Abstract
Background
A subset of developmental disorders (DD) is characterized by disease-specific genome-wide methylation changes. These episignatures inform on the underlying pathogenic mechanisms and can be used to assess the pathogenicity of genomic variants as well as confirm clinical diagnoses. Currently, the detection of these episignature requires the use of indirect methylation profiling methodologies. We hypothesized that long-read whole genome sequencing would not only enable the detection of single nucleotide variants and structural variants but also episignatures.
Methods
Genome-wide nanopore sequencing was performed in 40 controls and 20 patients with confirmed or suspected episignature-associated DD, representing 13 distinct diseases. Following genomic variant and methylome calling, hierarchical clustering and dimensional reduction were used to determine the compatibility with microarray-based episignatures. Subsequently, we developed a support vector machine (SVM) for the detection of each DD.
Results
Nanopore sequencing-based methylome patterns were concordant with microarray-based episignatures. Our SVM-based classifier identified the episignatures in 17/19 patients with a (likely) pathogenic variant and none of the controls. The remaining patients in which no episignature was identified were also classified as controls by a commercial microarray assay. In addition, we identified all underlying pathogenic single nucleotide and structural variants and showed haplotype-aware skewed X-inactivation evaluation directs clinical interpretation.
Conclusion
This proof-of-concept study demonstrates nanopore sequencing enables episignature detection. In addition, concurrent haplotyped genomic and epigenomic analyses leverage simultaneous detection of single nucleotide/structural variants, X-inactivation, and imprinting, consolidating a multi-step sequential process into a single diagnostic assay.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-024-01419-z.
Keywords: Long-read sequencing, Developmental disorders, Methylation, Methylome, Episignatures, Nanopore sequencing, X-inactivation, Support vector machine
Background
The epigenome plays a central role in regulating differential gene expression. Epigenetic variation is therefore involved in several adaptative but also pathological processes. Repeat expansion-associated modifications of promotor methylation have long been recognized as a cause of developmental disorders (DD) [1, 2]. Imprinting defects are also known to disturb development, growth, and metabolism [3]. More recently, methylome studies in patients with unexplained DD revealed rare epigenetic changes in 23% of patients with a 2.8-fold excess of de novo epivariants [4], suggesting that these may be involved in their etiology. Yet, apart from a handful of targeted methylation assays for well-known imprinted regions, epigenomic analyses are currently not routinely performed in the diagnostic work-up of patients with DD.
Besides localized epigenetic variants, genome-wide methylation studies in DD have recently revealed multi-loci, disorder-specific methylation disturbances, called episignatures. Episignatures have initially been identified in disorders caused by known chromatin and/or methylation regulatory gene disruptions [5]. An increasing number of disorders are characterized by episignatures and recognizable methylation pattern changes have now been detected in over 60 diseases [6]. For some, such as certain microdeletion syndromes [7], the mechanism causing reproducible genome-wide methylation changes remains unclear. Mapping the methylome in DD will therefore improve our understanding of pathogenic mechanisms and might leverage new insights in the origins of phenotypic variability. In addition, episignatures have a high diagnostic value in assessing the pathogenicity of genomic variants of unknown significance (VUS) and in confirming clinical diagnoses in patients for which the underlying genomic variant could not be identified [8].
The current standard method used to identify episignatures in DD is based on bisulfite conversion of methylated cytosines followed by methylation analysis using Illumina Epic/Infinium methylation microarrays [6]. Microarray-based episignature analysis is proposed on the publicly available EpiGenCentral platform for a subset of diseases [9] and a larger panel of episignatures can be evaluated through the commercialized EpiSign assay [10]. A clinical implementation study of this method reported the detection of an episignature in 11% of patients with DD without conclusive genetic finding and in 35% of patients with a VUS in a gene for which an episignature is described [10], demonstrating its diagnostic value. One drawback of this assay is that it represents an extra step in the diagnostic odyssey of patients which comes with additional diagnostic delays and increased costs.
With the advent of long-read sequencing technologies, detection of single nucleotide variants (SNVs), structural variants (SVs), and base modifications in a single assay becomes reality [11]. Both prevailing long-read methodologies, nanopore (Oxford Nanopore Technologies) and single molecule real-time sequencing (PacBio), enable methylation detection from the native DNA strands without amplification, bisulfite, or enzymatic conversion biases [12]. Moreover, the generated long reads enable haplotyping [13], allowing the phasing of genetic and epigenetic variation. Recent studies showed the ability of targeted nanopore sequencing to detect X-chromosome inactivation (XCI) [14], methylation disturbances at imprinted loci [15], and short tandem repeats causing DD [16] as well as the concurrent detection of specific SNVs, SVs, and methylation changes for tumor classification [17].
Considering the potential of nanopore sequencing to detect localized methylation levels and disease states, we reasoned it should be possible to map methylome-wide disturbances in chromatinopathies. Hence, we explored the potential to concurrently detect episignatures and underlying pathogenic variants. We performed long-read whole genome nanopore sequencing (lrWGS) of 20 patients representing 13 different rare DD with known episignatures, as well as 40 controls. We report non-inferiority of nanopore sequencing for detecting episignatures and demonstrate concurrent detection of underlying SNVs or SVs. We also confirm the detection of imprinting as well as haplotype-aware skewed XCI in an untargeted setting. In addition, our results illustrate some limitations of current episignatures. Overall, our study underscores the diagnostic value of lrWGS which might ease rare diseases diagnosis by offering simultaneous genome and epigenome analysis.
Methods
Study cohort
Twenty patients with a confirmed or a suspected DD for which an episignature is described (Table 1, Additional file 1: Table S1), as well as 40 healthy controls, from the Center of Human Genetics of Leuven were prospectively recruited. For the first phase of our study, we included 11 patients (EPI_01 to EPI_11) representing four diseases as well as five controls. Seven patients were initially sequenced: two with Kabuki, Sotos, and Wiedemann-Steiner syndrome, and one with Cornelia de Lange syndrome. Subsequently, one additional patient was recruited for each of these four diseases. The samples of these four extra individuals were blinded by the recruiting physician for subsequent analyses. For the second phase, nine additional patients (EPI_12 to EPI_20) with different confirmed or suspected diseases were included to increase the amount of evaluated episignatures as well as 35 other controls. Two of the patients were selected based on the presence of a VUS and not a (likely) pathogenic variant, as evaluation and illustration of one of the major applications of episignature detection. The genomic variants of all patients had been identified by chromosomal microarray, targeted Sanger sequencing, or trio-based exome sequencing (WES) (Additional file 1: Table S1).
Table 1.
