Abstract
Oligonucleotide-based DNA microarrays are becoming increasingly useful for the analysis of gene expression and single nucleotide polymorphisms. Here we report a systematic study of the sensitivity, specificity and dynamic range of microarray signals and their dependence on the labeling and hybridization conditions as well as on the length, concentration, attachment moiety and purity of the oligonucleotides. Both a controlled set of in vitro synthesized transcripts and RNAs from biological samples were used in these experiments. An algorithm is presented that allows the efficient selection of oligonucleotides able to discriminate a single nucleotide mismatch. Critical parameters for various applications are discussed based on statistical analysis of the results. These data will facilitate the design and standardization of custom-made microarrays applicable to gene expression profiling and sequencing analyses.
INTRODUCTION
DNA microarrays hold the promise of becoming a revolutionary tool for large-scale parallel analyses of genome sequence and gene expression (1–4). Current applications range from global analyses of transcriptional programmes in yeast or mammals (5,6) to establishment of novel criteria for the classification and evaluation of clinical course of tumors (7–10) and to accelerated discovery of drug targets (11,12).
Methods for microarray fabrication include spotting of DNA onto nylon membranes or glass slides by robots with pins or ink jet printers (13,14). The DNA spotted corresponds to fragments of genomic DNA, cDNAs, PCR products or chemically synthesized oligonucleotides (15). cDNA arrays are often used in RNA expression analysis, while oligonucleotide arrays are additionally used for sequence analyses. Oligonucleotides can also be synthesized in situ on the surface of the array by means of light-directed combinatorial synthesis (photolithography) or ink jet technologies, which allow microarrays of higher density. Current state-of-the-art technology allows the inclusion of more than 400 000 sequences representing up to 13 000 genes and expressed sequence tags (see below) on a surface of 1.6 cm2 (16).
Oligonucleotide-based microarrays offer a number of advantages over cDNA microarrays, including (i) more controlled specificity of hybridization, which makes them particularly useful for the analysis of single nucleotide polymorphisms (17) or mutational analysis (18,19); (ii) versatility to address subtle questions about transcriptome composition such as the presence and prevalence of alternatively spliced or alternatively polyadenylated transcripts (20,21); (iii) capacity to systematically screen whole genomic regions for gene discovery (22,23); and (iv) the fact that only sequence information (not biological samples or cDNA collections) is required to generate custom-made microarrays.
Despite the predictable impact and widespread use of oligonucleotide-based microarrays, there is a paucity of publicly available information regarding the design and use of this technology. In addition there is a need for standardization that will facilitate comparison of microarray data (24). Basic questions such as the number of oligonucleotides required for reliable detection of an RNA can have profound practical consequences for the quality and financial feasibility of specific experiments or projects.
Early reports using in situ photolithographic synthesis employed 300 probe pairs (match and single mismatch control) of 15 nt in length per gene studied. Statistical analysis of the hybridization signals allowed the detection of 2-fold changes in the levels of cytokine mRNAs in T lymphocytes under a variety of physiological stimuli (25). Improved photolithographic in situ synthesis resulting in reliable longer oligonucleotides, together with statistical analyses of the data, allowed these investigators to reduce the number of probes required per gene to 20 oligonucleotide pairs of 25 nt in length (8,26), and more recently to 16 oligonucleotide pairs per gene (16).
Ink jet procedures have allowed in situ synthesis of longer oligos. A single 60 nt-long oligo per gene has rendered results comparable with those obtained using cDNA microarrays, and has allowed functional annotation of complete chromosomes under dozens of experimental conditions (22). One disadvantage of long oligonucleotide probes, shared with cDNA microarrays, is the difficulty of generating reliable mismatch controls that will assess the specificity of hybridization.
Here we report results of experiments designed to optimize the selection of oligonucleotides and the performance of oligonucleotide-based microarrays. Specificity, sensitivity and dynamic range of the signals were analyzed with regard to characteristics of the oligonucleotide (e.g. length and purity), hybridization conditions, labeling method and other parameters both in a controlled system composed of in vitro transcribed RNAs and using mRNA from mammalian cells.
MATERIALS AND METHODS
Oligonucleotide selection
Oligonucleotides were selected using modified Gene Skipper software and selection rules modified from published criteria (25). The algorithm used applies the following set of hierarchical conditions. (i) Exclusion of oligonucleotides with ‘adverse’ base composition: total number of As or Ts less than 10; total number of Cs or Gs less than six; no more than six As or Ts in a row; a palindrome score (a measure of probe self-complementarity) of <7 nt. (ii) Selection of sets of oligonucleotides with homogeneously high melting temperature. (iii) Exclusion of oligonucleotides with perfect complementarity to other sequences present in the set of genes to be analyzed. (iv) Exclusion of oligonucleotides with ability to form hairpin loops. The program also selects the corresponding mismatch control oligonucleotides, containing a single transversion in the central position. This software is freely available by email request to schwager@embl-heidelberg.de.
Spotting and attachment to glass slides
Unless indicated, HPLC-purified oligonucleotides containing an amino group and six carbon spacer at the 5′ end were spotted onto aminosilane-coated glass slides using either a GMS 417 spotter (Affymetrix) or a SDDC Microarray spotter (Engineering Systems Inc.), with equivalent results. Fifty picoliters of oligonucleotide solutions were spotted at concentrations of 30–100 µM. Attachment was achieved by incubating the coated glass slides with the spotted oligos for 4 h at 60°C and 10 min at 120°C. Alternative attachment protocols, e.g. overnight incubation at 37°C, resulted in decreased sensitivity.
Attachment onto epoxy surfaces or after acid treatment of glass slides was as described previously (27).
Preparation and labeling of in vitro transcripts
Templates for transcription of selected genes were generated by PCR from the corresponding cDNAs using oligonucleotides containing a T7 promoter in the (–) strand. Standard 25 µl in vitro transcription reactions were set up containing 100 µM fluorescently labeled nucleotides [cyanine 5-CTP (Cy5) or cyanine 3-CTP (Cy3); NEN], 200 µM CTP, 500 µM ATP, UTP, GTP (Amersham Pharmacia), 200 ng of template DNA and 1.6 U/µl T7 RNA polymerase (Promega). After incubation for 2 h at 37°C, the DNA template was digested with 10 U of DNase I (Promega) at 37°C for 30 min. The samples were then purified using Chroma-Spin columns (Clontech) and stored at –20°C.
