID derivatives
The plasmid pET32-Pcf11 was a gift from Dr. Claire Moore (Tufts University, Boston, MA). The sequence encoding the first 140 amino acids of Pcf11 was cloned into the pET-28-(a) expression vector (Novagen). Oligonucleotide-directed mutagenesis was used to make the mutations described in Figure 2. Plasmids were transformed in E. coli BL21(DE3), and 6-histidine-tagged versions of each protein were purified on Ni-NTA-agarose according to the manufacturer's instructions (QIAGEN).
Purification of GST-CTD fusion protein
The plasmid encoding GST-CTD with a 6-histidine tag at the C terminus (pGST-yCTDH6) was obtained from Dr. Arno Greenleaf (Duke University, Durham, NC). E. coli BL21 cells containing plasmids encoding glutathione S-transferase (GST) or GST-CTD were grown in LB containing 100 μg/mL Ampicillin at 37°CtoanOD600 of 0.5–0.8, and protein expression was induced by the addition of IPTG to 1.0 mM. After 3 h at 37°C, cells were harvested and lysed, and proteins were purified by affinity chromatography on glutathione-Sepharose 4B (Amersham Biosciences) according to the manufacturer's instructions.
Formation and dismantling of ECs
ECs were formed on a tailed template as previously described (Zhang et al. 2004) with the following changes: 0.6 μg of highly purified yeast Pol IIA was used in each reaction and transcription reactions were done at 30°C for 30 min. For dismantling reactions, ECs in solution or immobilized on magnetic beads were treated with 3 μg of Pcf11 for 30 min prior to further analysis. When used, immobilized complexes were washed prior to treatment with Pcf11. Analysis of ECs on native gels was done as previously described (Zhang et al. 2004).
Conversion of Pol IIA to Pol IIB
Pol IIA was converted to Pol IIB by treating 0.6 μg of purified Pol II in 15 μL of transcription buffer with 0.5 μg/mL chymotrypsin at 30°C for 30 min. PMSF was added to a final concentration of 2 mM to inactivate the chymotrypsin. For controls, the order of addition for PMSF and chymotrypsin was reversed.
GST-CTD pull-down analysis for Pcf11 binding
For this analysis, 200 ng of GST or GST-CTD was bound to 15 μL of glutathione-Sepharose 4B (Amersham Biosciences) in binding buffer [150 mM KCl, 25 mM HEPES at pH 7.6, 50 mM (NH4)2SO4, 12% glycerol, 1.0 mM MnCl2, 0.5 mM Dithiothreitol (DTT), 0.1 mM PMSF] for 1 h at 4°C. Beads were washed five times with the binding buffer containing 200 mM KCl, and resuspended in 15 μL of binding buffer. One-hundred nanograms of each Pcf11 derivative was incubated with GST-CTD beads for 1 h at 4°C. Each resin was washed four times with the binding buffer containing 200 mM KCl for 5 min at 4°C. Bound material was analyzed by Western blotting with antibody against the 6-histidine tags on each Pcf11 derivative and on GST-CTD.
UV cross-linking analysis of Pcf11-RNA binding
A 330-nt, radiolabeled RNA was produced by transcribing a restriction-cut plasmid with T7 polymerase in the presence of [α-32P]UTP and other nonradioactive nucleotides. RNA was isolated by phenol extraction and ethanol precipitation. For the UV cross-linking experiments, 1 μg of each CID derivative was incubated with 300,000 cpm of RNA at room temperature for 15 min in 150 mM KCl, 25 mM HEPES (pH 7.6), 1.0 mM MnCl2, 12% glycerol, and 0.5 mM DTT. Samples were irradiated with UV light from a short-wavelength transilluminator for 2 min. RNA was digested with 2 μg of RNase A for 30 min at 37°C. The entire sample was electrophoresed on an SDS–polyacrylamide gel. Radioactively tagged proteins were detected with a PhosphorImager.
Pcf11-mediated pull-down of RNA by immobilized GST-CTD
Five micrograms of GST-CTD was incubated with 20 μL of glutathione Sepharose at room temperature for 30 min in 150 mM KCl, 25 mM HEPES (pH 7.6), 50 mM (NH4)2SO4, 1.0 mM MnCl2, 12% glycerol, and 0.5 mM DTT and then extensively washed. Beads were then incubated with 3 μg of CID derivatives or no protein for 15 min followed by extensive washing. Finally, each sample was incubated with 300,000 cpm of RNA at room temperature for 20 min followed by extensive washing to remove unbound material. RNA was recovered from the beads by incubating the beads at 42°C for 30 min in 100 μL of 20 mM EDTA (pH 8), 200 mM NaCl, 1% SDS, 250 μg/mL yeast RNA, and 0.1 mg/mL proteinase K. RNA was phenol-extracted, ethanol-precipitated, and analyzed on a denaturing polyacrylamide gel.
