Abstract
BLASTX alignment between 189.5 Mb of rice genomic sequence and translated Arabidopsis thaliana annotated coding sequences (CDS) identified 60 syntenic regions involving 4–22 rice orthologs covering ≤3.2 cM (centiMorgan). Most regions are <3 cM in length. A detailed and updated version of a table representing these regions is available on our web site. Thirty-five rice loci match two distinct A.thaliana loci, as expected from the duplicated nature of the A.thaliana genome. One A.thaliana locus matches two distinct rice regions, suggesting that rice chromosomal sequence duplications exist. A high level of rearrangement characterizing the 60 syntenic regions illustrates the ancient nature of the speciation between A.thaliana and rice. The apparent reduced level of microcollinearity implies the dispersion to new genomic locations, via transposon activity, of single or small clusters of genes in the rice genome, which represents a significant additional effector of plant genome evolution.
INTRODUCTION
Rice is one of the most important crops in the world, accounting for 50–80% of the daily diet of approximately half the world’s population (1), with an annual production of approximately half a billion tons (http://apps.fao.org/). It therefore has a key role to play as a genome model in the monocots, as for Arabidopsis in the dicot family. It has the smallest genome among the Graminae family, estimated to be ∼440 Mb divided into 12 pairs of chromosomes (2,3). The International Rice Genome Sequencing Project (IRGSP) is an international collaboration to sequence the rice genome, with each chromosome being sequenced by one or a few nations (4). By November 15, 2001, ∼189.5 Mb from 1394 BAC/PAC clones had been released into the public domain and the project should be completed by the end of 2004 (http://www. rgp.dna.affrc.jp/).
The rice genome is 3.5 times the size of the dicotyledonous model genome Arabidopsis thaliana. The complete Arabidopsis sequence was released in December 2000 by the Arabidopsis Genome Initiative (AGI) and covers 115.4 Mb with 25 498 predicted genes encoding proteins from 11 000 families (5). Gene density and structure have been well studied with an average of one gene every 4.5 kb. In order to understand the mechanisms that have led to present day genome structures, we assume that Arabidopsis and all higher plants have inherited gene order and content, with modifications, through common ancestry. Thus, the individual genes in modern day plant species can be used to reconstruct ancestral genome structure. These assumptions have already been tested and largely verified between Arabidopsis and species within the dicot family, especially its closest relatives, the cultivated Brassica species (6–10), but also tomato (11,12) and soybean (13). Similarly, in cereals, comparison of high density genetic maps, mostly restriction fragment length polymorphism (RFLP) markers using cDNA probes, led to the conclusion that significant collinearity (i.e. macro-synteny) exists among cereals and has been maintained over an evolutionary period as long as 60 million years. A model in which the synteny between cereals is illustrated by concentric circles has been proposed, involving all linkage groups of each cereal genome involved: rice, foxtail millet (Setaria italica), sugar cane, sorghum, maize, Triticeae and oats (14,15).
In contrast to studies within monocot and dicot families, very few studies have been performed on synteny between the two groups which diverged from a common ancestor between 120 and 200 million years ago (MYA) (16). Paterson et al. (17), using a model based on an estimated rate of structural chromosomal mutation, predicted that within a region averaging 3 cM in length ∼50% of the genes would be maintained by chance between monocot and dicot genomes, although the evidence for such conservation is not clear. Recent studies by Vision et al. (18) on Arabidopsis genome duplication suggest that any regions which are homologous between Arabidopsis and genomes that diverged from a common ancestor >100 MYA will be <10 cM (centiMorgan). Devos et al. (19) in a study based on similarities between Arabidopsis genomic sequences (five BACs in a contig on chromosome II) and mapped rice cDNAs [expressed sequence tags (ESTs)] concluded that synteny between Arabidopsis and rice has been eroded during evolution so that it is no longer detectable using a comparative mapping strategy. However, hundreds of genes may lie between any pair of adjacent markers so that macro-synteny (involving mapped cDNAs or ESTs) does not necessarily imply micro-synteny, i.e. conservation of local gene order, and orientation. Conversely, conserved micro-synteny can exist between species that lack obvious signs of large macro-synteny, as suggested by Devos et al. (19).
If recombination has not broken up the local order of plant genes through genome evolution, ancestral homeologous relationships can be revealed. A more recent study based on BLASTN alignment of Arabidopsis genomic sequence against mapped rice ESTs (20) identified regions of conserved structure between rice and Arabidopsis. These contain five homologous genes characterized by a single inversion between the two genomes and are 194 and 219–301 kb in size on Arabidopsis chromosome IV and rice chromosome II, respectively. More recently the same group sequenced 340 kb from this region of rice chromosome II, revealing 56 putative protein coding genes (21). They confirmed conservation of gene content and order between the two genome segments and identified four additional segments of the Arabidopsis genome that show similar conservation. In all, 22 of the 56 genes initially identified in the rice genome segment were represented in this set of five Arabidopsis genome segments, with at least five genes present in conserved order in each segment. However, as pointed out by Devos et al. (19), the alignment of conserved domains in non-orthologous genes in BLAST queries and the identification of different members of multigene families may complicate the interpretation of comparisons by simple BLAST alignment. This could provide apparent support for conserved relationships. Unless care is taken to avoid identification of artifactual syntenic regions through the alignment of similar but non-orthologous sequences and to test the statistical validity of the results obtained, the value of alignment-based studies is questionable.
Here we have developed an automated BLASTX strategy, aligning rice genomic sequences against translated annotated coding sequences (CDS) on Arabidopsis chromosomes, to allow comparison of the ongoing rice genome sequence with the complete, annotated Arabidopsis genomic sequence. The validity of the results obtained has been checked manually and neighboring BACs concatenated into longer contigs. Using 189.5 Mb rice sequences available in the public domain, we have identified 60 small-scale syntenic regions between those two genomes involving 4–22 annotated genes covering ≤3.2 cM. The limited conservation of gene order suggests that the identification of genes related to valuable agronomic traits in cereal crop plants through map based cloning approaches will not be possible using this approach. However, our results will provide valuable information allowing a better understanding of the molecular mechanisms that have shaped present day plant genomes since the separation of the common ancestor of monocots and dicots some 200 MYA.
MATERIALS AND METHODS
The search for syntenic regions between Arabidopsis and rice is a four-step process. (i) Detection of rice genes by comparison with translated annotated Arabidopsis CDS. (ii) Identification of syntenic loci between rice and Arabidopsis BAC clones. (iii) Test of permutation for the statistical accuracy of the method. (iv) Extension of syntenic loci based on overlapping rice BAC clones.
Gene identification on the rice genome
A local library was constructed corresponding to all non-redundant annotated Arabidopsis CDS in order on the five chromosomes. A BLASTX (22) search was carried out between the rice genomic sequences released in the public domain and this translated Arabidopsis CDS library. For each rice BAC clone we searched for regions encoding similar proteins by looking for significant high-scoring segment pairs (HSPs) after BLASTX analysis. BLAST results were treated using the modules of BioPerl (http://www.bioperl.org).
Contigs of HSPs were constructed with the following parameters. (i) HSPs must be collinear on the same strand. (ii) No overlap exceeding 15 amino acids to avoid repetitive domains within a protein. (iii) Maximum distance of 4000 bp between two HSPs in order to avoid fusing two identical genes.
Further, single HSPs (or HSP contigs displaying the above characteristics) were considered as potential genes in the rice genome only when the alignment length exceeded 80 amino acids or was ≥50% of the Arabidopsis peptide length in order to exclude similarities limited to well-conserved domains. Single HSPs and HSP contigs built on the rice genome are hereafter referred to as genes, although the true, complete gene structure may be slightly different. Genes were not considered if they showed similarity to transposons. This system links each identified potential rice gene to all potential Arabidopsis homologs.