To precisely measure the amount of RNA synthesized, an aliquot of the reaction was spiked with a trace of [α-32P]GTP. The transcripts were quantified after fractionation by denaturing polyacrylamide gel electrophoresis, excising the corresponding band and measuring radioactivity with a liquid scintillation counter.
Preparation and labeling of RNA from HeLa cells
Total RNA was extracted from HeLa cells using the RNAeasy kit (Qiagen) and the concentration estimated by measuring optical density at 260 nm. Poly(A)+ purification was carried out using olig-dT cellulose columns (28).
For direct cDNA labeling, 2 µg of poly(A)+ RNA was incubated in a 25 µl reaction containing 200 pmol of 14-nt random primers, 200 pmol oligo-dT (12–18 nt in length), which was heated at 70°C for 10 min and then left on ice for 5 min. cDNA synthesis was carried out in 55 µl reactions containing 400 U Superscript II Reverse Transcriptase (Invitrogen), 100 µM Cy5-dUTP (New England Nuclear), 200 µM dTTP, 500 µM dATP, dCTP, dGTP (Amersham Pharmacia), and the buffer conditions recommended by the manufacturer. After 2 h incubation at 42°C the reaction was stopped by incubating at 65°C for 10 min in the presence of 50 mM NaOH and 1 mM EDTA in a final volume of 58 µl. Labeled cDNA was purified using Chroma-Spin columns +STE-10 (Clontech) and stored at –20°C.
For cRNA labeling, either 5–20 µg of total HeLa RNA or 2 µg of poly(A)+ RNA was used. Signals obtained using 20 µg of total HeLa RNA and 2 µg of poly(A)+ RNA were comparable. RNA was incubated with 8 µM T7-dT(24) primer in a 25 µl volume at 70°C for 10 min and then incubated at 4°C for 5 min. First strand synthesis was carried out in a 41.7 µl reaction containing 420 U Superscript II Reverse Transcriptase (Invitrogen), 500 µM dNTP mix (Amersham Pharmacia) and 10 mM DTT, under the buffer conditions recommended by the manufacturer. After 1 h incubation at 37°C the reaction mixture was chilled on ice for 5 min and then the second strand was synthesized in a 75 µl reaction mix containing 20 U DNA polymerase I, 5 U DNA ligase, 5 U RNase H (all three enzymes from Invitrogen), 200 µM dNTP mix, under the buffer conditions recommended by the manufacturer, incubated for 2 h at 16°C in a Thermocycler, and the reaction stopped with 60 mM EDTA in a total final volume of 85 µl. After phenol/chloroform extraction and ethanol precipitation in the presence of 20 µg glycogen (Roche), the pellet was washed twice with 70% ethanol, dried and resuspended in 6 µl of distilled water. T7 transcription was carried out overnight at 37°C in 25 µl using one-fourth of cDNA, 160 U T7 Polymerase (Promega), 1 mM DTT, 500 µM ATP, UTP, GTP, 250 µM CTP and 100 µM Cy5-CTP. After digestion with 1 U of RNase-free DNase for 30 min, labeled cRNA was purified twice using Chroma-Spin columns (Clontech).
Fragmentation of labeled cRNA was achieved by incubation of 15 µg of cRNA in 20 mM Tris–acetate pH 8.1, 50 mM potassium acetate, 15 mM magnesium acetate for 15 min at 94°C.
Hybridization and washing
Slides were incubated in a glass chamber for 45 min at 42°C with pre-warmed pre-hybridization buffer (6× SSC, 0.5% SDS, 1% BSA) and subsequently quickly washed with distilled water pre-warmed at the same temperature and dried by short centrifugation.
Poly(A)+ (5 µg) and 1 µg of human Cot DNA were added to the sample, dried in a speed vac at 45°C and redissolved in 12 µl (for a 24 × 24 mm cover slip) of hybridization buffer (50% formamide, 6× SSC, 0.5% SDS, 5× Denhardt’s solution; 58% formamide for RNA from HeLa cells). Hybridization was carried out in a humid chamber for 16 h. Washings were performed twice at 42°C inside a glass chamber containing 0.1× SSC, 0.1% SDS for 5 min and twice more in 0.1× SSC for 5 min. Washes at higher temperatures (47, 55 and 62°C) or lower concentrations of SSC (0.03× SSC, 0.01× SSC or water) resulted in very significant losses in fluorescent signals. Slides were subsequently dried by brief centrifugation.
Data analyses
Microarrays were scanned using either a GMS 418 array scanner (Affymetrix) or a Gene Pix 4000B simultaneous dual wave-length scanner (Axon Instruments Inc.). The data obtained were analyzed using Chip Skipper software. This software is freely available by email request to schwager@embl-heidelberg.de.
The intensity values per spot were determined by creating a circle adjusted to the size of the spot (diameter ∼15 µm centered on the spot) and integrating the intensity value per pixel in that area. Background was extracted by determining the median values of pixels located on the perimeter of a square surrounding the circle centered on the same position. Intensity values were normalized using as spotting controls a mix of oligonucleotides of known concentration labeled with Cy5 and Cy3.
RESULTS
RNAs corresponding to the antisense sequence of five eukaryotic RNA binding proteins were transcribed in vitro in the presence of fluorescent Cy5- or Cy3-labeled nucleotides. The rationale for the use of antisense transcripts was to allow direct comparisons with hybridization of labeled cDNAs or cRNAs generated in subsequent experiments aimed to analyze mRNAs from biological samples (see below). A trace of radioactive nucleotides was used in the transcription reactions to allow the quantification of the yield of purified RNAs. The RNAs were hybridized to an oligonucleotide microarray where HPLC-purified 5′-amino modified oligos of 25, 30 or 35 nt in length were printed on the activated surface of a glass slide (see Materials and Methods). Figure 1A indicates the layout of oligos corresponding to one of the genes. Fifteen non-overlapping oligos corresponding to each length, complementary in sequence to the corresponding transcript, were selected according to the algorithm described in Materials and Methods, and printed in triplicate. Mismatch controls containing a single transversion change in the middle position of each oligo were printed in triplicate next to the corresponding perfect match oligo. The sequences of oligonucleotides used for the analysis of two of the genes are provided as Supplementary Material. After hybridization to in vitro transcribed RNA and washing, fluorescent signals were detected using a confocal microarray scanner.
