Abstract
Emerging rodent-borne hantaviruses cause severe diseases in humans with no approved vaccines or therapeutics. We recently isolated a monoclonal broadly neutralizing antibody (nAb) from a Puumala virus–experienced human donor. Here, we report its structure bound to its target, the Gn/Gc glycoprotein heterodimer comprising the viral fusion complex. The structure explains the broad activity of the nAb: It recognizes conserved Gc fusion loop sequences and the main chain of variable Gn sequences, thereby straddling the Gn/Gc heterodimer and locking it in its prefusion conformation. We show that the nAb’s accelerated dissociation from the divergent Andes virus Gn/Gc at endosomal acidic pH limits its potency against this highly lethal virus and correct this liability by engineering an optimized variant that sets a benchmark as a candidate pan-hantavirus therapeutic.
INTRODUCTION
Infections by rodent-borne hantaviruses are associated with more than 50,000 annually diagnosed cases of zoonotic disease worldwide. At least 50 distinct hantaviruses are known (1) and are classified into two clades on the basis of their sequence, geographic distribution, and natural reservoir (2). “Old World” hantaviruses (OWH), including Hantaan virus (HTNV), Dobrava-Belgrade virus, and Puumala virus (PUUV), are endemic in regions of Europe, Asia, or worldwide (Seoul virus) and cause hemorrhagic fever with renal syndrome (HFRS) with case fatality rates of up to 15% (2). The “New World” hantaviruses (NWH), including Andes virus (ANDV), Sin Nombre virus (SNV), and Choclo virus, are the major etiologic agents of hantavirus cardiopulmonary syndrome (HCPS) in the Americas. Although much less prevalent, HCPS has case fatality rates of up to 40% (3). The capacity of ANDV to spread through human-to-human contact (4, 5) and the essentially unknown but growing potential for hantavirus emergence suggest that the public health risks posed by these agents have been underestimated (6). The lack of Food and Drug Administration (FDA)–approved, European Medicines Agency-approved, or emergency use–authorized vaccines and therapeutics constitutes a critical gap in our preparedness for a large hantavirus outbreak.
Hantaviruses are enveloped viruses with a trisegmented negative-strand RNA genome that encodes three structural proteins: a polymerase, a nucleocapsid protein, and a membrane-anchored surface glycoprotein precursor that is proteolytically processed into N-terminal (Gn) and C-terminal (Gc) subunits. Gn/Gc complexes associate to form square-shaped hetero-tetrameric (Gn/Gc)4 spikes (7, 8) that mediate all steps in viral entry, including interaction with cellular attachment factors and receptors (9–11), as well as fusion of viral and endosomal membranes after virion endocytosis (12–14). Structural analyses of virus particles (10, 11, 15, 16) revealed that Gn/Gc tetramers associate laterally to form (often incomplete) lattices of spikes at the virion surface. Each tetramer is formed by four Gn subunits in the center and four Gc subunits at the periphery. The Gn ectodomain comprises two independently folded regions: the head (GnH), predicted to interact with cellular receptors, and the base (GnB), which provides tetrameric contacts stabilizing the spike. As a class-II fusion protein, Gc folds into three structured β sheet–rich domains (I, II, and III), mediates lateral contacts between spike tetramers, induces the membrane curvature required for budding of nascent virions, and catalyzes viral fusion with an endosomal membrane during entry (17). Gn interacts cotranslationally with Gc to form a metastable heterodimeric prefusion complex (15), in which the tip of domain II is partially buried by the “capping loop,” a stretch of 11 to 14 residues projecting from Gn. After virion internalization, the acidic endosomal milieu triggers conformational changes in the glycoprotein shell, initiating membrane fusion. As the prefusion complex dissociates, the tip of domain II reorganizes to form a hydrophobic surface [target membrane–interacting region (TMIR)] that inserts into the endosomal membrane, and domain III relocates to bring the transmembrane segment closer to the TMIR. These structural rearrangements lead to the formation of an energetically favored postfusion trimer while driving the merger of viral and endosomal membranes (14). Gn/Gc spikes are thus the primary target for neutralizing antibodies (nAbs) capable of blocking one or more steps in hantavirus entry.
Monoclonal nAbs isolated from convalescent individuals (18–20) and immunized animals (21, 22) hold promise as anti-hantavirus therapeutics. However, most highly potent nAbs identified to date recognize variable surface-exposed epitopes in Gn and display a narrow spectrum of activity against evolutionarily divergent hantaviruses. Conversely, Gc-specific nAbs recognize more conserved (but cryptic) epitopes and exhibit greater antiviral breadth but at the expense of potency. We recently described ADI-42898, a rare nAb isolated from a PUUV-convalescent donor that bridges this gap between breadth and potency, likely by recognizing a quaternary epitope spanning Gn and Gc (20). ADI-42898 neutralized a large panel of divergent OWHs and NWHs and protected against viral infection in a bank vole reservoir model of PUUV challenge and against infection and lethal disease in an ANDV Syrian hamster model (20). Given that ADI-42898 is a promising candidate for broad anti-hantavirus prophylaxis and therapy, understanding its molecular mechanism of action is a high priority and may inform the design of universal hantavirus vaccines.
RESULTS
X-ray structure of ADI-42898 bound to the Gn/Gc fusion complex
To uncover the structural basis of ADI-42898:Gn/Gc recognition, we crystallized a complex formed by a single-chain, variable domain fragment (scFv) of the antibody and the PUUV glycoprotein complex (table S1). We generated the latter by joining the GnH (amino acid residues 20 to 381) and the Gc ectodomain (residues 659 to 1093) with a linker as previously reported for ANDV and Maporal virus (MPRLV) (15). In the crystal structure, PUUV GnH/Gc forms a prefusion heterodimer resembling those of ANDV and MPRLV, with GnH making extensive contacts with Gc at the tip of domain II (Fig. 1, A and B). ADI-42898 simultaneously binds both glycoprotein subunits and engages three discontinuous regions: the capping loop in GnH and the bc and cd loops in Gc domain II (Fig. 1C and fig. S1). The total surface area buried by the antibody is 745 Å2, distributed in a 40:60 ratio between Gn and Gc (Fig. 1D). In line with the quaternary nature of the epitope, dissociation of the GnH/Gc complex at acidic pH abrogated binding by ADI-42898, but not by a Gc binding nAb ADI-42885 (Fig. 1E). The Gc and Gn portions of the ADI-42898 epitope are framed by the complementarity-determining regions (CDRs) H1 and H3 of the antibody’s heavy chain (HC) and by CDRs L1 and L2 of its light chain (LC), such that (i) T28H1 and S30H1 are hydrogen-bonded with side chain atoms of the Gc bc loop; (ii) R98H3 and Y99H3 interact with a cleft between the Gc bc and cd loops; and (iii) Y30L1 and Y53L2 contact main chain atoms in the Gn capping loop (Fig. 1C and fig. S2). We exchanged each of these contact residues for alanine (A) and measured the variants’ binding activities to recombinant vesicular stomatitis virus (rVSV) pseudotypes bearing PUUV or ANDV Gn/Gc using enzyme-linked immunosorbent assays (ELISA) (fig. S3). Whereas substitution of T28H1 and S30H1 had no effect on Gn/Gc engagement, exchanges of Y53L2, R98H3, and Y30L1 each substantially reduced binding, with Y30L1 having the strongest impact [reduction by about 70-fold (ANDV) and about 90-fold (PUUV)]. Exchange of Y99H3 to A rendered the antibody completely inactive. We conclude that contacts between CDR-H3 and the cd loop in Gc and between CDR-L1 and CDR-L2 and the Gn capping loop drive ADI-42898:Gn/Gc recognition.
Fig. 1. ADI-42898 binds a pH-sensitive quaternary epitope at the Gn/Gc interface.