Concurrent detection of episignatures (SVM-based) and underlying genomic variants. The seven first columns of the table describe patient characteristics and (likely) pathogenic genomic variants or VUS (marked by a “?”) detected by standard of care methods. The next column indicates the result of the SVM-based classifier for each sample. Patients for which no episignature is identified by the classifiers are classified as control (italic). The last column specifies the ranking of the underlying genomic variant in the fast track analysis of a phenotype-drive prioritization tool (Geneyx) [18]. For the variants prioritized in a subanalysis, both the subanalysis (structural variant (SV) or X-linked (XL)) and the ranking in the subanalysis are provided. *Manual curation of the gene was needed for the identification of this mosaic variant given Clair3 was targeting heterozygous SNVs
| Study ID | Age | Sex | Gene | Transcript | Variant | Syndrome | SVM classifier result | SV/SNV ranking |
|---|---|---|---|---|---|---|---|---|
| EPI_01 | 12 | F | KMT2D | NM_003482.4 | c.8311C > T p.(Arg2771*) | Kabuki | Kabuki | 1 |
| EPI_02 | 24 | F | KMTD2 | NM_003482.4 | c.8304_8307del p.(Ser2768Argfs*18) | Kabuki | Kabuki | 1 |
| EPI_03 | 7 | M | NSD1 | NM_022455.5 | c.6188 T > C p.(Leu2063Pro) | Sotos | Sotos | 1 |
| EPI_04 | 6 | M | NSD1 | NM_022455.5 | c.3659_3660del p.(Glu1220Alafs*5) | Sotos | Sotos | 1 |
| EPI_05 | 11 | M | KMT2A | NM_001197104.2 | c.6571C > T p.(Arg2191*) | Wiedemann-Steiner | Wiedemann-Steiner | 1 |
| EPI_06 | 22 | M | KMT2A | NM_001197104.2 | c.10168C > T p.(Gln3390*) | Wiedemann-Steiner | Wiedemann-Steiner | 1 |
| EPI_07 | 14 | F | SMC3 | NM_005445.4 | c.720_722del p.(Asp240del) | Cornelia de Lange | Cornelia de Lange | 3 |
| EPI_08 | 46 | M | KMT2D | NM_003482.4 | c.16517_16518del p.(Glu5506Glyfs*5) | Kabuki | Kabuki | 1 |
| EPI_09 | 2 | M | NSD1 | NM_022455.5 | c.4905C > G p.(Cys1635Trp) | Sotos | Sotos | 1 |
| EPI_10 | 5 | M | KMT2A | NM_001197104.2 | c.3460C > T p.(Arg1154Trp) | Wiedemann-Steiner | Wiedemann-Steiner | 1 |
| EPI_11 | 5 | M | SMC3 | NM_005445.4 | c.2166_2167insCCCGAG p.(Glu722_Thr723insProGlu) | Cornelia de Lange | Cornelia de Lange | 4 |
| EPI_12 | 13 | F | SMARCA2 | NM_003070.5 | c.3476G > A p.(Arg1159Gln) | Nicolaides-Baraitser | Nicolaides-Baraitser | 1 |
| EPI_13 | 15 | F | PHF6 | NM_001015877.2 | c.306del p.(Tyr103Thrfs*40) | Börjeson-Forssman-Lehmann | control | 1 |
| EPI_14 | 14 | F | KANSL1 | NM_015443.4 | c.1419dup p.(Arg474Thrfs*3) | Koolen-De Vries | Koolen-De Vries | 1 |
| EPI_15 | 2 months | M | EHMT1 | NM_024757.5 | c.3717-2A > G | Kleefstra | Kleefstra | 2 |
| EPI_16 | 3 | M | chr7:g.72900001_76430000del | Williams-Beuren | Williams Beuren | SV: 1 | ||
| EPI_17 | 4 | M | EP300 | NM_001429.4 | g.41168065_41171580del | Rubinstein-Taybi | Rubinstein-Taybi | SV: 1 |
| EPI_18 | 3 | M | ZNF711 | NM_001330574.2 | c.1361A > G p.(His454Arg) | MRX97? | control | 4 |
| EPI_19 | 3 | M | KMT2D | NM_003482.4 | c.3002_3005del p.(Leu1001Profs*15) | Kabuki | Kabuki | Absent* |
| ATRX | NM_000489.6 | c.7263_7265del p.(Gln2425del) | ATRX? | XL: 1 | ||||
| EPI_20 | 5 | M | UBE2A | NM_003336.4 | c.19C > T p.(Arg7Trp) | MRXSN | control | 2 |
Long-read sequencing
At inclusion, one blood sample (EDTA) was collected from the participants and fresh or frozen (− 80 °C) blood was used for HMW DNA extraction (Promega Wizard®, Monarch®). For four patients and two controls, DNA extracted with the automated Revvity—Chemagic® workflow and stored at − 20 °C was retrieved (Additional file 2: Table S2). Libraries were prepared from 3 µg of DNA using the Oxford Nanopore Technologies (ONT) Ligation Sequencing Kit (SQK-LSK110). Each sample was loaded on a R.9.4.1 Flow Cell and reloaded once after 24–48 h, for a total sequencing period of 72–96 h on a PromethION®. From HMW DNA extracted from fresh blood, we obtained a median output of 95.6 GB and a median N50 of 40.4 kb. Using frozen blood, the median output was 116.25 GB, and the median N50 31.9 kb. Using stored non-HMW DNA, the median output and N50 were 113.7 GB and 17 kb, respectively (Additional file 2: Table S2).
For patient EPI_13, additional urine and buccal swab samples were collected. DNA extraction of these tissues was performed using the Qiagen® and MagCore® chemistries, respectively. Using the SQK-LSK110 chemistry and R.9.4.1 Flow Cells, we generated an output of 100 GB for the urine (N50 = 9.79 kb) and 146.7 GB for the buccal swab (N50 = 7.25 kb).
(Modified) basecalling, mapping, SVs and SNVs calling
To extract methylation information from raw ONT data, we performed modified basecalling with Dorado (v0.3.0) using the dna_r9.4.1_e8_sup@v3.3_5mCG model [17]. After basecalling, reads were aligned to hg38 with minimap2 (v2.24) [19]. Subsequently, bedMethyl files, used in downstream analysis, were generated from aligned BAM files using modkit (v0.1.13 with pileup –preset traditional –only-tabs) [20]. Methylation was visualized with Methylartist [21] and Integrative Genomics Viewer (IGV) [22].
To evaluate concurrent detection of the underlying genomic variants, SNVs were called with Clair3 (v1.0.4) [23], which was also used to generate haplotagged BAM files. SVs were called using Sniffles2 (v2.0.2) [24, 25]. To improve the sensitivity for large copy number variants (> 50 kb), known to be difficult to detect with current nanopore SV callers [25], we applied QDNAseq (v1.3.8) [26], a read-depth based software. Presence of the genomic variant was verified in the Geneyx platform v6.0. [18], enabling phenotype-driven as well as ACMG classification based variant prioritization to perform a fast analysis in the absence of parental data.