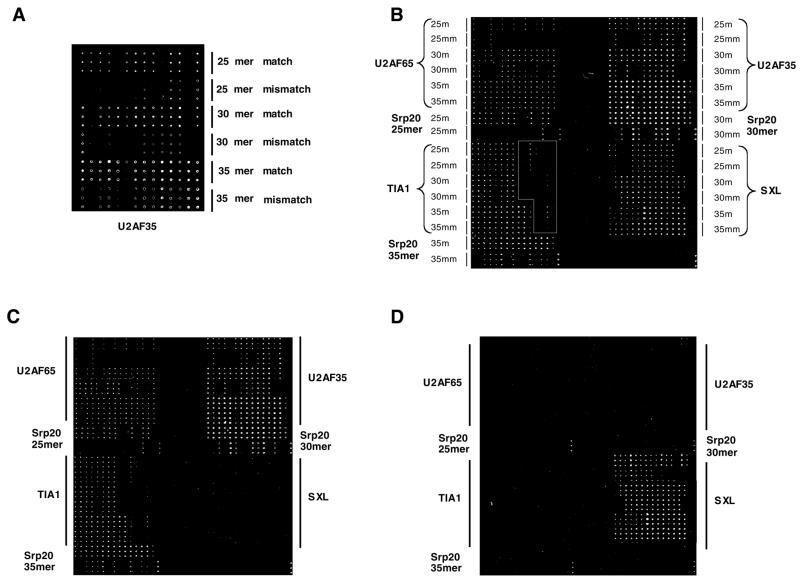
Figure 1.
Hybridization of in vitro transcribed RNAs to oligonucleotide microarrays. (A) Layout of oligos corresponding to one gene, and fluorescent scan of the hybridization data corresponding to oligos for one of the genes under study (U2AF35). (B) Microarray layout for the five genes under study and fluorescent scan of hybridization data to the complete set of in vitro transcribed RNAs. The layout of oligo lengths and M/MM controls is indicated. The white box surrounds an area where the oligos spotted correspond to regions of the mRNA not present in our in vitro transcripts. (C) As in (B), without RNA corresponding to the gene SXL. (D) As in (B), with only SXL RNA.
Specificity and sensitivity optimization
The results shown in Figure 1A indicate that the signals associated with perfect match oligos were stronger than those associated with their mismatch controls.
Figure 1B shows similar results for the five genes studied. Triplets of fluorescent signals present at the bottom-right position of each quarter correspond to fluorescent markers used as spotting controls. As a first test of specificity, one of the RNAs (SXL) was omitted from the hybridization mix. A reduction in the fluorescent signals corresponding to SXL oligonucleotides was observed (compare Fig. 1B and C). As a second test, only SXL transcripts were hybridized to the microarray. Figure 1D shows that little fluorescence was detected associated with oligos corresponding to genes different from SXL. A third specificity test was built in the design of the microarray. Oligos corresponding to sequences in the 3′-untranslated region (3′-UTR) of some of the genes were present in the microarray, whereas labeled in vitro synthesized RNAs were limited to the open reading frames. Hybridization to oligos complementary to the UTR regions was undetectable for most probes (positions inside the white rectangles in Fig. 1B).
Taken together, the results of Figure 1 indicate that hybridization to a significant fraction of the oligonucleotides selected is specific, as shown by the discrimination of single nucleotide mismatches.
Table 1 summarizes quantitative information obtained from at least three independent experiments carried out as in Figure 1B. RNAs labeled with Cy-5 and RNAs labeled with Cy-3 were used in each experiment, thus providing a duplicate read out of each result. As signals for each oligo were obtained in triplicate, the figures in Tables 1 and 2 correspond to the average and standard deviation of at least 270 independent measurements for each gene and oligo length. The data were further validated by results from more than 40 hybridization experiments.
Table 1. Variation of microarray sensitivity and specificity with oligo length: M/MM ratios for oligos corresponding to the different genes and oligonucleotide lengths.
| Length | M/MM | % M/MM >2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| U2AF65 | Srp20 | U2AF35 | TIA1 | SXL | Average | Median | Average | Median | |
| 25mer |
3.3 ± 0.6 |
11.1 ± 1.3 |
3.6 ± 0.1 |
4.6 ± 0.1 |
4.4 ± 0.4 |
5.4 |
4.4 |
77% |
75% |
| 30mer |
2.7 ± 0.6 |
9.7 ± 0.4 |
4.3 ± 0.2 |
2.6 ± 0.4 |
2.2 ± 0.2 |
4.3 |
2.7 |
79% |
85% |
| 35mer | 1.8 ± 0.2 | 8.2 ± 0.4 | 2.8 ± 0.2 | 1.8 ± 0.4 | 1.7 ± 0.1 | 3.3 | 1.8 | 74% | 60% |
Average and median M/MM values, as well as percentage of oligos with a M/MM discrimination >2, are indicated.
Table 2. Variation of microarray sensitivity and specificity with oligo length: ratios between fluorescent intensities of different lengths of oligonucleotides corresponding to the indicated genes.
| Length | Signal intensity ratios | ||||||
|---|---|---|---|---|---|---|---|
| U2AF65 | Srp20 | U2AF35 | TIA1 | SXL | Average | Median | |
| 35/25 |
2.2 ± 0.6 |
3.6 ± 0.5 |
2.8 ± 0.5 |
2 ± 0.1 |
2.5 ± 0.1 |
2.6 ± 0.6 |
2.5 |
| 35/30 |
0.6 ± 0.1 |
0.6 ± 0.1 |
1.3 ± 0.4 |
1.1 ± 0.1 |
1.2 ± 0.1 |
1 ± 0.3 |
1.1 |
| 30/25 | 2.9 ± 0.4 | 5.8 ± 0.6 | 1.6 ± 0.1 | 1.7 ± 0.2 | 2.1 ± 0 | 2.8 ± 1.7 | 2.1 |
Average, standard deviation and median M/MM values are also shown.