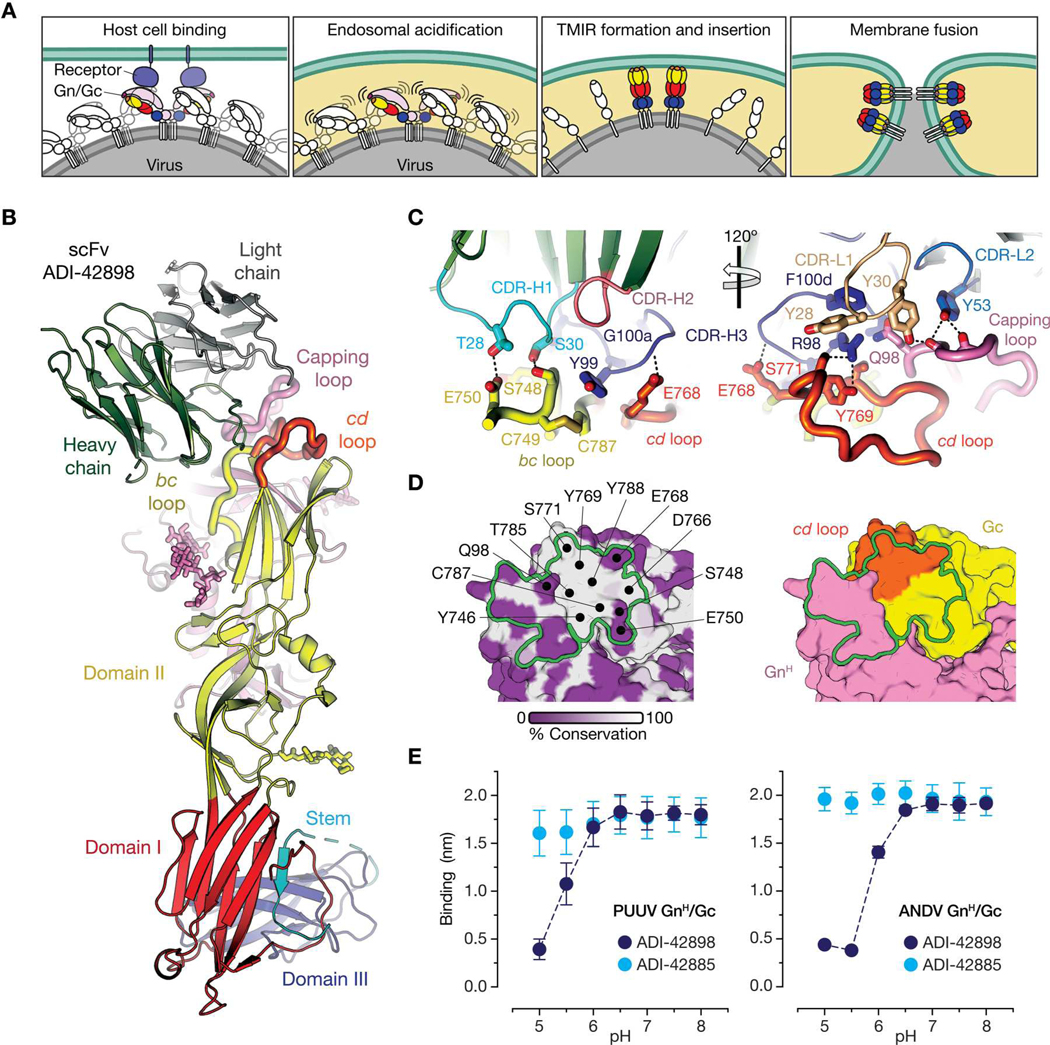
(A) Schematic mechanistic model of hantavirus membrane fusion. Gn, light pink; Gc domain I, red; Gc domain II, yellow; Gc domain III, blue. TMIR, target membrane–interacting region. (B) Ribbon representation of x-ray structure of PUUV GnH/Gc in complex with an scFv fragment of ADI-42898. ADI-42898 heavy chain, green; ADI-42898 light chain, gray; Gc domains I to III are colored as in (A). ADI-42898’s epitope is shaped by three loops: Gn capping loop, pink; Gc bc loop, yellow; Gc cd loop, orange. (C) The ADI-42898:GnH/Gc contact surface in (B) was enlarged for clarity to depict amino acid residues involved in interface formation. ADI-42898 residues are numbered according to the Kabat scheme. CDR-H and CDR-L, complementarity-determining regions of the heavy and light chains, respectively. Dashed lines indicate hydrogen bonds. (D) Surface-shaded representation of the PUUV GnH/Gc complex with the ADI-42898 contact surface indicated as a green outline. Each amino acid residue was shaded according to the degree of sequence variability among hantavirus Gn/Gc proteins at that position based on a multiple sequence alignment (white to purple, 100 to 0% sequence conservation) (left). Amino acid side chains of GnH/Gc involved in ADI-42898 binding are colored according to their domain organization (right). (E) GnH/Gc binding to biolayer interferometry (BLI) sensors coated with ADI-42898 or ADI-42885 at the indicated pH values. Data are presented as averages ± SD, n = 3 from three independent experiments.
Of the above six contact residues in the ADI-42898 paratope, only Y53L2 resulted from a somatic hypermutation (SHM) (fig. S4A). To determine whether any other SHMs contribute to ADI-42898 activity, we reverted the HC and LC chains to their inferred germline (IGL) ancestors (fig. S4) but retained the mature CDR-H3, because its germline sequence could not be predicted with confidence. We generated three germline-reverted antibodies, the IGL ancestor (HCIGL:LCIGL) and two chimeras containing the IGL of one chain and the mature [wild-type (WT)] version of the other (HCIGL:LCWT and HCWT:LCIGL), and assessed their binding properties as above (fig. S4, B, C, and E). Concordant with our findings above indicating a key role for the LC in Gn/Gc binding, only the chimera containing the mature LC (HCIGL:LCWT) and CDR-H3 retained partial binding activity. A closer examination of the structure suggests that the SHM Y49S (fig. S4D) located in the framework region 2 of the LC might be required to preorganize H3 into a conformation favorable for epitope recognition. Consistent with this hypothesis, introduction of the SHM Y49S into HCWT:LCIGL rescued ADI-42898’s binding activity (fig. S4, C to E).
To identify critical binding determinants in ADI-42898’s quaternary Gn/Gc-spanning epitope, we assessed the effects of natural sequence variations in Gn/Gc on antibody neutralization activity. Specifically, we surveyed rVSVs bearing Gn/Gc derived from eight divergent, rodent-borne hantaviruses (fig. S5). Although rVSVs bearing NWH Gn/Gc were generally less susceptible to ADI-42898 neutralization than their OWH counterparts, rVSV bearing the Laguna Negra virus (LANV) Gn/Gc was a notable exception to this trend. The enhanced susceptibility of rVSV-LANV-Gn/Gc can be tentatively attributed to residue 98Gn (Q/S in OWH and LANV, N in NWH), the only residue in the capping loop whose side chain is directly contacted by ADI-42898 (figs. S1, S2, and S5). The key role for 98Gn in susceptibility to ADI-42898 neutralization, regardless of viral clade and the length of the capping loop, is also supported by our previous observation that rVSV-PUUV-Gn/GcQ98R escapes ADI-42898 neutralization (20).
N-linked glycosylation of the Gn capping loop affords viral escape from ADI-42898 neutralization
To uncover additional Gn/Gc residues important for ADI-42898 recognition, we selected escape mutants with rVSV-ANDV- and -SNV-Gn/Gc, both of which bear an asparagine at residue 98Gn. Three mutations at distinct sites in the capping loop—A96TGn in ANDV Gn/Gc and S88NGn and T90NGn in SNV Gn/Gc—afforded resistance to ADI-42898 (fig. S6). All variations introduced an N-glycosylation motif (sequon; N-X-S/T) into the capping loop (fig. S6, A and B). rVSV-ANDV- and -SNV-Gn/Gc thus appear to “resurface” their capping loops to sterically occlude antibody recognition. To obtain experimental evidence for the acquisition of a new Gn glycosylation site by these escape mutants, we subjected virus-like particles (VLPs) bearing ANDV Gn/Gc or ANDV Gn/GcA96T to liquid chromatography–coupled tandem mass spectrometry (LC-MS/MS) (fig. S7). Consistent with our predictions, LC-MS/MS analysis of tryptic glycopeptides revealed near-full N-glycan occupancy at amino acid residue N94Gn but no detectable N-glycosylation at the same position in its WT Gn/Gc counterpart. These findings reinforce the conclusion that contacts with the polypeptide backbone of the highly variable Gn capping loop in divergent hantaviruses drive ADI-42898 recognition. Furthermore, our failure to isolate ADI-42898–mediated escape mutants located in Gc, which accounts for most of the buried surface area at the epitope-paratope interface (Fig. 1D), may reflect the viral fitness cost exacted by such mutations. In addition, N-glycosylation sequons have not been observed previously in the capping loop sequences of hantavirus strains (23, 24).
ADI-42898 blocks viral membrane fusion by locking Gn/Gc in its prefusion conformation
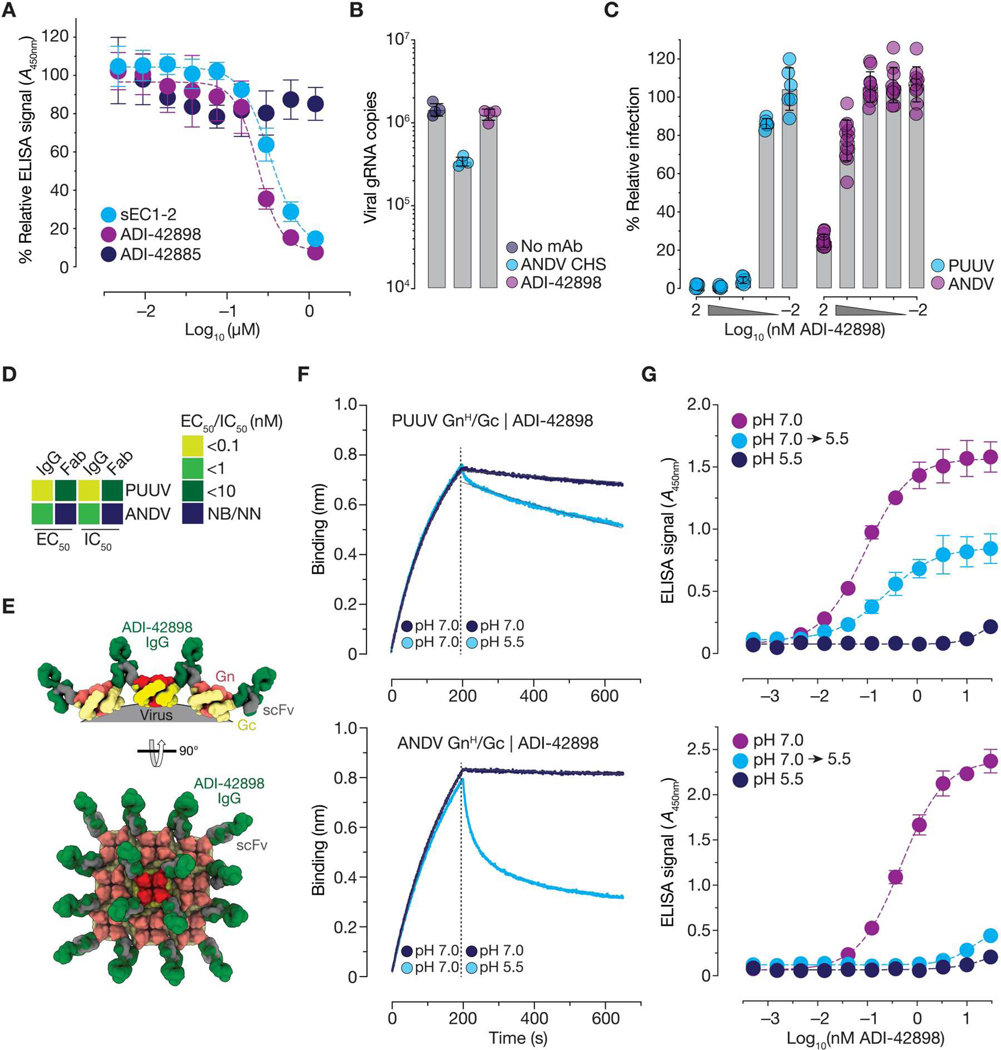
These structural studies suggested multiple non-mutually exclusive mechanisms by which ADI-42898 could block hantavirus entry. First, the apical location of its epitope in the Gn/Gc tetramer raised the possibility that ADI-42898 could affect virus-receptor binding. Consistent with this hypothesis, ADI-42898 prevented capture of rVSV-ANDV-Gn/Gc by a soluble form of the NWH clade–specific hantavirus receptor, protocadherin-1 (PCDH1) (9) in a dose-dependent manner, whereas a broadly neutralizing Gc-targeting monoclonal antibody (mAb), ADI-42885, had no effect (Fig. 2A). Despite this activity, ADI-42898 could not prevent viral attachment to PCDH1-bearing endothelial cells, likely because virions can exploit alternative attachment factors in cultured cells, as observed previously (Fig. 2B) (9). Thus, the inhibition of virus-receptor interaction cannot fully explain the potent neutralizing activity of ADI-42898, at least in cell culture.
