Abstract
HTT1a has been identified in human and mouse HD brain as the pathogenic exon 1 mRNA generated from aberrant splicing between exon 1 and 2 that contributes to aggregate formation and neuronal dysfunction (Sathasivam et al., 2013). Detection of the HTT exon 1 protein (HTTex1p) has been accomplished with surrogate antibodies in fluorescence-based reporter assays (MSD, HTRF), and immunoprecipitation assays, in HD postmortem cerebellum and knock-in mice but direct detection by SDS-PAGE and western blot assay has been lacking. Here proteins in subcellular fractions prepared from human and mouse HD brain were separated by SDS-PAGE and probed by western blot with neo-epitope monoclonal antibodies (P90–1B12 and 11G2) directed to the C-terminal 8 residues of HTTex1p. In human HD putamen and cortex, HTTex1p migrated at 56–60 kD and at higher molecular masses (HMM) consistent with the presence of CAG repeat expansion in HTT1a. HTTex1p in control brain was low or undetectable. Immunofluorescence labeling of human HD cortex using P90–11G2 revealed small aggregates that sparsely populated the neuropil in layers 3 and 5. In caudate putamen of 6 month old HD knock-in mice (Q50, Q80, Q111, Q140 and Q175) HTTex1p migration was inversely correlated with CAG repeat length and appeared as a SDS soluble high molecular mass (HMM) smear in HD Q111, Q140 and Q175 mice but not in Q50 and Q80 mice indicating a CAG repeat size threshold for detecting HTTex1p aggregation. Migration of HTTex1p and HMM smear changed with age in caudate putamen of Q111, Q175 and YAC128 mice. Treating HD Q111 mice with siRNA to MSH3, a modifier of CAG repeat expansion, significantly reduced levels of the HMM smear indicating that the effects of curbing CAG repeat expansion was quantifiable. These results show that P90 antibodies can be used in western blot assays and immunostaining to track and quantify HTTex1p levels, subcellular localization, and solubility.
Introduction
An mRNA arising from aberrant splicing between exon 1 and 2 in the Q150 HD mouse model generates a polyadenylated mRNA named HTT1a that is CAG repeat length dependent and also detected in HD patient brain and HD patient fibroblasts (Sathasivam et al., 2013;Neueder et al., 2017). HTT1a mRNA was proposed to give rise to the toxic HTT exon 1 protein (HTTex1p) expressed in the R6/2 transgenic mouse model (Mangiarini et al., 1996;Davies et al., 1997). R6/2 mice are more severely affected than HD knock in mice because they exhibit phenotypes at an earlier age including large nuclear aggregates in the striatum, behavioral deficits, and early death at about 16–20 weeks. The MW8 antibody made to C-terminal 8 amino acids of exon 1 AEEPLHRP has been used as a surrogate marker of HTTex1p in immunoprecipitation assays, Meso Scale Discovery (MSD) and homogeneous time resolved fluorescence (HTRF) assays (Sathasivam et al., 2013;Landles et al., 2020). Antibody MW8 paired with antibody 4C9 or 2B7 had been used in HTRF assays to detect soluble and aggregated forms respectively of HTTex1p (Smith et al., 2023;Landles et al., 2024). MW8 also detects other N-terminal HTT fragments (Baldo et al., 2024) and by immunostaining shows a non-specific signal for aggregates in human control brain tissue (Landles et al., 2020). S830 is a sheep polyclonal made to exon 1–53Q and along with MW8 mouse monoclonal was considered the most sensitive for detecting aggregates by immunostaining (Bayram-Weston et al., 2016;Smith et al., 2023) and was paired with MW8 for use in immunoprecipitation assays to detect HTTex1p in Q150 knock-in mice and in human cerebellum (Neueder et al., 2017). Fluorescence-based assays are sensitive high throughput bioassays that can be costly and labor intensive. Also, detection of HTTex1p by HTRF assay requires considerable mouse brain tissue (Landles et al., 2024). A sensitive and direct detection of HTTex1p by SDS-PAGE and western blot would provide a rapid, cost-effective method that utilizes smaller tissue samples.
HTTex1p has been identified in HD knock-in mice and YAC128 transgenic mice using HTRF, MSD and immunoprecipitation assays (Landles et al., 2010;Neueder et al., 2017;Fienko et al., 2022;Landles et al., 2024) but direct detection in the human HD brain by western blot and immunostaining has been elusive. Monoclonal neo-epitope antibodies (P90 1B12 and 11G2) directed to the C-terminal 83–90 of HTTex1p (based on HTT with 23 Qs) recently developed and characterized (Baldo et al., 2024;Missineo et al., 2024), that are highly sensitive for detecting purified HTTex1p by western blot compared to other antibodies directed to N-terminal HTT fragments, such as MW8, offered the possibility of detecting HTTex1p using SDS-PAGE and western blot. Here we tested the sensitivity of P90 neo-epitope antibodies to identify HTTex1p by SDS-PAGE and western blot in brain samples from human postmortem adult onset and juvenile onset HD patient, YAC128 transgenic mouse, and HD knock-in mice from an allelic series Q50, Q80, HD Q111, Q140 and Q175 and compared the results with R6/2 transgenic HD mice which express exon 1. We found that neo-epitope P90 antibodies detected HTTex1p in putamen and cortex from human HD brain as a band at 56–60 kD and also appeared in small neuropil aggregates in HD cortex by immunofluorescence. In HD knock-in mouse models HTTex1p appeared as a band and as a high molecular mass (HMM) smear measurable by densitometry. The migration and solubility of HTTex1p in SDS-PAGE depended on CAG repeat length, subcellular compartment, and age of mice. Silencing MSH3 mRNA which encodes a mismatch repair protein that drives CAG expansion in Q111 mice attenuated HTTex1p smear in caudate putamen, suggesting P90 antibody is a sensitive quantifiable readout for HTTex1p aggregation.
Methods
Sources of Human and Mouse Brain Tissue
Table 1 lists the WT and HD brain tissue, regions, sources, and references to our prior publications where brain tissues were used (Aronin et al., 1995;Sapp et al., 2012;Iuliano et al., 2021). All human tissues were de-identified and stored frozen at −80° C and age, sex, and CAG repeat length determined from PCR analysis of brain tissue are noted. HD grade information is included in the table when available.
Table 1.
Control and HD brains used for western blot analysis of HTTex1p.
