Abstract
For a long time, snakes were presented as a textbook example of a group with gradual differentiation of homologous ZZ/ZW sex chromosomes. However, recent advances revealed that the ZZ/ZW sex chromosomes characterize only caenophidian snakes and certain species of boas and pythons have nonhomologous XX/XY sex chromosomes. We used genome coverage analysis in four non-caenophidian species to identify their sex chromosomes, and we examined the homology of sex chromosomes across phylogenetically informative snake lineages. We identified sex chromosomes for the first time in 13 species of non-caenophidian snakes, providing much deeper insights into the evolutionary history of snake sex chromosomes. The evolution of sex chromosomes in snakes is more complex than previously thought. Snakes may have had ancestral XX/XY sex chromosomes, which are still present in a blind snake and some boas, and there were several transitions to derived XX/XY sex chromosomes with different gene content and two or even three transitions to ZZ/ZW sex chromosomes. However, we discuss more alternative scenarios. In any case, we document that (1) some genomic regions were likely repeatedly co-opted as sex chromosomes in phylogenetically distant lineages, even with opposite types of heterogamety; (2) snake lineages differ greatly in the rate of differentiation of sex chromosomes; (3) snakes likely originally possessed sex chromosomes prone to turnovers. The sex chromosomes became evolutionarily highly stable once their differentiation progressed in the megadiverse caenophidian snakes. Snakes thus provide an ideal system for studying the evolutionary factors that drive unequal rates of differentiation, turnovers and stability of sex chromosomes.
Keywords: DNA-seq, genomics, reptiles, sex chromosomes, sex determination, qPCR
Introduction
Snakes are a group of squamate reptiles that appeared in the fossil record in the Jurassic period (Caldwell et al. 2015). They represent approximately one third of the total species diversity of extant non-avian reptiles (Uetz et al. 2024) and have colonized all continents, except Antarctica, and the vast majority of terrestrial and aquatic habitats, including oceans. More than 4,000 currently recognized species are classified in about 30 families (Pyron et al. 2013, 2014; Zheng and Wiens 2016; Uetz et al. 2024). Snakes show unique morphological traits and have emerged as a popular model group for a variety of scientific fields. Prominent research in snakes includes the evolution of venom and its application in medicine (Koh et al. 2006; Chan et al. 2016; Oliveira et al. 2022), the loss of limbs and the adaptation to fossorial and aquatic lifestyles (Simoes et al. 2015; Leal and Cohn 2018; Silva et al. 2018), the adaptation to the marine environment (Brischoux and Shine 2011), but they also played an important role in the field of the evolution of sex chromosomes (Ohno 1967; Bull 1980; Singh et al. 1980).
The ZZ/ZW sex chromosomes were identified decades ago in many caenophidian snakes (Beçak et al. 1962, 1964; Beçak and Beçak 1969; Singh et al. 1968; Singh 1972; Mengden and Stock 1980), i.e. in the group containing over 80% of all extant species of snakes (Uetz et al. 2024). The sex chromosomes in caenophidian snakes can be detected by both conventional and molecular cytogenetic methods, because they are large (typically the 4th or 5th largest chromosome pair), often heteromorphic with heterochromatic W (Singh et al. 1968; Beçak and Beçak 1969; Mengden and Stock 1980) and rich in repeats (Singh et al. 1980; Augstenová et al. 2018b; Rovatsos et al. 2018). Later, molecular and genomic studies confirmed that the caenophidian snakes possess homologous and highly differentiated ZZ/ZW sex chromosomes, which evolved likely at least 69 MYA (Vicoso et al. 2013; Rovatsos et al. 2015; Matsubara et al. 2016; Yin et al. 2016; Kumar et al. 2017).
In contrast to caenophidian snakes, sex chromosomes were not detected by cytogenetic methods in other snake lineages with the exception of ZZ/ZW sex chromosomes in the Madagascar boa Acrantophis sp. cf. dumerili (Sanziniidae) (Mengden and Stock 1980; Augstenová et al. 2018a) and putatively in the long-nosed worm snake, Myriopholis macrorhyncha (Leptotyphlopidae), where a heterochromatic chromosome, interpreted as a W chromosome, was reported in a single female (Matsubara et al. 2019). It was assumed for a long time that all snakes share homologous ZZ/ZW sex chromosomes, which are homomorphic and at an early stage of differentiation in the snakes outside Caenophidia (Beçak et al. 1964). The striking contrast between caenophidian and non-caenophidian snakes inspired Susumu Ohno (1967) to formulate the now classical scenario on progressive, step-by-step differentiation of sex chromosomes. This view was further reproduced and expanded by a series of influential studies, despite the fact that there was little evidence on sex chromosomes in snakes outside the clade Caenophidia (Ray-Chaudhuri et al. 1971; Bull 1980; Jones and Singh 1985; Marshall Graves and Shetty 2001; Ezaz et al. 2006; Matsubara et al. 2006, 2016; Bellot and Page 2009; Pokorná and Kratochvíl 2009; O’Meally et al. 2010, 2012; Vicoso et al. 2013; Yin et al. 2016).
In contrast, all-female, largely homozygous progeny produced by occasional (facultative) parthenogenesis in several species of boas and pythons (namely, in Boa constrictor, Chilabothrus angulifer, Chilabothrus subflavus, Epicrates cenchria, Epicrates maurus, Eunectes murinus, Malayopython reticulatus, and Python regius) (Booth et al. 2011a, 2011b, 2014; Kinney et al. 2013; Shibata et al. 2017; Seixas et al. 2020; Bailey et al. 2024) indicated that these snakes might possess male heterogamety. Similarly, a colour mutation in the ball python Python regius showed an inheritance pattern consistent with a XX/XY sex determination system (Mallery and Carrillo 2014). The verification of the male heterogamety in two species of boas (Boa constrictor and B. imperator) and one species of python (Python bivittatus) was provided by Gamble et al. (2017) based on restriction site-associated DNA sequencing (RAD-seq). Notably, the XX/XY sex chromosomes of the python share partial gene content with the caenophidian ZZ/ZW chromosomes, while the sex chromosomes of the boas evolved from a different genomic region (Gamble et al. 2017). However, the heteromorphic ZZ/ZW sex chromosomes in the boa Acrantophis sp. cf. dumerili, which is phylogenetically positioned between the python and the boas with known XX/XY sex chromosomes, suggest that the evolution of sex determination in snakes is even more variable (Augstenová et al. 2018a).
To better reconstruct the evolution of sex chromosomes in snakes, we sequenced genomes of both sexes in four non-caenophidian species with phylogenetically informative positions. Namely, we studied the boa Acrantophis sp. cf. dumerili, the Kenyan sand boa Eryx colubrinus (Erycidae), the Cuban wood snake Tropidophis melanurus (Tropidophiidae) and the Eurasian blind snake Xerotyphlops vermicularis (Typhlopidae). We selected these species as (1) Acrantophis sp. cf. dumerili is the only snake outside Caenophidia known to have heteromorphic and cytogenetically easily detectable sex chromosomes, although cytogenetic analyses, including molecular cytogenetic techniques, have been performed in many non-caenophidian lineages (Beçak et al. 1964, 1969; Mengden and Stock 1980; Oguiura et al. 2009; Mezzasalma et al. 2016; Viana et al. 2016; Augstenová et al. 2018a, 2019; Charvát et al. 2022); (2) E. colubrinus is a member of the family Erycidae, phylogenetically placed among boas with known male and female heterogamety; (3) T. melanurus is a member of the family Tropidophiidae, representing a clade sister to all snakes other than blind snakes and thread snakes; (4) X. vermicularis as a representative of the blind snakes (Pyron et al. 2013; Zheng and Wiens 2016), a group of snakes poorly studied using cytogenetic or genomic approaches. We identified the gene content of sex chromosomes in these four species by comparative gene coverage analysis and tested the homology of their sex chromosomes across other snakes by a comparison of sex-specific gene copy numbers using quantitative real-time PCR (qPCR).
