Abstract
Background
Since Mycobacterium avium subspecies paratuberculosis (MAP) was isolated from intestinal tissue of a human patient suffering Crohn's disease, a controversial discussion exists whether MAP have a role in the etiology of Crohn's disease or not. Raw milk may be a potential vehicle for the transmission of MAP to human population. In a previous paper, we have demonstrated that MAP are found in raw milk samples obtained from a defined region in Switzerland. The aim of this work is to collect data about the prevalence of MAP specific IS900 insertion sequence in bulk-tank milk samples in different regions of Switzerland. Furthermore, we examined eventual correlation between the presence of MAP and the somatic cell counts, the total colony counts and the presence of Enterobacteriaceae.
Results
273 (19.7%) of the 1384 examined bulk-tank milk samples tested IS900 PCR-positive. The prevalence, however, in the different regions of Switzerland shows significant differences and ranged from 1.7% to 49.2%. Furthermore, there were no statistically significant (p >> 0.05) differences between the somatic cell counts and the total colony counts of PCR-positive and PCR-negative milk samples. Enterobacteriaceae occur as often in IS900 PCR-positive as in PCR-negative milk samples.
Conclusion
This is the first study, which investigates the prevalence of MAP in bulk-tank milk samples all over Switzerland and infers the herd-level prevalence of MAP infection in dairy herds. The prevalence of 19.7% IS900 PCR-positive bulk-milk samples shows a wide distribution of subclinical MAP-infections in dairy stock in Switzerland. MAP can therefore often be transmitted to humans by raw milk consumption.
Background
MAP belongs to the slow growing mycobacteria in the genus Mycobacterium. The cell envelope of mycobacteria contains a high proportion of lipids that make them resistant to dye extraction by acid-alcohol (acid-fast in Ziehl-Neelsen stain). This high proportion of lipids plays a crucial role in the adaptation of mycobacteria to intracellular growth in macrophages as well as in drug resistance [1]. Various mycobacterial species contain a number of highly specific insertion sequences (IS) integrated in the genome [1]. Some of those genetic elements can be used to disthiguish species of mycobacteria [2-4].
MAP belongs to the M. avium-complex and has specific characteristics: mycobactin-dependence very slow growth (incubation of the cultures for 16 weeks), and a tendency to form large clumps [5]. The specific insertion sequence of MAP is the IS900 element [6-8].
In ruminants, infection by MAP causes paratuberculosis, a granulomatous enteritis which is responsible for important economic losses through production losses and premature culling [9-12]. Paratuberculosis is prevalent in domestic ruminants worldwide and occurs also in other domesticated, wild and zoo animals including primate [13]. It is principally a subclinical infection with a long incubation period. Clinical signs are only seen in a minority of infected animals [14]. MAP is shed intermittently by subclinically and clinically infected cows into manure and within macrophages in milk [15,16]. Subclinical shedders are the main problem because they have no clinical signs and contaminate the environment with MAP, which represents a source for new infections.
Crohn's disease, a chronic granulomatous, incurable ileocolitis of humans, has significant similarity to the paucimicrobial form of paratuberculosis in animals [17]. Chiodini et al. [10] isolated MAP from intestinal tissue of a human patient suffering Crohn's disease. Since then, a controversial discussion exists whether MAP plays a role in the etiology of Crohn's disease or not [4,17-19]. Recently, research is making advances in understanding MAP-infections, and is indicating more and more that at least some cases of Crohn's disease may be caused by this organism [20].