Identification of syntenic regions between Arabidopsis and rice
Because the rice genome is still being sequenced, we first identified pairs of collinear rice and Arabidopsis BAC clones. When neighboring rice genes showed BLAST hits to the same Arabidopsis CDS they were considered as having the same function and to represent tandem duplications. Thus, syntenic regions are defined by a number of similar functions rather than similar genes to avoid artifactual detection of regions containing tandemly duplicated genes. Starting from individual genes, a second one was added to a syntenic block if the pairs of identified genes were sufficiently close in the two genomes and did not correspond to a transposon or a cluster. The distance between two hits is not measured in base pairs but in gene or CDS numbers to avoid further problems due to distinct gene densities between Arabidopsis and rice and is, in any case, a better criterion for synteny. Syntenic blocks were retained if the number of different functions is ≥3 and duplicated genes have copy numbers <3.
Accuracy
In order to validate this method a dotter analysis was conducted on a set of 30 previously identified syntenic loci. None of these regions were rejected after this manual inspection. It was important to test the accuracy of the method before performing the permutation statistical test.
Permutation statistical test
The permutation tests allow an evaluation of the significance of the detected syntenic sites. The null hypothesis which was tested is that the number of syntenic sites detected on the genome could be the result of a random distribution of the genes. For each permutation the rice genes were randomized, syntenic sites defined and the number of sites counted. The probability is the fraction of the randomized data that are equal to or greater than the number of sites detected with the real data. This provides a measure of the reliability of sites detected.
Extension of syntenic loci
The syntenic regions previously identified as statistically accurate were enlarged when overlapping rice BACs were available in each considered region. Rice BACs were aligned against Arabidopsis CDS using BLASTX to extend and/or merge syntenic blocks. Selected syntenic regions obtained using BLASTX results were checked by a dotter (23) analysis.
Identification of ESTs
A classical BLASTN approach between EST libraries and rice or Arabidopsis genomic sequences allowed us to identify ESTs involved in those syntenic regions with a threshold of ≥90% sequence identity.
RESULTS
Identification and statistical validity of syntenic regions between rice and Arabidopsis
Whereas rice genomic sequence is accumulating rapidly in international databases, the correct annotation of this sequence will require considerable time. In order to compare gene order and conservation between the complete Arabidopsis sequence and the ongoing rice project, we developed an approach to detect regions of the rice genome whose conceptual translation products show significant similarity to annotated proteins from the complete Arabidopsis sequence. Despite the known limits of the current annotation of the Arabidopsis genome (24), the vast majority of genes have been identified, although detailed exon–intron structures may be incorrect. As our strategy is based on identification of BLAST HSPs, which generally correspond to individual exons, errors in overall gene structure do not prevent detection of orthologous sequences. Furthermore, this approach allows a regular update of the results to be carried out as raw rice genomic sequence is submitted to public databases.
A BLASTX alignment was performed between rice BAC sequences (available in the public domain on November 15, 2001) and a library corresponding to all translated annotated Arabidopsis CDS ordered on each of the five chromosomes. Identification of significant sequence alignments is usually carried out using a cut-off BLAST probability score (Expect or e value). However, this approach is generally used in gene annotation, in which the aim is to propose possible functions through identification of conserved motifs. It does not distinguish between proteins showing similarity over most or all of their length and shorter, conserved regions (for example, kinase or ring finger motifs). This is exacerbated by the fact that raw scores in BLAST output are those of the highest-scoring HSP. We therefore decided to sum similarities, using criteria based on length and percent identity, to identify pairs of sequences showing similarity over most or all of their length. We tested several values to determine the optimum BLAST parameter to use to detect truly orthologous sequences.
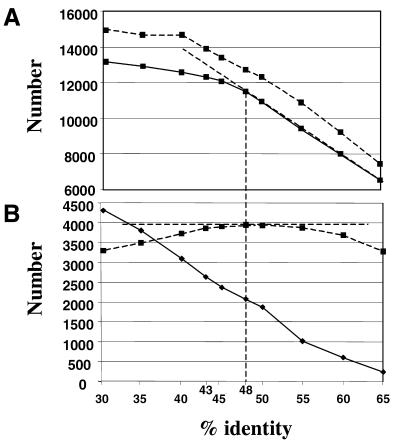
Figure 1 shows a plot of the number of rice genes, transposons and different functions detected by BLASTX alignment for varying percentages of identity. It can be clearly seen that, from 65 to 48% identity, there is a linear relationship between the percentage and the number of genes, up to approximately 11 500. Below 48%, the number of identified genes tends towards a plateau at around 13 000. In fact, this slight increase is largely due to alignment with transposon and transposon-like sequences, as can be clearly seen in Figure 1. The number of different functions detected reaches a maximum at 48% identity, then decreases as, at lower percentage identities, an increasing number of genes are considered as having identical functions. Consequently, each of these would be considered as a potential ortholog of all others in our subsequent analysis, leading inevitably to artifactual syntenic points. Using a cut-off of 48% therefore optimizes detection of rice genes while minimizing the definition of large gene families based solely on shorter conserved sequences.
Figure 1.
Detection of rice genes as a function of BLAST similarity values. (A) BLAST alignments were carried out as described in Materials and Methods. The number of rice genes identified (solid line) and the number of Arabidopsis CDS translations detected (dotted line) are shown for values from 33 to 65% identity. (B) The number of transposons (solid line) and the number of different functions (dotted line) are plotted for values between 33 and 65% identity.
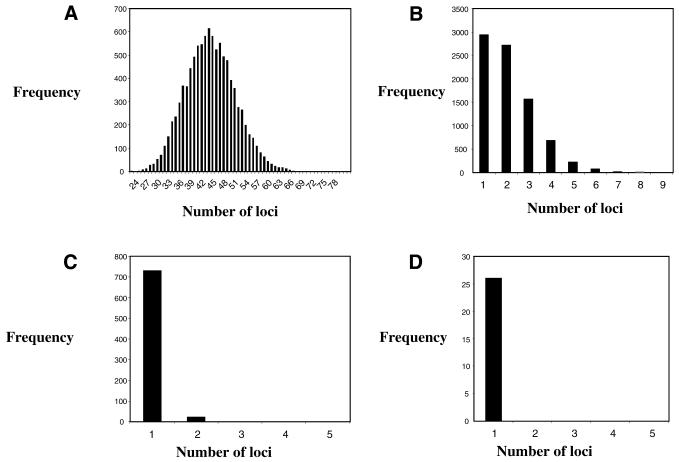
Using a 48% cut-off score, we found three to six consecutive or close Arabidopsis CDS on 303 (22%) of the 1394 rice BACs analyzed (233, 47, 16 and 7 BACs involving 3, 4, 5 and 6 Arabidopsis CDS, respectively). In order to determine the statistical significance of these observations, we analyzed a set of 10 000 permutations for these four cases. Figure 2 shows that the average number of syntenic regions involving 3, 4, 5 or 6 Arabidopsis CDS among the random permutations is 42.4, 1.77, 0.076 and 0.002, respectively. Thus, although for each category the results are statistically significant, syntenic blocks of three genes should be considered with caution. On the other hand, the permutation statistical test shows that the 70 syntenic regions involving four or more CDS are highly significant. A further indication of the validity of our analysis is that we found a similar number of syntenic regions on chromosomes 7 and 3 while about twice as much data are available for the former compared with the latter (Table 1). If our results had been due to chance, we would expect the number of syntenic regions to increase with the amount of sequence available.