Materials and Methods
Studied Material and DNA Isolation
We collected samples from 20 species of snakes from 9 non-caenophidian families (Boidae, Calabariidae, Candoiidae, Erycidae, Pythonidae, Sanziniidae, Tropidophiidae, Typhlopidae, and Xenopeltidae) and 5 caenophidian species from 5 families (Acrochordidae, Colubridae, Pareidae, Viperidae, and Xenodermidae) (supplementary table S1, Supplementary Material online). Total DNA was isolated from fresh blood or tissue samples, using the DNeasy Blood and Tissue Kit (Qiagen), according to the manufacturer's protocol. The quality and the concentration of isolated DNA samples were estimated by NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific Inc.).
DNA-Seq and Coverage Analysis
In order to identify X- or Z-specific genes, we used a comparative gene coverage analysis, which was previously applied successfully in other reptiles (Vicoso et al. 2013; Kostmann et al. 2021; Pensabene et al. 2023). The method assumes that Illumina sequencing is relatively unbiased, and the Illumina reads are proportional to copies of DNA molecules used for the preparation of the sequencing libraries. X-specific single-copy genes have two copies per cell in the genome of XX females and a single copy in XY males, while autosomal genes should mostly have an equal number of copies per genome in both sexes. This difference should lead to different coverage in XY males and XX females in the map-to-reference assemblies of the X-specific genes. Similar logic applies to Z-specific genes. Therefore, we expect a normalized male-to-female gene coverage ratio of 0.5 for X-specific, 1.0 for autosomal or pseudoautosomal, and 2.0 for Z-specific genes.
The chromosome-level assemblies of the western terrestrial garter snake Thamnophis elegans (GCA_009769535.1), the green anole Anolis carolinensis (GCA_000090745.2; Alföldi et al. 2011), the common wall lizard Podarcis muralis (GCA_004329235.1; Andrade et al. 2019), and the chicken Gallus gallus (GCA_000002315.3; Warren et al. 2017) were used for genome-wide cross-species comparisons of genes and assessment of sex chromosome homology.
Total DNA from both sexes of Acrantophis sp. cf. dumerili, E. colubrinus, T. melanurus, and X. vermicularis was sequenced by Novogene (Cambridge, UK) on Illumina HiSeq2500 platform with 150 base pairs (bp) paired-end option and 350 bp library size (DNA-seq). Low-quality raw reads and sequencing adaptors were trimmed by Trimmomatic (Bolger et al. 2014), and reads shorter than 50 bp were removed from further analysis. Trimmed reads were checked in FASTQC (Andrews 2010) and MULTIQC (Ewels et al. 2016) for quality control. The raw Illumina reads are deposited in the NCBI database under BioProject PRJNA1076544. The NCBI database was also used to obtain the Illumina sequences from two caenophidian species, Naja naja (SRX7124244; Suryamohan et al. 2020) and Crotalus viridis (SRX7273744; SRX7273711; Schield et al. 2019) for comparison.
The trimmed Illumina reads from each sample were mapped independently using Geneious Prime 2023.0.4 (https://www.geneious.com) to a reference data set consisting of 174,674 exons from the Thamnophis elegans genome project (GCA_009769535.1). The average coverage was estimated for each gene from the assembly report. Subsequently, we removed exons with unexpectedly high or low coverage (i.e. 3-fold difference from the average read coverage of each specimen) and we calculated the male-to-female gene coverage ratio in each species, normalized to the average coverage for each specimen. Genes with known homology and annotation in chicken and garter snake chromosomes (further abbreviated as GGA and TEL, respectively) were used for cross-species comparisons.
Validation of Z-Specific/X-Specific Genes and Test of Homology of Sex Determination by qPCR
The differences in copies of X- or Z-specific genes between males and females can be detected by qPCR (Rovatsos et al. 2014a), analogously as in the comparative gene coverage analysis. We designed primers from a random subset of gene exons using Primer-Blast software (Ye et al. 2012) for the putative X-/Z-specific genes revealed by the comparative gene coverage analysis in Acrantophis sp. cf. dumerili, E. colubrinus, X. vermicularis, and T. melanurus. All designed primers were tested by standard PCR to exclude primers with secondary products. Additionally, we used primers for the genes adarb2 (GGA2), immt (GGA4), and tanc2 (GGA27), which are Z-specific in caenophidian snakes, as controls and mecom (GGA9) for normalization of the qPCR values (Rovatsos et al. 2015). The qPCR tests were performed in triplicates for each primer and specimen according to the protocol described in Rovatsos et al. (2014a) using the LightCycler II 480 (Roche Diagnostics). From the normalized qPCR quantification values, we calculated the male-to-female gene copy ratio which is expected to be 0.5 for X-specific, 1.0 for autosomal or pseudoautosomal, and 2.0 for Z-specific genes (Rovatsos et al. 2014a; Rovatsos and Kratochvíl 2017).
Genes proven to be Z-/X-specific in A. sp. cf. dumerili, E. colubrinus, T. melanurus, and X. vermicularis by both gene coverage analysis and the qPCR test were further tested for Z-/X-specificity in additional 21 snake species (supplementary tables S2 and S3, Supplementary Material online).
Estimation of Sex Chromosome Age and Degree of Differentiation
The minimum age of each uncovered sex chromosome system was estimated as the age of the basal split of lineages with likely homologous sex chromosomes as reported by Kumar et al. (2017). Quantification of sex chromosome differentiation cannot be done from chromosome morphology, as the degree of heteromorphism of sex chromosomes does not correlate with sequence divergence (Pokorná et al. 2011). We follow Charlesworth (2021) in attempting to estimate this parameter from the estimation of the size of sex-linked regions from evidence of gene loss. The degree of differentiation could be estimated by comparing the number of genes lost from the Y (or W) chromosome with the number of genes retained on the X (or Z) chromosome, ideally taking into an account a number of genes in the region before it became a sex chromosome. In snakes, this estimation could now be made in caenophidian snakes, but not for the other snake sex chromosome systems due to the lack of chromosome-level genome assemblies. Therefore, we quantified the degree of sex chromosome differentiation by the number of hemizygous genes detected by our cross-genome comparison in each species relative to the total number of mapped genes in a given species.
Results
Identification of Z-Specific Loci by Genome Coverage Analysis in A. sp. cf. dumerili
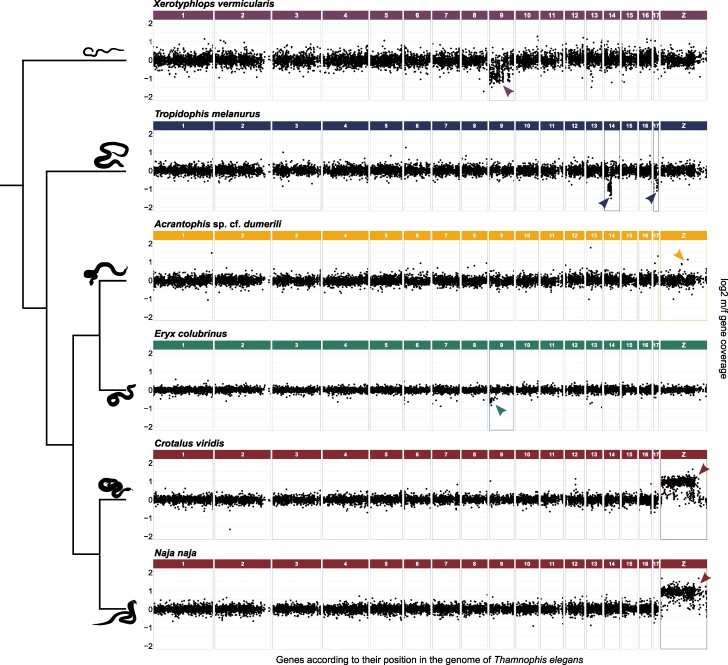
The comparative gene coverage analysis resulted in 11,106 genes in A. sp. cf. dumerili with known homologs in the chicken and garter snake genomes. In total, 47 genes have male-to-female read coverage between 1.43 and 3.33, which is the expected range for Z-specificity (Fig. 1; Table 1; supplementary table S2, Supplementary Material online). Their homologs are located mainly on the chicken chromosomes 1 (GGA1; 6 genes) and 2 (GGA2; 7 genes) and on the garter snake Z chromosome (TELZ; 6 genes).