Fig. 2. Mechanistic basis of viral entry blockade by ADI-42898.
(A) Capacity of Gn/Gc-specific mAbs to block rVSV-ANDV-Gn/Gc capture by a soluble protein comprising the first two extracellular cadherin repeats of the NWH cell surface receptor PCDH1 (sEC1–2) was measured by ELISA. Competitive inhibition by sEC1–2 was used as a positive control. Data are presented as averages ± SD, n = 4 to 6 from two or three independent experiments. (B) Capacity of ADI-42898 to block rVSV-ANDV-Gn/Gc attachment to human umbilical vein endothelial cells (HUVECs). Bound viral particles were enumerated by quantitative reverse transcription PCR (RT-qPCR) detecting rVSV genomic RNA (gRNA). CHS, convalescent hamster serum. Data are presented as averages ± SD, n = 4 from four independent experiments. (C) Capacity of ADI-42898 to inhibit PUUV or ANDV Gn/Gc–mediated fusion-infection. rVSV particles were preincubated with ADI-42898, followed by fusion-infection on HUVECs. Data are presented as averages ± SD, n = 6 to 12 from two to four independent experiments. (D) Heatmap of EC50 (from PUUV and ANDV VLP dose-response binding curves) and IC50 values (from rVSV-PUUV and -ANDV-Gn/Gc dose-response neutralization curves) derived by nonlinear regression analysis (see fig. S11). Data points are colored according to binding and neutralization potency. IgGs or Fabs with EC50 or IC50 values of >100 nM were designated as non-binding (NB) or non-neutralizing (NN). Also see data file S2 for EC50 and IC50 values. (E) Models of ADI-42898 IgG engagement of a local (Gn/Gc)4 lattice based on the PUUV Gn/GcH:ADI-42898 structure (Fig. 1) are shown in orthogonal (top) and en face (bottom) views. scFv, gray; ADI-42898, green; Gn, red; Gc, yellow. (F) BLI sensorgrams for association of PUUV (top) and ANDV (bottom) GnH/Gc to ADI-42898 at pH 7.0, followed by dissociation of GnH/Gc at pH 7.0 and 5.5 (pH shift indicated by dotted line). Data are presented as averages, n = 4 from four independent experiments are shown. (G) Capacity of ADI-42898 to bind VLPs bearing PUUV (top) and ANDV (bottom) Gn/Gc at different pHs was measured by ELISA. Where indicated, Gn/Gc:ADI-42898 complexes were shifted from pH 7.0 to pH 5.5. Data are presented as averages ± SD, n = 4 from two independent experiments.
We next considered a second mechanism of action: inhibition of the acid-induced Gn/Gc rearrangements required for viral membrane fusion through ADI-42898’s recognition and stabilization of the Gn/Gc prefusion complex (Fig. 1, A to D). To test this hypothesis, we measured the impact of ADI-42898 in a “fusion-infection” assay, in which rVSV-Gn/Gc particles were forced to infect cells by fusing their viral membranes with the plasma membranes of endothelial cells (Fig. 2C). ADI-42898 inhibited fusion-infection by rVSVs bearing both OWH and NWH Gn/Gc in a dose-dependent manner, suggesting two non-mutually exclusive mechanisms of action: (i) ADI-42898 directly blocks viral membrane fusion, or (ii) it elutes bound virions from the cell surface. We found no evidence that ADI-42898 enhanced viral elution from cells in a quantitative polymerase chain reaction (qPCR) assay for viral genomes (fig. S8). Therefore, we conclude that viral neutralization by ADI-42898 is primarily driven by its capacity to lock the Gn/Gc complex in a prefusion conformation, thereby preventing its acid-triggered dissociation, formation of the TMIR, and insertion of the TMIR into the endosomal membrane (Fig. 1A and fig. S9).
Cross-linking of neighboring Gn/Gc tetramers contributes to ADI-42898’s neutralization activity
Although ADI-42898 was broadly active against hantavirus Gn/Gc in the fusion-infection experiments, it was much more potent against PUUV than against ANDV Gn/Gc–initiated fusion (Fig. 2C), consistent with its relative neutralizing activities against viruses bearing these glycoproteins (fig. S5). We surmised that these differences arise from the reduced binding affinity of ADI-42898 for ANDV Gn/Gc. Consistent with this hypothesis, an ADI-42898 monovalent antibody-binding fragment (Fab) bound much less well to ANDV GnH/Gc and ANDV VLPs than to its PUUV counterparts in biolayer interferometry (BLI) (fig. S10) and ELISA-based assays, and it suffered greater losses in neutralizing activity against ANDV than against PUUV (Fig. 2D and fig. S11). Comparisons of Fabs with immunoglobulin Gs (IgGs) in these experiments reinforced the conclusion that bivalent binding is critical to the activity of ADI-42898 against both PUUV and ANDV, but especially against the latter. Modeling of ADI-42898 IgG molecules bound to Gn/Gc tetramers [Protein Data Bank: 6ZJM, (15)] and to lattices of tetramers [Electron Microscopy Data Bank: 11236, (15)] suggested that ADI-42898 cannot bind bivalently to a single tetramer and must instead cross-link neighboring tetramers in the virion lattice (Fig. 2E).
PUUV and ANDV Gn/Gc:ADI-42898 complexes are differentially labile to endosomal acid pH
Our above observations that ADI-42898’s reduced binding affinity for ANDV Gn/Gc could be largely overcome through IgG-dependent avidity suggested that binding differences at the neutral pH of the extracellular milieu may not alone explain ADI-42898’s reduced neutralization potency against ANDV. Accordingly, we next considered that ADI-42898:Gn/Gc immunocomplexes might be differentially labile at the acidic pH of late endosomes (pH 5.5), where hantavirus membrane fusion is triggered and where ADI-42898 presumably deploys its fusion-blocking activity. To investigate this possibility, we dipped ADI-42898–coated BLI sensors into PUUV or ANDV GnH/Gc solutions at pH 7.0 to form immunocomplexes and then monitored their dissociation at pH 5.5 (Fig. 2F). Although both PUUV and ANDV GnH/Gc dissociated more rapidly from ADI-42898 at pH 5.5 than at pH 7.0, the ANDV GnH/Gc:ADI-42898 complex was markedly less stable at acidic pH than its PUUV counterpart, with a half-life (t1/2) of 519 s versus 1083 s for ANDV and PUUV GnH/Gc, respectively (Fig. 2F), possibly as a function of its lower binding affinity for ANDV Gn/Gc. Qualitatively similar results were obtained in a distinct assay format in which we assessed the pH-dependent release of ADI-42898 from immobilized ANDV and PUUV VLPs by ELISA (Fig. 2G). Together, these findings support the following model for ADI-42898’s mechanism of action: IgGs engage PUUV and ANDV virions efficiently in the extracellular space and transit with them into late endosomes. IgGs that remain bound to Gn/Gc upon endosomal acidification affect virus neutralization by locking the viral glycoprotein spikes in their prefusion conformation. The potency of ADI-42898 against ANDV is greatly diminished relative to PUUV because a larger proportion of the antibody molecules bound to ANDV virions are released from their Gn/Gc targets in endosomes, thereby allowing more Gn/Gc spikes to undergo acid-triggered, fusion-driving conformational changes.