Raw milk may be a potential vehicle for the transmission of MAP to human population. Sweeney et al. [21] found, that MAP is shed into milk by 12% of subclinically infected cows in a concentration of 2 to 8 CFU per 50 ml milk. Moreover, results of laboratory pasteurization tests of whole milk spiked with MAP showed that MAP was capable of surviving pasteurization of raw milk [22-25]. Nevertheless, literature data are very divergent. Sung et al. [26] reported a D-value (decimal reduction time) for MAP in milk of 11 sec at 71°C, so there is a possibility for MAP to survive HTST (high temperature short time) pasteurization if the initial number is >101 cfu ml-1 milk. Pearce et al. [27], however, found a D-value of 2.03 sec at 72°C, which is equivalent to a >7 log10 reduction at 72°C for 15 sec (HTST pasteurization). In a study of whole-pasteurized cows milk obtained from retail outlets in England 7% of these samples tested MAP-positive (IS900-positive) by PCR [28]. Grant et al. [29] pasteurized raw cows' milk naturally infected with MAP and found, that MAP is capable of surviving commercial scale pasteurization at 73°C for 15 s and 25 s with and without prior homogenization if the bacterial cells are present in sufficient numbers before heat treatment. They also tested commercially pasteurized cows' milk samples from dairy processing establishments for the present of MAP by culture. 1.8% of the tested pasteurized milk samples were MAP-positive [30]. Furthermore, the production process for hard and semihard raw milk cheese seems not to completely reduce MAP. Spahr and Schafroth [31] found D-values of 27.7 days for hard and 45.5 days for semihard cheese manufactured of artificially contaminated raw milk.
The aim of this study was to collect data about the prevalence of the MAP specific IS900 insertion sequence in bulk-tank milk samples from different regions of Switzerland. Furthermore, we examined eventual correlation between the presence of MAP and the somatic cell counts, the total colony counts and the presence of Enterobacteriaceae in bulk-tank milk samples.
Results
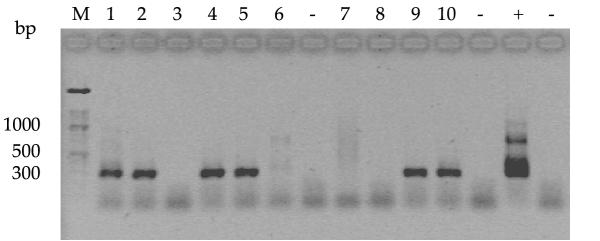
273 (19.7%) of the 1384 milk samples were IS900 PCR-positive. PCR amplification of the MAP specific insertion sequence IS900 and subsequent agarose gel analysis of the amplified products showed a single band of 298 bp for each of the positive milk samples (Fig. 1). The PCR products, however, differed in intensity.
Figure 1.
Agarose gel electrophoresis of PCR products obtained from different bulk-tank milk samples following IS900 nested PCR. M: 100 bp molecular wight ladder (Roche Diagnostics, Rotkreuz, Switzerland). Lanes 1, 2, 4, 5, 9, 10: positive samples with a specific amplification product of 298 bp. Lanes 3, 6, 7, 8: negative samples. +: positive control with IS900 nested product of 298 bp in length and additional higher molecular weight amplification products at 350 bp, 400 bp and 700 bp. -: negative control.
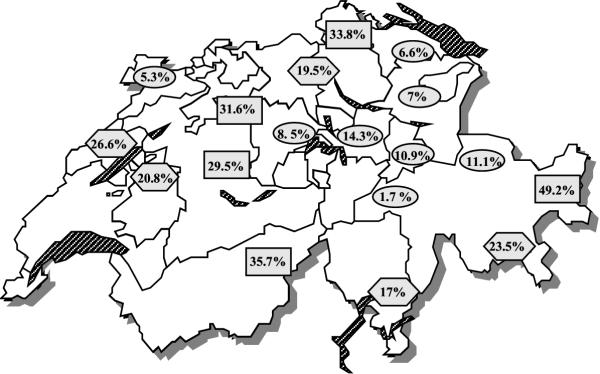
The prevalence of IS900-containing milk samples in the 18 regions of Switzerland were different (Fig. 2). In five regions in different parts of Switzerland, prevalence ranging from 29.5% to 49.2%, in eight regions, situated in the center, the east and one of them in the northwest part of Switzerland, only prevalence from 1.7% to 14.3% were found.
Figure 2.
Prevalence of the MAP specific IS900 insertion sequence in bulk-tank milk samples in different regions of Switzerland ( high prevalence;
high prevalence;  medium prevalence;
medium prevalence;  low prevalence)
low prevalence)
The results of somatic cell counts ranged from 11'000 cells ml-1 to 981'000 cells ml-1, with mean value of 130'670 cells ml-1. In 42 (3.5%) milk samples, the somatic cell counts were higher than the Swiss legal limit of 350'000 cells ml-1.