| CAG | Category* | Age | Sex | Brain region | Source (see footnote) | Other name | Publication | |
|---|---|---|---|---|---|---|---|---|
| 1060 | 17/17 | Control | 55 | m | cortex | 1 | C1 | Sapp et al., 2012, Aronin et al., 1995 |
| 1063 | 17/22 | Control | 93 | f | cortex | 1 | C2 | Aronin et al., 1995 |
| 1070 | 9/17 | Control | 68 | m | cortex | 1 | C8 | Sapp et al., 2012, Aronin et al., 1995 |
| 206 | NA | Control | 49 | m | cortex | 2 | Sapp et al., 2012 | |
| A136 | NA | Control | NA | NA | post. parietal cortex | 3 | ||
| Z006 | 17/18 | Control | 74 | m | putamen | 1 | ||
| Z021 | 12/17 | Control | 68 | f | putamen | 1 | Iuliano et al., 2021 | |
| Z060 | 19/25 | Control | 76 | f | putamen | 1 | Iuliano et al., 2021 | |
| Z125 | 15/19 | Control | 64 | m | putamen | 1 | Iuliano et al., 2021 | |
| 1254 | NA | Control | 93 | f | cortex | 4 | ||
| 1067 | 16/43 | HD3 | 62 | f | cortex | 1 | A2 | Aronin et al., 1995 |
| 1069 | 17/43 | HD2 | 57 | m | cortex | 1 | A3 | Sapp et al., 2012, Aronin et al., 1995 |
| 1065 | 25/40 | HD2 | 40 | f | cortex | 1 | A11 | Sapp et al., 2012, Aronin et al., 1995 |
| 1429 | 18/41 | HD | 72 | m | cortex | 4 | Iuliano et al., 2021 | |
| 1484 | 25/44 | HD | 78 | f | cortex | 4 | Iuliano et al., 2021 | |
| 253 | 20/43 | HD1 | 45 | f | cortex | 2 | ||
| 210 | 43/48 | HD | 43 | f | cortex | 2 | Sapp et al., 2012 | |
| A143 | NA | HD1 | NA | NA | post. parietal cortex | 3 | ||
| A122 | NA | HD4 | NA | NA | post. parietal cortex | 3 | ||
| Z008 | 18/41 | HD | 61 | f | putamen | 1 | Iuliano et al., 2021 | |
| Z063 | 24/44 | HD | 59 | f | putamen | 1 | ||
| Z076 | 18/53 | HD | 29 | m | putamen | 1 | ||
| 3053 | 27/42 | HD3 | 66 | f | caudate or putamen | 5 | A12 | Sapp et al., 2012, Aronin et al., 1995 |
| 1057 | 17/42 | HD1 | 32 | f | cortex | 1 | A4 | Sapp et al., 2012, Aronin et al., 1995 |
| 1068 | 15/69 | HD3 | 28 | f | cortex | 1 | J5 | Sapp et al., 2012, Aronin et al., 1995 |
| 1066 | 17/105 | HD4 | 12 | f | cortex | 1 | J6 | Aronin et al., 1995 |
| 4501 | NA | HD4 | 8 | f | cortex | 2 | ||
| 3123 | 17/83 | HD | 16 | f | putamen | 5 | J11 | Aronin et al., 1995 |
Massachusetts General Hospital Neuropharmacology Laboratory Brain Bank
New York Brain Bank at Columbia University
Anton Reiner, University of Tennessee
Massachusetts Alzheimer’s Disease Resource Center
Harvard Brain Tissue Resource Center
Number represents HD grade based on Vonsattel grading system when neuropathological classification was performed (Vonsattel et al., 1985); NA=not available.
Mouse brain lysates prepared fresh and remaining from prior studies were used (Alterman et al., 2019;Sapp et al., 2020;O’Reilly et al., 2023;Yamada et al., 2024). The animal protocols for these studies were approved by the MGH Subcommittee on Research Animal Care (SRAC)-OLAW protocol #2004N000248 and University of Massachusetts Chan Medical School Institution Animal Care and Use Committee (IACUC PROTO202000010). All procedures conform to the USD Animal Welfare Act, the ‘ILAR Guide for the Care and Use of Laboratory Animals and followed institutional guidelines. HD knock-in mice (Q50, Q80, Q111, Q140 and Q175) were obtained from Jackson laboratories. CAG repeat determined from tail DNA was done in house or was provided by the supplier at the time of delivery. Mice were euthanized and the brain was removed and fresh-frozen or a cardiac perfusion with PBS was performed before brain removal and freezing at −80°C. The cortex from an R6/2 mouse was provided by Dr. Vanita Chopra.
For analysis of MSH3 silencing, samples of mouse caudate putamen from a prior study (O’Reilly et al., 2023) and from a new cohort of mice that were injected bilaterally (125 μg per ventricle) in the lateral ventricle with di-valent siRNA directed to a non-targeting control (NTC) or MSH3–1000 at three months and sacrificed at 5 months. Sequence and chemistry of the siRNAs was previously published (O’Reilly et al., 2023). Needle punches of the caudate putamen were taken from brain slices to make crude homogenates (CH).
Preparation of lysates for SDS-PAGE from human and mouse brains.
Lysates from brain tissue were prepared using one of two basic methods reported in our prior studies (Kegel et al., 2000;Sapp et al., 2020). Pieces of human putamen or cortex or mouse caudate putamen (1–2 mm3) were homogenized in 10mM HEPES pH7.2, 250mM sucrose, 1mM EDTA + Protease inhibitor tablet (Roche) + 1mM NaF + 1mM Na3VO4. An aliquot of the CH was removed, and, in some cases, the sample was centrifuged at 800×g for 15 minutes at 4°C. The supernatant (S1) from this spin corresponds to the cytoplasmic fraction and the pellet (P1), which was resuspended in the same buffer, includes nuclear proteins. For most experiments reported here the CH was used as indicated in figure legends.
A second method of preparation of brain lysates required a different buffer. In brief, both striata per mouse brain were homogenized in 3 ml of 0.32M sucrose, 10mM DTT + Protease inhibitor tablet (Roche) and 100 μl was removed as a CH. Some lysates that had been prepared in this manner were used in a published study of 6-month-old WT, Q50, Q80, Q111, Q140 and Q175 mice (Sapp et al., 2020). The remaining samples that were stored frozen were used in the current study.
To solubilize aggregated HTT, two methods were tested including our previously published protocol (Sapp et al., 2012). Specifically, 20 μg of CH prepared in 10mM HEPES pH7.2, 250mM sucrose, 1mM EDTA as described above from cortex of control and HD human brain, cortex of R6/2mice, and caudate putamen of 6-month-old WT and Q175/Q7 mice were incubated in 100 mM DTT and 8M urea for 30 minutes at room temperature with vortexing every 5 minutes before adding LDS sample buffer (Invitrogen) and separating by SDS-PAGE. A second method described by Landles et al. (2010) uses a sequential treatment with SDS followed by formic acid (Landles et al., 2010). 50 μg of R6/2 cortex crude homogenate was centrifuged at 13000 ×g for 15 min at 4°C and the pellet was resuspended in 50 μl 1× SDS buffer (2% SDS, 5% beta-mercaptoethanol, 15% glycerol). 50 μg of P1 fraction from HD and juvenile HD patient putamen (3053 and 3123, respectively) was diluted to 25 μl with H20 and 25 μl 2× SDS buffer (4% SDS, 10% beta-mercaptoethanol, 30% glycerol) was added. All samples were boiled for 10 minutes, sonicated for 20 seconds, centrifuged at 13000×g for 15 min at 4°C and the supernatant was removed as the SDS soluble fraction. 100 μl formic acid was added to the pellets and the samples were incubated at 37°C with shaking at 250 rpm for 1 hour, then dried overnight under vacuum in a desiccator. Dried formic acid pellets were neutralized with 1M Tris pH8.6 before adding LDS sample buffer, boiled for 10 minutes and the entire sample was separated by SDS-PAGE as described below.
SDS-Page and western blot
Protein concentration was determined using the Bradford assay (BioRad kit). Equal amounts of protein were prepared in 1X LDS buffer (Invitrogen) + 100mM DTT, boiled for 5 minutes, separated by SDS-PAGE using 3–8% Tris-acetate (NuPAGE 15-well, 1.5mm, Invitrogen or Criterion 26-well, 1.0mm, BioRad) or 4–12% Bis-tris gels (NuPAGE 10-well, 1.5mm, Invitrogen) at 120V until the dye-front reached the bottom of the gel (~1.5 hours), then gels were soaked in Tris-glycine transfer buffer + 0.1% SDS for 5 minutes before transfer. We compared wet transfers at 100V for 1 hour and 35V overnight to our normal transfer using TransBlot Turbo (BioRad) and did not see an appreciable change in the detection of the high molecular weight protein, so we transferred proteins to nitrocellulose with the TransBlot Turbo apparatus at high molecular weight setting (25V for 10 minutes) throughout this study. Blots were incubated in blocking buffer (5% blotting-grade blocker (BioRad) in TBST) for 1 hour at room temperature, then in primary antibody diluted in blocking buffer at 4°C overnight with agitation.