Detection of differentially methylated regions: imprinting and X-inactivation
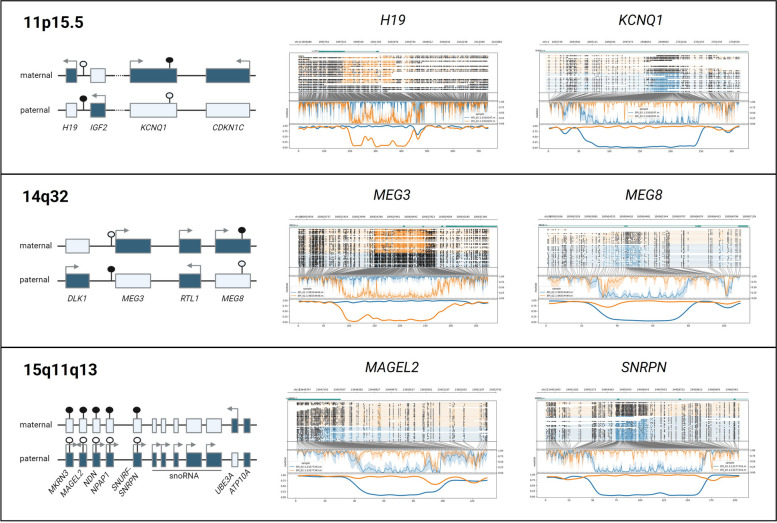
Imprinting was assessed by quantifying and visualizing methylation of both haplotypes at six loci located in three imprinted regions associated with DD (11p15.5, 14q32, and 15q11-q13). The following CpGs were assessed: 242 CpGs at the H19 (chr11:1,997,509–2003349, hg38), 194 CpG at the KCNQ1 (chr11:2,698,155–2,701,028, hg38), 188 CpG at the MEG3 (chr14:100,824,185–100827640, hg38), 45 CpG at the MEG8 (chr14:100,904,325–100905081, hg38), 116 CpG at the SNRPN (chr15:24,954,564–24,956,828, hg38), and 52 CpG at the MAGEL2 (chr15:23,647,178–23,648,424, hg38) imprinted loci.
To evaluate XCI, we assessed methylation at two validated loci, also recently targeted by CRISPR-Cas9 nanopore sequencing for this purpose: 115 CpGs at the AR gene loci (chrX:67,543,761–67,546,170, hg38) and 57 CpGs at the RP2 gene loci (chrX:46,836,539–46,837,273, hg38) [14]. In addition, 99 CpGs in the promotor region of PHF6 were evaluated (chrX:134,372,110–134374891, hg38).
Differential methylation at these loci was quantified and visualized in the six females of our cohort. Loci with < 10 × total coverage or < 5 × coverage for one of the haplotypes were excluded from subsequent analyses (16/120 loci for imprinting evaluation and 3/18 loci for XCI analyses).
For the mothers of EPI_18 and EPI_19, XCI was evaluated by the standard of care method, i.e., comparing the relative amount of PCR amplification products of both AR gene CAG-repeat haplotypes after methylation-sensitive restriction enzyme digestion.
Distinction of episignatures by dimensional reduction and clustering
Thirty-four disease-specific episignatures have been identified through microarrays and published by Aref-Eshghi et al. [5]. The most significant microarray probes for each disease and methylation levels at these loci have been made available. We extracted the DNA methylation beta values from this published dataset at all episignatures probes’ loci for the 34 diseases and the included control. This extraction provided, for every episignature locus, one array-based example of the episignature in presence of the disorder and 34 examples of methylation levels at these loci in the absence of the disorder. Using pyliftover [27], all the probes (loci) encoding episignatures were lifted from hg19 to hg38. During the liftover, only a single CpG locus in the SBBYSS episignature was lost. Methylation levels were extracted from the nanopore sequencing data at the episignature loci using hg38 coordinates. During this process, the percentage of methylated reads was extracted from bedMethyl files. Using python (v3.9.15), three approaches were used to determine the similarity between array- and nanopore-based episignatures: UMAP (umap-learn_v0.5.4 [28], n_neighbors = 2) and t-SNE (sklearn_v1.2.2 [29], n_components = 2, perplexity = 2) for dimensional reduction, and hierarchical clustering (scipy_v1.9.3 [30], seaborn_v0.12.2 [31]). These three approaches were first applied using seven samples, representative of four diseases (EPI_01 to EPI_07) as well as five healthy controls, and repeated including four blinded samples (EPI_08 to EPI_11).
Development of a SVM classifier
We adopted the one-vs-all approach and trained 34 individual support vector machines (SVMs) using disorder episignature loci DNA methylation median beta values [5]. A linear kernel (sklearn v1.2.2 [29]) was used to train the SVMs, as it demonstrated strong performance with the given dataset. All parameters, excluding class_weight, were set to their default values as provided by sklearn v.1.2.2 [29]. SVMs were trained to predict the presence of a specific disorder episignature considering all the other cases as controls. Each SVM was trained using 35 array derived episignatures [5]: one “positive” episignature representing the disorder of interest and the 34 “negative” episignatures, representative of the other disorders as well as a control dataset, all being used as control. While this approach guaranteed one and 34 examples of methylation values in the presence and in the absence of the disorder, respectively, it also introduced an unbalance between the positive (disorder presence) and negative (disorder absence) classes that could affect the SVM training. To overcome this issue, we adjusted the weight of the classes during the training process. Due to the limited number of positive samples (only one per SVM), alternative methods for addressing class imbalance, such as data augmentation or sampling strategies, were not feasible. Therefore, class weights were adjusted, with the positive class weight set to ten, and the negative class weight set to one. The value of ten for the positive class was chosen heuristically after experimenting with different values.
To predict each sample’s class, we ran all the 34 SVM classifiers on the 20 patient samples and 40 control samples and assigned the sample to the class with the highest confidence score. Confidence scores represent the decision function indicating the signed distance of a sample from the separating hyperplane. If none of the classifiers returned a positive confidence score or all the confidence scores were < 0.30, the sample was classified as control.
Results
Imprinting detection
To verify the haplotype-aware methylation detection, six loci localized in three imprinted regions (11p15.5, 14q32, and 15q11-q13) were investigated in six patients (Fig. 1, Additional file 4: Fig. S1). Visualizing both haplotypes’ methylation, we observe consistent methylation of most CpGs of one allele in contrast to the absence of methylation of most CpGs of the other haplotype at the different loci in all patients. The H19 imprinted region is less covered than the other imprinted loci after phasing in several individuals, probably because of its location in a repetitive genomic region. Further quantification of methylation at these six imprinted loci shows mean percentages of methylation between 89.3 and 96.3% for the methylated haplotypes and 4.1 to 16.5% for the unmethylated alleles (Additional file 4: Fig. S1, Additional file 3: Table S3A). These results are concordant with expectations of mono-allelic expression at imprinted loci and illustrate direct allelic methylation measurement as comparison to standard technologies where disturbances of one allele’s methylation are inferred from total methylation levels.
Fig. 1.