Figure 2.
MonteCarlo statistical test. Analyses were carried out as described in Materials and Methods. Horizontal axes present the number of loci obtained among 10 000 permutations for a specific number of Arabidopsis CDS. Vertical axes present the frequency observed. (A) Syntenic region involving three Arabidopsis CDS (mean 42.4 ± 6.75). (B) Syntenic region involving four Arabidopsis CDS (mean= 1.77 ± 0.07). (C) Syntenic region involving five Arabidopsis CDS (mean = 0.076). (D) Syntenic region involving six Arabidopsis CDS (mean = 0.002).
Table 1. Status of the International Rice Genome Sequencing Project.
| Chromosome | Site of genome center | Web site | Location | Published BAC/PAC sequences (Mb) | Number of syntenic regions |
|---|---|---|---|---|---|
| 1 | Korea Rice Genome Research Program (KRGRP) | http://bioserver.myongji.ac.kr/ricemac.html | Korea | 374 (52.1 Mb) | 18 |
| Rice Genome Research Program (RGP) | http://rgp.dna.affrc.go.jp/ | Japan | |||
| 2 | Rice Genome Research Program (RGP) | http://rgp.dna.affrc.go.jp/ | Japan | 153 (18.8 Mb) | 4 |
| John Innes Centre (John Innes Centre) | http://jic.bbsrc.ac.uk | UK | |||
| 3 | Clemson University (CUGI) | http://www.genome.clemson.edu/ | USA | 93 (13.2 Mb) | 7 |
| Cold Spring Harbor Laboratory (CSHL) | http://nucleus.cshl.org/riceweb/ | USA | |||
| Washington University School of Medicine (GSC) | http://genome.wustl.edu/gsc/ | USA | |||
| Plant Genome Initiative at Rutgers (PGIR) | http://mbclserver.rutgers.edu/pigr/ | USA | |||
| The Institute for Genomic Research (TIGR) | http://www.tigr.org/tdb/rice | USA | |||
| 4 | National Center for Gene Research Chinese Academy of Sciences (NCGR) | http://www.ncgr.ac.cn/who/index.html | China | 145 (20.7 Mb) | 4 |
| 5 | Academia Sinica Plant Genome Center (ASPGC) | http://biometrics.sinica.edu.tw/genome | Taiwan | 42 (5.3 Mb) | 4 |
| 6 | Rice Genome Research Program (RGP) | http://rgp.dna.affrc.go.jp/ | Japan | 146 (21.0 Mb) | 5 |
| 7 | Rice Genome Research Program (RGP) | http://rgp.dna.affrc.go.jp/ | Japan | 166 (20.3 Mb) | 6 |
| 8 | Rice Genome Research Program (RGP) | http://rgp.dna.affrc.go.jp/ | Japan | 89 (10.7 Mb) | 4 |
| 9 | National Center for Genetic Engineering and Biotechnology (BIOTEC) | http://www.cs.ait.th/nstda/biotec.biotec.html | Thailand | 8 (1.1 Mb) | 0 |
| 10 | Clemson University (CUGI) | http://www.genome.clemson.edu/ | USA | 169 (24.9 Mb) | 7 |
| Cold Spring Harbor Laboratory (CSHL) | http://nucleus.cshl.org/riceweb/ | USA | |||
| Washington University School of Medicine (GSC) | http://genome.wustl.edu/gsc/ | USA | |||
| Plant Genome Initiative at Rutgers (PGIR) | http://mbclserver.rutgers.edu/pigr/ | USA | |||
| The Institute for Genomic Research (TIGR) | http://www.tigr.org/tdb/rice | USA | |||
| 11 | Clemson University (CUGI) | http://www.genome.clemson.edu/ | USA | 0 | 0 |
| Cold Spring Harbor Laboratory (CSHL) | http://nucleus.cshl.org/riceweb/ | USA | |||
| Washington University School of Medicine (GSC) | http://genome.wustl.edu/gsc/ | USA | |||
| Plant Genome Initiative at Rutgers (PGIR) | http://mbclserver.rutgers.edu/pigr/ | USA | |||
| The Institute for Genomic Research (TIGR) | http://www.tigr.org/tdb/rice | USA | |||
| Genome Center of Wisconsin | http://www.gcow.wisc.edu/ | USA | |||
| 12 | Genoscope (Genoscope) | http://www.genoscope.fr | France | 9 (1.4 Mb) | 1 |
| TOTAL | 1394 (189.5 Mb) | 60 |
A total of 1394 rice BAC/PAC sequences (189.5 Mb, ∼40% of the rice genome) had been released on November 15, 2001. The last column of the table presents the 60 syntenic regions distributed on the 12 rice chromosomes.
Visual inspection of the original BLAST alignments of individual BACs showed numerous additional, apparently syntenic regions, but which involve multigene families in clusters such as receptor-like kinases and for which no accurate interpretation was possible. We therefore concentrated our study on regions containing four or more CDS from different gene families on each rice BAC for further analysis.
Extension of the syntenic regions
The results obtained for each BAC were controlled manually to check the validity of our approach and to construct contigs from neighboring sequences. Manual checking of the 70 blocks of at least four CDS allowed us to build contigs for a number of the original syntenic blocks. Each sequence within the blocks was also realigned against the Arabidopsis CDS library using the BLASTX program to identify potentially syntenic genes within these regions that had not been detected in the first step of our analysis. This allowed extension of some regions by identification of one or two conserved genes on neighboring BACs. As sequencing proceeds, this step will not be necessary, as it will be possible to carry out the same analysis using longer contigs and, eventually, whole chromosomes as overlapping chunks. Finally, 60 syntenic regions between Arabidopsis and rice were identified. Among these loci 18, 4, 7, 4, 4, 5, 6, 4, 7, 1 are located on rice chromosomes I–VIII, X, XII, respectively, as presented in Table 1. A detailed and updated table representing the syntenic regions between Arabidopsis and rice is available on our web site: http://gamay.univ-perp.fr/~salse/nar_supplement/.
The 60 syntenic regions involve 4–21 annotated CDS with an average P value of 1e–31. Extension of all regions is not possible. For example, 26 of them correspond to a single rice BAC clone for which no overlapping or genetically close rice BAC was found (as only ∼40% of the rice genomic sequence is available in the public domain). Thus, we expect to be able to extend other syntenic points as sequencing progresses, especially when a clear minimum tiling path is available for all rice chromosomes. We did not carry out an exhaustive check on strict gene order and orientation, as for many BACs sequencing is still in progress (the sequence is presented in several unordered pieces). However, visual inspection of syntenic blocks involving several completed BACs from chromosome 1 showed that strand and orientation are conserved between rice and Arabidopsis (data not shown). Nevertheless, the low level of conservation in these regions suggests that either gene evolution is heterogeneous within collinear regions (25,26) or intensive rearrangement (inversion, translocation and insertion) has taken place. For more detailed analysis we chose to concentrate our attention on regions of chromosomes I and X, for which sequencing is most advanced. All BACs presented here are available as single contigs, not ordered or unordered pieces and are placed in well-established minimal tiling paths by the corresponding sequencing groups.