Sources of antibodies used for western blot.
Antibodies and dilutions were as follows: anti-HTT antibody Ab1 (aa1–17, (DiFiglia et al., 1995), 1:2000, rabbit), MW8 (Developmental Studies Hybridoma Bank, University of Iowa, 1:500, mouse), S830 (generous gift from Dr. Gillian Bates, 1:6000, sheep), P90 antibodies (neo-epitope to aa83–90, clones 1B12 and 11G2, Coriel Institute, 5 μg/ml, rabbit), MSH3 (Santa Cruz BioTech #sc-271079, 1:500, mouse), GAPDH (MilliporeSigma #MAB374, 1:10000, mouse), B-actin (MilliporeSigma #A5441, 1:5000, mouse). Blots were washed in TBST, then incubated in peroxidase-labeled secondary antibodies diluted 1:2500 for rabbit IgG and 1:5000 for mouse and sheep IgG in blocking buffer for 1 hour at room temperature. Blots were washed in TBST, and bands were visualized using SuperSignal West Pico PLUS Chemiluminescent substrate (Thermo Scientific) and ChemiDoc XRS+ with Image Lab software (BioRad). Some blots were stripped for 30 minutes with Restore stripping buffer (Thermo Scientific) before washing, blocking and reprobing with a different antibody.
Pixel intensity quantification.
Image analysis was performed using ImageJ software. Bands were manually circled, and the area and average intensity were determined. The total intensity of each band was calculated by multiplying the area by the average intensity. For the Q111 mice treated with MSH3, the average intensity of the smear was measured in equal areas for each lane corresponding to the upper part of the smear down to the lower edge of the longest smear and the same for the bands a (see Fig. 6B where the dashed lines in the first lane of the western blot show the areas measured). The total intensity of the bands or average intensity of the smear was normalized to the loading control signal of B-actin or GAPDH. The intensity of HTTex1p smear in each mouse treated with NTC or siRNA MSH3-1000 was calculated as a ratio relative to levels of MSH3.
Fig. 6. Levels of HTTex1p in HD mice caudate putamen: Effects of protein concentration and treatment with siRNA to MSH3.
A. Signal for HTTex1p detected with P90 antibody is concentration dependent. S1 fraction from Q111 mouse 2751 was loaded at different protein concentrations for SDS-PAGE (5–40 μg) using 3–8% Tris-acetate 15-well gel. Western blot probed with P90 shows signal for 75 kD band (arrow) increases with increasing protein concentration (graph below, correlation co-efficient, R2 =0.9898, p=0.0051). HTTex1p smear at bracket is also concentration dependent. Similarly full length HTT detected with Ab1 (above) and GAPDH (below) increase with protein concentration. The 75 kD band at level of arrow was detected in all Q111 samples but not WT. B, C, D. Effects of reducing levels of MSH3 mRNA in Q111 mice on HTTex1p levels. Three experiments show that HTTex1p smear is reduced in most lanes where MSH3 levels are markedly reduced. Graphs show intensity of HTTex1p smear and 75 kD band (at arrow in B). Equal areas of the smear and 75 kD band as indicated by dashed lines in the first lane in B were measured by densitometry. Graphs below western blots show signal intensity of 75 kD band is not changed but HTTex1p smear is significantly reduced in lysates from siRNA MSH3-treated mice compared to NTC (D, *p=0.01813, unpaired t test, N=7 NTC and 7 MSH3. Data from mouse at open arrowhead is removed because the signal for HTTex1p and the loading control were not normal.). Note that there is 1 mouse in B and 1 in C that have a high HTTex1p signal indicated by arrowhead on western blots and graphs. If these mice are removed from the analysis, the levels of HTTex1p would be significantly less in the MSH3siRNA-treated mice in each experiment. Blots were stripped and re-probed with anti-HTT antibody Ab1 (aa1–17), anti-MSH3 and anti-B-actin antibodies. E. There is a significant correlation between MSH3 and HTTex1p signal intensities when the data from the 3 experiments were combined (R2 = 0.3040; ***p=0.0003, n= 19 NTC and 19 MSH3 mice). 20 μg crude homogenate samples were separated on 3–8% Tris-acetate 15-well gel in B and C and 26-well gel in D. HTTex1p was detected with P90 antibody 1B12.
Procedures on HD human tissue sections for double immunofluorescence.
Tissue sections were processed for immunofluorescence as previously described (Deng et al., 2021). Briefly sections were treated with 0.1 M sodium borohydride/0.1 M D-lysine in 0.1 M PB for 1 hour, 0.5% H2O2 PB for 1 hour then 1% nonfat milk PBTX for 2 hours. Tissue sections were incubated in primary antibody combinations at 4°C for 3 days then in secondary antibody combinations for 2 hours at room temperature. Antibody combinations were: Primary antibody 1: Mouse MW8 (IgG2a antibody). Secondary antibody: Goat anti-mouse IgG2a Alexa 594. Primary antibody 2: Rabbit P90 −11G2 or 1B12 (IgG 1k antibody). Secondary antibody: Goat anti-rabbit IgG Alexa 647. Tissue sections were then mounted on gelatin-coated slides, dried, treated with Sudan black B solutions and cover-slipped with antifade solutions.
Results
HTTex1p in human HD brain exhibits CAG repeat expansion
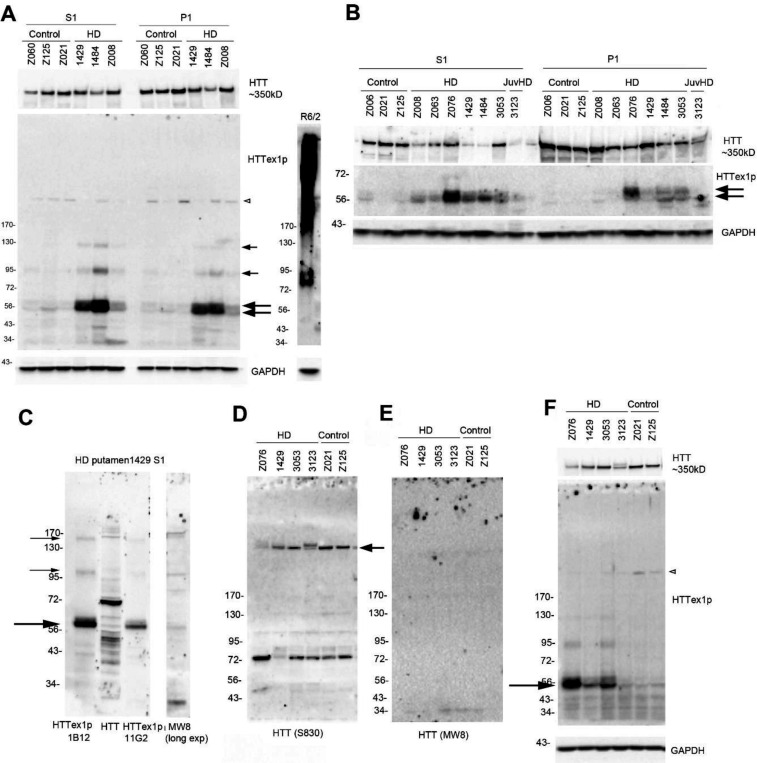
To detect HTTex1p, we examined subcellular fractions from human putamen and cortex of HD and control brains. In human HD putamen, HTTex1p appeared in S1 and P1 fractions as one or two bands migrating to about 56–60 kD (Fig. 1A, B, large arrows) and as higher molecular mass (HMM) proteins of about 95 kD and 125 kD (A, small arrows) consistent with CAG repeat expansions. In contrast to human brain, HTTex1p in cortex of a R6/2 mouse that expresses human exon 1 with about 115 CAG repeats migrated to about 85 kD and as a broad SDS soluble smear that extended from the top of the blot to about 170 kD (Fig. 1A). HTTex1p levels in human putamen were generally greater in S1 than P1 fractions (Fig. 1B) and were detected in lysates that had been stored frozen (Fig. 1A) or prepared fresh (Fig. 1B). Control human brain had no or very weak signal for HTTex1p at 56–60 kD. Blots re-probed with an antibody that detects HTT1–17 (Ab1) revealed the presence of full length HTT in all samples (Fig. 1A,B).