Fig. 1.
Log2-transformed male-to-female ratios of normalized read coverage per gene in Acrantophis sp. cf. dumerili, Crotalus viridis, Eryx colubrinus, Naja naja, Tropidophis melanurus, and Xerotyphlops vermicularis. Genes are illustrated based on the position of their orthologs in the genome of Thamnophis elegans. The X-specific genes are expected to show log2-transformed male-to-female read coverage ratio approx. −1.00, Z-specific genes approx. 1.00, and autosomal and pseudoautosomal genes approx. 0.00. All data are available in supplementary table S2, Supplementary Material online. Phylogenetic relationships follow Pyron et al. (2013) and Zheng and Wiens (2016).
Table 1.
Total number of mapped genes, hemizygous genes, their percentage in the genomes, and estimated age of the sex chromosome systems
| Species | No. of mapped genes with known homologs in TEL and GGA | No. of hemizygous genes | Percentage of hemizygous genes | Minimal estimated age of the sex chromosome system |
|---|---|---|---|---|
| Xerotyphlops vermicularis | 10,971 | 284 | 2.59 | … |
| Tropidophis melanurus | 11,075 | 113 | 1.02 | … |
| Acrantophis sp. cf. dumerili | 11,106 | 47 | 0.42 | 64 MY |
| Eryx colubrinus | 11,092 | 34 | 0.31 | 45 MY |
| Crotalus viridis | 11,104 | 621 | 5.59 | 69 MY |
| Naja naja | 11,113 | 713 | 6.42 | 69 MY |
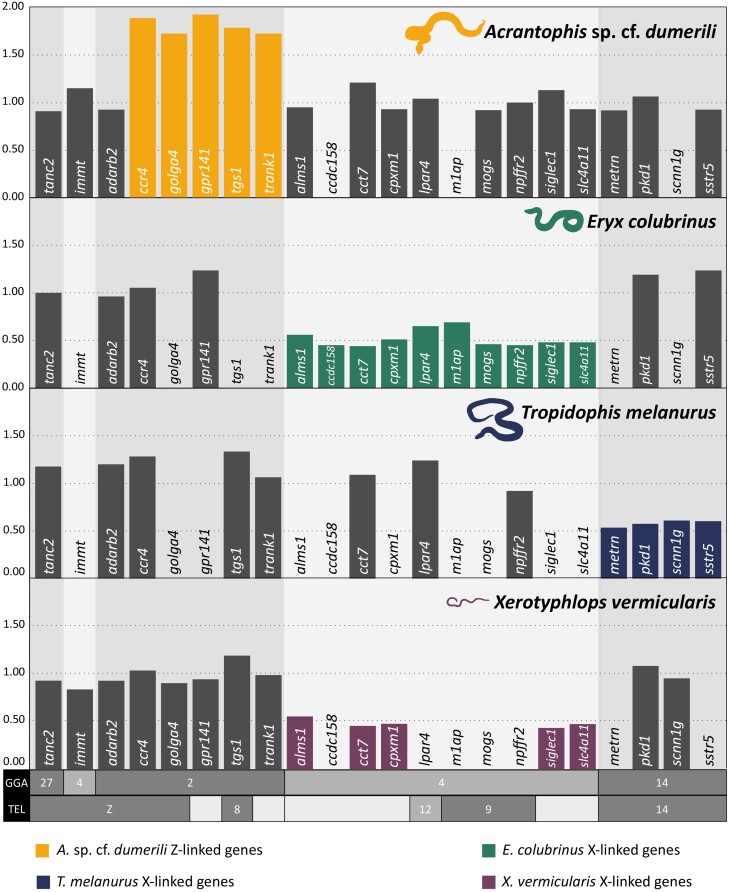
To validate the identification of sex chromosomes by the comparative gene coverage analysis, we successfully designed primers for five Z-specific candidate genes (ccr4, golga4, gpr141, tgs1, and trank1) (Fig. 2; supplementary table S3, Supplementary Material online) with known homology to GGA2. These genes showed the male-to-female gene copy ratio between 1.72 and 1.92, consistent with Z-specificity (Fig. 2; supplementary table S3, Supplementary Material online).
Fig. 2.
Normalized male-to-female gene dose ratios for 22 genes in Acrantophis sp. cf. dumerili, Eryx colubrinus, Tropidophis melanurus, and Xerotyphlops vermicularis. X-specific genes should show ratios approx. 0.5, Z-specific genes approx. 2.00, while autosomal and pseudoautosomal genes approx. 1.00. The missing bars indicate that the specific gene was not successfully amplified by qPCR. All data are available in supplementary table S3, Supplementary Material online.
Identification of X-Specific Loci by Genome Coverage Analysis in E. colubrinus
In E. colubrinus, we identified 11,092 genes with known homologs in the chicken and garter snake genomes. Only 34 genes have the male-to-female read coverage ratio between 0.3 and 0.7 expected for X-specificity (Fig. 1; Table 1; supplementary table S2, Supplementary Material online). These likely hemizygous genes were linked mainly to the chicken chromosome 4 (GGA4; 19 genes) and the garter snake chromosome 9 (TEL9; 19 genes).
We designed primers for ten candidate X-specific genes (alms1, ccdc158, cct7, cpxm1, lpar4, m1ap, mogs, npffr2, siglec1, and slc4a11), which showed the male-to-female gene copy ratio between 0.45 and 0.69, consistent with X-specificity (Fig. 2; supplementary table S3, Supplementary Material online).
Identification of X-Specific Loci by Genome Coverage Analysis in T. melanurus
A total of 11,075 genes with known homologs in the chicken and garter snake genomes were detected in T. melanurus, 113 of them with male-to-female read coverage ratio between 0.3 and 0.7 (Fig. 1; Table 1; supplementary table S2, Supplementary Material online). The homologs of these putative X-specific genes are linked mainly to the chicken chromosome 14 (GGA14; 65 genes) and the garter snake chromosome 14 (TEL14; 65 genes).
We designed primers for four candidate X-specific genes in T. melanurus with homologs linked to GGA14 (metrn, pkd1, scnn1g, and sstr5) (Fig. 2; supplementary table S3, Supplementary Material online). All four genes show the male-to-female gene copy ratio between 0.53 and 0.61 consistent with X-specificity (Fig. 2; supplementary table S3, Supplementary Material online).
Identification of X-Specific Loci by Genome Coverage Analysis in X. vermicularis
In X. vermicularis, 10,971 genes with known homologs in the chicken and garter snake genomes were found. Out of them, 284 genes have the male-to-female read coverage ratio between 0.3 and 0.7 (Fig. 1; Table 1; supplementary table S2, Supplementary Material online). The homologs of these candidate X-specific genes mapped mainly to the chicken chromosome 4 (GGA4; 168 genes) or garter snake chromosome 9 (TEL9; 167 genes).
We designed primers for five X-specific candidate genes of X. vermicularis (alms1, cct7, cpxm1, siglec1, and slc4a11) (Fig. 2; Table 1; supplementary table S3, Supplementary Material online), which showed the male-to-female gene copy ratio between 0.43 and 0.55, consistent with their X-specificity (Fig. 2; supplementary table S3, Supplementary Material online).
Identification of Z-Specific Loci by Genome Coverage Analysis in C. viridis
In this caenophidian snake, we identified homologs for 11,104 genes in the chicken and garter snake genomes. Many of them (621 genes) have male-to-female read coverage between 1.43 and 3.33, which is the expected range for Z-specificity (Fig. 1; Table 1; supplementary table S2, Supplementary Material online) as is expected for the caenophidian snakes with highly differentiated ZZ/ZW sex chromosomes.