Detection of differential methylation at imprinted loci. Haplotype aware methylation visualization [21] of EPI_02 at 6 imprinted loci (middle and right part). The maternal allele is represented in orange and paternal allele in blue. The upper part of each plot represents the reads, with methylated CpGs visualized as black circles. In the lower part of each plot, after translation from genome to CpG-only coordinate space, raw log-likelihood ratios are plotted above a smoothened graph of methylation fraction. The left part illustrates gene localization and expression at these 3 imprinted loci. Active genes are represented as full boxes with arrows, whereas untranslated genes are represented by empty boxes. Full circles represent methylated CpG islands whereas empty circles represent unmethylated CpG islands
Episignature distinction using microarray data as reference
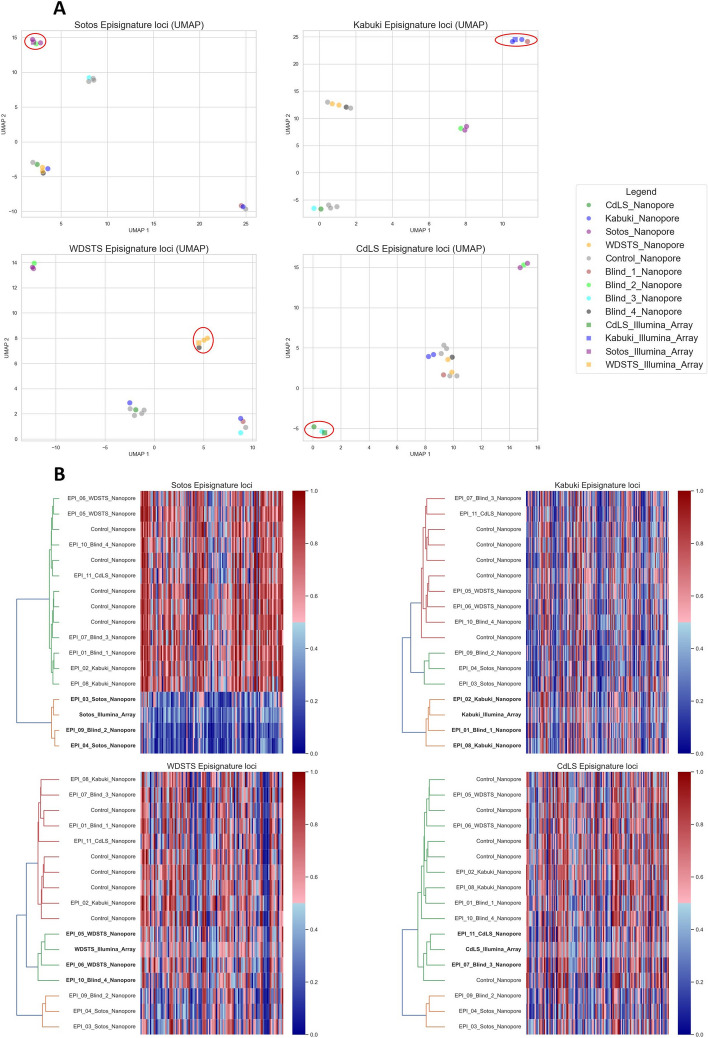
To evaluate the potential of nanopore sequencing-based episignature detection, seven patients representing four different diseases (Kabuki, Sotos, Wiedemann-Steiner, and Cornelia de Lange syndromes) were initially sequenced. Two-dimensional reduction analyses of episignatures (Fig. 2A, Additional file 5: Fig. S2), comparing our samples to the microarray-based disease reference as well as five healthy controls, co-located nanopore methylation values close to the reference microarray data for each disease sample at their disease-specific loci.
Fig. 2.
Nanopore methylation analysis enables to distinguish disease samples from controls and other diseases samples. UMAP scatter plots (A), hierarchical clustering (B, left part), and heatmaps (B, right part) based on methylation levels at Sotos, Kabuki, Wiedemann-Steiner (WDSTS), and Cornelia de Lange (CdLS) syndromes episignature loci. Nanopore sequencing data of five healthy controls and 11 (seven known and four blinded) patients with one of the four disorders are represented, along with the array-based reference for the respective episignature. Patients sequenced with nanopore cluster in close proximity to their respective array-based episignature reference. Blinded samples can be assigned the correct diagnosis
To further confirm the hypothesis that nanopore-based methylome analysis can be used to recognize episignatures, we sequenced an additional four blinded cases (one of each disease). Hierarchical clustering (Fig. 2B) of methylation levels at the disease’s episignature loci combined with the microarray methylome profiles used as a reference clustered all blinded samples with the disease reference and other samples with the same disorder. Similarly, the blinded samples clustered with samples with the same disease in the UMAP analysis (Fig. 2A). Methylation levels at other disease-specific loci are discriminative for some diseases (e.g., CdLS and WDSTS loci for Sotos), but not for all (e.g., Kabuki loci for Sotos), as was shown in the original publications [5]. Moreover, the heatmaps show how strong the methylation differences are in some diseases (e.g., Sotos), but subtle in others. Nonetheless, even in those diseases with small methylation variation, hierarchical clustering enables to recognize each sample’s disease.
SVM-based episignature detection
Our primary results showed, through different approaches, a high similarity between the microarray reference [5] and nanopore episignatures. However, methods like UMAP require interpretation upon visual representation, and in the case of hierarchical clustering, determining the cutoffs or number of clusters is not trivial. For this reason, we developed an automated and generic approach to screen samples for 34 shared episignatures [5] using SVM classifiers. Our classifiers were tested on the 11 patients assessed in our first analyses and nine additional samples, representing 13 diseases, as well as 40 controls. In 17/20 patients with a (suspected) DD, the classifiers recognized an episignature and assigned the sample to the right disease (Table 1). All healthy individuals were classified as controls.
For patient EPI_19, both a maternally inherited, hemizygous c.7263_7265del p.(Gln2425del) VUS in ATRX and a pathogenic but mosaic c.3002_3005del p.(Leu1001Profs*15) variant in KMT2D (present in 10–21% of cells according to WES and Sanger sequencing) were identified. The detection of the mosaic KMT2D variant was driven by clinical suspicion, but an additional ATRX syndrome could not be excluded solely based on the patient’s phenotype. The SVMs classified this patient as Kabuki syndrome, the syndrome caused by pathogenic variants in KMT2D, but not as ATRX syndrome. Interestingly, the confidence score value returned by the SVM was lower compared to other Kabuki patients: 0.70 in EPI_19 in contrast to 1.94, 1.78, and 2.30 for the three other Kabuki samples (Additional file 6: Table S4), likely as consequence of the mosaic status of the KMT2D variant. Similarly, EpiSign [10] also identified the Kabuki and not the ATRX signature. In addition, XCI testing was performed in the mother and showed a 41/59 inactivation ratio. Hence, there is no skewed XCI in maternal blood. Moreover, segregation analysis showed the presence of the ATRX c.7263_7265del p.(Gln2425del) variant in a healthy brother of the patient, further supporting the ATRX variant to be benign. Together with the result of our classifier, those results indicate the KMT2D c.3002_3005del p.(Leu1001Profs*15) variant to be pathogenic, but not the ATRX c.7263_7265del p.(Gln2425del) variant.
In three samples (EPI_13, EPI_18, and EPI20), the classifier did not recognize any of the reference episignatures (Table 1). For those cases, clinical testing using EpiSign also returned negative.