Affinity maturation of ADI-42898 enhances cross-clade neutralization potency by increasing immunocomplex stability at acid pH
To enhance ADI-42898’s potency against ANDV without compromising its cross-hantavirus clade breadth, we affinity-matured ADI-42898 against ANDV Gn/Gc in vitro (see Materials and Methods and fig. S12). We generated a panel of 1020 Fabs bearing single CDR mutations, screened for improved binding to ANDV GnH/Gc, and counterscreened for retention of binding to PUUV GnH/Gc. Twelve clones bearing eight distinct amino acid exchanges showed improved off-rates to ANDV GnH/Gc and improved or similar off-rates to PUUV GnH/Gc (fig. S12A). These variants were then screened in combination, yielding 26 progeny Fabs with improved and parent-like binding affinity for ANDV and PUUV GnH/Gc, respectively (fig. S12B and data file S1). Of these, two affinity-matured antibodies, ADI-65533 and ADI-65534, were prioritized because they displayed great enhancements over ADI-42898 in neutralization potency against VSVs bearing NWH Gn/Gc (ANDV) with no attendant loss in neutralization against their OWH counterparts (PUUV and HTNV) (fig. S12C). Last, because in vitro engineering can lead to polyreactivity with potential risks of off-target binding and accelerated clearance in vivo (25), we evaluated ADI-65533 and ADI-65534 in a polyreactivity assay that is predictive of serum half-life in humans (26, 27). Both mAbs lacked polyreactivity in this assay, indicating a low risk for poor pharmacokinetic behavior (fig. S12D). Accordingly, we selected these affinity-matured variants of ADI-42898 for further analysis.
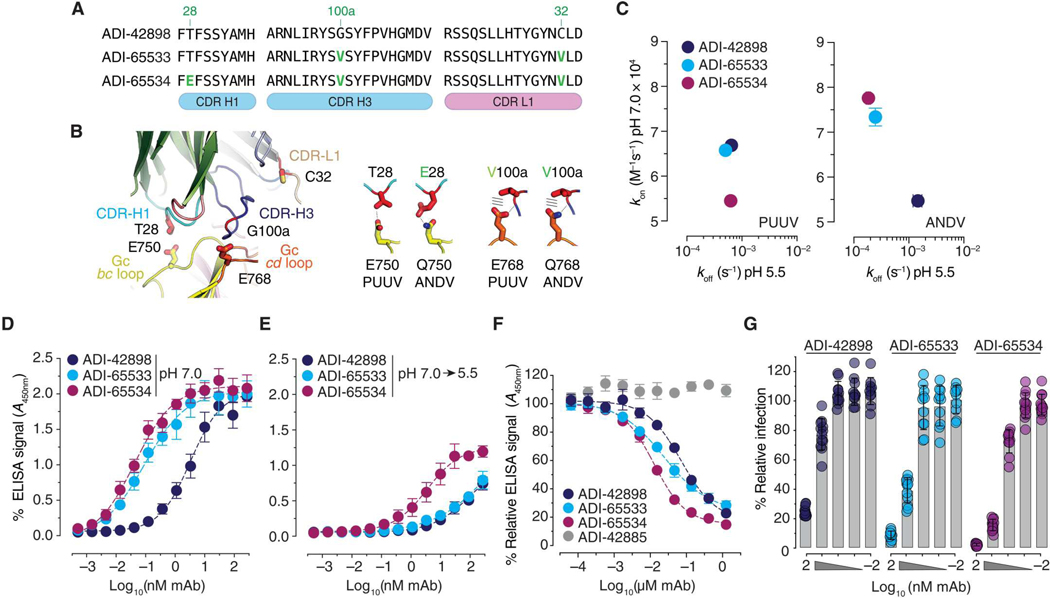
ADI-65533 and ADI-65534 share two amino acid exchanges, G100aH3V and C32L1V (Fig. 3A). The main chain of G100a forms a hydrogen bond with the side chain of E768 in PUUV Gc (Q768 in ANDV Gc). The introduction of a V at this position might allow the formation of additional van der Waals interactions with residue Q768 in ANDV Gn/Gc (represented as lines in Fig. 3B, right). Residue C32 does not contact Gn/Gc but is part of a cluster of hydrophobic amino acids that includes F100d in CDR-H3 and thus may indirectly stabilize the conformation of that loop. ADI-65534 contains one additional substitution, T28E in the CDR-H1. The side chains of T28 and E750 in PUUV Gc (Q750 in ANDV Gc) form a hydrogen bond; therefore, the mutation of T28 to the negatively charged E may afford the formation of two hydrogen bonds with Q750 and thereby strengthen ADI-65534’s interaction with ANDV Gn/Gc. However, this may come at the expense of charge repulsion between ADI-65534 (E28) and PUUV Gn/Gc (E750) (Fig. 3B, middle).
Fig. 3. Affinity-matured mAbs, ADI-65533 and ADI-65534, show improved inhibition of virus entry.
(A) Sequence alignment of regions in ADI-42898 and its affinity-matured variants containing amino acid substitutions (green indicates Kabat numbering). (B) ADI-42898:PUUV GnH/Gc interface highlighting the amino acid changes in (A) and their contact residues in Gc (left). Potential interactions between the changed amino acid in CDR-H1 (T28E) with residues in the bc loops of PUUV and ANDV Gc are shown (middle). Potential interactions between the changed amino acid in CDR-H3 (G100aV) with residues in the cd loops of PUUV and ANDV Gc are shown (right). (C) Graphical representation of association (kon at pH 7.0) and dissociation rate (koff at pH 5.5) constants for mAb interactions with PUUV and ANDV GnH/Gc. Rates were isolated from BLI sensorgrams in fig. S13; data are presented as averages ± SD, n = 4 from four independent experiments are shown. (D and E) Capacity of mAbs to bind ANDV Gn/Gc–decorated VLPs at pH 7.0 (D) or in a pH 7.0-to-pH 5.5 shift regime (E) was measured by ELISA. Data are presented as averages ± SD, n = 4 to 6 from two or three independent experiments. (F) Capacity of Gn/Gc-specific mAbs to block rVSV-ANDV-Gn/Gc capture by sEC1–2 was measured by ELISA. Data are presented as averages ± SD, n = 6 from three independent experiments. (G) Capacity of ADI-42898, ADI-65533, and ADI-65534 to inhibit ANDV Gn/Gc–mediated fusion-infection. rVSV-ANDV-Gn/Gc particles were preincubated with mAbs, followed by fusion-infection of HUVECs. Data are presented as averages ± SD, n = 10 to 12 from four independent experiments.
To investigate the impact of these amino acid substitutions on the antibodies’ mechanisms of action, we generated a kinetic map for mAb:GnH/Gc binding by plotting association (kon at pH 7.0) and dissociation (koff at pH 5.5) rates measured by BLI (Fig. 3C and fig. S13). Both affinity-matured mAbs bound better to ANDV GnH/Gc than did their ADI-42898 parent by virtue of their enhanced association at neutral pH and a 10-fold reduction in their dissociation at acidic pH (Fig. 3C, right). ADI-65533 bound to PUUV GnH/Gc as well as ADI-42898, but ADI-65534 bound less well at neutral pH, likely because of the repulsive effect of the E28/E750 pair discussed above (Fig. 3C, left).
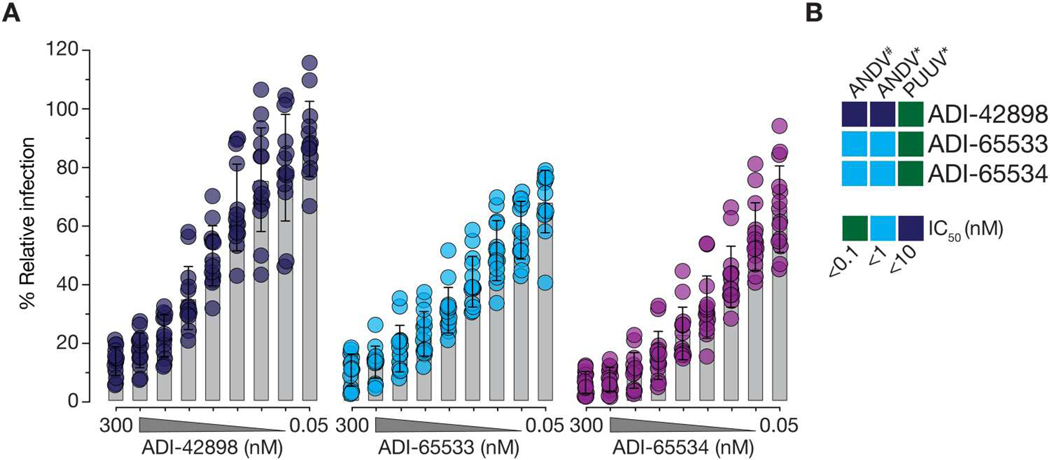
These findings were recapitulated in assays with viral particles: Both affinity-matured variants bound better to ANDV VLPs than their ADI-42898 parent in a direct-binding ELISA at neutral pH and outcompeted ADI-42898 for rVSV-ANDV-Gn/Gc binding in a competition ELISA with PCDH1 (Fig. 3, D to F). ADI-65534 also showed reduced acid pH–dependent release from immobilized ANDV VLPs (Fig. 3E). However, despite its slower dissociation from ANDV GnH/Gc in the BLI assay, ADI-65533 resembled ADI-42898 in its capacity to remain bound to ANDV VLPs at acidic pH, suggesting that additional variables associated with membrane-bound tetramers and tetramer lattices in viral particles influence the binding and dissociation of ADI-42898 and its variants. In line with its increased stability at neutral and low pH, ADI-65534 was superior to both ADI-65533 and ADI-42898 in its capacity to block ANDV Gn/Gc–mediated fusion-infection [half-maximal inhibitory concentration (IC50): 15.4 nM versus 2.3 nM for ADI-42898 and ADI-65534, respectively; Fig. 3G], whereas the affinity-matured mAbs had no advantage over ADI-42898 in blocking PUUV Gn/Gc–mediated fusion-infection (fig. S14). Concordant with their enhanced capacity to block viral membrane fusion by ANDV but not PUUV Gn/Gc, both affinity-matured mAbs selectively increased neutralization potency against authentic ANDV relative to their parent mAb (Fig. 4 and fig. S15). Together, these findings support the hypothesis that the mutations in the ADI-65534 variant increase its propensity to stay bound to and lock ANDV Gn/Gc in its prefusion conformation after viral internalization and endosomal acidification, thereby allowing for more efficient viral neutralization in endosomal compartments.