Oligo length
Two main conclusions can be drawn from these data. First, a decrease in match/mismatch (M/MM) ratio was observed with the increase in oligo length. This is particularly clear when the average median values are considered (from 4.4 for 25mers to 1.8 for 35mers). This trend is expected, as longer oligos are more likely to energetically accommodate a single nucleotide mismatch at a central position. Statistically, ∼75% of the oligonucleotides selected showed a M/MM discrimination >2-fold.
The second conclusion is that up to 4-fold differences in M/MM ratios could be observed for the different genes studied. These could not be attributed to overall differences in G+C content, or other obvious sequence features.
The effect of oligonucleotide length on the intensity of the fluorescent signals was also analyzed. Table 2 shows the ratios between the intensity of the signals for each gene and oligo length. While the signals for 30 and 35mer oligonucleotides were 2–5 times higher than 25mers, no significant difference was observed between oligos of 30 and 35 nt. Once again, differences in microarray performance were observed for different genes. Sensitivity measurements indicated that ≥0.1 ng (0.3 fmol) could be routinely detected. This level of sensitivity would enable the detection of one specific mRNA present at 0.01% in 1 µg of poly(A)+ RNA. This would be equivalent to detect low abundance mRNA species (e.g. PPAR-α, HMGcoA), but may represent difficulties to detect very low abundance transcripts (e.g. Fas or Insulin receptor) (29). Although the threshold of detection depended on the sensitivity of the scanner used, similar M/MM ratios were obtained with different scanners.
Oligo concentration
Tables 3 and 4 show the results obtained for one of the genes (SXL) with different amounts of oligonucleotides spotted. Equivalent results were obtained for the other four genes studied (data not shown). Only marginal increases in specificity and sensitivity were observed by increasing the concentrations of the oligo in the spotting solution from 20 to 50 pmol of oligo/µl. Although additional tests in a range of concentrations from 10 to 100 pmol/µl registered up to 10-fold differences in signal intensity, no significant differences were normally observed for concentrations between 30 and 100 pmol/µl. These observations indicate that the amount of oligonucleotide attached at spotting concentrations between 30 and 100 µM were not rate limiting for detection of fluorescent RNAs. Consistent with this conclusion, hybridization of higher amounts of target RNAs resulted in increased fluorescent signals (data not shown).
Table 3. Variation of microarray sensitivity and specificity with the concentration of oligos spotted: M/MM ratios.
| Oligo concentration (pmol/µl) | M/MM | ||
|---|---|---|---|
| 25mer | 30mer | 35mer | |
| 50 |
4.3 ± 0.6 |
2 ± 0.1 |
2 ± 0.2 |
| 30 |
3.6 ± 0.6 |
2 ± 0.1 |
1.5 ± 0.1 |
| 20 | 3.8 ± 0.1 | 2.2 ± 0.2 | 1.8 ± 0.1 |
Average and standard deviations are shown for oligos of the indicated lengths, spotted at the concentrations indicated.
Table 4. Variation of microarray sensitivity and specificity with the concentration of oligos spotted: fluorescent intensities for oligos of the indicated lengths, spotted at the concentrations indicated.
| Oligo concentration (pmol/µl) | Signal intensities (×106) | |||
|---|---|---|---|---|
| 25mer | 30mer | 35mer | Average | |
| 50 |
1.75 ± 0.07 |
2.49 ± 0.07 |
2.98 ± 0.08 |
2.41 ± 0.62 |
| 30 |
1.75 ± 0.05 |
2.38 ± 0.01 |
2.51 ± 0.05 |
2.21 ± 0.41 |
| 20 | 1.06 ± 0.09 | 2.09 ± 0.02 | 2.36 ± 0.07 | 1.84 ± 0.69 |
Average and standard deviation values for all lengths are shown.
The performance of different attachment chemistries was also tested. Higher sensitivity (between 10- and 100-fold) was observed with silanized coating compared with pan-epoxy coating or acid treatment of the glass surface (27). Although 5′ amino modification was not strictly required, 2–4-fold increases in detection levels were observed when amino-modified oligos were used.
Hybridization temperature and formamide concentration
Next, the effect of different hybridization temperatures and percentage of formamide on sensitivity and specificity of the signals were analyzed. Figure 2 shows the results obtained for one gene (SXL) and one oligo length (30mer), which were representative of the performance of other genes and oligonucleotide lengths. Hybridization at temperatures between 4 and 25°C resulted in poor microarray performance due to low signal intensities (4°C) or high background (25°C). Therefore, a range of temperatures between 30 and 42°C was tested. Figure 2A shows that while increasing the temperature from 30 to 35°C resulted in a 25% increase in M/MM ratio (up to 40% for other genes, and not higher for shorter oligos), a further increase to 42°C did not improve (in fact, decreased) discrimination.
Figure 2.
Variation of specificity and sensitivity of the microarray with hybridization temperature and formamide concentration. (A) Variation of M/MM ratios with temperature. Average M/MM ratios for 30-nt oligos corresponding to SXL at the indicated temperatures. Standard deviations are represented by vertical bars. Filled squares represent values at 50% formamide; triangles represent the value at 58% formamide. (B) Variation of signal intensities with temperature. Average fluorescent intensities for 30-nt oligos corresponding to SXL at the indicated temperatures. Standard deviations are represented by vertical bars. Filled squares represent values at 50% formamide; triangles represent the value at 58% formamide.
The reverse tendency was observed regarding hybridization intensities. Figure 2B shows a slight decrease in hybridization signals between 30 and 35°C, followed by an increase when the hybridization was carried out at 42°C.