Supporting evidence from phased X-inactivation analysis
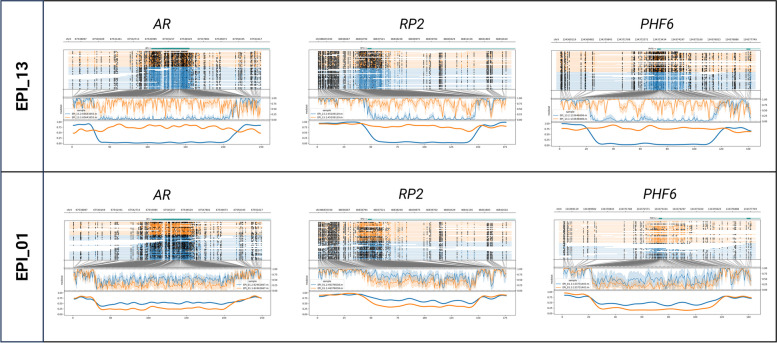
Patient EPI_13 had a diagnosis of Börjeson-Forssman-Lehmann syndrome (BFLS), after identification of a de novo pathogenic c.306del p.(Tyr103Thrfs*40) variant in PHF6. BFLS is an X-linked disorder with variable penetrance and expression in females. Functional mosaicism, a phenomenon where the allele on which the pathogenic variant is located is active in some but not all cells due to XCI, can explain, at least in part, this variation [32]. Hence, we hypothesize that the female patient with BFLS (EPI_13) might express the allele with the pathogenic variant in relevant tissues but have skewed XCI in blood with predominant inactivation of the abnormal allele. This could explain the absence of episignature in blood. Methylation analysis of the two AR, RP2, and PHF6 promotor haplotypes on the patient’s blood sample showed divergent methylation levels between both haplotypes: 75%:2% at the AR loci, 80%:5% at the RP2 loci, and 79%:10% at the PHF6 loci (coverage of 37 × for the three loci). These results point to skewing of XCI. In comparison, 26 to 68% of the reads of each of the two haplotypes are methylated at these loci in the five other females of the cohort (Fig. 3, Additional file 3: Table S3B). Evaluation of XCI of the patient’s urine and buccal swab samples also revealed an imbalanced inactivation, the percentages of methylation of both alleles being measured at 76%:19% (AR) and 75%:12% (RP2) for the urine sample and 71%:3% (AR) and 83%:3% (RP2) for the buccal swab sample (Additional file 4: Fig. S1).
Fig. 3.
Evaluation of X-inactivation. Phased nanopore sequencing methylation levels at AR and RP2 loci, as well as at the promotor of the PHF6 gene, in patient EPI_13 (top) and EPI_01 (bottom). Orange and blue represent the two different haplotypes of each sample. In the upper part of each plot, the genome coordinates, genes, and reads are illustrated. On each read, modified bases are represented by full, dark circles, unmodified bases by open circles. In the lower part of the plot, after translation from genome to CpG-only coordinate space, raw log-likelihood ratios are plotted above a smoothened graph of methylation fraction [21]. About half reads of each allele are methylated at the three loci in patient EPI_01 whereas in patient EPI_13 one allele (orange) is methylated in all reads in contrast to the other one which is unmethylated
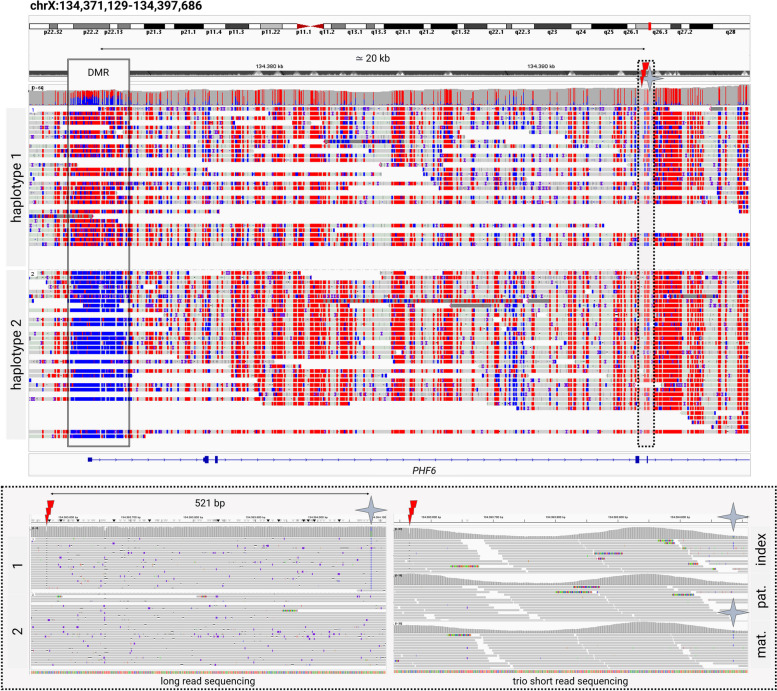
To further substantiate our hypothesis, we confirmed EPI_13’s PHF6 c.306del p.(Tyr103Thrfs*40) pathogenic variant to be localized on the hypermethylated inactive allele by haplotyping. Phasing is enabled by the long reads spanning the 5′UTR differentially methylated region to the pathogenic variant. Taking further advantage of this property, we could determine that the de novo PHF6 pathogenic variant occurred on the maternal allele. Available trio WES data showed an informative maternally inherited SNV 521 base pairs downstream of the pathogenic variant and nanopore long reads co-localized these two variants to be in cis (Fig. 4). The RP2 locus did not contain any informative SNV. The AR differentially methylated region, however, also contained an informative SNV which confirms the inactivation of the mutated maternal allele (Additional file 7: Fig. S3). These results support the hypothesis that this variant is pathogenic but, due to skewed X-inactivation, is not causing an epigenetic fingerprint in white blood cells.
Fig. 4.
Phasing of methylome and genome variation. Visualization of EPI_13’s Oxford Nanopore long-read sequencing and Illumina short read exome sequencing BAM files in IGV [22], focusing on the PHF6 gene. The long reads spanning the promotor differentially methylated region (DMR) and the pathogenic c.306delA variant (marked by the red thunderbolt sign) are shown in the upper image. Methylation is marked in red; unmethylated CpGs are represented in blue. The lower box is zooming into the region of the pathogenic variant (marked by a dotted line) and the closest informative SNV (marked by a blue star), showing both the long-read sequencing data of the index as well as trio short read exome sequencing data of the index and her parents
Concurrent epigenetic and genetic diagnosis
Given that long-read sequencing offers genome in parallel with methylome sequencing, we wanted to evaluate and confirm the potential of long-read sequencing to enable concurrent episignature, SV, and SNV detection. Hence, we assessed whether the (likely) pathogenic variants (or VUS for EPI_18 and EPI_19) could be detected in the nanopore sequencing data. Eighteen of 19 underlying SNVs were detected using the standard bioinformatic analysis (see “Methods”). The mosaic KMT2D variant in EPI_19 was only present in 17% (5/29) of the nanopore reads and could not be detected. However, the variant allele frequency in previous short-read WES was also too low (22%, 38/174 reads) to be detected after default variant allele frequency-based filtering. Both short- and long-read methods did allow the KMT2D variant detection after manual curation driven by clinical suspicion. Two patients were heterozygous for a pathogenic SV and both were detected: the multi-exonic deletion in the EP300 gene in EPI_17 and the 7q11.23 microdeletion in EPI_16. Additionally, the long-read sequencing data allowed mapping the breakpoints of the EP300 deletion (NM_001429: g.41168065_41171580del, hg38). The Geneyx platform [18] was used to prioritize variants based on the patients’ phenotype and ACMG variant classification, in the absence of parental data. All (likely) pathogenic variants were ranked in the top 4 variants to evaluate, most of them being ranked as first (Table 1). The only variant that was not prioritized is the VUS in ATRX that was ranked as first variant in the X-linked analysis. Both SVs were also ranked first in the SV subanalysis of the tool.
Discussion
Episignatures are diagnostically important. Their presence provides evidence for the pathogenicity of VUS or can act as biomarker for a disease when the underlying genomic variant cannot be identified [6, 8]. This proof-of-concept study illustrates the potential of lrWGS for episignature detection. Despite known technological differences, we show that microarray reference data can be used for several diseases. Moreover, nanopore sequencing enables to concurrently identify the episignature inducing SNVs and SVs. In addition, we show that phased methylation maps inform about imprinting and skewed XCI. Altogether, long-read sequencing haplotype-aware mapping of genetic and methylation variation provides a comprehensive genome analysis, combining several analyses that currently require multiple technologies.