Identification of Z-Specific Loci by Genome Coverage Analysis in N. naja
Similar results were obtained also for the second analyzed caenophidian snake. We found homologs in the chicken and garter snake genomes for 11,113 genes of N. naja, 713 of them have male-to-female read coverage between 1.43 and 3.33, as expected for Z-specificity (Fig. 1; Table 1; supplementary table S2, Supplementary Material online).
Test of Homology of Sex Chromosomes by qPCR
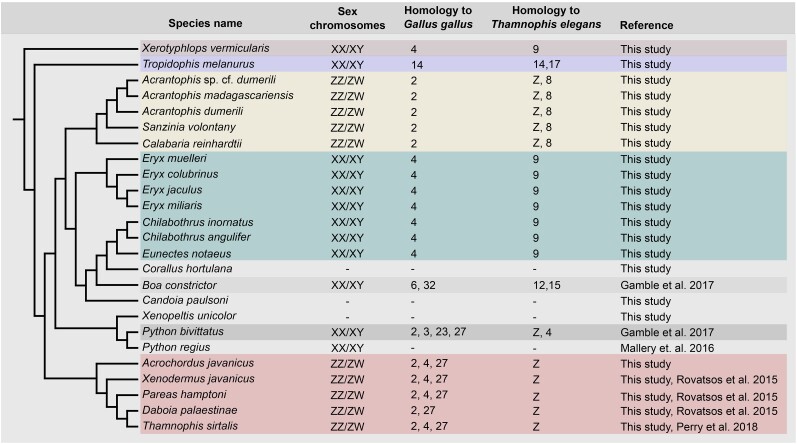
We tested Z-/X-specificity of 6 genes linked to GGA2 (adarb2, ccr4, golga4, gpr141, tgs1, and trank1), 11 genes linked to GGA4 (alms1, ccdc158, cct7, cpxm1, immt, lpar4, m1ap, mogs, npffr2, siglec1, and slc4a11), 4 genes linked to GGA14 (metrn, pkd1, scnn1g, and sstr5), and 1 to GGA27 (tanc2) across the phylogenetic spectrum of snakes (Fig. 3; supplementary table S3, Supplementary Material online). The qPCR showed that Acrantophis sp. cf. dumerili has homologous ZZ/ZW sex chromosomes with other tested species from the family Sanziniidae (A. dumerili, A. madagascariensis, and S. volontany) and with Calabaria reinhardtii (family Calabariidae). Some of the tested genes gave a pattern consistent with their Z-specificity in all five caenophidian snakes examined (Acrochordus javanicus, Daboia palaestinae, Pareas hamptoni, Thamnophis sirtalis, and Xenodermus javanicus). Furthermore, E. colubrinus has homologous XX/XY sex chromosomes with other tested species from the family Erycidae (E. mulleri, E. miliaris, and E. jaculus) and three species from the family Boidae (Eunectes notaeus, Chilabothrus angulifer, and Chilabothrus inornatus), but not with the closely related Boa constrictor and Corallus hortulana. In addition, hemizygosity of the same genes in the XX/XY sex chromosomes of E. colubrinus and of the phylogenetically distant X. vermicularis, uncovered by the comparative analyses, was further verified, as all of the X-specific genes of X. vermicularis tested by qPCR also show X-specific values in E. colubrinus. Notably, the XX/XY sex chromosomes of T. melanurus do not share X-specific genes with any other tested species. Finally, all tested genes seem to be autosomal or pseudoautosomal in the pythons Python bivittatus and Python regius, in Xenopeltis unicolor and in the boas Boa constrictor and Corallus hortulana, indicating that these species have either poorly differentiated sex chromosomes homologous to some of the other tested species or completely different sex determination systems.
Fig. 3.
Phylogenetic overview of the sex determination system and sex chromosome homology from analyzed species of snakes. Phylogenetic relationships follow Pyron et al. (2013) and Zheng and Wiens (2016). Information for sex chromosomes were collected from Gamble et al. 2017; Mallery et al. 2016; Rovatsos et al. 2015; Perry et al. 2018.
Estimated Age and Degree of Differentiation of Sex Chromosomes
The minimum age could be estimated for the ZZ/ZW sex chromosomes of caenophidian snakes as an estimated age of the split between the family Acrochordidae and other caenophidian snakes, the ZZ/ZW sex chromosomes of the families Sanziniidae and Calabaridae, and the XX/XY sex chromosomes of the genera Eryx, Chilabothrus, and Eunectes. Each of these sex chromosome systems is tens of millions of years old. However, they differ greatly in the degree of differentiation. We estimate that about 6% of the total number of genes in the genome are hemizygous in the examined caenophidian snakes, i.e. were lost from their well-known differentiated W sex chromosomes. Hundreds of genes representing around 1% and 2.5%, respectively, of the total number of mapped genes are hemizygous in the Cuban wood snake T. melanurus and the blind snake X. vermicularis. On the other hand, despite their age, we found only a few dozen of hemizygous genes (less than 0.5%) in the sand boa E. colubrinus and the Madagascar boa A. sp. cf. dumerili (Table 1).
Discussion
Unexpectedly Large Variability of Sex Chromosomes in Snakes
This study expands our knowledge of the variability of sex determination systems in snakes, an ecologically important and highly diversified group with over 4,000 recently recognized species, which have been historically used as one of the most important models for the early theories of sex chromosome evolution (Ohno 1967). We present four previously unknown snake sex chromosome systems and we identified sex chromosomes for the first time in 13 species of snakes. Now, we can recognize seven potentially distinct sex chromosome systems in snakes (Fig. 3):
Heteromorphic and highly differentiated ZZ/ZW sex chromosomes in caenophidian snakes syntenic with GGA2,4,27/TELZ (Rovatsos et al. 2015). These highly stable sex chromosomes are likely at least 69 million years old.
Poorly differentiated XX/XY sex chromosomes in the Burmese python Python bivittatus, with homologs of their X-linked genes to GGA2,3,23,27/TEL4,Z (Gamble et al. 2017).
Homomorphic (Viana et al. 2016) XX/XY sex chromosomes in the boas Boa constrictor and B. imperator including genes with homologs linked to GGA6,32/TEL12,15 (Gamble et al. 2017).
Mostly homomorphic and poorly differentiated (Augstenová et al. 2019), yet old (>64 million years) ZZ/ZW sex chromosomes in the Madagascar boas (family Sanziniidae) and the Calabar ground boa (family Calabaridae), with homologs of Z-specific genes linked to GGA2/TEL8,Z (current study).
XX/XY sex chromosomes with an extensive X-specific region in the Cuban wood snake T. melanurus, with X-specific genes homologous to those linked to GGA14/TEL14 (current study).
Homomorphic (Viana et al. 2016; Augstenová et al. 2019; Charvát et al. 2022) XX/XY sex chromosomes in the sand boas (genus Eryx), the Greater Antillean boas (genus Chilabothrus), and the anacondas (genus Eunectes), with homologs of the X-specific genes linked to GGA4/TEL9 (current study). The age of this system can be estimated to be approximately 45 million years.
Homomorphic (Mezzasalma et al. 2016) XX/XY sex chromosomes with an extensive X-specific region in the blind snake X. vermicularis, syntenic with the genomic regions GGA4/TEL9 (current study).
We exclude the reported putative ZZ/ZW sex chromosome system in the long-nosed worm snake Myriopholis macrorhyncha (family Leptotyphlopidae) from this list for now, as it is based only on the presence of one heterochromatic microchromosome, visualized by C-banding, in the karyotype of a single studied individual (Matsubara et al. 2019). Since this observation can also be explained by heterochromatin heteromorphism not linked to sex, which was, among many vertebrates, also reported in snakes (Charvát et al. 2022), we stress the need to examine more individuals of this species using cytogenetic or genomic methods in the future.