Fig. 4. Affinity-matured mAbs broadly neutralize hantaviruses.
(A) Potency of ADI-42898, ADI-65533, and ADI-65534 against ANDV infection of HUVECs. Data are presented as averages ± SD; n = 14 from seven independent experiments are shown. (B) Heatmap of IC50 values from ANDV and PUUV dose-response neutralization curves [(A) and fig. S15] derived by nonlinear regression analysis. Data points are colored according to mAb neutralization potency. Also see data file S2 for IC50 values. # Data from focus-reduction neutralization assays; *data from microneutralization assays.
ADI-65534 protects Syrian hamsters from lethal ANDV challenge when administered after the onset of viremia
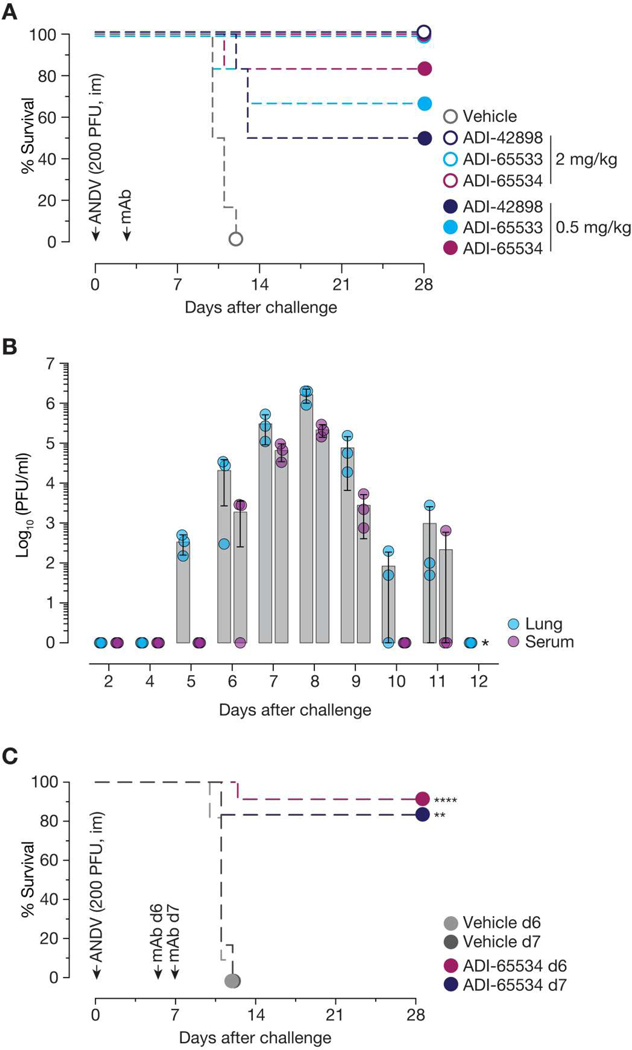
Last, we examined the capacity of ADI-42898 and both affinity-matured variants, ADI-65533 and ADI-65534, to protect Syrian hamsters against lethal ANDV challenge (Fig. 5). We previously showed that a single 6-mg/kg dose of ADI-42898 was highly protective when administered 3 days after virus challenge (20). Here, we found that all three antibodies were fully protective even at a lower 2-mg/kg dose, a testament to their cross-clade potency (Fig. 5A). However, by dosing down further, we could identify a breakthrough condition—0.5 mg/kg per animal—at which ADI-42898 provided only 50% protection but ADI-65534 afforded about 85% protection. Affinity maturation of ADI-42898 to ADI-65534 did not compromise efficacy against PUUV, because both antibodies afforded equivalent protection in a pre-exposure setting in a bank vole model of PUUV challenge (fig. S16).
Fig. 5. Affinity-matured mAbs confer post-exposure protection in Syrian hamsters.
(A) Syrian golden hamsters were challenged with ANDV [200 PFU, intramuscularly (im)], followed by treatment with a single dose of mAb [2 or 0.5 mg/kg, intraperitoneally (ip)] 3 days after virus exposure. Mortality of hamsters was monitored for 28 days. Averages from one experiment, n = 6 per group. (B) In a time course study, Syrian golden hamsters were challenged with ANDV (200 PFU, im). At the indicated days after virus challenge, animals were euthanized, and virus titers in serum samples and lung tissues were assessed by plaque assay. Averages from one experiment are shown, n = 3 per group. * indicates that virus titers could not be determined. (C) Syrian golden hamsters were challenged with ANDV (200 PFU, im), followed by treatment with a single dose of mAb (25 mg/kg, ip) 6 or 7 days after virus challenge, respectively. Averages are shown from two experiments (treatment on day 6), n = 9 (mAb) and n = 11 (vehicle) or from one experiment (treatment on day 7), n = 6 (mAb) and n = 6 (vehicle). Comparisons are shown between untreated and mAb-treated animals and were analyzed by Mantel-Cox test: **P = 0.0021; ****P < 0.0001.
A limitation of the preceding mAb protection experiments in the Syrian hamster model of ANDV challenge—one shared by nearly all such previously published studies—is that most animals were likely not yet viremic at the time of mAb treatment (3 days after virus challenge). To evaluate the protective efficacy of our lead mAb, ADI-65534, when administered after the onset of viral replication, we first performed a longitudinal study to establish the time course of ANDV replication in vivo. We inoculated hamsters intramuscularly with 200 plaque-forming units (PFU) of ANDV and measured viral titers in lung tissue and serum daily (Fig. 5B). Viral infectivity was first consistently detectable on day 5 in lung tissue (day 6 in serum) and peaked at about 106 and about 105 PFU/ml on day 8 after challenge in lung tissue and serum, respectively, with a sharp decline thereafter. Substantial viral replication could not be measured on day 3 after challenge. In a follow-up study, we treated hamsters with a single 25-mg/kg dose of ADI-65534 on day 6 or 7 after ANDV exposure, when viral replication was approaching its peak (Fig. 5, B and C). Although all animals in the vehicle group succumbed to viral challenge, ADI-65534 protected more than 80% of the hamsters treated on day 6 or 7 (Fig. 5C).
DISCUSSION
In sum, we have determined the structure of a unique hantavirus cross-clade nAb from a PUUV-experienced human donor, ADI-42898, bound to its quaternary epitope in the prefusion conformation of the viral spike complex. This structure explains the broad cross-clade neutralizing activity of ADI-42898, despite its recognition of a highly variable loop sequence in Gn. Mechanistic studies showed that ADI-42898 blocks viral membrane fusion by stapling together the spike’s Gn and Gc subunits, which must rearrange for Gc to insert into the target membrane at acidic pH. These experiments also uncovered a key property of ADI-42898 that limits its activity against ANDV: its rapid dissociation from the viral spike at the acidic pH of late endosomes where membrane fusion takes place. Correcting this liability through high-throughput antibody engineering yielded an affinity-optimized antibody variant, ADI-65534, with enhanced cross-clade neutralization potency. ADI-65534 demonstrated substantial in vivo efficacy, including in stringent post-exposure protection studies. This antibody is a promising candidate for prophylaxis and therapy of HFRS and HCPS and represents an important step toward preparedness for future hantavirus outbreaks caused by known and novel hantaviruses.
Other recent work has identified a human nAb, SNV-53, elicited by infection with an NWH (SNV), which selectively targets Gn/Gc complexes but not either subunit alone (28). Genetic and low-resolution structural evidence are consistent with the hypothesis that the nAb SNV-53 recognizes an antigenic site resembling that of ADI-42898 in comprising the GnH capping loop and the adjacent Gc fusion loops. These two hantavirus-specific nAbs also share functional properties, including their primary mechanism of action (membrane fusion blockade) and their requirement for bivalent binding to effect potent neutralization. Despite these similarities, however, ADI-42898 and SNV-53 appear to differ in their relative angle of approach to their epitopes. Specifically, SNV-53 engages its epitope from a steeper angle relative to the central axis of the spike than does ADI-42898, projecting its constant regions laterally and causing them to clash with adjacent Gn/Gc spikes in the higher-order surface lattice on viral particles (28). SNV-53’s lateral angle of attack, which resembles that of previously described Gc-targeting antibodies (23), raises the possibility that it can only engage lattice-free sites on virions, whereas ADI-42898’s more vertical angle of attack may allow it to bind Gn/Gc tetramers in both continuous and incomplete lattices. Although a high-resolution structure of an SNV-53:Gn/Gc complex is not yet available, current evidence also suggests that ADI-42898 contacts sequences in Gc with overall higher sequence conservation across hantaviruses than does SNV-53 (18, 28).