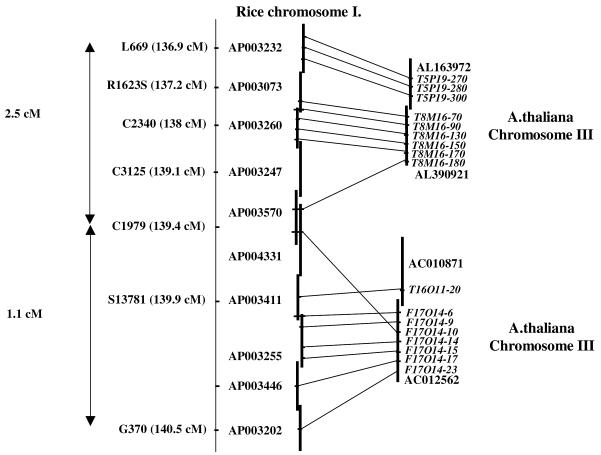
Figure 3 shows an example of one of these regions. It covers 3.6 cM on the rice genome and two separate regions on chromosome III of Arabidopsis separated by ∼19 Mb. Nine CDS out of 24 on two overlapping Arabidopsis chromosome III BAC clones are similar to nine genes on four overlapping rice chromosome I BAC clones covering ≤2.5 cM. Two other overlapping Arabidopsis chromosome III BAC clones show eight CDS out of 26 similar to genes on five overlapping chromosome I rice BAC clones covering ≤1.1 cM. Table 2 represents a detailed analysis of this region by reporting the BAC clone accession number, its genetic location, CDS accession numbers and putative functions, BLASTX expect value and matching ESTs. This result is typical of those we observe in that short, contiguous blocks of rice genes match widely separated blocks on the Arabidopsis genome. Tables corresponding to the 60 syntenic regions identified are available on our web site (http://lgdp.univ-perp.fr/~salse/nar_supplement).
Figure 3.
Conserved microstructure between a rice chromosome I locus and two Arabidopsis loci. Schematic representation of a syntenic region between a rice chromosome I locus and two loci on Arabidopsis chromosome III. Vertical lines show overlapping Arabidopsis and rice BAC clones. Conserved Arabidopsis annotated CDS and their rice orthologous counterparts are linked by solid lines.
Table 2. Detailed analysis of a syntenic region between a rice chromosome I locus and Arabidopsis chromosome III loci.
| Arabidopsis | Rice | ESTs | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis BAC Accession no. | Arabidopsis chromosomes | CDS Accession no. | Putative functions | Rice BAC Accession no. | Map location (cM) | Rice chromosomes | Score BlastP | Rice Accession no. | Arabidopsis Accession no. |
| AL163972 (21.4 Mb) | III | T5P19-270 | Nodulin-like protein | AP003232 | 136.9 | I | 1e–140 | BF430686 | – |
| T5P19-280 | Cytochrome P450-like protein | 6e–47 | – | – | |||||
| T5P19-300 | Hypothetical protein | 3e–42 | – | AV555810 | |||||
| AL390921 (21.5 Mb) | III | T8M16-70 | Putative protein | AP003073 | 137.2 | I | 4e–21 | – | AI994742 |
| T8M16-90 | Calcium-dependant protein kinase | 6e–58 | AU172459 | AI996306 | |||||
| T8M16-130 | Calmodulin-3 like protein | AP003260 | 138 | I | 3e–53 | AF090687 | AV549547 | ||
| T8M16-150 | Hypothetical protein | 1e–32 | – | – | |||||
| T8M16-170 | Putative glycerol-3-phosphate dehydrogenase protein | 2e–35 | BI305405 | AI999691 | |||||
| T8M16-180 | Promoter-binding factor-like protein | AP004331 | 139.4 | I | 6e–39 | AU166506 | AV560052 | ||
| AC012562 (2.6 Mb) | III | F17O14-6 | Putative 2,3-biphosphoglycerate-independent phosphoglycerate mutase | 1e–114 | AU100655 | AV545256 | |||
| F17O14-9 | Unknown protein | 3e–13 | D46400 | – | |||||
| F17O14-14 | Unknown protein | AP003255 | 139.9 | I | dot | – | – | ||
| F17O14-15 | Putative protein kinase | 9e–91 | – | AV537987 | |||||
| F17O14-17 | Putative ubiquitin-conjugating enzyme | AP003446 | 139.9–140.5 | I | 4e–16 | AU162581 | AV536792 | ||
| F17O14-23 | Putative protein kinase | AP003302 | 140.5 | I | 9e–39 | – | AV552421 | ||
| F17O14-10 | Unknown protein | AP004331 | 139.4 | I | 8e–74 | C74394 | AV558909 | ||
| AC010871 (2.6 Mb) | T16O11-20 | Unknown protein | AP003411 | 139.9 | I | 8e–54 | – | AI996977 |
The central block of this table presents overlapping rice BACs on chromsome I (Rice BAC accession number, map location, BLASTX score) syntenic to two distinct loci of Arabidopsis genome (Arabidopsis BAC accession number, Arabidopsis chromosome, CDS accession number, putative function). Rice and Arabidopsis ESTs are mentioned by their accession number.
Extensive rearrangement in the syntenic regions
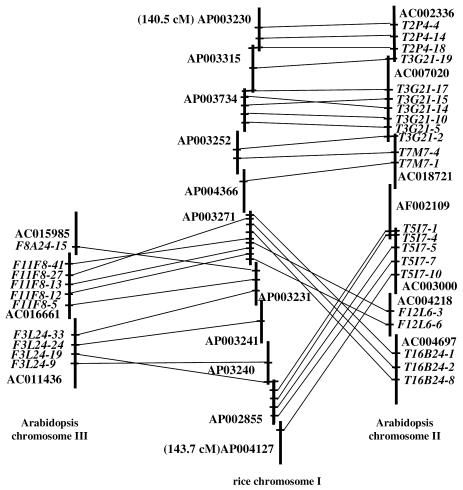
In addition to the limited length of detected syntenic blocks, our analysis shows that, within these regions, considerable rearrangements have occurred. Figure 4 illustrates a locus involving 11 overlapping rice BAC clones on chromosome I covering ≤3.2 cM. This picture is divided into two blocks. The first corresponds to the synteny between rice chromosome I and Arabidopsis chromosome II, the second to that with Arabidopsis chromosome III. The synteny with Arabidopsis chromosome II involves 22 CDS distributed over seven overlapping BAC clones, that with Arabidopsis chromosome III involves 10 CDS distributed over three overlapping BAC clones.
Figure 4.
Conserved microstructure between a rice chromosome I locus and two Arabidopsis loci. Schematic representation of a syntenic region between a rice chromosome I locus and loci on Arabidopsis chromosomes II and III. Vertical lines show overlapping Arabidopsis and rice BAC clones. Conserved Arabidopsis annotated CDS and their rice orthologous counterparts are linked by solid lines.
This region illustrates several levels of rearrangement generally encountered in all the 60 syntenic blocks. The first level of rearrangement takes place within each block of synteny. The gene order on Arabidopsis BAC clones is not strictly conserved on their rice orthologous counterparts. This is clearly the case for two CDS between rice chromosome I and Arabidopsis chromosome II (T3G21-15/T3G21-14) and between rice chromosome I and Arabidopsis chromosome III (F3L24-9/F3L24-19). The second level of rearrangement is at the macro-synteny level. This can be seen between rice chromosome I and Arabidopsis chromosome II, involving rice BAC clones AP003271 and AP002855 and Arabidopsis BAC clones AC003000, AC004218 and AC004697, which clearly show a macro-scale inversion. The final level of rearrangement takes place between syntenic blocks. Within the previously described macro-scale inversion, rice BAC clones AP003271, AP003231, AP003241 and AP003240 have orthologous counterparts on Arabidopsis chromosome III.