Reconstruction of the Evolution of Sex Chromosomes in Snakes
At the current stage of knowledge, it is difficult to reconstruct the phylogenetic history of sex determination in snakes in detail. As discussed below, the homology of sex chromosomes among several of the uncovered systems is difficult to identify. Moreover, the negative results of the qPCR tests in the members of the genera Xenopeltis, Candoia, and Corallus suggest that even more sex determination systems may occur in snakes. It is important to note that the applied genome coverage analysis and the qPCR comparison of gene copies between the sexes are more effective in systems with relatively differentiated sex chromosomes. Therefore, we cannot exclude the possibility that the members of the genera Xenopeltis, Candoia, and Corallus may have homologous sex chromosomes with other studied species, but with a lower degree of differentiation, below the sensitivity of the applied methods, or the tested genes are located in the pseudoautosomal regions in them. The extent of pseudoautosomal regions can be evolutionary quite plastic, as known for instance in otherwise conserved avian sex chromosomes (Zhou et al. 2014). More studies are clearly needed to identify sex chromosomes in these three and other unstudied snake lineages, particularly in additional species of blind snakes and thread snakes. Another complication for the reconstruction of the ancestral sex determination is that the synteny of sex chromosomes does not automatically mean the homology of sex determination (Kratochvíl et al. 2021). For example, the sex-determining locus was transposed to at least three different chromosomes among closely related salmonid species (Yano et al. 2013; Lubieniecki et al. 2015). The proper reconstruction of the evolutionary history of sex determination in snakes will thus require knowledge on sex-determining loci. Unfortunately, a sex-determining locus has not been identified yet in any non-avian reptile, although several putative sex-determining loci have been proposed, e.g. in anguimorphan reptiles, dragon lizards, skinks, and sphaerodactylid geckos (Deakin et al. 2016; Rovatsos et al. 2019; Pinto et al. 2023, 2024).
Having all these difficulties in mind, we think that some insights into the evolutionary history of snake sex determination can be made. Environmental sex determination has never been reported in any snake species (Valenzuela and Lance 2004), therefore, we assume that the common ancestor of all snakes had genotypic sex determination. Based on the phylogenetic distribution and known gene content (Figs. 3 and 4), it seems that XX/XY sex chromosomes evolved in snakes four or even five times. One XX/XY system evolved in the ancestor of T. melanurus. It might be an evolutionary novelty of this lineage, but more data is needed from other lineages to reconstruct the situation at the base of the snake phylogenetic tree. Two male heterogametic systems are now known in the boas: the XX/XY system of the genus Boa uncovered by Gamble et al. (2017), which might be its apomorphy, and tentatively the ancestral system of the boas present in the genera Eryx, Chilabothrus, and Eunectes. Intriguingly, the highly differentiated XX/XY sex chromosomes in the blind snake X. vermicularis include the same region as the differentiated region in the member of the genus Eryx. We cannot differentiate whether the sex chromosomes in the blind snake and the putative ancestral sex chromosomes of the boas represent independent co-option and differentiation of the same region, or whether it points that this region was part of the ancestral snake sex chromosomes still present in these lineages.
Fig. 4.
Phylogenetic overview of snakes with known sex determination and confirmed homology of their sex chromosomes. Phylogenetic relationships follow Pyron et al. (2013), Reynolds et al. (2014), and Zheng and Wiens (2016). Information for sex chromosomes were collected from Matsubara et al. 2019; Gamble et al. 2017; Rovatsos et al. 2015; Schield et al. 2019; Matsubara et al. 2006; Vicoso et al. 2013; Li et al. 2021; Singchat et al. 2020; Suryamohan et al. 2020; Perry et al. 2018.
The only other XX/XY system described in snakes was found in the python P. bivittatus. Interestingly, the XX/XY sex chromosomes of P. bivittatus seem to share a part of the same genomic region with the ZZ/ZW sex chromosomes of caenophidian snakes (Gamble et al. 2017) and of the boa families Sanziniidae and Calabariidae (Fig. 4). Do the sex chromosomes of these two families represent an ancestral situation in boas then? If yes, it would be possible that the last common ancestor of boas and the python and of caenophidian snakes had sex chromosomes homologous to the caenophidian sex chromosomes, which stayed poorly differentiated in Sanziniidae and Calabariidae and turned into XX/XY sex chromosomes in the ancestor of the python. This scenario would not be compatible with the ancestral snake XX/XY hypothesis described above, but would resemble the old hypothesis by Susumu Ohno (1967) that at least some non-caenophidian snakes share homologous, yet poorly differentiated sex chromosomes with caenophidian snakes. Alternatively, the sharing of the same genomic region by the ZZ/ZW sex chromosomes in Sanziniidae and Calabariidae, ZZ/ZW in caenophidian snakes and the XX/XY sex chromosomes in the python can reflect independent co-option of this region as sex chromosomes.
Both scenarios—i.e. the ancestral snake XX/XY sex chromosomes homologous to those of the blind snake and of some boas, and the homology of the caenophidian and boa ZZ/ZW sex chromosomes—assume that the shared genomic regions in sex chromosomes in some lineages have to occur due to their independent co-options. It was previously proposed that the gene content can “predispose some chromosomes to a specialized role in sex determination”, e.g. due to limited options of genes involved in gonad development (Marshall Graves and Peichel 2010; O’Meally et al. 2012). Such repeated co-options of the same regions were documented multiple times in other amniote lineages and certain genomic regions seem to be often nonrandomly involved in them; however, there are many unique systems among amniotes as well (Kratochvíl et al. 2021).
In any case, it is evident that sex chromosomes originating from the same region in snakes exhibited considerable divergence in their rates of differentiation. A notable example can be observed in the sex chromosomes of Caenophidia and those of the families Sanziniidae and Calabariidae, both of which can be likely traced back to the Cretaceous period (Table 1). Both these groups have female heterogametic systems, yet the degree of differentiation of their sex chromosomes varies dramatically (Fig. 1; Table 1). In a broader context, all findings allow us to conclude that sex chromosomes in snakes display a greater variability than previously assumed. The emerging pattern suggests that snakes may have ancestrally possessed sex chromosomes more susceptible to turnovers, even connected with a change in heterogamety, as known in several lineages of amphibians, teleost fishes, or plants (Myosho et al. 2015; Franchini et al. 2018; Jeffries et al. 2018; El Taher et al. 2021; Wang et al. 2022). However, once differentiation of sex chromosomes proceeded in the common ancestor of caenophidian snakes, they became highly evolutionarily stable. This scenario provides an explanation for the observed variability in some lineages, contrasted by the remarkable long-term stability of sex chromosomes in other lineages, not only within snakes but also in iguanian and scincomorphan reptiles (Rovatsos et al. 2014b; Altmanová et al. 2018; Kostmann et al. 2021; Kratochvíl et al. 2021).
Snakes as a Past and Current Model System for Sex Chromosome Evolution
In snakes, we can observe in a single lineage both fairly frequent turnovers of sex chromosomes and their astonishing stability. Over 80% of snake species belong to a group in which sex chromosomes have been documented to be highly differentiated and conserved (Rovatsos et al. 2014a, 2014b). Unlike many teleosts, such as cichlids, amphibians, and other lineages with frequent turnovers of often poorly differentiated sex chromosomes (Myosho et al. 2015; Franchini et al. 2018; Jeffries et al. 2018; El Taher et al. 2021), and unlike many diversified lineages with highly stable, differentiated sex chromosomes, such as iguanas, viviparous mammals, and birds (Marshall Graves 2016; Altmanová et al. 2018; Mazzoleni et al. 2021), snakes offer the opportunity to study both these aspects in a single lineage. Moreover, the same genes have been found to be linked to differentiated regions of sex chromosomes in different snake lineages (Fig. 4), allowing us to study the sex chromosome differentiation from the same genomic region. Like other reptiles, snakes have conserved chromosome synteny (Pinto et al. 2023); therefore, we can test convergence in differentiation (e.g. hemizygosity, divergence of gametologs, gene dose regulatory mechanisms) in sex chromosomes independently evolved from the regions with the same genes within a single lineage. In addition to the historical importance of snakes in sex chromosome research (Ohno 1967), we anticipate that the group will continue to contribute to our general knowledge of sex chromosomes and sex determination.