The mode of viral recognition used by ADI-42898 and SNV-53 places them in a class of nAbs elicited by divergent viruses bearing class-II membrane fusion glycoproteins, including phleboviruses and alphaviruses, which recognize a conserved interface spanning the membrane-interacting fusion loops of the fusion subunit and sequences in its companion protein that partially shield the fusion loops and regulate their exposure during viral entry (29, 30). Consequently, these nAbs appear to block infection primarily by preventing dissociation of the intersubunit interface and membrane insertion of the fusion loops triggered by endosomal acidic pH (with secondary effects on virus-receptor recognition or viral attachment, in some cases). Implicit in this mechanism is the requirement that antibody molecules stay bound to virions during viral internalization and endocytic trafficking for long enough and at high enough occupancies to prevent viral membrane fusion. We show that the potency of ADI-42898 against viruses bearing Gn/Gc from divergent hantaviruses is inversely correlated with the rates at which the respective ADI-42898:Gn/Gc complexes dissociate at endosomal acidic pH. Concordantly, the enhanced neutralization potency of affinity-matured ADI-42898 variants was at least in part associated with reduced immunocomplex dissociation at acidic pH. This principle may apply not only to other class-II glycoprotein-binding antibodies of the same type as ADI-42898 but also to more divergent classes of endosomal fusion-blocking nAbs. As a case in point, the most potent and broad Ebola virus–specific nAbs remain bound to their glycoprotein targets at endosomal acidic pH and after cleavage by endosomal proteases, whereas nAbs with lower neutralization potencies generally do not (31, 32). The acid-dependent stability of nAb:glycoprotein complexes may provide one useful metric to assess the potency of nAbs that block one or more endosomal steps in viral entry.
Neutralization escape from ADI-42898 in a VSV-based assay occurred through the acquisition of an N-linked glycan at different locations in the GnH capping loop masking this antigenic site, as also observed for SNV-53 (28). Such sequons in the capping loop have not been reported previously for rodent-borne hantaviruses (13), raising the possibility that they are not compatible with the formation of the highly interconnected (Gn/Gc)4 surface lattice thought to be required for the budding of hantavirus particles (as opposed to VSVs bearing hantavirus Gn/Gc, which encode their own budding machinery) (7). If true, then this would suggest that nAbs of the ADI-42898 class may be relatively resistant to neutralization escape by authentic hantaviruses. This property, together with the highly conserved nature of their antigenic site among hantaviruses, suggests that elicitation of ADI-42898–like antibodies could form the basis of a broadly protective vaccine strategy. However, such antibodies appear to arise only rarely as part of antibody responses to natural infection (providing an alternative explanation for the paucity of sequons observed in the GnH capping loop in available viral sequences). Further, DNA immunization with hantavirus M segments encoding for Gn/Gc elicits a largely virus- and clade-specific response (33, 34). Thus, the engineering of glycoprotein variants to stabilize the Gn/Gc interface in its prefusion conformation and to mask other non-interfacial Gn epitopes might be necessary to focus humoral recognition onto this class of quaternary epitopes. Similar structure-based design approaches stabilizing inherently metastable but highly immunogenic viral surface proteins yielded the generation of FDA-approved respiratory syncytial virus and severe acute respiratory syndrome coronavirus 2 vaccines (35, 36) as well as a vaccine candidate for dengue virus (37).
Specific and effective treatment for patients with HCPS and HFRS remains challenging. Early disease symptoms are largely nonspecific, resulting in delayed diagnosis and hospital admission of patients presenting with severe late-phase disease symptoms and high viremia. For patients with HCPS in particular, a sudden progression from early symptoms to shock and respiratory and cardiac failure has been reported, leaving only a short window for medical intervention (38). This long prodrome complicates attempts to extrapolate the results of antibody protection studies in the acute and highly lethal ANDV challenge model in Syrian hamsters to protective efficacy in humans. This is especially the case for nearly all of the studies reported to date, in which doses of antibody were administered no later than 3 days after infection (18, 19, 21, 28). In a single in vivo study, Syrian hamsters were administered two doses of an antibody cocktail at days 8 and 10 after a lethal intranasal ANDV challenge; this high-antibody dose treatment at a late stage of disease resulted in survival of only half of the animals (39). To evaluate ADI-65534 under a similarly stringent post-exposure treatment regime, we first performed a longitudinal study in ANDV-challenged Syrian hamsters. The earliest time point at which we could detect viral infection in lung tissue was day 5 after virus challenge. The capacity of the affinity-matured lead antibody, ADI-65534, to protect in this model when administered as a single dose at day 6 or 7, close to the peak of viral infection on day 8, thus advances a promising pan-hantavirus candidate therapeutic and represents an important step toward the development of a more meaningful animal model to evaluate hantavirus treatments.
One limitation of this study and others on therapeutic counter-measures against hantaviruses is the incomplete understanding of how protective efficacy in the Syrian hamster model translates to humans given the very different kinetic trajectories of disease observed in the two species. By substantially extending the therapeutic window for a single dose of a nAb in Syrian hamsters, we hope to inform the design of clinical trials aimed at evaluating mAbs in viremic human patients with HCPS and HFRS. Further, it remains unclear whether mAb monotherapy for hantavirus disease risks eliciting viral neutralization escape in vivo. Although more work is needed, it is conceivable that the nature of the intersubunit antigenic site targeted by ADI-65534 reduces the risk of escape in the context of authentic hantavirus particles.
MATERIALS AND METHODS
Study design
We initiated this study to determine how a protective pan-hantavirus human mAb, ADI-42898, engages a quaternary epitope in the viral surface glycoprotein inhibiting viral membrane fusion and infection. We crystallized a complex formed by an scFv of ADI-42898 bound to the PUUV glycoprotein complex Gn/Gc and identified critical binding determinants constituting the Gn/Gc:mAb interface. We performed mechanistic studies to evaluate ADI-42898’s cross-clade binding and membrane fusion inhibition activity. Through high-throughput antibody engineering, we yielded antibody variants with optimized glycoprotein affinity and evaluated their protective efficacy in Syrian hamster ANDV lethal challenge models for HCPS.
Syrian golden hamster (Mesocricetus auratus) animal experiments were conducted under Institutional Animal Care and Use Committee–approved protocols in compliance with the Animal Welfare Act, Public Health Service Policy, and other applicable federal statutes and regulations relating to animals and experiments involving animals. The animal biosafety level–4 (BSL-4) facility where these studies were conducted [U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID)] is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council. Further, all studies were performed in accordance with institutional policies for biosafety and biosecurity. Groups of six 5- to 10-month-old female animals were challenged per condition to provide greater than 80% power in a one-tailed Fisher’s exact test. This sample size would allow the experimenter to detect a minimum efficacy rate of 88% (five of six protected) in the treated group compared with 0% (zero of six not protected) in the control group at a 95% confidence level. Data randomization and blinding were not performed. Several hamsters designated as untreated were excluded from the dataset (see the “Syrian hamster challenge studies” section in the Supplementary Materials).
Statistical analysis
Statistical details for each experiment can be found in the respective figure legends. These include the number of replicates (n), measures of precision, and the statistical test used. Dose-response neutralization curves were subjected to nonlinear regression analysis to derive IC50 values (four-parameter, variable slope sigmoidal dose-response equation constraining bottom parameters to 0 to 100). Dose-response binding curves were subjected to nonlinear regression analysis to derive EC50 values (variable slope sigmoidal dose-response equation). Survival curves from the hamster challenge study (Fig. 5C) were compared by a Mantel-Cox (log-rank) test. Viral RNA load in animal tissue (fig. S16) was analyzed using a Kruskal-Wallis test with Dunn’s correction for multiple comparisons. All statistical analyses were carried out in GraphPad Prism.
Supplementary Material
Acknowledgments:
We thank I. Gutierrez, E. Valencia (Albert Einstein College of Medicine), and J. Wigren-Byström (Umeå University) for laboratory management and technical support. We acknowledge A. Haouz and the staff of the Protein Crystallogenesis Facility (Institut Pasteur) for help with crystallization trials and thank the staff of the PX1 and PX2 beamlines (Synchrotron SOLEIL) for beamline support. We thank S. Maki for virus cultivation and O. Vapalahti and the Viral Zoonosis Research Unit for help with BSL-3 animal work (Departments of Virology and Veterinary Biosciences, University of Helsinki). We appreciate the help of K. Cogliano for project management and administrative support; we also thank M. Loomis and S. Khanal for technical assistance with authentic hantavirus assays (USAMRIID). We thank M. Vasquez, E. Krauland, and J. Nett for helpful comments on the manuscript; Adimab’s High-Throughput Expression group for antibody sequencing, expression, and purification; Adimab’s Protein Analytics group for performing BLI characterizations of the affinity-matured progeny; and M. Brown for kinetics data analyses. We thank members of all of our groups and the Prometheus consortium for feedback on preliminary versions of the manuscript.
Funding:
This research was supported by NIAID of the NIH under award number U19AI142777 (Centers of Excellence in Translational Research) to K.C., L.M.W., J.M.D., A.S.H., S.B.B., M.N.E.F., and L.Z. M.N.E.F. was supported by a grant from the Swedish Research Council (no. 2020–06235). C.A. was supported by research grants from Region Västerbotten and Umeå University (RV-579011, RV-734361, and RV-965866). J.T. was supported by the Finnish Cultural Foundation (no. 00221066). R.K.J. was supported by grants from the NIH (R2AI156482, P20GM134974, and P20GM121307). R.G.U. was supported by the Bundesministerium für Bildung und Forschung (BMBF) within the Research Network Zoonotic Infectious Diseases (01KI1721A and 01KI2004A). T.S. was supported by the Academy of Finland (no. 321809). P.G.-C. was supported by a grant by the National French Research Agency (ANR-18-CE11–0011), and A.S. was supported with a fellowship by the French Fondation pour la Recherche Médicale (FDM20170638040). F.A.R. and P.G.-C. were supported by Labex IBEID (ANR-10-LABX-62-IBEID). The content is solely the responsibility of the authors and does not necessarily represent the official views of our institutions or funders. Opinions, conclusions, interpretations, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Department of the Army, the U.S. Department of Defense, or the U.S. Department of Health and Human Services.