Standard hybridization solutions include 50% formamide. Absence or lower concentrations of formamide (e.g. 40%) resulted in poor fluorescent signals. The effects of increasing formamide concentration to 58% were assessed, and are represented as triangles in Figure 2. While the increase in formamide concentration caused a slight increase in M/MM discrimination, it was accompanied by a more substantial decrease in hybridization signals. Use of higher (70–80%) formamide concentrations resulted in very poor sensitivity. These effects can be explained, at least qualitatively, by the more stringent hybridization conditions imposed by the presence of formamide.
Washing temperatures were also systematically tested. Temperatures of 47, 55 and 65°C resulted in progressive and significant loss of signals compared with 42°C. Temperatures of 37 or 25°C resulted in progressive loss of M/MM discrimination compared with 42°C.
Purity and source of oligonucleotides
A potentially important issue for large-scale microarray performance is the quality and source of oligonucleotides. To address this, seven selected oligonucleotides corresponding to one of the genes under study, chosen strategically to represent oligos with different levels of performance, were obtained from four different commercial providers. Both non-purified and HPLC-purified oligonucleotides were obtained from three of these providers. Tables 5 and 6 summarize the results of the comparison. Two conclusions can be drawn from these results. First, differences in performance of up to 70% between providers were observed, both regarding sensitivity and M/MM ratios. Secondly, while purified oligos could provide up to 5-fold better sensitivity, non-purified oligos showed higher M/MM discrimination. This could be due to an increased proportion of oligos shorter than full length in the non-purified preparations, resulting in lower sensitivity (Table 2) while showing higher M/MM discrimination (Table 1). Consistent with this possibility, the degree of full length oligo in non-purified preparations, assessed by electrophoresis on denaturing gels, correlated with performances more comparable with those of purified oligonucleotides (data not shown).
Table 5. Variation of microarray sensitivity and specificity with degree of purification among different commercial providers: average M/MM ratios for seven pairs of HPLC-purified versus non-purified oligos for four different commercial providers.
| Provider | 1 | 2 | 3 | 4a |
|---|---|---|---|---|
| Non-purified | 6.1 ± 0.4 | 4.6 ± 0.1 | 4.4 ± 0.5 | – |
| HPLC-purified | 3.9 ± 0.1 | 3.2 ± 0.1 | 4.1 ± 0.3 | 4.9 ± 0.3 |
aProvider 4 could not produce non-purified oligos.
Table 6. Variation of microarray sensitivity and specificity with degree of purification among different commercial providers: average relative signal intensities for HPLC-purified versus non-purified oligos for four different commercial providers.
| Provider | 1 | 2a | 3 | 4 |
|---|---|---|---|---|
| Non-purified | 1.3 ± 0.2 | 1 ± 0 | 0.8 ± 0.2 | – |
| HPLC-purified | 4.3 ± 0.2 | 3.5 ± 0.1 | 4.1 ± 0.2 | 3 ± 0.2 |
aThe value of non-purified oligos from provider 2 was set arbitrarily to 1.
Dynamic range
Microarray analyses often serve to compare the relative abundance of a set of RNA species between two samples. To address what was the dynamic range of sensitivity of our microarrays, experiments were carried out in which different amounts of Cy5- and Cy3-labeled RNA samples were hybridized together to the same microarray. The ratio between the signals obtained by scanning the slides at the wavelength characteristic of each fluorochrome was compared with the input ratio between the two RNAs. Table 7 shows statistical analyses of such comparisons for the five genes under study. Whereas approximately linear responses were observed for input ratios between 1 and 10, higher input ratios were underestimated by up to 3-fold. This indicates that changes in concentration >10-fold may not be accurately quantified. Interestingly, a 1:1 input ratio was measured in the microarray as a 1.6 Cy5:Cy3 ratio. This effect could be due to lower levels of Cy3 incorporation during transcription of the target RNAs or to less efficient detection of RNAs labeled with this fluorochrome. From a practical point of view, this emphasizes the need for reciprocal labeling in order to establish reliable comparisons between two samples.
Table 7. Comparison between input ratios of in vitro transcripts labeled with Cy5/Cy3 and the observed fluorescence values after hybridization.
| Input ratios | Observed ratios | |
|---|---|---|
| Median | Average | |
| 1 |
1.6 |
1.6 ± 0.1 |
| 2 |
2.3 |
2.5 ± 0.2 |
| 10 |
10.4 |
12.4 ± 2.7 |
| 30 |
20.2 |
24.1 ± 6 |
| 50 |
26.7 |
30.4 ± 6.4 |
| 100 | 30.1 | 37.8 ± 13.5 |
Average, standard deviation and median values correspond to oligos of 30 nt.
Analysis of HeLa mRNAs
The experiments described above were carried out using precise amounts of specific RNAs transcribed in vitro. To verify the performance of the microarrays with RNAs obtained from biological samples, poly(A)+ RNA was isolated from HeLa cells in culture and fluorescently labeled by a variety of procedures (see below).
Figure 3 shows results obtained under optimized hybridization conditions and indicate that, although the discrimination was reduced compared with the values obtained for the simplified system, most of the oligonucleotides still distinguished between the match and the single mismatch control sequence. Statistical analyses of the results indicated that: (i) ∼30% better M/MM discrimination was observed for 25-nt oligos compared with 35-nt oligos (average discrimination for 25mers was 1.8); (ii) 30- or 35-nt oligos had ∼40% better sensitivity than 25mers; and (iii) the intensity of signals associated with genes not present in the sample (SXL) was on average 100-fold lower than for genes expected to be expressed. If SXL RNA was spiked in the sample, however, signals associated with the corresponding oligos were of comparable intensity as when present in a simpler mix of RNAs (data not shown).
Figure 3.
Hybridization of fluorescently labeled HeLa mRNAs to oligonucleotide microarrays. Microarray layout for the five genes under study, and fluorescent scan of hybridization data to labeled HeLa mRNAs, obtained under optimized conditions.
Table 8 shows the percentage of oligonucleotides showing >2-fold M/MM discrimination for RNAs analyzed using a variety of sample labeling protocols (see Materials and Methods). The data indicate that a level of discrimination similar to that obtained for in vitro transcribed RNAs can also be achieved for the complex mixture of HeLa mRNAs using 25-nt oligos and oligo-dT-primed cDNA linearly amplified by transcription with T7 RNA polymerase. As expected, the fraction of oligos showing discrimination was reduced when oligos corresponding to SXL, a gene not expressed in HeLa cells, were considered (Table 8). The difference in discrimination between expressed genes and SXL was reduced for longer oligos, particularly when amplification was carried out using random primers.