Fig. 1. HTTex1p in HD putamen migrates at 56/60 kD and as higher molecular mass bands in S1 and P1 fractions.
A. S1 and P1 fractions were prepared from putamen of 3 HD brains (#1429, 1484 and Z008) with adult-onset HD and 3 control brains (Z060, Z125 and Z021) as described in methods and were stored frozen at - 80°C. Prominent signal for HTTex1p occurs at 56–60 kD in two HD cases, 1429 and 1484, and less so in Z008 (arrows). There is also a doublet seen at lower levels in the control brains. HMM bands (small arrows) migrate to about 95 kD and 125 kD in the S1 and P1 fractions of the HD brains. P90 antibody cross-reacts with a 200 kD protein present in control and HD brains (open arrowhead). Western blot membrane was re-probed with anti-HTT antibody Ab1 (aa1–17) to detect full-length HTT and is shown above. R6/2 mouse cortex was run on a separate gel and HTTex1p runs at about 85 kD with a broad SDS soluble smear that reaches the top of the gel which was not detected in human putamen. 20 μg samples were run on a 15-well, 3–8% Tris-acetate gel. B. Fresh S1 and P1 fractions were prepared from putamen of the same brains shown in A as well as 3 more adult onset (Z063, Z076, and 3053) and 1 juvenile HD patient (3123). HTTex1p is generally detected as stronger 56–60 kD bands in the HD S1 fraction compared to the P1 fraction and 2 separate bands can clearly be seen in some of the P1 fractions (arrows). 20 μg samples were run on a 26-well, 3–8% Tris-acetate gel. C. HTTex1p in HD putamen co-migrates with N-terminal HTT fragments detected with anti-HTT Ab1 and MW8 antibodies. Shown is HD putamen of case 1429 S1 fraction also shown in 1A. HTTex1p is detected with 2 P90 antibodies (1B12 and 11G2) and HTT proteins are detected with Ab1 (aa1–17) and MW8 (aa 83–90, exon 1 protein). HTTex1p migrates at about 60 kD with both P90 antibodies and appears as an intense band capped by a weaker intensity slight slower migrating band that may be HTTex1p with a longer CAG repeat or as a post translational modification. Ab1 and MW8 detect bands that co-migrate with the P90 bands (large arrow). Here antibody P90 detects a stronger signal for HTTex1p than does 11G2 and reacts with slower migrating proteins at 100 kD and 160 kD (small arrows), which may be due to CAG repeat expansion. 20 μg samples were run on a 10-well, 4–12% Bis-tris gel. D, E. 15 μg crude homogenate from 4 HD patient and 2 control putamen were run on 3–8% Tris-acetate 15-well gels and probed with anti-HTT antibodies S830 (D) and MW8 (E). S830 detects full length HTT at arrow in control and HD and a band at 72 kD. MW8 shows no signal except for a very weak band below 34 kD. F. The MW8 blot shown in E was stripped and re-probed with P90 antibody. HTTex1p specific band appears at 56–60 kD as in A and B and here is strongly detected in 3 of the 4 HD cases and weakly in the 2 control brains (arrow). P90 also reacted with a 200 kD protein in WT and HD putamen (open arrowhead). HTTex1p was detected with P90 antibody 1B12 except in C where noted.
To determine if HTTex1p detected by P90 antibody in HD putamen was also recognized by other anti-HTT antibodies, membranes with transferred proteins were cut into strips and probed with MW8 (aa83–90), Ab1 (aa1 −17) or P90 (1B12 and 1G11). Results showed that Ab1 and MW8 reacted with proteins that co-migrated with HTTex1p and with other N-terminal fragments not recognized by P90 (Fig. 1C). To confirm specificity of P90 for HTTex1p in human HD brain, proteins transferred to nitrocellulose membranes were probed with S830 antibody (Fig. 1D) and with MW8 (Fig. 1E) and then stripped and probed with P90 (Fig. 1F). Only P90 antibody detected the HTTex1p at 56–60 kD in human HD putamen (3 of the 4 HD samples at arrow).
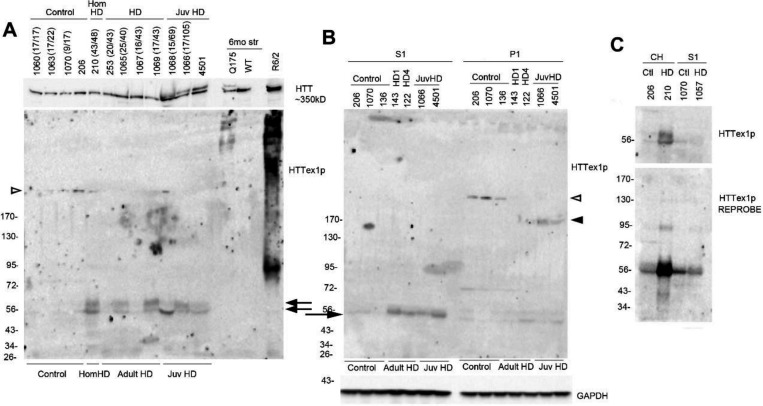
Next, we examined HTTex1p in postmortem cortex of adult and juvenile onset HD cases (for CAG repeat number see Table 1). In crude homogenates (CH) HTTex1p migrated to 56–60 kD (Fig. 2A, arrows). By comparison, HTTex1p in CH of cortex from R6/2 mice migrated to 85 kD and from 6-month Q175 mouse caudate putamen migrated at the top of the blot as a few HMM proteins. Treating CH with 8M urea before SDS PAGE had no effect on the solubility of HTTex1p in either human or mouse brain samples (Supp. Fig. 1). Applying a protocol by Landles et al. (Landles et al., 2010), in which the P1 fraction was treated with SDS followed by formic acid, the HMM smear in the R6/2 cortex CH was partially solubilized to a discrete band (Supp. Fig. 2A, small arrow) but there was no increase in HTTex1p signal at 85 kD (Supp. Fig. 2A, large arrow). Both bands were cleared with formic acid treatment. In the P1 fraction from the human HD putamen, there was no change in HTTex1p signal with SDS solubilization and the protein was not detected after treatment with formic acid (Supp. Fig 2B). HTTex1p migrated to 56–60 kD in the S1 fraction, and in the P1 fractions from two of four juvenile onset HD patients at 170 kD signifying CAG repeat expansion (Fig. 2B, arrowhead). HTTex1p was typically absent in western blots of human control cortex but appeared after re-probing with P90 antibody suggesting that HTTex1p occurred at low levels in control brain (Fig. 2C). P90 antibody reacted with a 200kD protein in samples of human control and HD brain (Figs. 1, 2, open arrowheads). Altogether, the results established presence of HTTex1p in human HD putamen and cortex and provided evidence for CAG repeat expansion of HTT1a mRNA. HD mice also expressed HTTex1p as a product migrating to about 85 kD and as a SDS soluble smear due to the presence of larger CAG repeats in exon 1 (115Q in R6/2 and 175Q in Q175 mice), and higher levels of protein expression from HTT exon1 in R6/2 mice.