Footnotes
Competing interests: K.C. is a member of the scientific advisory board of Integrum Scientific, LLC and holds equity in Eitr Biologics Inc. A.S.H. is a scientific advisor to and holds equity in Integrum Scientific, LLC. E.S.E., C.G.R., J.C.G., A.Z.W., and L.M.W. are current or former employees of Adimab LLC and may hold shares in Adimab LLC. L.M.W., A.Z.W., P.G.-C., F.A.R., E.M., K.C., S.B.B., D.M.A., C.A., M.N.E.F., A.S.H., and J.M.D. are listed as inventors on a provisional patent application entitled “Antibodies against Orthohantaviruses” (application no. 63/490,215).
Data and materials availability:
All data associated with this study are present in the paper or the Supplementary Materials. The coordinate files of the structures described in this manuscript have been submitted to the Protein Data Bank with ID 7QQB. The LC-MS/MS data described in this manuscript have been submitted to Chorus with ID 1815. Materials are available through material transfer agreement by contacting K.C. or P.G.-C.
REFERENCES AND NOTES
- 1.Laenen L, Vergote V, Calisher CH, Klempa B, Klingström J, Kuhn JH, Maes P, Hantaviridae: Current classification and future perspectives. Viruses 11, 788 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jonsson CB, Figueiredo LTM, Vapalahti O, A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev 23, 412–441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson DC, Sargianou M, Papa A, Chra P, Starakis I, Panos G, Epidemiology of Hantavirus infections in humans: A comprehensive, global overview. Crit. Rev. Microbiol 40, 261–272 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Martínez VP, Di Paola N, Alonso DO, Pérez-Sautu U, Bellomo CM, Iglesias AA, Coelho RM, López B, Periolo N, Larson PA, Nagle ER, Chitty JA, Pratt CB, Díaz J, Cisterna D, Campos J, Sharma H, Dighero-Kemp B, Biondo E, Lewis L, Anselmo C, Olivera CP, Pontoriero F, Lavarra E, Kuhn JH, Strella T, Edelstein A, Burgos MI, Kaler M, Rubinstein A, Kugelman JR, Sanchez-Lockhart M, Perandones C, Palacios G, “Super-Spreaders” and Person-to-Person Transmission of Andes Virus in Argentina. N. Engl. J. Med 383, 2230–2241 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Valdebenito C, Calvo M, Vial C, Mansilla R, Marco C, Palma RE, Vial PA, Valdivieso F, Mertz G, Ferrés M, Person-to-person household and nosocomial transmission of andes hantavirus, Southern Chile, 2011. Emerg. Infect. Dis 20, 1629–1636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klempa B, Hantaviruses and climate change. Clin. Microbiol. Infect 15, 518–523 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Huiskonen JT, Hepojoki J, Laurinmäki P, Vaheri A, Lankinen H, Butcher SJ, Grünewald K, Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol 84, 4889–4897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittler E, Dieterle ME, Kleinfelter LM, Slough MM, Chandran K, Jangra RK, Hantavirus entry: Perspectives and recent advances. Adv. Virus Res 104, 185–224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jangra RK, Herbert AS, Li R, Jae LT, Kleinfelter LM, Slough MM, Barker SL, Guardado-Calvo P, Román-Sosa G, Dieterle ME, Kuehne AI, Muena NA, Wirchnianski AS, Nyakatura EK, Fels JM, Ng M, Mittler E, Pan J, Bharrhan S, Wec AZ, Lai JR, Sidhu SS, Tischler ND, Rey FA, Moffat J, Brummelkamp TR, Wang Z, Dye JM, Chandran K, Protocadherin-1 is essential for cell entry by New World hantaviruses. Nature 563, 559–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Rissanen I, Zeltina A, Hepojoki J, Raghwani J, Harlos K, Pybus OG, Huiskonen JT, Bowden TA, A molecular-level account of the antigenic hantaviral surface. Cell Rep. 15, 959–967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rissanen I, Stass R, Zeltina A, Li S, Hepojoki J, Harlos K, Gilbert RJC, Huiskonen JT, Bowden TA, Structural transitions of the conserved and metastable hantaviral glycoprotein envelope. J. Virol 91, e00378–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acuña R, Bignon EA, Mancini R, Lozach P-Y, Tischler ND, Acidification triggers Andes hantavirus membrane fusion and rearrangement of Gc into a stable post-fusion homotrimer. J. Gen. Virol 96, 3192–3197 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Guardado-Calvo P, Rey FA, The surface glycoproteins of hantaviruses. Curr. Opin. Virol 50, 87–94 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Guardado-Calvo P, Bignon EA, Stettner E, Jeffers SA, Pérez-Vargas J, Pehau-Arnaudet G, Tortorici MA, Jestin J-L, England P, Tischler ND, Rey FA, Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc. PLOS Pathog. 12, e1005813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serris A, Stass R, Bignon EA, Muena NA, Manuguerra J-C, Jangra RK, Li S, Chandran K, Tischler ND, Huiskonen JT, Rey FA, Guardado-Calvo P, The hantavirus surface glycoprotein lattice and its fusion control mechanism. Cell 183, 442–456.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willensky S, Bar-Rogovsky H, Bignon EA, Tischler ND, Modis Y, Dessau M, Crystal Structure of Glycoprotein C from a Hantavirus in the Post-fusion Conformation. PLOS Pathog. 12, e1005948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guardado-Calvo P, Rey FA, The envelope proteins of the bunyavirales. Adv. Virus Res 98, 83–118 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Engdahl TB, Kuzmina NA, Ronk AJ, Mire CE, Hyde MA, Kose N, Josleyn MD, Sutton RE, Mehta A, Wolters RM, Lloyd NM, Valdivieso FR, Ksiazek TG, Hooper JW, Bukreyev A, Crowe JE Jr., Broad and potently neutralizing monoclonal antibodies isolated from human survivors of New World hantavirus infection. Cell Rep. 35, 109086 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrido JL, Prescott J, Calvo M, Bravo F, Alvarez R, Salas A, Riquelme R, Rioseco ML, Williamson BN, Haddock E, Feldmann H, Barria MI, Two recombinant human monoclonal antibodies that protect against lethal Andes hantavirus infection in vivo. Sci. Transl. Med 10, eaat6420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mittler E, Wec AZ, Tynell J, Guardado-Calvo P, Wigren-Byström J, Polanco LC, O’Brien CM, Slough MM, Abelson DM, Serris A, Sakharkar M, Pehau-Arnaudet G, Bakken RR, Geoghegan JC, Jangra RK, Keller M, Zeitlin L, Vapalahti O, Ulrich RG, Bornholdt ZA, Ahlm C, Rey FA, Dye JM, Bradfute SB, Strandin T, Herbert AS, Forsell MNE, Walker LM, Chandran K, Human antibody recognizing a quaternary epitope in the Puumala virus glycoprotein provides broad protection against orthohantaviruses. Sci. Transl. Med 14, eabl5399 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duehr J, McMahon M, Williamson B, Amanat F, Durbin A, Hawman DW, Noack D, Uhl S, Tan GS, Feldmann H, Krammer F, Neutralizing Monoclonal Antibodies against the Gn and the Gc of the Andes Virus Glycoprotein Spike Complex Protect from Virus Challenge in a Preclinical Hamster Model. mBio 11, e00028–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rissanen I, Krumm SA, Stass R, Whitaker A, Voss JE, Bruce EA, Rothenberger S, Kunz S, Burton DR, Huiskonen JT, Botten JW, Bowden TA, Doores KJ, Structural basis for a neutralizing antibody response elicited by a recombinant hantaan virus Gn immunogen. mBio 12, e0253120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rissanen I, Stass R, Krumm SA, Seow J, Hulswit RJ, Paesen GC, Hepojoki J, Vapalahti O, Lundkvist Å, Reynard O, Volchkov V, Doores KJ, Huiskonen JT, Bowden TA, Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein. eLife 9, e58242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bignon EA, Albornoz A, Guardado-Calvo P, Rey FA, Tischler ND, Molecular organization and dynamics of the fusion protein Gc at the hantavirus surface. eLife 8, e46028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sievers SA, Scharf L, West AP Jr., P. J. Bjorkman, Antibody engineering for increased potency, breadth and half-life. Curr. Opin. HIV AIDS 10, 151–159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu T-Y, Torrey J, Thomas J, Bobrowicz P, Vásquez M, Wittrup KD, Krauland E, Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: A FACS-based, high-throughput selection and analytical tool. Protein Eng. Des. Sel 26, 663–670 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, Cao Y, Lynaugh H, Brown M, Baruah H, Gray LT, Krauland EM, Xu Y, Vásquez M, Wittrup KD, Biophysical properties of the clinical-stage antibody landscape. Proc. Natl. Acad. Sci. U.S.A 114, 944–949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engdahl TB, Binshtein E, Brocato RL, Kuzmina NA, Principe LM, Kwilas SA, Kim RK, Chapman NS, Porter MS, Guardado-Calvo P, Rey FA, Handal LS, Diaz SM, Zagol-Ikapitte IA, Tran MH, McDonald WH, Meiler J, Reidy JX, Trivette A, Bukreyev A, Hooper JW, Crowe JE, Antigenic mapping and functional characterization of human New World hantavirus neutralizing antibodies. eLife 12, e81743 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman NS, Zhao H, Kose N, Westover JB, Kalveram B, Bombardi R, Rodriguez J, Sutton R, Genualdi J, LaBeaud AD, Mutuku FM, Pittman PR, Freiberg AN, Gowen BB, Fremont DH, Crowe JE Jr., Potent neutralization of Rift Valley fever virus by human monoclonal antibodies through fusion inhibition. Proc. Natl. Acad. Sci. U.S.A 118, e2025642118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou QF, Fox JM, Earnest JT, Ng T-S, Kim AS, Fibriansah G, Kostyuchenko VA, Shi J, Shu B, Diamond MS, Lok S-M, Structural basis of Chikungunya virus inhibition by monoclonal antibodies. Proc. Natl. Acad. Sci. U.S.A 117, 27637–27645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilchuk P, Kuzmina N, Ilinykh PA, Huang K, Gunn BM, Bryan A, Davidson E, Doranz BJ, Turner HL, Fusco ML, Bramble MS, Hoff NA, Binshtein E, Kose N, Flyak AI, Flinko R, Orlandi C, Carnahan R, Parrish EH, Sevy AM, Bombardi RG, Singh PK, Mukadi P, Muyembe-Tamfum JJ, Ohi MD, Saphire EO, Lewis GK, Alter G, Ward AB, Rimoin AW, Bukreyev A, Crowe JE Jr., Multifunctional Pan-ebolavirus antibody recognizes a site of broad vulnerability on the ebolavirus glycoprotein. Immunity 49, 363–374.