Table 8. Oligonucleotide discrimination for HeLa mRNAs labeled using different protocols: percentage of oligos with M/MM ratios >2 indicated for oligos of different lengths and different labeling methods.
| Labeling method | % M/MM >2 | |||
|---|---|---|---|---|
| Direct labeling | Amplification using odT | Amplification using random primers | Amplification using random primers + odT | |
| 25mer |
58 |
73 |
54 |
73 |
| 30mer |
50 |
63 |
50 |
50 |
| 35mer |
46 |
47 |
44 |
33 |
| |
SXL |
|
|
|
| 25mer |
17 |
10 |
27 |
13 |
| 30mer |
25 |
20 |
35 |
27 |
| 35mer | 20 | 18 | 33 | 40 |
The lower part of the table indicates the same values for oligos corresponding to SXL, a Drosophila gene whose transcripts are not present in HeLa cells.
As additional controls of specificity, 25-nt oligos corresponding to human β-actin, β-tubulin and the Arabidopsis genes mgd and fad were included in the microarray. While the proportion of oligos showing a M/MM ratio >1 was between 87 and 100% for genes expressed in HeLa cells (U2AF65, U2AF35, TIA-1, SRp20, β-actin and β-tubulin) this proportion was 50% or less for oligos corresponding to control genes (SXL, mgd and fad), as expected from random distribution of spurious hybridization. Accordingly, the median M/MM ratio for all oligo lengths was 1.6–1.7 for oligos corresponding to genes expressed in HeLa cells, whereas it was 1.0 for control genes (Table 9). These data suggest that M/MM discrimination does occur for the majority of the oligos that are able to hybridize to RNAs present in the sample, although often this ratio is ≤2-fold.
Table 9. Oligonucleotide discrimination for HeLa mRNAs labeled using different protocols: M/MM ratios for all genes and oligo lengths and different labeling methods, for oligos corresponding to genes expressed in HeLa cells versus control genes.
| Amplification method | HeLa genes | Control genes | ||
|---|---|---|---|---|
| Average | Median | Average | Median | |
| odT |
1.7 ± 0.8 |
1.6 |
1.0 ± 0.5 |
1.0 |
| Random primer | 1.4 ± 0.5 | 1.5 | 0.9 ± 0.6 | 1.0 |
Discrimination could not be improved further by using more stringent washing conditions. Amplification improved the sensitivity of detection by a factor of 10 compared with direct labeling by reverse transcription. RNA fragmentation of T7-derived transcripts, achieved by partial degradation at pH 8.1 in the presence of 15 mM magnesium, also increased sensitivity by 1.5–2.0-fold, although this was accompanied by moderate (1.5-fold) decreases in M/MM discrimination.
As an additional test for the specificity of the signals detected, HeLa cells were transfected with an expression vector encoding TIA-1, or the gene was knocked down in tissue culture by transfecting short double-stranded RNA oligos corresponding to TIA-1 sequences (30). RNA isolated from these cells was labeled with either Cy5 or Cy3 and compared with RNA from untransfected cells labeled with the other dye. As predicted, increases or decreases in hybridization signals specific for TIA-1 were detected depending on whether TIA-1 was overexpressed or its expression inhibited (data not shown).
Performance of longer oligonucleotides
Oligos significantly longer than 35 nt have been used in the literature (23). The rationale for the use of longer oligo microarrays is that their sensitivity could approach that of cDNA microarrays. To compare the performance of long versus short (25–35 nt) oligos, two 60-nt oligos were selected for each of the genes under study, which covered sequences that included a subset of the 25–35 nt-long oligos described above. 25–35- and 60-nt oligos were printed in the same slides. The results of hybridization experiments using in vitro transcribed RNAs indicated that hybridization signals associated to 60-nt oligos were 10-fold higher than the signals detected for the corresponding 25mers (Table 10). This ratio was reduced to 2-fold when signals obtained for 60mers and 30mers were compared. Hybridization of HeLa RNAs was also within a similar range of values (Table 10).
Table 10. Performance of 60-nt oligos compared with 25 and 30mers: average and median values of the ratios between fluorescent signals associated with oligos of different lengths, after hybridization of labeled RNAs either generated by in vitro transcription (IVT) or isolated from HeLa cells.
| 60/25 | 60/30 | |||
|---|---|---|---|---|
| Average | Median | Average | Median | |
| IVT | 10 ± 6.7 | 10.5 | 1.8 ± 1.2 | 1.3 |
| HeLa | 7.1 ± 3.3 | 7.1 | 2.7 ± 1.4 | 2.2 |
One difficulty associated with the use of long oligos is that the difference in hybridization stability between perfect match and single mismatch controls is predicted to be too low to permit discrimination, and therefore that hybridization specificity is more difficult to assess for each oligo. To address this question, in vitro transcribed fluorescent RNAs corresponding to the five genes analyzed in Figure 1 were hybridized to the microarray containing short and long oligos. Hybridization signals corresponding to these genes were on average 15.4 times higher than those associated to the four control genes (β-actin, β-tubulin, mgd and fad) (Table 11). This ratio was 20-fold when the performance of 25-nt oligos was compared between the same set of genes. We conclude that 60-nt oligos can provide adequate specificity and better sensitivity than shorter oligos in this experimental set up.
Table 11. Performance of 60-nt oligos compared with 25 and 30mers: average ratios between fluorescent signals associated with oligos complementary to sequences present in the labeled RNAs and signals from oligos complementary to control Drosophila and Arabidopsis genes.
| |
Sample/control |
|
|---|---|---|
| 25mers | 60mers | |
| IVT |
20.7 ± 7 |
15.4 ± 6 |
| HeLa | 11.9 ± 3 | 3.3 ± 1 |
The behavior of 60-nt oligos was also analyzed using HeLa cell RNAs as targets. Signals associated with 60mers were on average 7-fold higher than for 25mers and 2.7-fold higher than for 30mers (Table 10). Specificity was measured as the average ratio between signals associated with human genes versus SXL and Arabidopsis controls. This ratio was 12 for 25 nt-long oligos, while it was reduced to 3.3 for 60mers.