Supplementary Material
Acknowledgments
We would like to express our gratitude to Jana Thomayerová for technical assistance. Blood or tissue samples for this study were kindly provided by Tomáš Peš (Zoo Plzeň), Richard Viduna, Petra Škárková, Kateřina Kucírková (Zoo Jihlava), Jan Suchánek, and Jan Hříbal. Computational resources were provided by the e-INFRA CZ project (ID:90254), supported by the Ministry of Education, Youth and Sports of the Czech Republic.
Contributor Information
Tomáš Pšenička, Department of Ecology, Faculty of Science, Charles University, Prague, Czech Republic.
Barbora Augstenová, Department of Ecology, Faculty of Science, Charles University, Prague, Czech Republic.
Daniel Frynta, Department of Zoology, Faculty of Science, Charles University, Prague, Czech Republic.
Panagiotis Kornilios, Department of Biology, University of Patras, Rio, Greece.
Lukáš Kratochvíl, Department of Ecology, Faculty of Science, Charles University, Prague, Czech Republic.
Michail Rovatsos, Department of Ecology, Faculty of Science, Charles University, Prague, Czech Republic.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Author Contributions
T.P. and B.A. performed the molecular work; M.R. performed the bioinformatic analysis; M.R. and L.K. conceived the project; D.F. and P.K. offered material and consultations; T.P., B.A., L.K., and M.R. drafted the first version of the manuscript. All authors contributed to the final form of the manuscript and are responsible for its content.
Funding
This study was supported by the Czech Science Foundation (GAČR 23-07347S) and Charles University Grant Agency (GAUK 358522).
Ethics
Animal handling and collection of blood samples were performed by accredited researchers (T.P.: CZ04583, L.K.: CZ02535, MR: CZ03540). The experimental procedures were approved by the Ethics Committee of the Faculty of Science, Charles University, and the Committee for Animal Welfare of the Ministry of Agriculture of the Czech Republic (permits No. MSMT-8604/2019-7 and MSMT-22486/2023-4).
Data Availability
The Illumina DNA-seq reads from all studied individuals are submitted in the GenBank database, under the BioProject PRJNA1076544.
References
- Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011:477(7366):587–591. 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmanová M, Rovatsos M, Pokorná MJ, Veselý M, Wagner F, Kratochvíl L. All iguana families with the exception of basilisks share sex chromosomes. Zoology. 2018:126:98–102. 10.1016/j.zool.2017.11.007. [DOI] [PubMed] [Google Scholar]
- Andrade P, Pinho C, Pérez i de Lanuza G, Afonso S, Brejcha J, Rubin C-J, Wallerman O, Pereira P, Sabatino SJ, Bellati A, et al. Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc Natl Acad U S A. 2019:116(12):5633–5642. 10.1073/pnas.1820320116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data; 2010. [accessed 2024 Dec 19]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Augstenová B, Johnson Pokorná M, Altmanová M, Frynta D, Rovatsos M, Kratochvíl L. ZW, XY, and yet ZW: sex chromosome evolution in snakes even more complicated. Evolution. 2018a:72(8):1701–1707. 10.1111/evo.13543. [DOI] [PubMed] [Google Scholar]
- Augstenová B, Mazzoleni S, Kostmann A, Altmanová M, Frynta D, Kratochvíl L, Rovatsos M. Cytogenetic analysis did not reveal differentiated sex chromosomes in ten species of boas and pythons (Reptilia: Serpentes). Genes (Basel). 2019:10(11):934. 10.3390/genes10110934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augstenová B, Mazzoleni S, Kratochvíl L, Rovatsos M. Evolutionary dynamics of the W chromosome in caenophidian snakes. Genes (Basel). 2018b:9(1):5. 10.3390/genes9010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CA, Righton AL, Reeves A, Ray D, Krebs J, Louis EE Jr. Facultative parthenogenesis discovered for the first time in Jamaican boa (Chilabothrus subflavus) using novel microsatellite markers. Zoo Biol. 2024:43(5):499–504. 10.1002/zoo.21852. [DOI] [PubMed] [Google Scholar]
- Beçak W, Beçak ML. Cytotaxonomy and chromosomal evolution in Serpentes. Cytogenetics. 1969:8(4):247–262. 10.1159/000130037. [DOI] [PubMed] [Google Scholar]
- Beçak W, Beçak ML, Nazareth HRS. Karyotypic studies of two species of South American snakes (Boa constrictor amarali and Bothrops jararaca). Cytogenetics. 1962:1(6):305–313. 10.1159/000129740. [DOI] [PubMed] [Google Scholar]
- Beçak W, Beçak ML, Nazareth HRS, Ohno S. Close karyological kinship between the reptilian suborder serpentes and the class aves. Chromosoma. 1964:15(5):606–617. 10.1007/BF00319994. [DOI] [PubMed] [Google Scholar]
- Bellott DW, Page DC. Reconstructing the evolution of vertebrate sex chromosomes. Cold Spring Harb Symp Quant Biol. 2009:74:345–353. 10.1101/sqb.2009.74.048. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014:30(15):2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth W, Johnson DH, Moore S, Schal C, Vargo EL. Evidence for viable, non-clonal but fatherless Boa constrictors. Biol Lett. 2011a:7(2):253–256. 10.1098/rsbl.2010.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth W, Million L, Reynolds RG, Burghardt GM, Vargo EL, Schal C, Tzika AC, Schuett GW. Consecutive virgin births in the New World boid snake, the Colombian rainbow boa, Epicrates maurus. J Hered. 2011b:102(6):759–763. 10.1093/jhered/esr080. [DOI] [PubMed] [Google Scholar]
- Booth W, Schuett GW, Ridgway A, Buxton DW, Castoe TA, Bastone G, Bennett C, McMahan W. New insights on facultative parthenogenesis in pythons. Biol J Linn Soc. 2014:112(3):461–468. 10.1111/bij.12286. [DOI] [Google Scholar]
- Brischoux F, Shine R. Morphological adaptations to marine life in snakes. J Morphol. 2011:272(5):566–572. 10.1002/jmor.10933. [DOI] [PubMed] [Google Scholar]
- Bull JJ. Sex determination in reptiles. Q Rev Biol. 1980:55(1):3–21. 10.1086/411613. [DOI] [Google Scholar]
- Caldwell MW, Nydam RL, Palci A, Apesteguía S. The oldest known snakes from the Middle Jurassic-Lower Cretaceous provide insights on snake evolution. Nat Commun. 2015:6(1):5996. 10.1038/ncomms6996. [DOI] [PubMed] [Google Scholar]
- Chan YS, Cheung RCF, Xia L, Wong JH, Ng TB, Chan WY. Snake venom toxins: toxicity and medicinal applications. Appl Microbiol Biotechnol. 2016:100(14):6165–6181. 10.1007/s00253-016-7610-9. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. The timing of genetic degeneration of sex chromosomes. Philos Trans R Soc Lond B Biol Sci. 2021:376(1832):20200093. 10.1098/rstb.2020.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvát T, Augstenová B, Frynta D, Kratochvíl L, Rovatsos M. Cytogenetic analysis of the members of the snake genera Cylindrophis, Eryx, Python, and Tropidophis. Genes (Basel). 2022:13(7):1185. 10.3390/genes13071185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Edwards MJ, Patel H, O'Meally D, Lian J, Stenhouse R, Ryan S, Livernois AM, Azad B, Holleley CE, et al. Anchoring genome sequence to chromosomes of the central bearded dragon (Pogona vitticeps) enables reconstruction of ancestral squamate macrochromosomes and identifies sequence content of the Z chromosome. BMC Genomics. 2016:17:447. 10.1186/s12864-016-2774-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Taher A, Ronco F, Matschiner M, Salzburger W, Böhne A. Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes. Sci Adv. 2021:7(36):eabe8215. 10.1126/sciadv.abe8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016:32(19):3047–3048. 