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wec AZ, Herbert AS, Murin CD, Nyakatura EK, Abelson DM, Fels JM, He S, James RM, de La Vega M-A, Zhu W, Bakken RR, Goodwin E, Turner HL, Jangra RK, Zeitlin L, Qiu X, Lai JR, Walker LM, Ward AB, Dye JM, Chandran K, Bornholdt ZA, Antibodies from a human survivor define sites of vulnerability for broad protection against Ebolaviruses. Cell 169, 878–890.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brocato RL, Josleyn MJ, Wahl-Jensen V, Schmaljohn CS, Hooper JW, Construction and nonclinical testing of a Puumala virus synthetic M gene-based DNA vaccine. Clin. Vaccine Immunol 20, 218–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hooper JW, Josleyn M, Ballantyne J, Brocato R, A novel Sin Nombre virus DNA vaccine and its inclusion in a candidate pan-hantavirus vaccine against hantavirus pulmonary syndrome (HPS) and hemorrhagic fever with renal syndrome (HFRS). Vaccine 31, 4314–4321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GBE, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang G-Y, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko S-Y, Todd J-P, Rao S, Graham BS, Kwong PD, Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342, 592–598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh C-L, Goldsmith JA, Schaub JM, DiVenere AM, Kuo H-C, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, Chou C-W, Byrne PO, Hjorth CK, Johnson NV, Ludes-Meyers J, Nguyen AW, Park J, Wang N, Amengor D, Lavinder JJ, Ippolito GC, Maynard JA, Finkelstein IJ, McLellan JS, Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouvinski A, Dejnirattisai W, Guardado-Calvo P, Vaney M-C, Sharma A, Duquerroy S, Supasa P, Wongwiwat W, Haouz A, Barba-Spaeth G, Mongkolsapaya J, Rey FA, Screaton GR, Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope. Nat. Commun 8, 15411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vial PA, Ferrés M, Vial C, Klingström J, Ahlm C, López R, Le Corre N, Mertz GJ, Hantavirus in humans: A review of clinical aspects and management. Lancet Infect. Dis 10.1016/S1473-3099(23)00128-7, (2023). [DOI] [PubMed] [Google Scholar]
- 39.Williamson BN, Prescott J, Garrido JL, Alvarez RA, Feldmann H, Barría MI, Therapeutic efficacy of human monoclonal antibodies against Andes virus infection in Syrian Hamsters. Emerg. Infect. Dis 27, 2707–2710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strandin T, Smura T, Ahola P, Aaltonen K, Sironen T, Hepojoki J, Eckerle I, Ulrich RG, Vapalahti O, Kipar A, Forbes KM, Orthohantavirus Isolated in Reservoir Host Cells Displays Minimal Genetic Changes and Retains Wild-Type Infection Properties. Viruses 12, 457 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinfelter LM, Jangra RK, Jae LT, Herbert AS, Mittler E, Stiles KM, Wirchnianski AS, Kielian M, Brummelkamp TR, Dye JM, Chandran K, Haploid genetic screen reveals a profound and direct dependence on cholesterol for hantavirus membrane fusion. mBio 6, e00801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slough MM, Chandran K, Jangra RK, Two point mutations in old world hantavirus glycoproteins afford the generation of highly infectious recombinant vesicular stomatitis virus vectors. mBio 10, e02372-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K, A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol 84, 163–175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dieterle ME, Solà-Riera C, Ye C, Goodfellow SM, Mittler E, Kasikci E, Bradfute SB, Klingström J, Jangra RK, Chandran K, Genetic depletion studies inform receptor usage by virulent hantaviruses in human endothelial cells. eLife 10, e69708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quiroz JA, Malonis RJ, Thackray LB, Cohen CA, Pallesen J, Jangra RK, Brown RS, Hofmann D, Holtsberg FW, Shulenin S, Nyakatura EK, Durnell LA, Rayannavar V, Daily JP, Ward AB, Aman MJ, Dye JM, Chandran K, Diamond MS, Kielian M, Lai JR, Human monoclonal antibodies against chikungunya virus target multiple distinct epitopes in the E1 and E2 glycoproteins. PLOS Pathog. 15, e1008061 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerkman PF, Dernstedt A, Tadala L, Mittler E, Dannborg M, Sundling C, Maleki KT, Tauriainen J, Tuiskunen-Bäck A, Wigren Byström J, Ocaya P, Thunberg T, Jangra RK, Román-Sosa G, Guardado-Calvo P, Rey FA, Klingström J, Chandran K, Puhar A, Ahlm C, Forsell MN, Generation of plasma cells and CD27−IgD− B cells during hantavirus infection is associated with distinct pathological findings. Clin. Transl. Immunology 10, e1313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundkvist A, Cheng Y, Sjölander KB, Niklasson B, Vaheri A, Plyusnin A, Cell culture adaptation of Puumala hantavirus changes the infectivity for its natural reservoir, Clethrionomys glareolus, and leads to accumulation of mutants with altered genomic RNA S segment. J. Virol 71, 9515–9523 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hooper JW, Larsen T, Custer DM, Schmaljohn CS, A lethal disease model for hantavirus pulmonary syndrome. Virology 289, 6–14 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Ali HS, Drewes S, Weber de V Melo, Schlegel M, Freise J, Groschup MH, Heckel G, Ulrich RG, Complete genome of a Puumala virus strain from Central Europe. Virus Genes 50, 292–298 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Sakharkar M, Rappazzo CG, Wieland-Alter WF, Hsieh C-L, Wrapp D, Esterman ES, Kaku CI, Wec AZ, Geoghegan JC, McLellan JS, Connor RI, Wright PF, Walker LM, Prolonged evolution of the human B cell response to SARS-CoV-2 infection. Sci. Immunol 6, eabg6916 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rappazzo CG, Tse LV, Kaku CI, Wrapp D, Sakharkar M, Huang D, Deveau LM, Yockachonis TJ, Herbert AS, Battles MB, O’Brien CM, Brown ME, Geoghegan JC, Belk J, Peng L, Yang L, Hou Y, Scobey TD, Burton DR, Nemazee D, Dye JM, Voss JE, Gunn BM, McLellan JS, Baric RS, Gralinski LE, Walker LM, Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science 371, 823–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD, Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney M-C, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, Girard-Blanc C, Petres S, Shepard WE, Desprès P, Arenzana-Seisdedos F, Dussart P, Mongkolsapaya J, Screaton GR, Rey FA, Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520, 109–113 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall III WB, Snoeyink J, Adams PD, Lovell SC, Richardson JS, Richardson DC, MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo W-P, Lin X-D, Wang W, Tian J-H, Cong M-L, Zhang H-L, Wang M-R, Zhou R-H, Wang J-B, Li M-H, Xu J, Holmes EC, Zhang Y-Z, Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLOS Pathog. 9, e1003159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robert X, Gouet P, Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haslwanter D, Lasso G, Wec AZ, Furtado ND, Raphael LMS, Tse AL, Sun Y, Stransky S, Pedreño-Lopez N, Correia CA, Bornholdt ZA, Sakharkar M, Avelino-Silva VI, Moyer CL, Watkins DI, Kallas EG, Sidoli S, Walker LM, Bonaldo MC, Chandran K, Genotype-specific features reduce the susceptibility of South American yellow fever virus strains to vaccine-induced antibodies. Cell Host Microbe 30, 248–259.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daniel Gietz R, Woods RA, Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Guide to Yeast Genetics and Molecular and Cell Biology - Part B, Guthrie C, Fink GR, Eds. (Methods in Enzymology, Elsevier, 2002), vol. 350, pp. 87–96. [DOI] [PubMed] [Google Scholar]
- 61.Wigren Byström J, Näslund J, Trulsson F, Evander M, Wesula Lwande O, Ahlm C, Bucht G, Quantification and kinetics of viral RNA transcripts produced in Orthohantavirus infected cells. Virol. J 15, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts A, Thomas WD, Guarner J, Lamirande EW, Babcock GJ, Greenough TC, Vogel L, Hayes N, Sullivan JL, Zaki S, Subbarao K, Ambrosino DM, Therapy with a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody reduces disease severity and viral burden in golden Syrian hamsters. J. Infect. Dis 193, 685–692 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials. The coordinate files of the structures described in this manuscript have been submitted to the Protein Data Bank with ID 7QQB. The LC-MS/MS data described in this manuscript have been submitted to Chorus with ID 1815. Materials are available through material transfer agreement by contacting K.C. or P.G.-C.