We conclude that while 60 nt-long oligos can provide significantly better sensitivity than 25 or 30mers, their specificity in complex mixtures of RNA is significantly lower than that obtained for 25mers.
DISCUSSION
The data presented in this manuscript will assist in the design of oligonucleotide-based DNA microarrays. The algorithm provided allows the selection of oligos of variable length with optimized uniform hybridization properties and with statistically significant discrimination between perfect match and a single nucleotide mismatch at a central position. Although at least part of the signal associated with mismatch control oligonucleotides is likely to be due to hybridization to the genuine target (31), we adopted the criterion of considering only those oligos showing at least 2-fold differences in hybridization ratios between match and mismatch. However, statistical analyses indicated that lower ratios could also be considered significant, as frequently assumed in the literature (21).
Conditions were found in which ∼75% of the oligos selected by our algorithm cleared the more stringent discrimination criteria. This corresponds to a 98.4% probability of obtaining at least one reliable measurement in a set of three selected oligonucleotides, a 99.0% probability of obtaining at least two reliable measurements in a set of five oligos, or a 93.76% probability of obtaining at least three reliable measurements in a set of five. These figures are significantly lower than the number of oligonucleotides utilized to assess gene expression in the literature (from 25 to 300 per RNA) (25), and could therefore significantly reduce the cost and simplify data processing of custom-made microarrays.
The next complementary step in oligonucleotide selection should involve extensive sequence comparisons (e.g. BLAST analyses) to minimize the chances that an oligo will hybridize to identical sequences present in two or more genes. This represents an intensive bioinformatic effort and selection of oligonucleotides with increased discrimination can only help to complement these efforts to improve microarray specificity.
An important conclusion of our results is that multiple aspects of microarray design contribute to their performance, from the choice of oligonucleotide provider or level of purification to hybridization temperature within narrow margins. Variations often work in opposite directions regarding sensitivity and specificity, and therefore an appropriate compromise may need to be reached for each experimental set up and application.
Let us consider the choice of oligonucleotide length. Oligos of 25 nt in length can provide optimal discrimination at the cost of significant losses in sensitivity, which may be very detrimental for detection of genes expressed at low levels. The data of Table 11 indicate that discrimination between genes expressed and not expressed in a biological RNA sample is 12-fold for oligos of 25 nt in length, but only 3.3-fold for 60 nt-long oligos covering similar sequences. In contrast, 60 nt-long oligos have higher sensitivity under the same conditions, but at the cost of more limited specificity, which in addition is difficult to quantify. The data of Table 10 indicate that 60-nt oligos are on average 7-fold more sensitive than 25mers. Therefore the 7-fold increase in sensitivity is accompanied by a ∼4-fold loss in specificity. Problems of specificity can be particularly serious considering that fewer oligos per gene are usually selected when longer oligos are used in the microarrays, thus providing a reduced number of independent measurements per gene.
The results of Tables 1 and 2 suggest that 30-nt oligos can represent an adequate compromise between sensitivity and specificity for optimal microarray performance in a system of limited RNA complexity. Similarly, the 3-fold lower sensitivity for RNAs from HeLa cells of 30mers compared with 60-nt oligos (Table 10) may be compensated by an increase in specificity, and by the possibility of assessing the degree of the specificity of each oligo by the use of mismatch controls.
Considered together, the data suggest that both the abundance in expression of the mRNAs to be studied and the degree of similarity to other RNAs present in the sample need to be taken into consideration for the choice of oligo length.
Rather subtle changes in hybridization conditions also affect microarray performance. Figure 2 shows an inverse correlation between signal intensity and M/MM discrimination in a range of temperatures between 30 and 42°C. While an increased M/MM ratio is expected from the lower thermal stability of imperfect duplexes under more stringent temperatures, the decrease observed at 42°C cannot be explained easily. Equally puzzling is the increase in hybridization signals between 35 and 42°C that follows the more expected decrease between 30 and 35°C. Increased signals at 42°C could be explained by the opening of secondary structures in the target cRNA, thereby facilitating hybridization to the microarray. If this is the case, it is conceivable that the reduction in discrimination observed at 42°C in Figure 2A could also be attributed to a general increase in the availability of target sequences for hybridization.
Finally, cost considerations can also play a relevant part in microarray design. Relatively small increases in sensitivity by the presence of expensive 5′ amino modifications or HPLC purification may be critical for some applications but not for others.
Concluding remarks
In this manuscript we have provided a quantitative and statistical analysis for the use of oligonucleotide-based microarrays that will aid in the selection of appropriate reagents and conditions for gene profiling and genotyping. Although these will vary depending on the specific application, we suggest that 30 nt-long oligos offer an adequate balance between sensitivity and specificity. Longer oligos can provide a slight increase in sensitivity at the cost of a significant decrease in specificity, which is also difficult to assess due to the absence of reliable mismatch controls. The higher cost of HPLC purification can be compensated by significant increases in sensitivity. Using the algorithm presented here, five oligonucleotides and their mismatch controls should be sufficient to provide statistically reliable quantification of signals corresponding to a gene or sequence feature.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank MWG Biotech AG for providing free oligonucleotides, George Dimopoulos, George K. Christophides, Thomas Preis, Vladimir Benes and members of the Ansorge and Valcárcel laboratories for technical help, reagents, discussions and critical reading of the manuscript. A.R. was the recipient of a Praxis XXI PhD fellowship from the Portuguese Ministry of Science and Technology. This work was supported in part by a grant from the Human Frontier Science Program Organization.