While this study opens the door for concurrent genomic and episignature assessment, it also illustrates limitations of current episignatures. In samples classified as control, no episignature was identified by the standard microarray-based assay neither, excluding inter-technology biases as the main cause for these negative results. For two of the genes of which the episignature could not be detected, UBE2A and PHF6, the reference microarray data we used is based on a limited sample set [5] without test nor validation cohort and might therefore be imprecise. Both genes, as well as ZNF711, are also listed among the genes for which the commercial microarray assay is reported to have a lower sensitivity [33]. Moreover, in the last years, sub-episignatures have been described for different types of variants in the same gene [34, 35]. While not yet been reported for UBE2A and ZNF711, such sub-episignatures could confound the classification of EPI 18 and EPI_20, which both harbor missense variants. Segregation, maternal XCI analyses, and functional analyses performed in a previous study have confirmed the pathogenicity of the c.19C > T p.(Arg7Trp) variant in UBE2A identified in EPI_20 [36]. Further segregation analyses could not be performed for the c.1223A > G p.(His408Arg) VUS in ZNF711 identified in EPI_18. However, further XCI analysis in the carrier mother of this individual, showing a random XCI, is an additional argument against the pathogenicity of the ZNF711 variant. In this case, the absence of the disease-associated episignature might therefore indicate the variant to be benign. However, one must carefully draw conclusions based on the absence of episignatures as a sub-episignature cannot be excluded. An evaluation of published episignatures also recently showed their high specificity but variable sensitivity [37].
Another constraint highlighted by this study is XCI. For the PHF6 c.306del p.(Tyr103Thrfs*40) variant (EPI_13), we hypothesized that skewed XCI in white blood cells might lead to predominant expression of the normal allele, resulting in a normal methylation pattern. Skewed XCI has already been reported in other females with BFLS [38]. Using haplotype-aware nanopore methylation mapping, we confirmed the unbalanced XCI in this patient and subsequently localized the variant on the inactivated allele. As the patient presents high phenotypic similarities with BFLS, we hypothesized that other tissues probably lack XCI skewing and might present the episignature. However, both urine and buccal swab samples also showed predominant inactivation of the affected allele. Other tissues could be investigated to substantiate our hypothesis further, but this requires more invasive procedures and might require tissue-specific epigenomic maps, which are currently lacking. Concurrent XCI and episignature evaluation in blood of carrier mothers of males with an X-linked disease episignature could, therefore, provide an indirect alternative to validate this hypothesis in the future.
An important potential pitfall in the diagnostic process of DD is mosaicism. Post-zygotic de novo pathogenic variants can lead to mosaic variants which often escape detection by the default variant calling algorithms. Interestingly, our classifier did detect the episignature in a mosaic heterozygous patient (EPI_19), albeit with a lower confidence score, reflecting the low-grade mosaicism for the pathogenic variant. Mosaicism detection can be improved by including mosaic samples in the training set and lowering the positive confidence score threshold [39]. Episignature evaluation was important in this boy as it contributed to classifying the X-linked ATRX variant as benign and reassuring the parents of low recurrence risk for future pregnancies. The ATRX episignature has indeed been reported as having a high sensitivity [37], and this conclusion was corroborated by segregation and maternal XCI analyses. Episignature detection can also potentially direct the clinical diagnostic laboratory to search for mosaic variants in specific genes, which would, as this KMT2D mosaic variant, be missed without targeted reanalysis of the data.
The correlation between microarray- and nanopore-based methylomes has been estimated at around 0.85 [40]. Here we show that despite these inter-assay biases and the subtle methylation changes that characterize the episignatures of some disorders, publicly available microarray-based methylation ratio profiles and/or positions [5] can be used for nanopore sequencing episignature detection for several disorders. Still, it is likely that comparing nanopore methylation to microarray reference data reduces the sensitivity. To further benchmark our approach, we explored the EpiGenCentral platform together with their episignatures to analyze a subset of our samples (Additional file 8: Table S5A). This comparison highlighted some challenges when using tools primarily developed for microarray data with lrWGS, and the classification outcomes were highly variable (Additional file 8: Table S5B). This underscores the importance of developing episignature tools that are specifically optimized for lrWGS technologies. Microarray references also have the disadvantage of being based on a restricted amount of CpGs, varying with the product’s version and each sample’s hybridization [41]. We envision that large-scale nanopore sequencing of patients and controls in the future will leverage high-resolution whole methylomes, enabling refinement of existing episignatures, potentially improving their sensitivity and specificity, and likely leading to the discovery of new episignatures.
Important drawbacks of lrWGS platforms have long been their lower accuracy and high costs. However, the costs of nanopore sequencing have been dropping and currently (for 30 × , genome-wide lrWGS) slightly exceed the costs of clinical episignature testing by microarrays in a large-scale setting. With the improving accuracy of lrWGS chemistries and analytical tools, the cost-effectiveness balance might change as concurrent genetic and epigenetic testing would enable to replace the combination of WES and methylation-sensitive microarray analyses. The other side of the coin of all the analyses enabled by lrWGS remains the high amount of resources that are needed to store, process, and analyze the data.
Conclusion
lrWGS enables concurrent phased genome and methylome variant detection, opening the door to large-scale studies about genomic and epigenomic variation and their interactions. While this study has used nanopore sequencing, other sequencing platforms combining methylation and base calling will probably produce similar results. XCI and imprinting detection have already been demonstrated with single-molecule real-time sequencing too [42, 43]. With its improving SNV calling accuracy [11] and more comprehensive SV detection, it seems likely that long-read sequencing will be implemented as the first-tier test for the diagnosis of DD in the future. Beyond known and potential new episignatures, we envision that shedding light on secondary epivariation will improve our understanding of genomic, especially non-coding, variants, and give us new insights into molecular mechanisms underlying diseases. In addition, an excess of de novo primary epivariants has been identified in patients with unexplained DD [4], and the presence of such epivariants has been associated with outlier gene expression [1]. Further exploration of the whole epigenome might, therefore, help us solve another part of the remaining unexplained DD.
Supplementary Information
Additional file 1: Table S1 Patients’ phenotypes and variant classification. Description of the phenotypes of the included patients, the underlying genomic (potential) pathogenic variants or VUS, the test that was previously used to identify this variant, and the classification of these variants according to the ACMG criteria [46, 47].
Additional file 2: Table S2 DNA extraction and sequencing summary. Description of the used methodologies for DNA extraction and sequencing as well as summary of the data output for each sample (estimated and basecalled bases as well as read length N50).
Additional file 3: Table S3 Differentially methylated regions: quantification. A- Imprinted loci: mean methylation levels at six imprinted loci; B- XCI: mean methylation levels at the AR, RP2, and PHF6 differentially methylated loci for each haplotype of the six female patients’ blood samples (as well as the urine and buccal swab samples of EPI_13).
Additional file 4: Fig. S1 Differentially methylated regions: visualization. Phased nanopore sequencing methylation levels at the same 6 imprinted loci in blood of all patients, as well as the AR, RP2, and PHF6 genes, in blood of all female patients (including the urine and buccal swab samples of EPI_13 for the loci on chromosome X).