Intragenome duplications
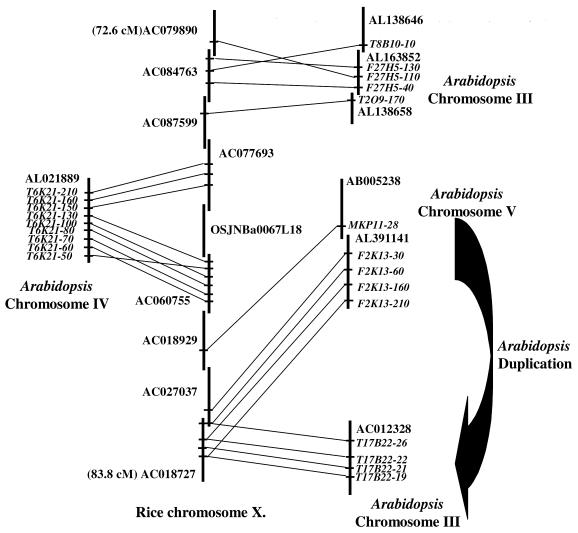
Duplicated Arabidopsis segments involved in multiple syntenic regions. Internal duplications in the Arabidopsis genome have been well documented (5,18,27). Figure 5 presents the results obtained for a region involving nine overlapping rice BACs on chromosome X. Five CDS annotated on three overlapping Arabidopsis BACs on chromosome III are syntenic to three rice BACs. Five CDS annotated on two overlapping Arabidopsis BACs on chromosome V are syntenic to three rice BACs. Nine CDS annotated on one Arabidopsis BAC on chromosome IV are syntenic to two rice BACs. Clearly, rice BAC clones AC027037 and AC018727 are syntenic to two distinct Arabidopsis loci on chromosomes III and V. Interestingly, the gene pair F2K13-60/T17B22-26 is the only one that is also one of the gene pairs duplicated between the two regions of the Arabidopsis genome. DNA-based sequence alignment revealed 24 large duplicated segments of ≥100 kb, comprising 65.6 Mb or 58% of the Arabidopsis genome. Many Arabidopsis duplications appear to have undergone further shuffling, such as local inversions after the duplication event. Among the 60 identified regions, 35 display synteny with duplicated Arabidopsis segments and we have indicated the Arabidopsis duplicated counterpart according to the data of Vision et al. (19; http://www.igd.cornell.edu/~tvision/arab/science_supplement.html) on our web site. The widespread occurrence of gene duplication and the consequent proliferation of large gene families in plants leads to difficulties in determining orthology between species. Thus, orthology can be defined unambiguously only if a high degree of collinearity is observed in the flanking regions of putative orthologs from different multigene families.
Figure 5.
Conserved microstructure between a rice chromosome X locus and multiple duplicated segments of the Arabidopsis genome. Schematic representation of a syntenic region between a rice chromosome X locus and loci on Arabidopsis chromosomes III, IV and V. Vertical lines show overlapping Arabidopsis and rice BAC clones. Conserved Arabidopsis annotated CDS and their rice orthologous counterparts are linked by solid lines.
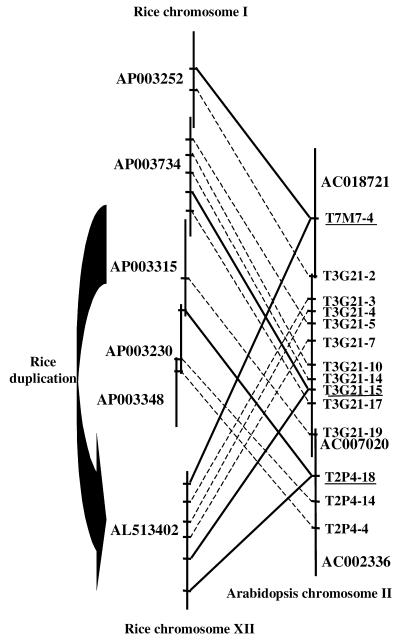
Evidence for local sequence duplications in the rice genome. Current evidence for duplications in the rice genome is limited. A high-density genetic linkage map involving 2275 molecular markers using a single F2 population obtained from a single cross between the japonica variety Nipponbare and the indica variety Kasalath clearly showed a duplication of distal ends between the short arms of chromosomes 11 and 12 in the rice genome (28). Understanding rice genome microstructural evolution necessitates the identification of local or large supplementary chromosomal duplications if they do exist. Figure 6 presents two syntenic regions between rice and Arabidopsis, involving 14 Arabidopsis genes. Among these, 11 are syntenic to a region of rice chromosome I, six to a region on chromosome XII and three are common to the two regions. These two distinct rice loci syntenic to a single Arabidopsis locus therefore reveal a putative local chromosome duplication in the rice genome. Interestingly, the region of Arabidopsis chromosome 2 is one of the rare regions that are not involved in intragenome duplications.
Figure 6.
Evidence for a local duplication in the rice genome. Schematic representation of a syntenic region between two rice on chromosomes I and XII loci and a single Arabidopsis chromosome II locus. Vertical lines show overlapping Arabidopsis and rice BAC clones. Conserved Arabidopsis annotated CDS and their rice orthologous counterparts are linked by solid lines. Pairs of genes conserved in only one or other of the regions are linked by dotted lines.
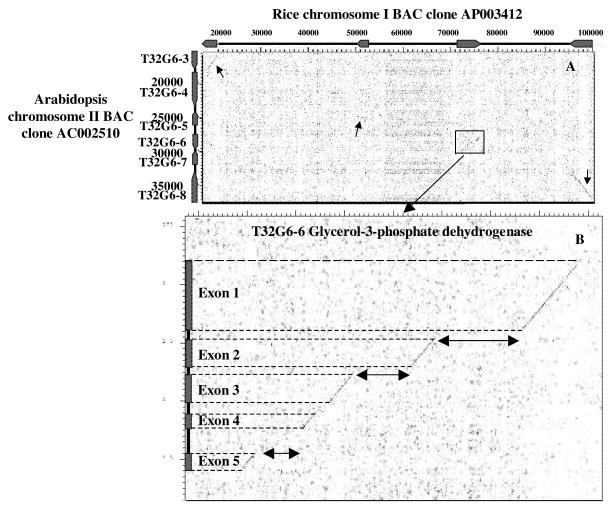
Fine scale analysis. We report in Figure 7A and B a dotplot analysis of another region where rice BAC AP003412 is syntenic to Arabidopsis BAC AC002510. Figure 7A shows a dot analysis between 80 kb of rice chromosome I BAC AP003412 (horizontal axis) and 25 kb of the Arabidopsis chromosome II BAC clone AC002510 (vertical axis). The six annotated CDS on the Arabidopsis sequence are schematically represented in boxes T32G6-3 to T32G6-8. The BLASTX approach showed four conserved CDS with high statistical P value (in parentheses): a putative cytokinin oxidase T32G6-3 (1e–102), an esterase T32G6-5 (4e–41), a glycerol-3-phosphate dehydrogenase T32G6-6 (1e–88) and a Ca2+ ATPase T32G6-8 (0.0). Diagonals represent conserved nucleotide sequences between the two BACs. It appears clearly on the dotplot graphical output that conserved nucleotide sequences correspond exclusively to these four CDS. In this case the BLASTX and dotplot analyses correspond perfectly. Surprisingly, all conserved genes identified by this approach showed conservation not only of the amino acid but also the nucleotide sequence, despite the fact that similarities were detected at the protein level.
Figure 7.
Fine scale study of sequence conservation. (A) Dot plot alignment between rice BAC AP003412 (horizontal) and Arabidopsis BAC AC002510 (vertical). Annotated CDS on Arabidopsis BAC AC002510 are represented as boxes (T32G6-3 to T32G6-8). Diagonals (arrows) on the dotplot output represent conserved CDS (or rice orthologs) on rice BAC AP003412. (B) Fine scale analysis of one conserved CDS, T32G6-6. Vertical boxes represent annotated exons for CDS T32G6-6 on Arabidopsis BAC clone AC002510. The orthologous sequence on rice BAC clone AP003412 is on the horizontal axis. Diagonals on the dotplot represent conserved sequences that match unambiguously to Arabidopsis exons.