Results of total colony counts ranged from 1.2 × 102 CFU ml-1 to 1.2 × 106 CFU ml-1, with mean value of 2 × 104 CFU ml-1. Enterobacteriaceae could be found in 237 (52.7%) of 450 investigated milk samples with colony counts ranging from 1 CFU ml-1 to 2 × 103 CFU ml-1.
Discussion
Positive IS900 PCR results provide presumptive evidence of the presence of MAP. The prevalence of 19.7% IS900 PCR-positive bulk-milk samples shows a wide distribution of subclinical MAP-infections in dairy stock in Switzerland. Since not all animals which are subclinically infected with MAP shed the organism in the milk and moreover this excretion is intermittent, it is probable that there are stocks with subclinically infected animals which haven't at the time of sample collection shed MAP in the milk. It is therefore possible that the prevalence of subclinically infected dairy stocks is even higher than we have found.
Moreover, there are distinct regional differences (Fig. 2) which are statistically (chi square-test) significant (p < 0.05). However, there is no explanation for these regional differences, even no difference between the mountain and the plains could be found. Conspicuous regional differences in the distribution of prevalence have previously been described in other countries [32,33].
The differences in the intensity of the PCR-amplicons can be explained by different concentrations of MAP in the bulk-tank milk samples.
For Switzerland, actually no other comparative data on the occurrence of MAP in milk samples exist. In two studies ELISA serology test results of Swiss milk cows are available. Meylan et al. [34] described a seroprevalence of 4.9% by examining 595 animals in one defined region of Switzerland. This study, however, concern with animal-level prevalence which is generally lower compared to herd-level prevalence. Stärk et al. [35] found a herd-level prevalence of 8.0% by investigating 113 stocks. In there study, however, no information is given about the geographic distribution of positive herds.
From some other countries herd-level data are available too (Table 1). Similar to our own study, Nielsen et al. [36,37] examined bulk-tank milk samples from six different regions in Denmark. They performed an ELISA test on 900 bulk-tank milk samples and detected a herd-level prevalence of 47% (423 MAP-antibody-positive samples). Grant et al. [30] performed an immunomagnetic PCR method on 244 bulk-tank milk samples from the United Kingdom and detected a herd-level prevalence of 7.8% (19 PCR-positive samples). In all other studies MAP antibodies were proofed in serum with ELISA-systems. The prevalence range from 6.96% to 54.7% in Europe, and from 43.3% to 55.4% in North America. These results show, that MAPis widely spread in northern Europe and in North America.
Table 1.
Herd-level prevalence of MAP in dairy stock (literature data)
| origin | material | method | number of investigated herds | herd prevalences | reference |
| Denmark | Bulk-tank milk samples | ELISA | 900 | 47% | [36,37] |
| UK | Bulk-tank milk samples | PCR | 244 | 7.8% | [30] |
| Austria | Serum samples | ELISA | 2757 | 6.96%a | [33] |
| Belgian | Serum samples | ELISA | 464 | 18% | [41] |
| Netherlands | Serum samples | ELISA | 378 | 54.7% | [42] |
| Canada | Serum samples | ELISA | 90 | 43.3% | [43] |
| Missouri | Serum samples | ELISA | 89 | 47% | [44] |
| Wisconsin | Serum samples | ELISA | 158 | 50%b | [32] |
| Michigan | Serum samples | ELISA | 121 | 55.4% | [45] |
a 4–13% in different regions. b more infected herds in the southern and western districts
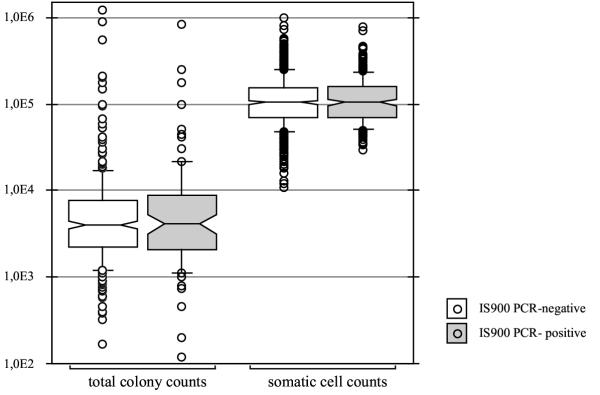
There were no statistically significant (p >> 0.05) differences between the somatic cell counts and the total colony counts of PCR-positive and PCR-negative milk samples by the Mann-Whitney-test (Fig. 3). Therefore, there is no correlation between the presence of MAP, the somatic cell counts or total colony counts. Enterobacteriaceae occur as often in IS900 PCR-positive milk as in PCR-negative samples. McNab et al. [38] and Nordlund et al. [39] found similar results.