REFERENCES
- 1.Noordewier M.O. and Warren,P.V. (2001) Gene expression microarrays and the integration of biological knowledge. Trends Biotechnol., 19, 412–415. [DOI] [PubMed] [Google Scholar]
- 2.Young R.A. (2000) Biomedical discovery with DNA arrays. Cell, 102, 9–15. [DOI] [PubMed] [Google Scholar]
- 3.Mills J.C., Roth,K.A., Cagan,R.L. and Gordon,J.I. (2001) DNA microarrays and beyond: completing the journey from tissue to cell. Nature Cell Biol., 3, E175–178. [DOI] [PubMed] [Google Scholar]
- 4.Bassett D.E. Jr, Eisen,M.B. and Boguski,M.S. (1999) Gene expression informatics–it’s all in your mine. Nature Genet., 21, 51–55. [DOI] [PubMed] [Google Scholar]
- 5.Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 6.Lee C.K., Klopp,R.G., Weindruch,R. and Prolla,T.A. (1999) Gene expression profile of aging and its retardation by caloric restriction. Science, 285, 1390–1393. [DOI] [PubMed] [Google Scholar]
- 7.Golub T.R., Slonim,D.K., Tamayo,P., Huard,C., Gaasenbeek,M., Mesirov,J.P., Coller,H., Loh,M.L., Downing,J.R., Caligiuri,M.A. et al. (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science, 286, 531–537. [DOI] [PubMed] [Google Scholar]
- 8.Alon U., Barkai,N., Notterman,D.A., Gish,K., Ybarra,S., Mack,D. and Levine,A.J. (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl Acad. Sci. USA, 96, 6745–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hippo Y., Taniguchi,H., Tsutsumi,S., Machida,N., Chong,J.M., Fukayama,M., Kodama,T. and Aburatani,H. (2002) Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res., 62, 233–240. [PubMed] [Google Scholar]
- 10.Liotta L. and Petricoin,E. (2000) Molecular profiling of human cancer. Nature Rev. Genet., 1, 48–56. [DOI] [PubMed] [Google Scholar]
- 11.Clarke P.A., te Poele,R., Wooster,R. and Workman,P. (2001) Gene expression microarray analysis in cancer biology, pharmacology, and drug development: progress and potential. Biochem. Pharmacol., 62, 1311–1336. [DOI] [PubMed] [Google Scholar]
- 12.Debouck C. and Goodfellow,P.N. (1999) DNA microarrays in drug discovery and development. Nature Genet., 21, 48–50. [DOI] [PubMed] [Google Scholar]
- 13.Granjeaud S., Bertucci,F. and Jordan,B.R. (1999) Expression profiling: DNA arrays in many guises. Bioessays, 21, 781–790. [DOI] [PubMed] [Google Scholar]
- 14.Hughes T.R., Mao,M., Jones,A.R., Burchard,J., Marton,M.J., Shannon,K.W., Lefkowitz,S.M., Ziman,M., Schelter,J.M., Meyer,M.R. et al. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol., 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 15.Southern E., Mir,K. and Shchepinov,M. (1999) Molecular interactions on microarrays. Nature Genet., 21, 5–9. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart D.J. and Barlow,C. (2001) Expressing what’s on your mind: DNA arrays and the brain. Nature Rev. Neurosci., 2, 63–68. [DOI] [PubMed] [Google Scholar]
- 17.LaForge K.S., Shick,V., Spangler,R., Proudnikov,D., Yuferov,V., Lysov,Y., Mirzabekov,A. and Kreek,M.J. (2000) Detection of single nucleotide polymorphisms of the human murine opioid receptor gene by hybridization or single nucleotide extension on custom oligonucleotide gelpad microchips: potential in studies of addiction. Am. J. Med. Genet., 96, 604–615. [DOI] [PubMed] [Google Scholar]
- 18.Hacia J.G. (1999) Resequencing and mutational analysis using oligonucleotide microarrays Nature Genet., 21, 42–47. [DOI] [PubMed] [Google Scholar]
- 19.Drobyshev A., Mologina,N., Shik,V., Pobedimskaya,D., Yershov,G. and Mirzabekov,A. (1997) Sequence analysis by hybridization with oligonucleotide microchip: identification of beta-thalassemia mutations. Gene, 188, 45–52. [DOI] [PubMed] [Google Scholar]
- 20.Modrek B. and Lee,C. (2001) A genomic view of alternative splicing. Nature Genet., 30, 13–19. [DOI] [PubMed] [Google Scholar]
- 21.Hu G.K., Madore,S.J., Moldover,B., Jatkoe,T., Balaban,D., Thomas,J. and Wang,Y. (2001) Predicting splice variant from DNA chip expression data. Genome Res., 11, 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoemaker D.D., Schadt,E.E., Armour,C.D., He,Y.D., Garrett-Engele,P., McDonagh,P.D., Loerch,P.M., Leonardson,A., Lum,P.Y., Cavet,G. et al. (2001) Experimental annotation of the human genome using microarray technology. Nature, 409, 922–927. [DOI] [PubMed] [Google Scholar]
- 23.Hughes T.R. and Shoemaker,D.D. (2001) DNA microarrays for expression profiling. Curr. Opin. Chem. Biol., 5, 21–25. [DOI] [PubMed] [Google Scholar]
- 24.Brazma A., Hingamp,P., Quackenbush,J., Sherlock,G., Spellman,P., Stoeckert,C., Aach,J., Ansorge,W., Ball,C.A., Causton,H.C. et al. (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genet., 29, 365–371. [DOI] [PubMed] [Google Scholar]
- 25.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. et al. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 26.Wodicka L., Dong,H., Mittmann,M., Ho,M.H. and Lockhart,D.J. (1997) Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol., 15, 1359–1367. [DOI] [PubMed] [Google Scholar]
- 27.Call D.R., Chandler,D.P. and Brockman,F. (2001) Fabrication of DNA microarrays using unmodified oligonucleotide probes. Biotechniques, 30, 368–372, 374, 376, passim. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Zhang J., Day,I. and Byrne,C. (2002) A novel medium throughput quantitative competitive PCR technology to simultaneously measure mRNA levels from multiple genes. Nucleic Acids Res., 30, e20–.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 31.Chudin E., Walker,R., Kosaka,A., Wu,S.X., Rabert,D., Chang,T.K. and Kreder,D.E. (2002) Assessment of the relationship between signal intensities and transcript concentration for Affymetrix GeneChip(R) arrays. Genome Biol., 3, RESEARCH0005–.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.