Additional file 5: Fig. S2 Nanopore methylation analysis allows to distinguish disease samples from controls and other diseases samples (t-SNE). t-SNE scatter plot from methylation levels at Sotos, Kabuki, Wiedemann-Steiner (WDSTS), and Cornelia de Lange (CdLS) syndromes episignature loci.
Additional file 6: Table S4 SVM classifiers results. A- SVM confidence scores: confidence score for the 20 cases and 40 controls. B- SVM classification simplified: highest confidence score and assigned disease for the 20 cases and 40 controls. Samples without recognized episignature (confidence score < 0.3) are classified as control.
Additional file 7: Fig. S3 Phasing of methylome and genome variation. Screenshots of the visualization of EPI_13’s Oxford Nanopore long read sequencing BAM files in IGV [22] at the AR gene locus.
Additional file 8: Table S5 Analysis with EpiGenCentral. A- Analysis description: short description of the analysis performed on EpiGenCentral platform [9]. B- Classification results: classification results from the EpiGenCentral platform.
Additional file 9: Table S6 Methylation values at known episignature loci: reference array data [5] and ONT data. The excel sheet contains a tab for every evaluated disorder. For every disorder (tab), the first 2 columns indicate the chromosome and the end position of the CpG loci of the disorder specific episignature.
Acknowledgements
The authors thank the patients and their families who participated in this research. We thank Jonas Demeulemeester for sharing his expertise about nanopore sequencing methylation calling at the start of the project and also thank Anne Bassett and Donna McDonnald-McGuinn for their critical review of the manuscript. Part of Fig. 1 was created with BioRender.com.
Authors’ contributions
Conceptualization: MG, BH, JRV; data curation: MG, BH, ES; formal analysis: MG, BH; funding acquisition: KDVB, JRV; investigation: MG; methodology: MG, BH; project administration: KVDB, JRV; resources: MG, JB, KDV, HP, GVB, HVE, KDVB; software: BH, ES; supervision: KVDB, JRV; validation: MG, BH, ES; visualization: MG, BH, ES; writing—original draft: MG, BH, KVDB, JRV; writing—review and editing: MG, BH, ES, JB, KDV, HP, GVB, HVE, KVDB, JRV. All authors read and approved the final manuscript.
Funding
This work has been made possible by FWO-TBM grant T-003819N, FWO grant G0A2622N, and KU Leuven grants C1-C14/18/092 and C14/22/125 to JRV. BH was supported by a Collaborative Doctoral Partnership Agreement of the Joint Research Center F.7/KU LEUVEN B&G 35332. JB is supported by a senior clinical investigator fellowship of the FWO Flanders.
Data availability
Methylation values at used genomic positions are shared in Additional file 9: Table S6. The used data and code are available on GitHub: JorisVermeeschLab/NSBEpi: Nanopore sequencing-based episignature detection (github.com) [44]. Fastq files of the 20 cases are available upon request using the European Genome-Phenome Archive (EGA) platform with study number EGAS50000000719 [45].
Declarations
Ethics approval and consent to participate
This study was approved by the Ethical committee research UZ/KU Leuven (S65304–S65991) and informed consent was obtained from all participants to participate. This study conformed with the principles of the Helsinki Declaration.
Consent for publication
Written informed consent was obtained from all participants for publication.
Competing interests
MG presented her research at the ACLF conference 2023 for Oxford Nanopore Technologies. Travel costs were reimbursed by the company. The remaining authors declare that they do not have any competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mathilde Geysens and Benjamin Huremagic are joint first authors.
References
- 1.Garg P, et al. A survey of rare epigenetic variation in 23,116 human genomes identifies disease-relevant epivariations and CGG expansions. Am J Hum Genet. Oct.2020;107(4):654–69. 10.1016/j.ajhg.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Depienne C, Mandel JL. 30 years of repeat expansion disorders: what have we learned and what are the remaining challenges? Cell Press. 2021;108(5):764–85. 10.1016/j.ajhg.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monk D, Mackay DJG, Eggermann T, Maher ER, Riccio A. Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat Rev. 2019;20:235–48. 10.1038/s41576-018-0092-0. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa M, et al. Identification of rare de novo epigenetic variations in congenital disorders. Nat Commun. 2018;9(1):1–11. 10.1038/s41467-018-04540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aref-Eshghi E, et al. Evaluation of DNA methylation episignatures for diagnosis and phenotype correlations in 42 Mendelian neurodevelopmental disorders. Am J Hum Genet. 2020;106(3):356–70. 10.1016/j.ajhg.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy MA, et al. Novel diagnostic DNA methylation episignatures expand and refine the epigenetic landscapes of Mendelian disorders. HGG Adv. 2022;3(1):100075. 10.1016/j.xhgg.2021.100075. [DOI] [PMC free article] [PubMed]
- 7.Rooney K, et al. Identification of a DNA methylation episignature in the 22q11.2 deletion syndrome. Int J Mol Sci. 2021;22:8611. 10.3390/ijms22168611. [DOI] [PMC free article] [PubMed]
- 8.Sadikovic B, Levy MA, Aref-Eshghi E. Functional annotation of genomic variation: DNA methylation episignatures in neurodevelopmental Mendelian disorders. Hum Mol Genet. 2020;29:1–27. 10.1093/hmg/ddaa144. [DOI] [PubMed] [Google Scholar]
- 9.Turinsky AL, et al. EpigenCentral: portal for DNA methylation data analysis and classification in rare diseases. Hum Mutat. Oct.2020;41(10):1722–33. 10.1002/humu.24076. [DOI] [PubMed] [Google Scholar]
- 10.Sadikovic B, et al. Clinical epigenomics: genome-wide DNA methylation analysis for the diagnosis of Mendelian disorders. Genet Med. 2021;23(6):1065–74. 10.1038/s41436-020-01096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolmogorov M, et al. Scalable nanopore sequencing of human genomes provides a comprehensive view of haplotype-resolved variation and methylation. Nat Methods. 2023;20(10):1483–92. 10.1038/s41592-023-01993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev. 2020;21:597–614. 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebert P, et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 2021;372:6537. 10.1126/SCIENCE.ABF7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson J, et al. A novel quantitative targeted analysis of X-chromosome inactivation (XCI) using nanopore sequencing. Sci Rep. 2023;13(1):12856. 10.1038/s41598-023-34413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada M, et al. Diagnosis of Prader-Willi syndrome and Angelman syndrome by targeted nanopore long-read sequencing. Eur J Med Genet. 2023;66:2. 10.1016/j.ejmg.2022.104690. [DOI] [PubMed] [Google Scholar]
- 16.Giesselmann P, et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat Biotechnol. 2019;37:1478–81. 10.1038/s41587-019-0293-x. [DOI] [PubMed] [Google Scholar]
- 17.Patel A, et al. Rapid-CNS2: rapid comprehensive adaptive nanopore-sequencing of CNS tumors, a proof-of-concept study. Acta Neuropathol. 2022;143(5):609–12. 10.1007/s00401-022-02415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahary D, et al. Genome analysis and knowledge-driven variant interpretation with TGex. BMC Med Genomics. 2019;12(1):200. 10.1186/s12920-019-0647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oxford Nanopore Technologies. modkit. Github. 2023. https://github.com/nanoporetech/modkit.