Oligonucleotide blocking experiment
The nascent transcript associated with the ECs has the sequence 5′-UGAUCUUAUCCUCUCCUCACCUCUCCCUCC UCUAUCUCCCCCCCUCACACUCAUUUCUCAUUCCACU CCC-3′. The DNA oligonucleotides tested for blocking were oligo 1, 5′-GAGAGGTGAGGAGAGGATAAGATCA-3′ (complementary to nucleotides 1–25 of the transcript); oligo 2, 5′-GAGTGTGAGGGGGGGAGATAGAGGAGG-3′ (complementary to nucleotides 26–52 of the transcript); and oligo 3, 5′-GGG AGTGGAATGAGAAATGAGTGTG-3′ (complementary to nucleotides 42–70 of the transcript). Each oligonucleotide was added to a final concentration of 0.5 nM and incubated with the ECs for 10 min prior to addition of CID.
Acknowledgments
We thank Claire Moore and Arno Greenleaf for providing plasmids and members of the Gilmour lab for several insightful comments. This work was supported by research grant GM47477 from the NIH to D.S.G.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1296305.
References
- Ackerman S., Bunick, D., Zandomeni, R., and Weinmann, R. 1983. RNA polymerase II ternary transcription complexes generated in vitro. Nucleic Acids Res. 11: 6041–6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M., Nissen, R.M., Kanazawa, S., Jabrane-Ferrat, N., and Peterlin, B.M. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8: 327–337. [DOI] [PubMed] [Google Scholar]
- Barilla D., Lee, B.A., and Proudfoot, N.J. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 98: 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. 1995. Regulation of transcriptional elongation by RNA polymerase II. Curr. Opin. Genet. Dev. 5: 210–216. [DOI] [PubMed] [Google Scholar]
- ____. 2002. The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14: 336–342. [DOI] [PubMed] [Google Scholar]
- Birse C.E., Minvielle-Sebastia, L., Lee, B.A., Keller, W., and Proudfoot, N.J. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301. [DOI] [PubMed] [Google Scholar]
- Blau J., Xiao, H., McCracken, S., O'Hare, P., Greenblatt, J., and Bentley, D. 1996. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16: 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.A., Imbalzano, A.N., and Kingston, R.E. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes & Dev. 10: 1479–1490. [DOI] [PubMed] [Google Scholar]
- Buratowski S. 2003. The CTD code. Nat. Struct. Biol. 10: 679–680. [DOI] [PubMed] [Google Scholar]
- Bushnell D.A. and Kornberg, R.D. 2003. Complete, 12-subunit RNA polymerase II at 4.1-Å resolution: Implications for the initiation of transcription. Proc. Natl. Acad. Sci. 100: 6969–6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick R.L. and Chamberlin, M.J. 1985. Studies on transcription of 3′-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry 24: 2245–2253. [DOI] [PubMed] [Google Scholar]
- Dezelee S., Wyers, F., Sentenac, A., and Fromageot, P. 1976. Two forms of RNA polymerase B in yeast. Proteolytic conversion in vitro of enzyme BI into BII. Eur. J. Biochem. 65: 543–552. [DOI] [PubMed] [Google Scholar]
- Edwards A.M., Darst, S.A., Feaver, W.J., Thompson, N.E., Burgess, R.R., and Kornberg, R.D. 1990. Purification and lipid-layer crystallization of yeast RNA polymerase II. Proc. Natl. Acad. Sci. 87: 2122–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg M.B., Baltimore, D., and Frankel, A.D. 1991. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. 88: 4045–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt A., Fu, J., and Kornberg, R.D. 1997. Formation and crystallization of yeast RNA polymerase II elongation complexes. J. Biol. Chem. 272: 30799–30805. [DOI] [PubMed] [Google Scholar]
- Gross S. and Moore, C. 2001a. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. 98: 6080–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001b. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 21: 8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Wind, M., and Reines, D. 1996. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc. Natl. Acad. Sci. 93: 6935–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R., Selby, C.P., Liu, M., Price, D.H., and Sancar, A. 1999. Human transcription release factor 2 dissociates RNA polymerases I and II stalled at a cyclobutane thymine dimer. J. Biol. Chem. 274: 24779–24786. [DOI] [PubMed] [Google Scholar]
- Izban M.G. and Luse, D.S. 1991. Transcription on nucleosomal templates by RNA polymerase II in vitro: Inhibition of elongation with enhancement of sequence-specific pausing. Genes & Dev. 5: 683–696. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Liu, M., Spencer, C.A., and Price, D.H. 2004. Involvement of transcription termination factor 2 in mitotic repression of transcription elongation. Mol. Cell 14: 375–385. [DOI] [PubMed] [Google Scholar]
- Kessler M. and Mathews, M.B. 1992. Premature termination and processing of human immunodeficiency virus type 1-promoted transcripts. J. Virol. 66: 4488–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.J. and Martinson, H.G. 2003. Poly(A)-dependent transcription termination: Continued communication of the poly(A) signal with the polymerase is required long after extrusion in vivo. J. Biol. Chem. 278: 41691–41701. [DOI] [PubMed] [Google Scholar]
- Kim M., Ahn, S.H., Krogan, N.J., Greenblatt, J.F., and Buratowski, S. 2004a. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23: 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Krogan, N.J., Vasiljeva, L., Rando, O.J., Nedea, E., Greenblatt, J.F., and Buratowski, S. 2004b. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522. [DOI] [PubMed] [Google Scholar]