Fig. 2. HTTex1p in HD human postmortem cortex.
A. Crude homogenates from postmortem control and HD cortex and from caudate putamen of 6-month-old of HD Q175 mice and cortex of a R6/2 mouse were probed with anti-HTTex1p antibody P90. Two bands at 56kD-60kD (arrows) are detected in 6 of the 8 HD cases and not in the controls and do not migrate in relation to CAG repeat which is indicated at the top of the figure for each case that was known. For comparison R6/2 mice express prominent HTTex1p at 80 kD and as a HMM smear which was not detected in human cortex. Lysates from HD Q175 mice express a few HMM bands for HTTex1p. Blot was re-probed with anti-HTT antibody Ab1. 20 μg samples were separated on a 3–8% Tris-acetate 26-well gel. B. HTTex1p in S1 and P1 fractions from postmortem adult and juvenile HD cortex. S1 and P1 fractions were prepared as described in methods and 20 μg samples were separated on a 3–8% Tris-acetate 26-well gel. Western blot shows presence of 56–60 kD band in S1 fraction of the adult and juvenile onset HD cases and at much lower level in the P1 fraction (arrow) as well as a HMM band at 170 kD in the P1 fraction of two juvenile HD cases (arrowhead). Samples of postmortem brains 136 (WT), and 143 and 122 (HD) are A7 posterior parietal cortex from A. Reiner collection where P90-positive aggregates were found detected by IHC. Note the presence of a 200 kD band in control brain (open arrowhead). C. HTTex1p in HD cortex that is revealed with re-probe of western blot. Top blot: CH or S1 fractions are from postmortem brain of controls (206,1070) homozygote (210, CAG repeat is 43/48) and presymptomatic HD patient (1057, grade 1, CAG repeat 17/42). HTTex1p in homozygote 210 is detected as two bands at 56–60 kD but signal is low or absent in all the other samples. The lower blot is a re-probe with P90 antibody. The signal for HTTex1p in cortex of homozygote (210) increases and is also detected in lysates of presymptomatic (1057) and WT controls (206, 1070). 20 μg samples were separated on a 3–8% Tris-acetate 26-well gel. HTTex1p was detected with P90 antibody 1B12.
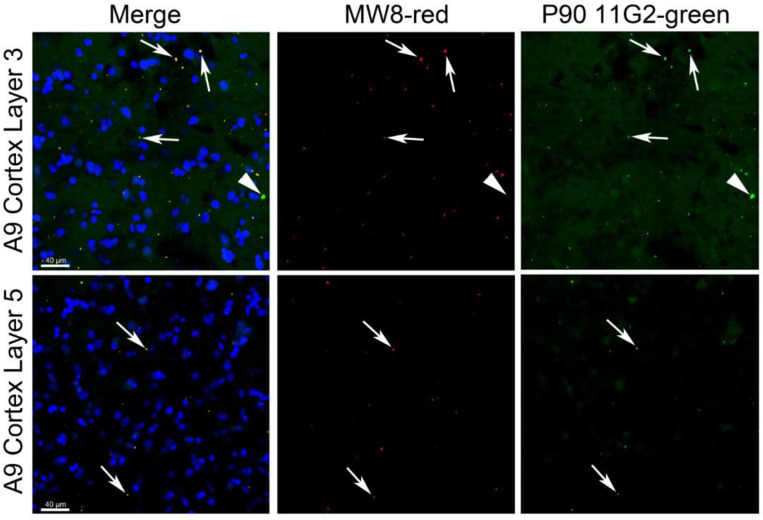
HTTex1p immunofluorescence labeling with P90 11G2 antibody reveals neuropil aggregates in layers 3 and 5 of human HD cortex
Western blot results described above showed HTTex1p in human HD cortex and putamen. To further support the presence of HTTex1p in human HD brain, we performed immunofluorescence staining with P90–11G2 antibody in cortex samples, Brodmann areas 3 and 9, from an adult-onset HD grade 1 patient (case 143 shown in Fig. 2B). Results revealed a sparse distribution of P90–11G2 labeled aggregates in layers 3 and 5 which are enriched in pyramidal neurons (Fig. 3). The NeuroTrace signal used to identify neuronal perikaryal showed that the P90–11G2 positive aggregates were located in the neuropil. Httex1p immunoreactive aggregates varied in size, some being relatively large, and they were partially co-labeled with MW8 antibody which by western blot detected a band that co-migrated with HTTex1p in human HD putamen (Fig. 1E) and has been used in HTRF and MSD based aggregation assays to identify HTTexp1 in cortex of HD knock-in mice (Landles et al., 2024). Thus, the immunofluorescence data support our western blot findings for the presence of HTTex1p in human HD cortex.
Fig. 3. Immunofluorescence staining of HTTex1p with P90 antibody.
Layer 3 (top panel) and layer 5 (bottom panel) of A9 cortex from grade 1 HD patient A143 stained with MW8 (red, merge and middle), P90 11G2 (green, merge and right) and NeuroTrace (blue, merge). Arrows indicate co-labeling of aggregates with both MW8 and P90. Arrowhead on right in upper panel shows a larger aggregate that is positive for P90 but does not colocalize with MW8. Scale bar in merge images is 40 μm.