Figure 7B also illustrates a general tendency observed in all 60 syntenic regions described in this work. The conserved nucleotide sequence between rice (horizontal) and Arabidopsis T32G6-6 (vertical) perfectly matches the annotated exons of the Arabidopsis CDS (schematically represented as boxes). Conservation in exon–intron structure as in exon sequences has been already shown between orthologous genes in the sh2-a1 homologous region of rice and sorghum (29). Moreover, this graphical representation clearly shows a general expansion of intronic regions in the rice genome compared with the corresponding introns on the annotated Arabidopsis CDS.
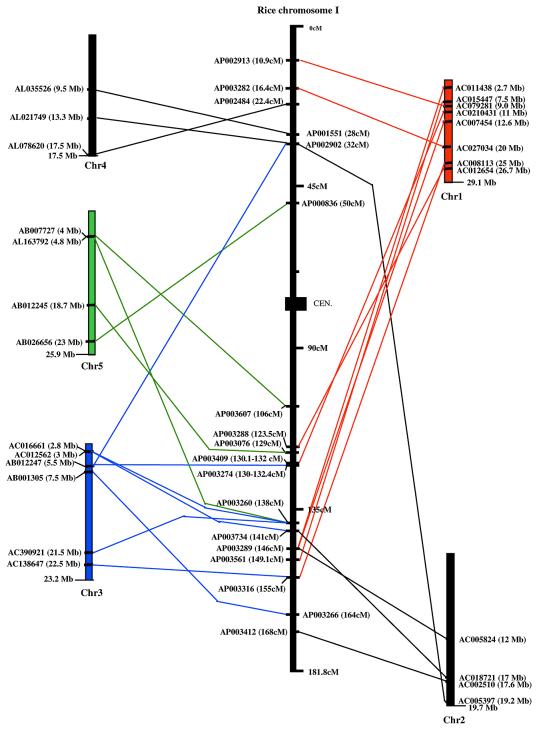
Detailed analysis of rice chromosome I. Figure 8 represents the distribution of the 18 syntenic regions involving four or more CDS between the five Arabidopsis chromosomes and rice chromosome I. Of the sequence of rice chromosome I, 97% has been released into the public domain, i.e. ∼52 Mb. No minimum tiling path is available yet, especially because numerous BAC clones from chromosome I are still in progress, i.e. in unordered pieces. Only six loci are found on the 50 cM of the north arm of the chromosome, mostly syntenic with Arabidopsis chromosomes I and IV, whereas the other 12 regions are found at the distal end of the south arm of the same chromosome. Moreover, Arabidopsis chromosomes are not represented with the same frequency in the 18 syntenic regions. Among these, eight have their Arabidopsis counterpart on chromosome I whereas only three and four rice regions have their counterpart on Arabidopsis chromosomes IV and V, respectively. In addition, six of the eight regions syntenic to Arabidopsis chromosome I are located within a region of ~30 cM on the south arm.
Figure 8.
Distribution of 18 syntenic regions between Arabidopsis and rice chromosome I. Localizations of syntenic points on rice chromosome 1 and the five Arabidopsis chromosomes are shown. When syntenic loci involve overlapping rice and Arabidopsis BACs, only one BAC from each region is mentioned in the figure.
DISCUSSION
Several private or public rice genomic sequences have been announced or published (30). Although the quality of the resulting sequences is sufficient for identification of individual genes, only the high quality, complete chromosome sequences currently being generated by the international rice programme will be useful in detailed molecular studies on genome structure and evolution. However, a complete, polished, fully annotated genomic sequence will only be available in several years. Uniform annotation of the Arabidopsis genome sequence (5) is still under way (http://www.tigr.org/tdb/e2k1/ath1/ath1.shtml). We present here a method aimed at using the ongoing IRGSP sequence to detect syntenic regions between the rice and completed Arabidopsis genomes, providing information on comparative genome structure between monocots and dicots. It has been designed to be easily and regularly updated (http://lgdp.univ-perp.fr/~salse/nar_supplement) as the complete rice genome sequence becomes available as ordered, overlapping BACs. This is made possible by the use of a BLASTX approach. The validity of the results obtained using a relatively low cut-off score for BLAST alignments, high sequence identity in HSPs and selection of the overall length of summed HSPs to avoid matching of functional motifs in proteins is shown by the results of the permutation test demonstrating statistical significance of blocks containing at least three matches in a limited number of Arabidopsis genes.
Early studies using comparative mapping strategies suggested considerable synteny between dicot (5–7) or grass (14,15,31) species. However, more recent sequence-based analyses between dicots have tended to demonstrate that, whereas micro-collinearity is detectable (10–12), syntenic blocks are limited to a few genes, although Grant et al. (13) described synteny covering almost complete soybean and Arabidopsis chromosomes. Between tomato and Arabidopsis, gene sequence conservation varies from 5/5 genes (12) to 12/17 (11). These two species diverged from a common ancestor an estimated 112–156 MYA and the last common ancestor of Arabidopsis and soybean is dated ∼90 MYA, both of which closely follow the divergence of dicot from monocot families (120–200 MYA; 16). Although comparative mapping studies suggest that many grass genomes have retained extensive collinearity over 60 million years or more (14), confirmation at the molecular level will require genomic sequence. A recent comparison of limited regions of barley and rice (32) or sorghum and rice (29) genomic clones showed 4/4 conserved genes, although more sequence information will be necessary to obtain reliable data. Gaut (33) has suggested that current comparative cereal maps do not adequately represent the complexity of collinearity, at least for maize.
Very little information is available on conserved gene order between monocots and dicots. Devos et al. (19), exploiting partial Arabidopsis genomic sequence and available rice EST sequences, used alignment at the nucleotide level to define orthologs as they found that sequences showing identity only at the amino acid level were too dissimilar to be considered true orthologs. Their conclusion that only limited regions of collinearity would be found is supported by more recent results (21), based on comparison of annotated Arabidopsis and rice genomic sequences, which suggested conservation of gene order and content only in short genome segments, with 22/56 genes (39%) found in conserved segments of the two genomes. A recent study using a limited number of annotated rice BACs led to similar conclusions (34). The results presented here refine and considerably extend the comparison of rice and Arabidopsis sequences, although our results suggest that conservation is in fact significantly lower, with ∼17% gene conservation in homeologous segments. One possible explanation is that the presence of members of multigene families may have led to artifactual identification of syntenic regions. Earlier studies did not clearly address the possible contribution of members of multigene families in both genomes and commonly used BLAST cut-off probabilities only rarely distinguish between individual members of gene families. A number of the syntenic blocks indicated by Mayer et al. (21), which are detected in our initial BLAST alignments, are rejected in the permutation analysis. This is probably because these authors used a particularly high cut-off probability (e–5), which leads to several tens or even hundreds of potential orthologs for some genes in each of the two genomes.
In the light of studies within the dicot family, the clearly reduced level of microcollinearity between the genomes of Arabidopsis and rice at the whole genome level is a surprising observation, raising the question as to why microsynteny has been eroded to such an extent over a time frame that is not much greater. Among the 60 syntenic loci, the majority of regions involving overlapping rice BAC clones are <5 cM in length, in agreement with the prediction of Paterson et al. (17). However, individual gene sequence conservation is far lower than the 50% suggested by these authors. Possibly, a very high level of genome rearrangement may have occurred in dicots very early after their divergence from monocots, resulting in a low level of collinearity, or high rates of rearrangement may have occurred specifically in the ancestors of Arabidopsis and its close relatives. Comparative genetic mapping studies have suggested that Arabidopsis may have had an unusually high frequency of chromosomal rearrangement during evolution (17).