- Kireeva M.L., Komissarova, N., Waugh, D.S., and Kashlev, M. 2000. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 275: 6530–6536. [DOI] [PubMed] [Google Scholar]
- Kireeva M.L., Hancock, B., Cremona, G.H., Walter, W., Studitsky, V.M., and Kashlev, M. 2005. Nature of the nucleosomal barrier to RNA polymerase II. Mol. Cell 18: 97–108. [DOI] [PubMed] [Google Scholar]
- Komissarova N., Becker, J., Solter, S., Kireeva, M., and Kashlev, M. 2002. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell 10: 1151–1162. [DOI] [PubMed] [Google Scholar]
- Krumm A., Meulia, T., and Groudine, M. 1993. Common mechanisms for the control of eukaryotic transcriptional elongation. BioEssays 15: 659–665. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Bartsch, I., and Grummt, I. 1990. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature 344: 559–562. [DOI] [PubMed] [Google Scholar]
- Lang W.H., Morrow, B.E., Ju, Q., Warner, J.R., and Reeder, R.H. 1994. A model for transcription termination by RNA polymerase I. Cell 79: 527–534. [DOI] [PubMed] [Google Scholar]
- Licatalosi D.D., Geiger, G., Minet, M., Schroeder, S., Cilli, K., McNeil, J.B., and Bentley, D.L. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9: 1101–1111. [DOI] [PubMed] [Google Scholar]
- Lis J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 63: 347–356. [DOI] [PubMed] [Google Scholar]
- Lis J.T., Mason, P., Peng, J., Price, D.H., and Werner, J. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes & Dev. 14: 792–803. [PMC free article] [PubMed] [Google Scholar]
- Luo W. and Bentley, D. 2004. A ribonucleolytic rat torpedoes RNA polymerase II. Cell 119: 911–914. [DOI] [PubMed] [Google Scholar]
- McCracken S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Greenblatt, J., Patterson, S.D., Wickens, M., and Bentley, D.L. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. [DOI] [PubMed] [Google Scholar]
- Meinhart A. and Cramer, P. 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430: 223–226. [DOI] [PubMed] [Google Scholar]
- Noble C.G., Hollingworth, D., Martin, S.R., Ennis-Adeniran, V., Smerdon, S.J., Kelly, G., Taylor, I.A., and Ramos, A. 2005. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat. Struct. Mol. Biol. 12: 144–151. [DOI] [PubMed] [Google Scholar]
- Orozco I.J., Kim, S.J., and Martinson, H.G. 2002. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J. Biol. Chem. 277: 42899–42911. [DOI] [PubMed] [Google Scholar]
- Park N.J., Tsao, D.C., and Martinson, H.G. 2004. The two steps of poly(A)-dependent termination, pausing and release, can be uncoupled by truncation of the RNA polymerase II carboxyl-terminal repeat domain. Mol. Cell. Biol. 24: 4092–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20: 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N.J. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16: 272–278. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Renner D.B., Yamaguchi, Y., Wada, T., Handa, H., and Price, D.H. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276: 42601–42609. [DOI] [PubMed] [Google Scholar]
- Richardson J.P. 2002. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta 1577: 251–260. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Sadowski M., Dichtl, B., Hubner, W., and Keller, W. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 22: 2167–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A., Tahiri-Alaoui, A., West, S., Thomas, B., Ramadass, A., Martianov, I., Dye, M., James, W., Proudfoot, N.J., and Akoulitchev, A. 2004. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature 432: 526–530. [DOI] [PubMed] [Google Scholar]
- Wada T., Takagi, T., Yamaguchi, Y., Ferdous, A., Imai, T., Hirose, S., Sugimoto, S., Yano, K., Hartzog, G.A., Winston, F., et al. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 12: 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S., Gromak, N., and Proudfoot, N.J. 2004. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Wada, T., and Handa, H. 1998. Interplay between positive and negative elongation factors: Drawing a new view of DRB. Genes Cells 3: 9–15. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Takagi, T., Wada, T., Yano, K., Furuya, A., Sugimoto, S., Hasegawa, J., and Handa, H. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97: 41–51. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Inukai, N., Narita, T., Wada, T., and Handa, H. 2002. Evidence that Negative Elongation Factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22: 2918–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankulov K., Blau, J., Purton, T., Roberts, S., and Bentley, D.L. 1994. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77: 749–759. [DOI] [PubMed] [Google Scholar]
- Zehring W.A., Lee, J.M., Weeks, J.R., Jokerst, R.S., and Greenleaf, A.L. 1988. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc. Natl. Acad. Sci. 85: 3698–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wu, C.H., and Gilmour, D.S. 2004. Analysis of Pol II elongation complexes by native gel electrophoresis: Evidence for a novel CTD-mediated termination mechanism. J. Biol. Chem. 279: 23223–23228. [DOI] [PubMed] [Google Scholar]