Solubility of HTTex1p protein in brain of HD knock in mice and YAC128 transgenic mice is dependent on age and CAG repeat length
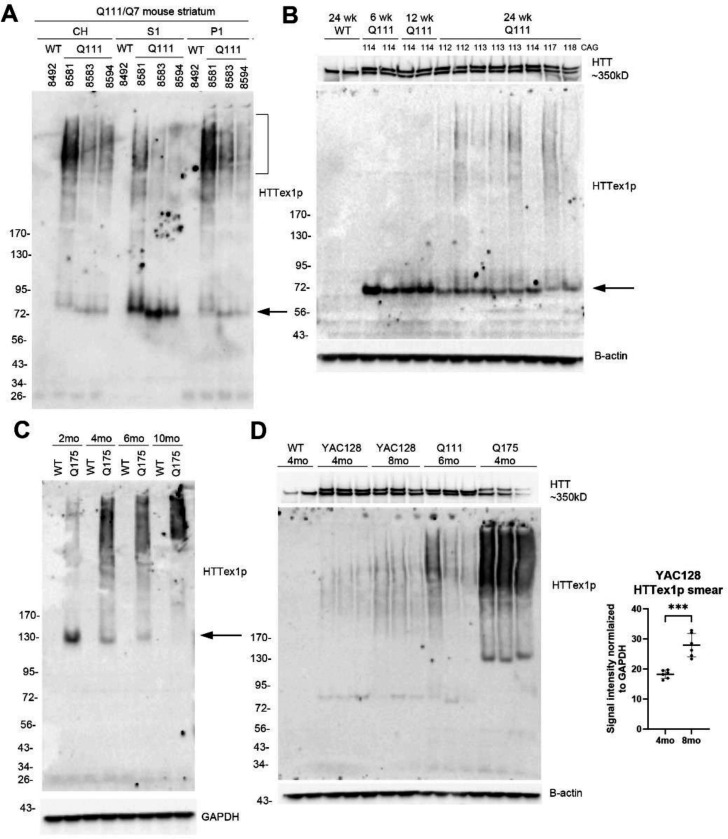
CAG repeat expansion in HTT1a drives aggregation of HTTex1p and nuclear localization (Dragileva et al., 2009;Fienko et al., 2022;Landles et al., 2024). To further address HTTex1p solubility, we examined subcellular fractions from HD knock-in mice Q111 and Q175, which belong to an allelic series that have human exon 1 inserted into the mouse exon 1 locus. The presence of HTTex1p in crude homogenates, S1, and P1 fractions of caudate putamen from 6-month-old Q111 mice were analyzed together by western blot (Fig. 4A). HTTex1p migrated to about 75 kD in S1 fractions and as a smear in crude homogenates and P1 fractions (Fig. 4A). In S1 fractions from 6-, 12- and 24-week-old Q111 mice compared in the same western blot, HTTex1p migrated to about 75 kD (Fig. 4B, arrow), but levels diminished with age to about 50% at 24 weeks when HTTex1p solubility diminished and was replaced by a HMM smear (Fig. 4B). No smear appeared in WT mice lysates at 24 weeks. HTTex1p was examined in Q175 mice of different ages where it migrated to about 130 kD (arrow) and decreased in intensity with age between 2 months and 10 months coinciding with a marked rise in SDS soluble smear (Fig. 4C). HTTex1p smear was also significantly increased at 8 months compared to 4 months in lysates from YAC128 mice (Fig. 4D). Altogether these data show that HTTex1p migration by western blot was age- dependent migrating more slowly by SDS-PAGE with increased CAG repeat expansion, although other factors affecting HTTex1p migration cannot be ruled out.
Fig. 4. Effects of age on HTTex1p expression in HD Q111, Q175 mice and YAC128 mice. Changes in levels, molecular mass and SDS solubility.
A. Crude homogenates (CH) as well as S1 and P1 fractions from the same mice were separated on a 3–8% Tris acetate 26-well gel and probed with P90 antibody. HTTex1p migrates to 75 kD (arrow) and is most expressed in the S1 fraction whereas it appears as a high molecular mass smear (HMM) (bracket) in CH and P1 fraction. B. Western blot of S1 fractions of caudate putamen from WT mouse at 24 weeks and Q111 mice at 6, 12 and 24 weeks. 20 μg samples were separated on a 15-well 3–8% Tris-acetate gel. HTTex1p migrates to about 75 kD (arrow), and intensity decreases progressively between 6 weeks and 24 weeks. Note that at 24 weeks HTTex1p smear appears in Q111 mice but not in WT mouse. CAG repeat was determined from tail DNA and is indicated at the top of the blot. Blot was re-probed with anti-HTT antibody Ab1 to detect full-length HTT. C. Western blot of crude homogenates of WT and Q175 mice at 2, 4, 6 and 10 months. 20 μg samples were separated on a 3–8% Tris-acetate 26-well gel. HTTex1p band migrates to about 130 kD (arrow), and intensity decreases with increased age from 2 months to 10 months. In contrast presence of HMM HTTex1p smear increases with age. HTTex1p is not detected in the WT mice. D. Western blot of CH prepared from caudate putamen of 4- and 8-month-old YAC128 mice show a significant increase in the HTTex1p smear from 4 to 8 months (***p<0.001, unpaired t test, n=8). Images for quantification are shown in Supp. Fig. 3. The HTTex1p band in the YAC128 mice runs at about 80 kD which is slightly larger than in the Q111 mice, as expected. Note that the loading controls, which are B-actin for B and D and GAPDH for C, show equal signal intensities indicating equal protein loading. HTTex1p was detected with P90 antibody 1B12.
We also compared HTTex1p expression in HD knock-in mice that had different CAG repeats in exon 1. (Fig. 5A). CH from caudate putamen of 6-month-old WT, Q50, Q80, Q111, Q140 and Q175 mice showed differences in HTTex1p migration. HTTex1p was ~65 kD in Q80 mice, 75 kD in Q111 mice, 100 kD in Q140 mice and 130 kD in Q175 mice (Fig. 5A small arrows). The intensity of HTTex1p also significantly increased with CAG repeat size (graphs in Fig. 5A). A HTTex1p smear appeared in Q111, Q140 and Q175 mice and increased in signal intensity with increase in CAG repeat number (graphs in Fig. 5A). HTTex1p was not detected in WT or Q50 mice. N-terminal HTT1–17 antibody Ab1 detected full length WT and mutant HTT at about 350 kD in all mice. In Q175 mice full-length HTT levels were significantly lower suggesting that the marked HTTex1p accumulation seen as a smear on western blot in these mice may affect levels of full length HTT (Fig. 5A, top).
Fig. 5. Intensity, size, and solubility of HTTex1p in caudate putamen of 6-month-old HD knock-in mice allelic series.
A. SDS-page and western blot probed with P90 in crude homogenates from caudate putamen of Q50, Q80, Q111, Q140 and Q175 mice. 15 μg samples were separated on 3–8% Tris-acetate 26-well gels. Numbers at top are CAG repeats determined from tail DNA. Large blot shows HTTex1p probed with P90 and the strip below is loading control for GAPDH. Migration of HTTex1p (arrows) slows with increasing CAG repeat from about 65 kD in HD Q80 to about 130 kD in HD Q175 mice. HTTex1p is not visible in WT or Q50 mice. HTTex1p smear is seen in lysates from Q111, Q140 and Q175 mice. Strip above large blot is a re-probe with anti-HTT antibody Ab1 and shows WT and mutant HTT. Note decline in levels of full-length WT and mutant HTT in Q175 samples. At right top graphs show intensity of HTTex1p and HTTex1p smear normalized to GAPDH. Bottom graph shows levels of WT and Mutant HTT normalized to GAPDH (*p<0.05, **p<0.01, ***p<0.01, One-way ANOVA with Tukey’s multiple comparison test, n=3 per group). B-D. Comparison of antibodies S830, MW8 and P90. 15 μg crude homogenates from n=1 each WT, Q50, Q80, Q111, Q140, Q175 striatum and R6/2 cortex were separated on 2 3–8% Tris-acetate 15-well gels, one probed with anti-HTT antibody S830 (B) and the other with anti-HTT antibody MW8 (C). The blot in C was stripped and re-probed with P90 antibody (D). In B, C, and D, all 3 antibodies detect 1–2 bands in R6/2 mouse cortex at ~90 and 100 kD and a prominent HMW SDS soluble smear. S830 detects full length HTT (arrow on left) in all cortex samples with a stronger signal for mutant than WT HTT. In C, MW8 detects the HTT exon 1 protein at ~60 kD in all HD knock-in mouse models. In D, P90 antibody detects HTTex1p which migrates in relation to its CAG repeat: at 65 kD for Q80, 75 kD in Q111, 100 kD in Q140 and 130 kD in Q175 (arrows on left). No CAG length dependent bands are detected in HD knock-in mice with S830 or MW8 (B, C). HTTex1p was detected with P90 antibody 1B12.