Figure 3.
Comparison of total colony counts and somatic cell counts of IS900 PCR-positive and IS900 PCR-negative bulk-tank milk samples (box plot representation).
Today it seems more and more probable that MAP could play a role in the etiology of Crohn's disease. But, many questions remain unanswered: minimal infection dose, concentration of MAP in the raw milk of subclinically infected cattle, heat resistance under commercial conditions, exact pathogenesis of Crohn's disease and paratuberculosis. Only the answers to these questions will allow an assessment of the risk associated with consumption of raw and pasteurised milk. It is especially important to investigate survive of MAP in the commercial pasteurization process by using naturally infected milk. Under laboratory conditions, the milk is in a quiescent state and not subjected to the turbulent flow. MAPwhich were used in the experiments were mostly present as single cells and in high concentrations. In naturally infected milk, however, MAP occur in low concentration and in clumps, which are enclosed in macrophages.
Conclusions
This is the first study, which investigates the herd-level prevalence of MAP all over Switzerland. Positive IS900 PCR results do provide presumptive evidence of the presence of MAP. The prevalence of 19.7% IS900 PCR-positive bulk-milk samples shows a wide distribution of subclinical MAP-infections in dairy stock in Switzerland. MAP can therefore often be transmitted to humans by raw milk consumption. Furthermore, there were no statistically significant (p >> 0.05) differences between the somatic cell counts and the total colony counts of PCR-positive and PCR-negative milk samples Enterobacteriaceae occur as often in IS900 PCR-positive as in PCR-negative milk samples.
Materials and Methods
Material
Between February and June 2001, 1384 bulk-tank milk samples were collected from 18 different regions all over Switzerland, with 41 to 100 samples per region. The samples were taken aseptically from diverse milk inspection services and brought cooled to the laboratory. Total colony counts and Enterobacteriaceae counts were done directly. For the detection of MAP, 20 ml milk was stored at -20°C up to investigation. Conservation of milk samples for somatic cell counts was done by adding 100 μl 4% Bronopol (Fluka, Buchs, Switzerland) to 10 ml milk.
Mycobacterial lysis
10 ml of each milk sample were mixed with 100 μl Triton® X-100 (Calbiochem, Darmstadt, Germany) and centrifuged at 4500 rpm for 30 min. Cream and whey were discarded, the pellet was frozen, transferred to a blue ribolyser tube (Hybaid, Ashford, United Kingdom), resuspended in 400 μl mycobacterial lysis buffer (86.6 ml H2O, 8 ml 5 M NaCL, 2 ml 2 M TrisHCl (pH 8), 3 ml 20% SDS, 400 μl 0,5 M EDTA, 220 μl 15.6 mg/ml proteinase K (Roche Diagnostics AG, Rotkreuz, Switzerland)), and incubated overnight at 37°C. MAP DNA was released by mechanical disruption of the lysate at 6.5 msec-1 for 45 sec in a Ribolyser (Hybaid, Ashford). Afterwards, the blue ribolyser tubes were immediately put on ice for 15 min.