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaz T, Stiglec R, Veyrunes F, Marshall Graves JA. Relationships between vertebrate ZW and XY sex chromosome systems. Curr Biol. 2006:16(17):R736–R743. 10.1016/j.cub.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Franchini P, Jones JC, Xiong P, Kneitz S, Gompert Z, Warren WC, Walter RB, Meyer A, Schartl M. Long-term experimental hybridisation results in the evolution of a new sex chromosome in swordtail fish. Nat Commun. 2018:9(1):5136. 10.1038/s41467-018-07648-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T, Castoe TA, Nielsen SV, Banks JL, Card DC, Schield DR, Schuett GW, Booth W. The discovery of XY sex chromosomes in a boa and python. Curr Biol. 2017:27(14):2148–2153. 10.1016/j.cub.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Jeffries DL, Lavanchy G, Sermier R, Sredl MJ, Miura I, Borzée A, Barrow LN, Canestrelli D, Crochet P-A, Dufresnes C, et al. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat Commun. 2018:9(1):4088. 10.1038/s41467-018-06517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KW, Singh L. Snakes and the evolution of sex chromosomes. Trends Genet. 1985:1:55–61. 10.1016/0168-9525(85)90024-1. [DOI] [Google Scholar]
- Kinney ME, Wack RF, Grahn RA, Lyons L. Parthenogenesis in a Brazilian rainbow boa (Epicrates cenchria cenchria). Zoo Biol. 2013:32(2):172–176. 10.1002/zoo.21050. [DOI] [PubMed] [Google Scholar]
- Koh DCI, Armugam A, Jeyaseelan K. Snake venom components and their applications in biomedicine. Cell Mol Life Sci. 2006:63(24):3030–3041. 10.1007/s00018-006-6315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostmann A, Kratochvíl L, Rovatsos M. Poorly differentiated XX/XY sex chromosomes are widely shared across skink radiation. Proc Biol Sci. 2021:288(1943):20202139. 10.1098/rspb.2020.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochvíl L, Gamble T, Rovatsos M. Sex chromosome evolution among amniotes: is the origin of sex chromosomes non-random? Philos Trans R Soc Lond B Biol Sci. 2021:376(1833):20200108. 10.1098/rstb.2020.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 2017:34(7):1812–1819. 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Leal F, Cohn MJ. Developmental, genetic, and genomic insights into the evolutionary loss of limbs in snakes. Genesis. 2018:56(1):e23077. 10.1002/dvg.23077. [DOI] [PubMed] [Google Scholar]
- Li A, Wang J, Sun K, Wang S, Zhao X, Wang T, Xiong L, Xu W, Qiu L, Shang Y, et al. Two reference-quality sea snake genomes reveal their divergent evolution of adaptive traits and venom systems. Mol Biol Evol. 2021:38(11):4867–4883. 10.1093/molbev/msab212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubieniecki KP, Lin S, Cabana EI, Li J, Lai YY, Davidson WS. Genomic instability of the sex-determining locus in Atlantic salmon (Salmo salar). G3 (Bethesda). 2015:5(11):2513–2522. 10.1534/g3.115.020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery CS Jr, Carrillo MM. A case study of sex-linkage in Python regius (Serpentes: Boidae), with new insights into sex determination in Henophidia. Phyllomedusa. 2016:15(1):29–42. 10.11606/issn.2316-9079.v15i1p29-42. [DOI] [Google Scholar]
- Marshall Graves JA. Did sex chromosome turnover promote divergence of the major mammal groups?: De novo sex chromosomes and drastic rearrangements may have posed reproductive barriers between monotremes, marsupials and placental mammals. BioEssays. 2016:38(8):734–743. 10.1002/bies.201600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Graves JA, Peichel CL. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 2010:11(4):205. 10.1186/gb-2010-11-4-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Graves JA, Shetty S. Sex from W to Z: evolution of vertebrate sex chromosomes and sex determining genes. J Exp Zool. 2001:290(5):449–462. 10.1002/jez.1088. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Kumazawa Y, Ota H, Nishida C, Matsuda Y. Karyotype analysis of four blind snake species (Reptilia: Squamata: Scolecophidia) and karyotypic changes in Serpentes. Cytogenet Genome Res. 2019:157(1–2):98–106. 10.1159/000496554. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Nishida C, Matsuda Y, Kumazawa Y. Sex chromosome evolution in snakes inferred from divergence patterns of two gametologous genes and chromosome distribution of sex chromosome-linked repetitive sequences. Zool Lett. 2016:2(1):19. 10.1186/s40851-016-0056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad U S A. 2006:103(48):18190–18195. 10.1073/pnas.0605274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoleni S, Němec P, Albrecht T, Lymberakis P, Kratochvíl L, Rovatsos M. Long-term stability of sex chromosome gene content allows accurate qPCR-based molecular sexing across birds. Mol Ecol Resour. 2021:21(6):2013–2021. 10.1111/1755-0998.13381. [DOI] [PubMed] [Google Scholar]
- Mengden GA, Stock AD. Chromosomal evolution in Serpentes; a comparison of G and C chromosome banding patterns of some colubrid and boid genera. Chromosoma. 1980:79(1):53–64. 10.1007/BF00328472. [DOI] [Google Scholar]
- Mezzasalma M, Andreone F, Glaw F, Petraccioli A, Odierna G, Guarino FM. A karyological study of three typhlopid species with some inferences on chromosome evolution in blindsnakes (Scolecophidia). Zool Anz. 2016:264:34–40. 10.1016/j.jcz.2016.07.001. [DOI] [Google Scholar]
- Myosho T, Takehana Y, Hamaguchi S, Sakaizumi M. Turnover of sex chromosomes in celebensis group medaka fishes. G3 (Bethesda). 2015:5(12):2685–2691. 10.1534/g3.115.021543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguiura N, Ferrarezzi H, Batistic RF. Cytogenetics and molecular data in snakes: a phylogenetic approach. Cytogenet Genome Res. 2009:127(2–4):128–142. 10.1159/000295789. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex-linked genes. Berlin: Springer-Verlag; 1967. [Google Scholar]
- Oliveira AL, Viegas MF, da Silva SL, Soares AM, Ramos MJ, Fernandes PA. The chemistry of snake venom and its medicinal potential. Nat Rev Chem. 2022:6(7):451–469. 10.1038/s41570-022-00393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meally D, Ezaz T, Georges A, Sarre SD, Marshall Graves JA. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res. 2012:20(1):7–19. 10.1007/s10577-011-9266-8. [DOI] [PubMed] [Google Scholar]
- O’Meally D, Patel HR, Stiglec R, Sarre SD, Georges A, Marshall Graves JA, Ezaz T. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 2010:18(7):787–800. 10.1007/s10577-010-9152-9. [DOI] [PubMed] [Google Scholar]
- Pensabene E, Yurchenko A, Kratochvíl L, Rovatsos M. Madagascar leaf-tail geckos (Uroplatus spp.) share independently evolved differentiated ZZ/ZW sex chromosomes. Cells. 2023:12(2):260. 10.3390/cells12020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry BW, Card DC, McGlothlin JW, Pasquesi GIM, Adams RH, Schield DR, Hales NR, Corbin AB, Demuth JP, Hoffmann FG, et al. Molecular adaptations for sensing and securing prey and insight into amniote genome diversity from the garter snake genome. Genome Biol Evol. 2018:10(8):2110–2129. 10.1093/gbe/evy157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorná M, Kratochvíl L. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool J Linn Soc. 2009:156(1):168–183. 10.1111/j.1096-3642.2008.00481.x. [DOI] [Google Scholar]