We compared HTTex1p detection with P90 antibody to two other anti-HTT antibodies S830 and MW8 in crude homogenates from mice with different CAG lengths, including R6/2 mice. All 3 antibodies detected a prominent HMM smear in the R6/2 mice and 1 or 2 bands ~90–100 kD (Fig. 5B, C, D). S830 also detected a very faint HMM smear in the Q111, Q140 and Q175 mice but no bands were dependent on CAG repeat length (Fig. 5B). There was no CAG-dependent HMM smear in these mice detected with MW8 antibody (Fig. 5C). Altogether these findings suggest that P90 neo-epitope antibody detects a combination of features of HTTex1p in the HD knock in mice that are not seen with other N-terminal anti-HTT antibodies.
HTTex1p smear decreases in caudate putamen of Q111 mice after lowering levels of MSH3, a mismatch repair protein.
Results above in HD knock in mice showed that the detection of HTTex1p by western blot was dependent on its subcellular compartment, age of mice, and CAG repeat length. To determine if HTTex1p detected with P90 antibody could be quantified by western blot assay, we compared different protein concentrations (5, 10, 20, and 40 μg) from the same Q111 mouse brain (2751) and performed densitometry. Results showed a concentration dependent increase in signal for HTTex1p, signifying the antibody was sufficiently sensitive to use for a quantitative assay (Fig. 6A).
The mismatch repair protein MSH3 is required for CAG repeat expansion and nuclear localization of mutant HTT (Dragileva et al., 2009). Previously we showed that treating 3-month old Q111 mice with MSH3–1000, a divalent siRNA targeting MSH3, lowered MSH3 protein by 54% and inhibited CAG repeat expansion at 5 months compared to treatment with a non-targeting control (NTC) (O’Reilly et al., 2023). In lysates from caudate putamen of mice treated with MSH3–1000 from this study along with two other cohorts (N=6–7 mice/study), there was a significant depletion in levels of HTTex1p smear that positively correlated with reduction in MSH3 protein levels (Fig. 6B–E, R2=0.3040, p=0.0003, n=19 mice per group from 3 studies). HTTex1p smear was conspicuously lowered in mice where MSH3 protein was reduced by 60–75% (Fig. 6B–D). In the same mice, there was no change in levels of full-length HTT. These results demonstrate that the changes in HTTex1p smear detected by P90 results from CAG repeat expansion in HTT1a and can be used to quantify the effects of treating HD Q111 mice with agents that inhibit somatic expansion.
Discussion
Aberrant splicing between exon 1 and exon 2 of HTT due to the presence of a cryptic polyadenylation in intron 1 produces an exon 1 mRNA named HTT1a that terminates at a proline residue (aa 90, based on 23 CAGs) (Sathasivam et al., 2013;Neueder et al., 2017). This fragment was identified as translating the endogenously generated HTT exon 1 protein product that may nucleate and recruit aggregates. The splicing anomaly is more frequent in human than in mouse HTT (Franich et al., 2019;Fienko et al., 2022). HTT1a mRNA has been detected in human HD brain, in a wide range of human peripheral tissues and in HD knock-in mouse models that express human exon 1 and its levels of expression are CAG repeat length dependent (Neueder et al., 2017;Fienko et al., 2022;Hoschek et al., 2024). HTTex1p levels in human and mouse HD brain has been assessed using immunoprecipitation and HTRF assays with N-terminal HTT antibodies that individually are not specific for the endogenous protein (Figure 4 in (Neueder et al., 2017)). Here we examined lysates from HD human, transgenic (R6/2, YAC128) and knock-in mouse brain lysates by SDS-PAGE and western blot using neo-epitope specific monoclonal antibody P90 made to the predicted C-terminal 8 amino acids of the HTT exon 1 protein (Baldo et al., 2024;Missineo et al., 2024). HTTex1p appeared in human HD putamen and cortex as 56–60 kD bands and in some HMM HTTex1p products consistent with the presence of CAG repeat expansion in HTT1a. In contrast to human samples, six-month-old mice from the “allelic series” of HD knock-in mouse models resolved HTTex1p as discrete bands and as prominent HMM smears that varied in migration and signal intensity in relation to CAG repeat length and age of the mice. The smears appeared in 6-month old Q111, Q140 and Q175 mice, consistent with the presence of aggregates that are prevalent by this age with immunostaining (Wheeler et al., 2000;Wheeler et al., 2002;Hickey et al., 2012;Smith et al., 2023) and in 4- and 8-month old YAC128 mouse striatum concurrent with loss of striatal volume at 3 months of age (Carroll et al., 2011) and appearance of EM48-positive nuclear aggregates by immunostaining at 2 months (Van Raamsdonk et al., 2005). Moreover, densitometry showed that the HTTex1p smear was significantly reduced in caudate putamen of 5-month-old Q111 mice treated at 3 months with siRNA targeting MSH3. O’Reilly and colleagues showed there was no difference in the instability index between the baseline 3-month mice and those treated with MSH3 siRNA for 2 months whereas mice treated with NTC had a significantly higher instability index suggesting that further expansion was blocked (O’Reilly et al., 2023). These findings support the presence of HTTex1p in human and mouse HD brain and the feasibility of using western blot detection of HTTex1p in HD mice as a biomarker to test the effects of agents that limit CAG repeat expansion.
The 56 kD HTTex1p detected in human HD cortex and putamen brain was more prevalent than the 60 kD band and co-migrated with bands detected with N-terminal anti-HTT antibodies Ab1 (HTT aa1–17) and MW8 (HTT aa83–90). MW8 has been used in HTRF and MSD aggregation assays to detect HTT1a protein in mouse HD cortex (Landles et al., 2024). Thus the presence of immunoreactive puncta we observed in human HD grade 1 (case 143) cortex that are co-labeled with P90–11G2 and MW8 antibodies supports the aggregate nature of these structures and is consistent with the aggregation prone nature of exon 1 protein in vitro (Scherzinger et al., 1997) and the formation of aggregated mutant huntingtin in dystrophic neurites in human HD cortex (DiFiglia et al., 1997). It is conceivable that the more prevalent 56 kD HTTex1p detected by western blot is the species that initiates the nucleation of aggregates which has been proposed as a role for HTTexp1 (see Fig 5 of (Neueder et al., 2017)). The slower migrating 60 kD seen in some of the human brains may be a CAG expanded form of HTTexp1 or a post-translational modification of HTTex1p, although the latter has not been reported. The low but distinct signal for HTTex1p in some control human brain samples fits with reports of HTT1a mRNA in control brain and other human tissues using a sensitive qPCR assay (Hoschek et al., 2024). Also, HTTex1p was weakly detected by immunoprecipitation assay in human cerebellum with 22 CAG repeats, using antibodies 3B5H10 in combination with S830 or MW8 (Neueder et al., 2017).