DNA extraction
For the DNA extraction, 400 μl phenol were added, the tubes vortexed for 20 sec, centrifuged at 13000 rpm for 2 min at 4°C, and the aqueous layer was pipetted to a new tube. The same steps were repeated with phenol/chloroform/isoamylalcohol (25:24:1) and chloroform/isoamylalcohol (24:1). To the last aqueous layer, 40 μl 3 M Sodium acetat and 800 μl ethanol abs. were pipetted. After careful mixing, the DNA was precipitated at -70°C, 30 min. Then, the tubes were centrifuged for 20 min at 13000 rpm and 4°C and the supernatant was discarded. The pellet was washed with 800 μl ice cold 70% ethanol, the tubes centrifuged for 5 min at 13000 rpm and 4°C, and the supernatant discarded. The pellet was dried in a concentrator (Vaudaux-Eppendorf, Basel, Switzerland) and resuspended in 50 μl TE-buffer.
IS900 nested PCR
5 μl of the resuspended DNA was used for an IS900 nested PCR specific to MAP. According to Hermon-Taylor et al. [4], the primers p90 5'-GAA GGG TGT TCG GGG CCG TCG CTT AGG-3' and p91 5'-GGC GTT GAG GTC GAT CGC CCA CGT GAC-3' were used for the first 30 amplification cycles. The PCR mix consisted of 50 μl reaction volume containing a final concentration of 2 μM of each primer, 2.5 mM MgCl2, 100 μM dNTP (Promega, Madison, USA), 2 U Taq polymerase (Promega, Madison) in 1× reaction buffer (Promega, Madison). Reactions were cycled as follows: 94°C for 5 min (1 cycle); 94°C for 1 min, 58°C for 1 min, 72°C for 3 min (30 cycles); 72°C for 7 min (1 cycle).
2 μl from the primary amplification were then used with the primers AV1 5'-ATG TGG TTG CTG TGT TGG ATG G-3' and AV2 5'-CCG CCG CAA TCA ACT CCA G-3' for the nested-PCR. The PCR mix consisted of 50 μl reaction volume containing a final concentration of 2 μM of each primer, 1.5 mM MgCl2, 100 μM dNTP (Promega, Madison), 2 U Taq polymerase (Promega, Madison) in 1× reaction buffer (Promega, Madison). Reactions were cycled as follows: 94°C for 5 min (1 cycle); 94°C for 1 min, 58°C for 1 min, 72°C for 3 min (40 cycles); 72°C for 7 min (1 cycle). PCR products were visualized on 1.5% agrose gels (Eurobio, Les Ulis Cedex, France). The IS900 nested product is 298 bp in length but a strong signal will also give non-specific bands at 350 bp, 400 bp and 700 bp [4]. For each batch of processed samples negative process controls and positive bacterial controls were included.
Total colony counts and Enterobacteriaceae counts
Additionally to MAP, total colony counts and Enterobacteriaceae counts were determined of 450 milk samples according to standard methods on plate count agar (Oxoid, Basingstoke, United Kindom) and violet red bile glucose agar (BBL, Cockeysville, USA), respectively.
Somatic cell counts
Of 1212 milk samples somatic cell counts were determined by a fluorescenceoptic method with a Fossomatic 90 system (Foss Electronic, Hillerod, Danmark) according to the manufacture's instruction.
Statistical analysis
Total colony counts and somatic cell counts were transformed to log10 values and depicted in box plots. By illustrating results in such a way median values, 50%, 75%, 80% and 90% ranges and extreme values can be analysed more conveniently [40]. Statistical significance was determined by the chi square-test and the Mann-Whitney-test.
Authors' contributions
SC carried out the experimental part of the study and drafting of the manuscript. RS made the initial observations and carried out the conception and design of the study
Both authors participated in producing and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors would like to thank to J. Hermon-Taylor and T. Bull, St. George's Hospital Medical School, London, for letting their nested IS900 PCR method be used; the different Swiss milk inspection services for their help in collecting the milk samples, and the milk inspection service of the North-East Switzerland for determination of the somatic cell counts.
Contributor Information
Sabrina Corti, Email: scorti@gmx.net.
Roger Stephan, Email: stephanr@fsafety.unizh.ch.