The most widely held view is that plant genome evolution occurs primarily by large-scale translocations and inversions, but this model is not sufficient to explain our results. It has been suggested that most eukaryotes are derived from an ancient polyploid, which had a tendency to evolve to a diploid state through sequence diversification and chromosomal rearrangement (13). This hypothesis is largely supported by studies on genomic sequences not only from plants but also from yeast and vertebrates (reviewed in 16). Polyploidy is particularly widespread among extant plant species relative to other eukaryotes. As a consequence, a common occurrence during plant evolution seems to be polyploidization followed by sequence and organizational divergence of the consequent homoeologous chromosomes (14,35). Chromosomal duplications are believed to occur by similar molecular mechanisms in all organisms and are important in genome evolution either in dicots (18) or monocots (14). Goldblatt (36) estimated that ≤70% of plant species may have evolved through polysomy and RFLP studies in a broad range of crop plants confirm that chromosome doubling has been a common event in plant genome evolution (37–40). One would expect that this mechanism would result in multiple, apparently homologous regions being identified in one genome relative to another. This was not observed in our results or during previous analyses (21). One explanation of our observations would be that both dicots and monocots have undergone at least one cycle of polyploidization since the two families separated. Subsequent deletion and/or translocation of genes, which has been clearly demonstrated in Arabidopsis (5,27), with arbitrary loss or translocation of genes from one or other of the resulting copies, would have led to the current situation, in which few genes are involved in limited syntenic regions between Arabidopsis and rice.
The results presented here, while detecting a large number of the known duplications in the Arabidopsis genome, also identify a duplicated region in the rice genome, although this was not the main aim of this study. Little information is available on duplications in the rice genome, apart from that of distal ends between the short arms of chromosomes 11 and 12 (28,41). In the latter study, 35 markers including 21 cDNA clones detected duplicated loci arrayed strictly in the same order along the two genomic regions with an expected length of ∼2.5 Mb. The degree of conservation based on these physical and genetic analyses demonstrates a single event of long range chromosomal duplication in the rice genome. However, it is possible that the density of markers used in this study (approximately one every 200 kb) was insufficient to detect duplication of more limited regions. Only three genes are common to all three regions shown in Figure 5, suggesting that most duplicated genes have been lost as previously suggested (42–44), although empirical studies suggest that most duplicated genes may retain function (45). Because very few BAC contigs on chromosome XII are available in the public database, extension of the chromosomal duplication was impossible. As soon as a complete overview of this duplication has been obtained when more sequence on chromosome XII is available, comparison of phylogenetic trees involving each duplicated gene (homologs) in rice and Arabidopsis genomes with orthologs from more distantly related grass genomes will reveal the nature of the evolutionary event of the chromosome duplication observed in the rice genome.
Several other short segments of the Arabidopsis sequence identified two regions of the rice genome. No examples of duplications common to both Arabidopsis and rice were found, which would be expected from the datation of the Arabidopsis duplication event. These distant tandem duplications, where a few genes from a collinear set appear duplicated, may be the legacies of tandem duplications of large segments or may indicate a cis preference for gene movement. The mobility of genes within plant genomes has long been recognized (46) via transposons, which are widespread in the rice genome and responsible for ‘exon shuffling’ (47). Regardless of their origin, these duplications can severely interfere with anchoring BAC clones for the physical mapping and map-based gene cloning. Deletion, rearrangement and dispersion to new genomic locations of single or small clusters of genes probably represents a significant effector of rice genome evolution. Intragenome comparison in rice will be necessary to determine the exact extent of duplication in this genome.
Interestingly, although only very short syntenic segments are detected in this study, Figure 7 clearly shows a non-random distribution of the syntenic points between chromosome I of rice and the five Arabidopsis chromosomes. A greater density of syntenic points is found on the south arm of the rice chromosome. Preliminary analyses (J.Salse and B.Piégu, unpublished results) indicate that transposon density is higher on the north arm, which could indicate that ancestral gene order has been disrupted to a greater extent in this region, leading to detection of fewer points. In addition, almost half of the points are found on Arabidopsis chromosome I and all but two of these are limited to a short segment of the rice chromosome. This apparent large-scale synteny may indicate that current chromosome structure may have evolved rather by translocation and/or rearrangement of single genes or limited regions of ancestral chromosomes. It may also explain the apparent contradiction between early mapping studies, often using widely separated markers and which suggested considerable intergenome synteny, and more recent, sequence-based analyses which show a much lower degree of conservation.
A fuller picture of the precise relationship between the Arabidopsis and rice genomes, as well as clearer evidence of rice chromosomal duplication, will emerge as more sequence data become available. It already seems clear that the limited level of synteny associated with a high rate of rearrangement within syntenic regions, as identified in our study, has limited applications in map-based cloning strategies. Although such a low level of microstructure conservation makes the use of a positional approach to integrate functional and structural genomic information from monocot and dicot species hazardous, detailed analysis of the complete genomes of these two model species will provide valuable information on plant genome structure and evolution.
Acknowledgments
ACKNOWLEDGEMENTS
We thank two anonymous referees for constructive comments. This work was supported by grants from the Centre National de la Recherche Scientifique. This work was based largely on the publicly available Arabidopsis and rice genomic sequences from the AGI and the IRGSP, respectively.