- Pokorná M, Kratochvíl L, Kejnovský E. Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and Lacertidae: Eremias velox). BMC Genet. 2011:12:90. 10.1186/1471-2156-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto BJ, Gamble T, Smith CH, Wilson MA. A lizard is never late: squamate genomics as a recent catalyst for understanding sex chromosome and microchromosome evolution. J Hered. 2023:114(5):445–458. 10.1093/jhered/esad023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto BJ, Nielsen SV, Sullivan KA, Behere A, Keating SE, van Schingen-Khan M, Nguyen TQ, Ziegler T, Pramuk J, Wilson MA, et al. It's a trap?! Escape from an ancient, ancestral sex chromosome system and implication of Foxl2 as the putative primary sex-determining gene in a lizard (Anguimorpha; Shinisauridae). Evolution. 2024:78(2):355–363. 10.1093/evolut/qpad205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron RA, Burbrink FT, Wiens JJ. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 2013:13:93. 10.1186/1471-2148-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron RA, Reynolds RG, Burbrink FT. A taxonomic revision of boas (Serpentes: Boidae). Zootaxa. 2014:3846(2):249–260. 10.11646/zootaxa.3846.2.5. [DOI] [PubMed] [Google Scholar]
- Ray-Chaudhuri SP, Singh L, Sharma T. Evolution of sex-chromosomes and formation of W-chromatin in snakes. Chromosoma. 1971:33(3):239–251. 10.1007/BF00284942. [DOI] [PubMed] [Google Scholar]
- Reynolds RG, Niemiller ML, Revell LJ. Toward a tree-of-life for the boas and pythons: multilocus species-level phylogeny with unprecedented taxon sampling. Mol Phylogenet Evol. 2014:71:201–213. 10.1016/j.ympev.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Rovatsos M, Altmanová M, Johnson Pokorná M, Augstenová B, Kratochvíl L. Cytogenetics of the Javan file snake (Acrochordus javanicus) and the evolution of snake sex chromosomes. J Zool Syst Evol Res. 2018:56(1):117–125. 10.1111/jzs.12180. [DOI] [Google Scholar]
- Rovatsos M, Altmanová M, Pokorná M, Kratochvíl L. Conserved sex chromosomes across adaptively radiated Anolis lizards. Evolution. 2014a:68(7):2079–2085. 10.1111/evo.12357. [DOI] [PubMed] [Google Scholar]
- Rovatsos M, Kratochvíl L. Molecular sexing applicable in 4000 species of lizards and snakes? From dream to real possibility. Methods Ecol Evol. 2017:8(8):902–906. 10.1111/2041-210X.12714. [DOI] [Google Scholar]
- Rovatsos M, Pokorná M, Altmanová M, Kratochvíl L. Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol Lett. 2014b:10(3):20131093. 10.1098/rsbl.2013.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos M, Rehák I, Velenský P, Kratochvíl L. Shared ancient sex chromosomes in varanids, beaded lizards, and alligator lizards. Mol Biol Evol. 2019:36(6):1113–1120. 10.1093/molbev/msz024. [DOI] [PubMed] [Google Scholar]
- Rovatsos M, Vukić J, Lymberakis P, Kratochvíl L. Evolutionary stability of sex chromosomes in snakes. Proc Roy Soc B Biol Sci. 2015:282(1821):20151992. 10.1098/rspb.2015.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schield DR, Card DC, Hales NR, Perry BW, Pasquesi GM, Blackmon H, Adams RH, Corbin AB, Smith CF, Ramesh B, et al. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 2019:29(4):590–601. 10.1101/gr.240952.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas F, Morinha F, Luis C, Alvura N, Pires MA. DNA-validated parthenogenesis: first case in a captive Cuban boa (Chilabothrus angulifer). Salamandra. 2020:56(1):83–86. [Google Scholar]
- Shibata H, Sakata S, Hirano Y, Nitasaka E, Sakabe A. Facultative parthenogenesis validated by DNA analyses in the green anaconda (Eunectes murinus). PLoS One. 2017:12(12):e0189654. 10.1371/journal.pone.0189654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva FM, Prudente ALC, Machado FA, Santos MM, Zaher H, Hingst-Zaher E. Aquatic adaptations in a neotropical coral snake: a study of morphological convergence. J Zool Syst Evol Res. 2018:56(3):382–394. 10.1111/jzs.12202. [DOI] [Google Scholar]
- Simoes BF, Sampaio FL, Jared C, Antoniazzi MM, Loew ER, Bowmaker JK, Rodriguez A, Hart NS, Hunt DM, Partridge JC, et al. Visual system evolution and the nature of the ancestral snake. J Evol Biol. 2015:28(7):1309–1320. 10.1111/jeb.12663. [DOI] [PubMed] [Google Scholar]
- Singchat W, Ahmad SF, Sillapaprayoon S, Muangmai N, Duengkae P, Peyachoknagul S, O’Connor RE, Griffin DK, Srikulnath K. Partial amniote sex chromosomal linkage homologies shared on snake W sex chromosomes support the ancestral super-sex chromosome evolution in amniotes. Front Genet. 2020:11:948. 10.3389/fgene.2020.00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh L. Evolution of karyotypes in snakes. Chromosoma. 1972:38(2):185–236. 10.1007/BF00326193. [DOI] [PubMed] [Google Scholar]
- Singh L, Purdom IF, Jones KW. Sex chromosome associated satellite DNA: evolution and conservation. Chromosoma. 1980:79(2):137–157. 10.1007/BF01175181. [DOI] [PubMed] [Google Scholar]
- Singh L, Sharma T, Ray-Chaudhuri SP. W chromosome in the Indian water snake (checkered keel back) Natrix piscator (Colubridae). Experientia. 1968:24(1):79–80. 10.1007/BF02136807. [DOI] [PubMed] [Google Scholar]
- Suryamohan K, Krishnankutty SP, Guillory J, Jevit M, Schröder MS, Wu M, Kuriakose B, Mathew OK, Perumal RC, Koludarov I, et al. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat Genet. 2020:52(1):106–117. 10.1038/s41588-019-0559-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Freed P, Aguilar R, Hošek J. The Reptile Database; 2024. [accessed 2024 Dec 19]. http://www.reptile-database.org
- Valenzuela N, Lance VA. Temperature-dependent sex determination. In: Reptilian incubation: environment, evolution and behaviour. Nottingham: Nottingham University Press; 2004. p. 211–227. [Google Scholar]
- Viana PF, Ribeiro LB, Souza GM, Chalkidis HM, Gross MC, Feldberg E. Is the karyotype of neotropical boid snakes really conserved? Cytotaxonomy, chromosomal rearrangements and karyotype organization in the Boidae family. PLoS One. 2016:11(8):e0160274. 10.1371/journal.pone.0160274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013:11(8):e1001643. 10.1371/journal.pbio.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Li Y, Li M, Yang W, Ma X, Zhang L, Wang Y, Feng Y, Zhang Y, Zhou R, et al. Repeated turnovers keep sex chromosomes young in willows. Genome Biol. 2022:23(1):200. 10.1186/s13059-022-02769-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Hillier LW, Tomlinson C, Minx P, Kremitzki M, Graves T, Markovic C, Bouk N, Pruitt KD, Thibaud-Nissen F, et al. A new chicken genome assembly provides insight into avian genome structure. G3 (Bethesda). 2017:7(1):109–117. 10.1534/g3.116.035923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A, Nicol B, Jouanno E, Quillet E, Fostier A, Guyomard R, Guiguen Y. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol Appl. 2013:6(3):486–496. 10.1111/eva.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012:13:134. 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Wang Z-J, Li Q-Y, Lian J-M, Zhou Y, Lu B-Z, Jin L-J, Qiu P-X, Zhang P, Zhu W-B, et al. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat Commun. 2016:7(1):13107. 10.1038/ncomms13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wiens JJ. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol Phylogenet Evol. 2016:94(Pt B):537–547. 10.1016/j.ympev.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Zhang J, Bachtrog D, An N, Huang Q, Jarvis ED, Gilbert MTP, Zhang G. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science. 2014:346(6215):1246338. 10.1126/science.1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina DNA-seq reads from all studied individuals are submitted in the GenBank database, under the BioProject PRJNA1076544.