References
- Rastogi N, Legrand E, Sola C. The mycobacteria: an introduction to nomenclature and pathogenesis. Rev Sci Tech. 2001;20:21–54. doi: 10.20506/rst.20.1.1265. [DOI] [PubMed] [Google Scholar]
- Moss MT, Green EP, Tizard ML, Malik ZP, Hermon-Taylor J. Specific detection of Mycobacterium paratuberculosis by DNA hybridisation with a fragment of the insertion element IS900. Gut. 1991;32:395–398. doi: 10.1136/gut.32.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull TJ, Hermon-Taylor J, Pavlik II, El-Zaatari F, Tizard M. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology. 2000;146:2185–2197. doi: 10.1099/00221287-146-9-2185. [DOI] [PubMed] [Google Scholar]
- Hermon-Taylor J, Bull TJ, Sheridan JM, Cheng J, Stellakis ML, Sumar N. Causation of Crohn's disease by Mycobacterium avium subspecies paratuberculosis. Can J Gastroenterol. 2000;14:521–539. doi: 10.1155/2000/798305. [DOI] [PubMed] [Google Scholar]
- Legrand E, Sola C, Rastogi N. Mycobacterium avium-intracellulare complex: phenotypic and genotypic markers and the molecular basis for interspecies transmission. Bull Soc Pathol Exot. 2000;93:182–192. [PubMed] [Google Scholar]
- McFadden JJ, Butcher PD, Chiodini R, Hermon-Taylor J. Crohn's disease-isolated mycobacteria are identical to Mycobacterium paratuberculosis, as determined by DNA probes that distinguish between mycobacterial species. J Clin Microbiol. 1987;25:796–801. doi: 10.1128/jcm.25.5.796-801.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DM, Gabric DM, de Lisle GW. Identification of a repetitive DNA sequence specific to Mycobacterium paratuberculosis. FEMS Microbiol Lett. 1989;51:175–178. doi: 10.1016/0378-1097(89)90503-X. [DOI] [PubMed] [Google Scholar]
- Green EP, Tizard ML, Moss MT, Thompson J, Winterbourne DJ, Mc Fadden JJ, Hermon-Taylor J. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 1989;17:9063–9073. doi: 10.1093/nar/17.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkal RS, Larsen AB, Booth GD. Analysis of the effect of inapparent bovine paratuberculosis. Am J Vet Res. 1975;36:837–838. [PubMed] [Google Scholar]
- Chiodini RJ, Van Kruiningen HJ, Merkal RS. Ruminant paratuberculosis (Johne's disease): the current status and future prospects. Cornell Vet. 1984;74:218–262. [PubMed] [Google Scholar]
- Wilson DJ, Rossiter C, Han HR, Sears PM. Association of Mycobacterium paratuberculosis infection with reduced mastitis, but with decreased milk production and increased cull rate in clinically normal dairy cows. Am J Vet Res. 1993;54:1851–1857. [PubMed] [Google Scholar]
- Kennedy DJ, Benedictus G. Control of Mycobacterium avium subsp. paratuberculosis infection in agricultural species. Rev Sci Tech. 2001;20:151–179. doi: 10.20506/rst.20.1.1274. [DOI] [PubMed] [Google Scholar]
- Cocito C, Gilot P, Coene M, de Kesel M, Poupart P, Vannuffel P. Paratuberculosis. Clin Microbiol Rev. 1994;7:328–345. doi: 10.1128/cmr.7.3.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy D, Holmström A, Plym Forshell K, Vindel E, Suarez Fernandez G. On-farm management of paratuberculosis (Johne's disease) in dairy herds. Bulletin of the IDF. 2001;362:18–31. [Google Scholar]
- Streeter RN, Hoffsis GF, Bech-Nielsen S, Shulaw WP, Rings DM. Isolation of Mycobacterium paratuberculosis from colostrum and milk of subclinically infected cows. Am J Vet Res. 1995;56:1322–1324. [PubMed] [Google Scholar]
- Sweeney RW. Transmission of paratuberculosis. Vet Clin North Am Food Anim Pract. 1996;12:305–312. doi: 10.1016/s0749-0720(15)30408-4. [DOI] [PubMed] [Google Scholar]