REFERENCES
- 1.Sasaki T. and Burr,B. (2000) International rice genome sequencing project: the effort to completely sequence the rice genome. Curr. Opin. Plant Biol., 3, 138–141. [DOI] [PubMed] [Google Scholar]
- 2.Fukui K. and Lijima,K. (1991) Somatic chromosome map of rice by imaging methods. Theor. Appl. Genet., 81, 589–596. [DOI] [PubMed] [Google Scholar]
- 3.Arumuganathan K. and Earle,E.D. (1991) Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep., 9, 208–218. [Google Scholar]
- 4.Delseny M., Salse,J., Cooke,R., Sallaud,C., Regad,F., Lagoda,P., Guiderdoni,E., Ventelon,M., Brugidou,C. and Ghesquiere,A. (2001) Rice genomics: Present and Future. Plant Physiol. Biochem., 39, 323–334. [Google Scholar]
- 5. The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 6.Kowalski S.P., Lan,T.H., Feldmann,K.A. and Paterson,A.H. (1994) Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals island of conserved organization. Genetics, 138, 449–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavell A.C., Lydiate,D.J., Parkin,I.A.P., Dean,C. and Trick,M. (1998) Collinearity between a 30 centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome, 41, 62–69. [PubMed] [Google Scholar]
- 8.Lagercrantz U. (1998) Comparative mapping between Arabidopsis thaliana and Brassica napus indicated that brassica genomes have evolved through extensive genome replication accompanied by chromosome fusion and frequent rearrangement. Genetics, 150, 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan T.H., DelMonte,T.A., Reischmann,K.P., Hyman,J., Kowalski,S.P., McFerson,J., Kresovich,S. and Paterson,A.H. (2000) An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Res., 10, 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt R., Acarkan,A. and Boivin,K. (2001) Comparative structural genomics in the Brassicaceae family. Plant Physiol. Biochem., 39, 253–262. [Google Scholar]
- 11.Ku H.M., Vision,T., Liu,S. and Tanksley,S.D. (2000) Comparative sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl Acad. Sci. USA, 97, 9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossberg M., Theres,K., Acarkan,A., Herrero,R., Schmitt,T., Schumacher,K., Schmitz,G. and Schmidt,R. (2001) Comparative sequence analysis reveals extensive microcolinearity in the lateral suppressor regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell, 13, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant D., Cregan,P. and Shoemaker,R.C. (2000) Genome organisation in dicots: genome duplications in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl Acad. Sci. USA, 97, 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore G., Devos,K.M., Wang,Z. and Gale,M.D. (1995) Grasses, line up and form a circle. Curr. Biol., 5, 737–739. [DOI] [PubMed] [Google Scholar]
- 15.Feuillet C. and Keller,B. (1999) High gene density is conserved at syntenic loci of small and large grass genome. Proc. Natl Acad. Sci. USA, 96, 8665–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe K.H., Gouy,M., Yang,Y.W., Sharp,P.M. and Li,W.H. (1989) Date of the monocot–dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl Acad. Sci. USA, 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paterson A.H., Lan,T.H., Reischmann,K.P., Chang,C., Lin,Y.R., Liu,S.C., Burow,M.D., Kowalski,S.P., Katsar,C.S., DelMonte,T.A., Feldmann,K.A., Schertz,K.F. and Wendel,J.F. (1996) Towards a unified genetic map of higher plants, transcending the monocot–dicot divergence. Nature Genet., 14, 380–382. [DOI] [PubMed] [Google Scholar]
- 18.Vision T.J., Brown,D.G. and Tanksley,S.D. (2000) The origins of genomic duplications in Arabidopsis. Science, 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- 19.Devos K.M., Beales,J., Nagamura,Y. and Sasaki,T. (1999) Arabidopsis–Rice: will colinearity allow gene prediction across the Eudicot–Monocot divide? Genome Res., 9, 825–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Dodeweerd A.M., Hall,C.R., Bent,E.G., Johnson,S.J., Bevan,M.W. and Bancroft,I. (1999) Identification and analysis of homoeologous segments of the genomes of rice and Arabidopsis thaliana. Genome, 42, 887–892. [PubMed] [Google Scholar]
- 21.Mayer K., Murphy,G., Tarchini,R., Wambutt,R., Volckaert,G., Pohl,T., Dusterrhoft,A., Stiekema,W., Entian,K.D., Terryn,N., Lemcke,K., Haase,D., Hall,C.R., Van Dodeweerd,A.M., Tingey,S.V., Mewes,H.W., Bevan,M.W. and Bancroft,I. (2001) Conservation of microstructure between a sequenced region of rice and multiple segments of the genome of Arabidopsis thaliana. Genome Res., 11, 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnhammer E.L.L. and Durbin,R. (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene, 167, 1–10. [DOI] [PubMed] [Google Scholar]
- 24.Pavy N., Rombauts,S., Dehais,P., Mathe,C., Ramana,D.V., Leroy,P. and Rouze,P. (1999) Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences. Bioinformatics, 11, 887–899. [DOI] [PubMed] [Google Scholar]
- 25.Leister D., Kurth,J., Laurie,D.A., Yano,M., Sasaki,T., Devos,K., Graner,A. and Schulze-Lefert,P. (1998) Rapid reorganization of resistance gene homologues in cereal genomes. Proc. Natl Acad. Sci. USA, 95, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallego F., Feuillet,C., Messmer,M., Penger,A., Graner,A., Yano,M., Sasaki,T. and Keller,B. (1998) Comparative mapping of the two wheat leaf rust resistance loci lri and lr10 in rice and barley. Genome, 41, 328–336. [DOI] [PubMed] [Google Scholar]
- 27.Blanc G., Barakat,A., Guyot,R., Cooke,R. and Delseny,M. (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell, 12, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harushima Y., Yano,M., Shomura,A., Sato,M., Shimano,T., Kuboki,Y., Yamamoto,T., Lin,S.Y., Antonio,B.A., Parco,A., Kajiya,H., Huang,N., Yamamoto,K., Nagamura,Y., Kurata,N., Khush,G.S. and Sasaki,T. (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics, 148, 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen M., SanMiguel,P. and Bennetzen,J.L. (1998) Sequence organization and conservation in sh2/a1 homologous regions of sorghum and rice. Genetics, 148, 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J., Hu,S., Wang,J., Li,S., Wong,K.G., Liu,B., Deng,Y., Dai,L., Zhou,Y., Zhang,X. et al. (2001) A draft sequence of the rice (Oryza sativa ssp. indica) genome. Chinese Science Bulletin, 46, 1937–1942. [Google Scholar]
- 31.Keller B. and Feuillet,C. (2000) Colinearity and gene density in grass genomes. Trends Plant Sci., 5, 246–251. [DOI] [PubMed] [Google Scholar]
- 32.Dubcovsky J., Ramakrishna,W., SanMiguel,P.J., Busso,C.S., Yan,L., Shiloff,B.A. and Bennetzen,J.L. (2001) Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes. Plant Physiol., 125, 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaut B.S. (2001) Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res., 11, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H., Sachidanandam,R. and Stein,L. (2001) Comparative genomics between rice and Arabidopsis shows scant collinearity in gene order. Genome Res., 12, 2020–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagercrantz U. and Lydiate,D.J. (1996) Comparative genome mapping in Brassica. Genetics, 144, 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldblatt P. (1979) Polyploidy in angiosperms: monocotyledons. Basic Life Sci., 13, 219–239. [DOI] [PubMed] [Google Scholar]
- 37.Reinisch A.J., Dong,J., Brubaker,C.L., Stelly,D.M., Wendel,J.F. and Paterson,A.H. (1994) A detailed RFLP map of cotton, Gossypium hitsutum x Gossypium barbadense: chromosome organisation and evolution in a disomic polyploid genome. Genetics, 138, 829–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoemaker R.C., Polzin,K., Labate,J., Specht,J., Brummer,E.C., Olson,T., Young,N., Concibido,V., Wilcox,J., Tamulonis,J.P., Kochert,G. and Boerma,H.R. (1996) Genome duplication in soybean (Glycine subgenus soja). Genetics, 144, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King J.J., Braden,J.M., Bark,O., McCallum,J.A. and Harvey,M.J. (1998) A low density genetic map of onion reveals a role for tandem duplication in the evolution of an extremely large diploid genome Theor. Appl. Genet., 96, 52–62. [Google Scholar]
- 40.Helentjaris T., Weber,D. and Wright,S. (1988) Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphism (RFLP). Genetics, 118, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J., Tanoue,H., Shimokawa,T., Umehara,Y., Yano,M. and Sasaki,T. (1998) Physical mapping of duplicated genomic regions of two chromosomes in rice. Genetics, 150, 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nei M. and Roychoudhury,A.K. (1973) Probability of fixation of nonfunctional genes at duplicate loci. Am. Nat., 107, 362–372. [Google Scholar]
- 43.Takahata N. and Maruyama,T. (1979) Polymorphism and loss of duplicate gene expression: a theoretical study with application to tetraploid fish. Proc. Natl Acad. Sci. USA, 76, 4521–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walsh J.B. (1995) How often do duplicated genes evolve new function? Genetics, 139, 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Force A., Lynch,M., Pickett,F.B., Amores,A., Yan,Y.L. and Postlethwait,J. (1999) Preservation of duplicate genes by complementary degenerative mutations. Genetics, 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClintock B. (1948) Mutable loci in maize. Carnegie Institute of Washington Year Book, Vol. 47, pp. 155–169.
- 47.Hirochika H. (1997) Retrotransposons in rice: their regulation and use for genome analysis. Plant Mol. Biol., 35, 231–240. [PubMed] [Google Scholar]