- 21.Cheetham SW, Kindlova M, Ewing AD. Methylartist: tools for visualizing modified bases from nanopore sequence data. Bioinformatics. 2022;38(11):3109–12. 10.1093/bioinformatics/btac292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhenxian Z, et al. Symphonizing pileup and full-alignment for deep learning-based long-read variant calling. Nat Comput Sci. 2022;2:797–803. 10.1038/s43588-022-00387-x. [DOI] [PubMed] [Google Scholar]
- 24.Sedlazeck FJ, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15(6):461–8. 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuenca-Guardiola J, et al. Improvement of large copy number variant detection by whole genome nanopore sequencing. J Adv Res. 2023;50:145–58. 10.1016/j.jare.2022.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheinin I, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–32. 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tretyakov K. pyliftover: Pure-python implementation of UCSC liftOver genome coordinate conversion. Github. 2023. https://github.com/konstantint/pyliftover.
- 28.McInnes L, Healy J, Saul N, Großberger L. UMAP: uniform manifold approximation and projection. J Open Source Softw. 2018;3(29):861. 10.21105/JOSS.00861.
- 29.Pedregosa F, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12(85):2825–30. http://jmlr.org/papers/v12/pedregosa11a.html.
- 30.Virtanen P, et al. SciPy 1.0: fundamental algorithms for scientific computing in python. Nat Methods. 2020;17(3):261–72. 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed]
- 31.Waskom ML. Seaborn: statistical data visualization. J Open Source Softw. 2021;6(60):3021. 10.21105/JOSS.03021.
- 32.Gerber CB, et al. Further characterization of Borjeson-Forssman-Lehmann syndrome in females due to de novo variants in PHF6. Clin Genet. 2022;102(3):182–90. 10.1111/CGE.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amsterdam UMC. EpiSign complete. 2023. https://genoomdiagnostiek.nl/en/product/episigncomplete/.
- 34.Bend EG, et al. Gene domain-specific DNA methylation episignatures highlight distinct molecular entities of ADNP syndrome. Clin Epigenetics. 2019;11(1):1–17. 10.1186/S13148-019-0658-5/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Jaarsveld RH, et al. Delineation of a KDM2B-related neurodevelopmental disorder and its associated DNA methylation signature. Genet Med. 2023;25(1):49–62. 10.1016/j.gim.2022.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haddad DM, et al. Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol Cell. 2013;50(6):831–43. 10.1016/j.molcel.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Husson T, et al. Episignatures in practice: independent evaluation of published episignatures for the molecular diagnostics of ten neurodevelopmental disorders. Eur J Hum Genet. 2023;32(2):190–9. 10.1038/s41431-023-01474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zweier C, et al. A new face of Borjeson-Forssman-Lehmann syndrome? De novo mutations in PHF6 in seven females with a distinct phenotype. J Med Genet. 2013;50:838–47. 10.1136/jmedgenet-2013-101918. [DOI] [PubMed] [Google Scholar]
- 39.Oexle K, et al. Episignature analysis of moderate effects and mosaics. Eur J Hum Genet. 2023;31(9):1032–9. 10.1038/s41431-023-01406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva C, et al. Whole human genome 5’-mC methylation analysis using long read nanopore sequencing. Epigenetics. 2022;17(13):1961–75. 10.1080/15592294.2022.2097473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giuili E, et al. Comprehensive evaluation of the implementation of episignatures for diagnosis of neurodevelopmental disorders (NDDs). Hum Genet. 2023;142(12):1721–35. 10.1007/s00439-023-02609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vollger MR et al. Synchronized long-read genome, methylome, epigenome, and transcriptome for resolving a Mendelian condition. bioRxiv preprint. 2023. 10.1101/2023.09.26.559521.
- 43.Olivia Tse OY, et al. Genome-wide detection of cytosine methylation by single molecule real-time sequencing. PNAS. 2021;118(5). 10.1073/pnas.2019768118/-/DCSupplemental. [DOI] [PMC free article] [PubMed]
- 44.Huremagic B, Geysens M. NSBEpi: nanopore sequencing-based episignature detection. GitHub. 2024. https://github.com/JorisVermeeschLab/NSBEpi.
- 45.Geysens M, Huremagic B. Clinical evaluation of long read sequencing-based episignature detection in developmental disorders. EGA. 2024. https://ega-archive.org/studies/EGAS50000000719. [DOI] [PMC free article] [PubMed]
- 46.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. 10.1038/GIM.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riggs ER, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22(2):245–57. 10.1038/S41436-019-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1 Patients’ phenotypes and variant classification. Description of the phenotypes of the included patients, the underlying genomic (potential) pathogenic variants or VUS, the test that was previously used to identify this variant, and the classification of these variants according to the ACMG criteria [46, 47].
Additional file 2: Table S2 DNA extraction and sequencing summary. Description of the used methodologies for DNA extraction and sequencing as well as summary of the data output for each sample (estimated and basecalled bases as well as read length N50).
Additional file 3: Table S3 Differentially methylated regions: quantification. A- Imprinted loci: mean methylation levels at six imprinted loci; B- XCI: mean methylation levels at the AR, RP2, and PHF6 differentially methylated loci for each haplotype of the six female patients’ blood samples (as well as the urine and buccal swab samples of EPI_13).
Additional file 4: Fig. S1 Differentially methylated regions: visualization. Phased nanopore sequencing methylation levels at the same 6 imprinted loci in blood of all patients, as well as the AR, RP2, and PHF6 genes, in blood of all female patients (including the urine and buccal swab samples of EPI_13 for the loci on chromosome X).
Additional file 5: Fig. S2 Nanopore methylation analysis allows to distinguish disease samples from controls and other diseases samples (t-SNE). t-SNE scatter plot from methylation levels at Sotos, Kabuki, Wiedemann-Steiner (WDSTS), and Cornelia de Lange (CdLS) syndromes episignature loci.
Additional file 6: Table S4 SVM classifiers results. A- SVM confidence scores: confidence score for the 20 cases and 40 controls. B- SVM classification simplified: highest confidence score and assigned disease for the 20 cases and 40 controls. Samples without recognized episignature (confidence score < 0.3) are classified as control.
Additional file 7: Fig. S3 Phasing of methylome and genome variation. Screenshots of the visualization of EPI_13’s Oxford Nanopore long read sequencing BAM files in IGV [22] at the AR gene locus.
Additional file 8: Table S5 Analysis with EpiGenCentral. A- Analysis description: short description of the analysis performed on EpiGenCentral platform [9]. B- Classification results: classification results from the EpiGenCentral platform.
Additional file 9: Table S6 Methylation values at known episignature loci: reference array data [5] and ONT data. The excel sheet contains a tab for every evaluated disorder. For every disorder (tab), the first 2 columns indicate the chromosome and the end position of the CpG loci of the disorder specific episignature.
Data Availability Statement
Methylation values at used genomic positions are shared in Additional file 9: Table S6. The used data and code are available on GitHub: JorisVermeeschLab/NSBEpi: Nanopore sequencing-based episignature detection (github.com) [44]. Fastq files of the 20 cases are available upon request using the European Genome-Phenome Archive (EGA) platform with study number EGAS50000000719 [45].