- Chiodini RJ. Crohn's disease and the mycobacterioses: a review and comparison of two disease entities. Clin Microbiol Rev. 1989;2:90–117. doi: 10.1128/cmr.2.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kruiningen HJ. Lack of support for a common etiology in Johne's disease of animals and Crohn's disease in humans. Inflamm Bowel Dis. 1999;5:183–191. doi: 10.1097/00054725-199908000-00007. [DOI] [PubMed] [Google Scholar]
- Chamberlin W, Graham DY, Hulten K, El-Zimaity HMT, Schwartz MR, Naser S, Shafran I, El-Zaatari FAK. R: Mycobacterium avium subsp. paratuberculosis as one cause of Crohn's disease. Aliment Pharmacol Ther. 2001;15:337–346. doi: 10.1046/j.1365-2036.2001.00933.x. [DOI] [PubMed] [Google Scholar]
- Mason O, Rowe MT, Ball HJ. Is Mycobacterium paratuberculosis a possible agent in Crohn's disease? Implications for the dairy industry. Milchwiss. 1997;52:311–313. [Google Scholar]
- Sweeney RW, Whitlock RH, Rosenberger AE. Mycobacterium paratuberculosis cultured from milk and supramammary lymph nodes of infected asymptomatic cows. J Clin Microbiol. 1992;30:166–171. doi: 10.1128/jcm.30.1.166-171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodini RJ, Hermon-Taylor J. The thermal resistance of Mycobacterium paratuberculosis in raw milk under conditions simulating pasteurization. J Vet Diagn Invest. 1993;5:629–631. doi: 10.1177/104063879300500424. [DOI] [PubMed] [Google Scholar]
- Grant IR, Ball HJ, Neill SD, Rowe MT. Inactivation of Mycobacterium paratuberculosis in cows' milk at pasteurization temperatures. Appl Environ Microbiol. 1996;62:631–636. doi: 10.1128/aem.62.2.631-636.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant IR, Ball HJ, Rowe MT. Effect of high-temperature, short-time (HTST) pasteurization on milk containing low numbers of Mycobacterium paratuberculosis. Lett Appl Microbiol. 1998;26:166–10. doi: 10.1046/j.1472-765X.1998.00309.x. [DOI] [PubMed] [Google Scholar]
- Keswani J, Frank JF. Thermal inactivation of Mycobacterium paratuberculosis in milk. J Food Prot. 1998;61:974–978. doi: 10.4315/0362-028x-61.8.974. [DOI] [PubMed] [Google Scholar]
- Sung N, Collins MT. Thermal tolerance of Mycobacterium paratuberculosis. Appl Environ Microbiol. 1998;64:999–1005. doi: 10.1128/aem.64.3.999-1005.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LE, Tuan Truong H, Crawford RA, Yates GF, Cavaignac S, de Lisle GW. Effect of turbulent-flow pasteurisation on survival of Mycobacterium avium subsp. paratuberculosis added to raw milk. Appl Environ Microbiol. 2001;67:3964–3969. doi: 10.1128/AEM.67.9.3964-3969.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar D, Ford J, Sanderson J, Withey S, Tizard M, Doran T, Hermon-Taylor J. IS900 PCR to detect Mycobacterium paratuberculosis in retail supplies of whole pasteurized cows' milk in England and Wales. Appl Environ Microbiol. 1996;62:3446–3452. doi: 10.1128/aem.62.9.3446-3452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant IR, Hitchings EI, McCarteny A, Ferguson F, Rowe MT. Effect of commercial-scale high-temperature, short-time pasteurization on the viability of Mycobacterium paratuberculosis in naturally infected cows' milk. Appl Environ Microbiol. 2002;68:602–607. doi: 10.1128/AEM.68.2.602-607.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant IR, Ball HJ, Rowe MT. Incidence of Mycobacterium paratuberculosis in bulk raw and commercially pasteurized cows' milk from approved dairy processing establishments in the United Kingdom. Appl Environ Microbiol. 2002;68:2428–2435. doi: 10.1128/AEM.68.5.2428-2435.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahr U, Schafroth K. Fate of Mycobacterium avium subsp. paratuberculosis in Swiss hard an semihard cheese manufactured from raw milk. Appl Environ Microbiol. 2001;67:4199–4205. doi: 10.1128/AEM.67.9.4199-4205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MT, Sockett DC, Goodger WJ, Conrad TA, Thomas CB, Carr DJ. Herd prevalence and geographic distribution of, and risk factors for, bovine paratuberculosis in Wisconsin. J Am Vet Med Assoc. 1994;204:636–641. [PubMed] [Google Scholar]
- Gasteiner J, Wenzl H, Fuchs K, Jark U, Baumgartner W. Serological cross-sectional study of paratuberculosis in cattle in Austria. Zbl Vet. 1999;46:457–466. doi: 10.1046/j.1439-0450.1999.00256.x. [DOI] [PubMed] [Google Scholar]
- Meylan M, Nicolet J, Busato A, Burnens A, Martig J. The prevalence of paratuberculosis in the Plateau de Diesse region. Schweiz Arch Tierheilk. 1995;137:22–25. [PubMed] [Google Scholar]
- Stärk KD, Frei-Stäheli C, Frei P, Pfeiffer DU, Danuser J, Audige L, Nicolet J, Strasser M, Gottstein B, Kihm U. Frequency and cost of health problems in Swiss dairy cows and their calves (1993–1994). Schweiz Arch Tierheilk. 1997;139:343–353. [PubMed] [Google Scholar]
- Nielsen SS, Thamsborg SM, Houe H, Bitsch V. Bulk-tank milk ELISA antibodies for estimating the prevalence of paratuberculosis in Danish dairy herds. Prev Vet Med. 2000;44:1–7. doi: 10.1016/S0167-5877(00)00098-2. [DOI] [PubMed] [Google Scholar]
- Nielsen SS, Thamsborg SM, Houe H, Bitsch V. Corrigendum to "Bulk-tank milk ELISA antibodies for estimating the prevalence of paratuberculosis in danish dairy herds". Prev Vet Med. 2000;46:297. doi: 10.1016/S0167-5877(00)00164-1. [DOI] [PubMed] [Google Scholar]
- McNab WB, Meek AH, Martin SW. An epidemiological study of paratuberculosis in dairy cattle in Ontario: study design and prevalence estimates. Can J Vet Res. 1991;55:246–251. [PMC free article] [PubMed] [Google Scholar]
- Nordlund KV, Goodger WJ, Pelletier J, Collins MT. Associations between subclinical paratuberculosis and milk production, milk components, and somatic cell counts in dairy herds. J Am Vet Med Assoc. 1996;208:1872–1876. [PubMed] [Google Scholar]
- Eggenberger E, Thun R. Eine graphische Methode zur Darstellung von Messwerten. Schweiz Arch Tierheilk. 1984;126:199–205. [PubMed] [Google Scholar]
- Boelaert F, Walravens K, Biront P, Vermeersch JP, Berkvens D, Godfroid J. Prevalence of paratuberculosis (Johne's disease) in the Belgian cattle population. Vet Microbiol. 2000;77:269–281. doi: 10.1016/S0378-1135(00)00312-6. [DOI] [PubMed] [Google Scholar]
- Muskens J, Barkema HW, Russchen E, van Maanen K, Schukken YH, Bakker D. Prevalence and regional distribution of paratuberculosis in dairy herds in The Netherlands. Vet Microbiol. 2000;77:253–261. doi: 10.1016/S0378-1135(00)00310-2. [DOI] [PubMed] [Google Scholar]
- VanLeeuwen JA, Keefe GP, Tremblay R, Power C, Wichtel JJ. Seroprevalence of infection with Mycobacterium avium subspecies paratuberculosis, bovine leukemia virus, and bovine viral diarrhea virus in maritime Canada dairy cattle. Can Vet J. 2001;42:193–198. [PMC free article] [PubMed] [Google Scholar]
- Thorne JG, Hardin LE. Estimated prevalence of paratuberculosis in Missouri, USA cattle. Prev Vet Med. 1997;31:51–57. doi: 10.1016/S0167-5877(96)01138-5. [DOI] [PubMed] [Google Scholar]
- Johnson-Ifearulundu Y, Kaneene JB. Distribution and environmental risk factors for paratuberculosis in dairy cattle herds in Michigan. Am J Vet Res. 1999;60:589–596. [PubMed] [Google Scholar]