The HTTex1p smear seen in HD mice with P90 antibody may partly correspond to SDS insoluble products identified in R6/2 mice using other biochemical assays. HMM products migrating above 300 kD were found in nuclear fractions from R6/2 mouse brain by agarose gel electrophoresis for resolving aggregates (AGERA) and probed with antibodies 4C9 and MW8 (Landles et al., 2020). Also, in nuclear fractions from brain lysates of R6/2 mice SDS insoluble products remained at the top of a western blot probed with S830 antibody (Landles et al., 2020). Consistent with these protein studies, a screen of antibodies by Bayram-Weston et al. showed that MW8 and S830 antibodies were the most sensitive for identifying nuclear inclusions and diffuse staining in HD transgenic and knock in mice (Bayram-Weston et al., 2016). The loss of HTTex1p smear we observed by western blot after lowering MSH3 mRNA contrasts with limited effects on aggregate formation seen by immunostaining after silencing MSH3 mRNA in HD Q111 mice and Q175 mice (Aldous et al., 2024;Driscoll et al., 2024). This suggests the HTTexp1 smear detected by P90 in western blot is a fraction of the insoluble mutant HTT present in tissue.
The HTTex1p smear detected with P90 antibody in caudate putamen of HD mice, did not appear in WT mice or in human HD postmortem putamen and cortex, although multiple slower migrating HTTex1p bands possibly caused by CAG repeat expansion occurred in human HD putamen, especially in soluble S1 fractions. The marked loss of neurons in HD patient neostriatum and cortex compared to HD knock-in mouse and the proliferation of glial cells which are reported to have less somatic instability than neurons (Vonsattel and DiFiglia, 1998;Shelbourne et al., 2007) could partly account for the paucity of HTTex1p accumulation seen in human HD brain. Also, RNA seq analysis of human HD neostriatum suggests that very large somatic expansions are infrequent and short-lived (Handsaker et al., 2024). Like the HD putamen, HD cortex lacked HTTex1p smears by western blot (only the 56–60 kD doublet) including the P1 fraction, which is nuclear enriched, suggesting that HTTex1p aggregation was not at sufficient levels for detection or is trapped in a nuclear compartment that was inaccessible by western blot. In studies of HD cortex using single serial fluorescence-activated nuclear sorting (sFANS) and single nucleus RNA seq, layer 5a pyramidal neurons in HD postmortem cortex exhibit CAG repeat expansions (Handsaker et al., 2024;Pressl et al., 2024). Since these neurons were reported as lost early in disease, they may be sparse in our postmortem samples. In support of these observations, immunofluorescence labeled HTTex1p aggregates detected with P90 antibody were only scantly present in parietal cortex of two HD postmortem brains that were also examined by western blot with P90 (Fig. 1, HD 143, and HD 122).
In summary, our results showed use of P90 antibodies for western blot detection of HTTex1p in subcellular fractions from human and mouse HD brain and immunofluorescence labeling of small neuropil aggregates in human HD cortex. The sizes and solubility of HTTex1p in HD mice varied with subcellular compartment, age and CAG repeat expansion. Relatively small amounts of tissue are required for detection of HTTex1p by SDS-PAGE and western blot, allowing tissues for other assays to be collected from the same mouse brain. Moreover, the ability to quantify changes in levels of HTTex1p smear in western blots after inhibiting CAG repeat expansion will be a useful readout for preclinical studies.
Supplementary Material
Table 2.
Sizes of immunoreactive HTTex1p detected with P90 antibody in HD mice.
| Mouse model | CAG range | Size of HTTex1p |
|---|---|---|
| R6/2 | 115–150 | 85–90 kD |
| Q80 | 84 | 65 kD |
| Q111 | 108–118 | 72–80 kD |
| Q140 | ~140 | 100 kD |
| Q175 | 185–202 | 130–140 kD |
| YAC128 | ~128 | 80 kD |
Acknowledgements
This work was supported by the Dake family fund, CHDI Foundation (CHDI-6367), and the National Institutes of Health (NIH U01 NS114098).
References
- Aldous S.G., Smith E.J., Landles C., Osborne G.F., Canibano-Pico M., Nita I.M., Phillips J., Zhang Y., Jin B., Hirst M.B., Benn C.L., Bond B.C., Edelmann W., Greene J.R., and Bates G.P. (2024). A CAG repeat threshold for therapeutics targeting somatic instability in Huntington’s disease. Brain 147, 1784–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alterman J.F., Godinho B., Hassler M.R., Ferguson C.M., Echeverria D., Sapp E., Haraszti R.A., Coles A.H., Conroy F., Miller R., Roux L., Yan P., Knox E.G., Turanov A.A., King R.M., Gernoux G., Mueller C., Gray-Edwards H.L., Moser R.P., Bishop N.C., Jaber S.M., Gounis M.J., Sena-Esteves M., Pai A.A., Difiglia M., Aronin N., and Khvorova A. (2019). A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat Biotechnol 37, 884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronin N., Chase K., Young C., Sapp E., Schwarz C., Matta N., Kornreich R., Landwehrmeyer B., Bird E., Beal M.F., and et al. (1995). CAG expansion affects the expression of mutant Huntingtin in the Huntington’s disease brain. Neuron 15, 1193–1201. [DOI] [PubMed] [Google Scholar]
- Baldo B., Peladan J., Albers J., Temowski T.S., Menalled L., Doherty E.M., Macdonald D., and Somalinga B. (Year). “Development of a HTT exon1-selective MSD immunoassay with novel HTT P90 neo-epitope specific antibodies”, in: 19th Annual Huntington’s Disease Therapeutics Conference). [Google Scholar]
- Bayram-Weston Z., Jones L., Dunnett S.B., and Brooks S.P. (2016). Comparison of mHTT Antibodies in Huntington’s Disease Mouse Models Reveal Specific Binding Profiles and Steady-State Ubiquitin Levels with Disease Development. PLoS One 11, e0155834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J.B., Lerch J.P., Franciosi S., Spreeuw A., Bissada N., Henkelman R.M., and Hayden M.R. (2011). Natural history of disease in the YAC128 mouse reveals a discrete signature of pathology in Huntington disease. Neurobiol Dis 43, 257–265. [DOI] [PubMed] [Google Scholar]
- Davies S.W., Turmaine M., Cozens B.A., Difiglia M., Sharp A.H., Ross C.A., Scherzinger E., Wanker E.E., Mangiarini L., and Bates G.P. (1997). Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90, 537–548. [DOI] [PubMed] [Google Scholar]
- Deng Y., Wang H., Joni M., Sekhri R., and Reiner A. (2021). Progression of basal ganglia pathology in heterozygous Q175 knock-in Huntington’s disease mice. J Comp Neurol 529, 1327–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difiglia M., Sapp E., Chase K., Schwarz C., Meloni A., Young C., Martin E., Vonsattel J.P., Carraway R., Reeves S.A., and et al. (1995). Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron 14, 1075–1081. [DOI] [PubMed] [Google Scholar]
- Difiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., and Aronin N. (1997). Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993. [DOI] [PubMed] [Google Scholar]
- Dragileva E., Hendricks A., Teed A., Gillis T., Lopez E.T., Friedberg E.C., Kucherlapati R., Edelmann W., Lunetta K.L., Macdonald M.E., and Wheeler V.C. (2009). Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol Dis 33, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R., Hampton L., Abraham N.A., Larigan J.D., Joseph N.F., Hernandez-Vega J.C., Geisler S., Yang F.C., Deninger M., Tran D.T., Khatri N., Godinho B., Kinberger G.A., Montagna D.R., Hirst W.D., Guardado C.L., Glajch K.E., Arnold H.M., Gallant-Behm C.L., and Weihofen A. (2024). Dose-dependent reduction of somatic expansions but not Htt aggregates by di-valent siRNA-mediated silencing of MSH3 in HdhQ111 mice. Sci Rep 14, 2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fienko S., Landles C., Sathasivam K., Mcateer S.J., Milton R.E., Osborne G.F., Smith E.J., Jones S.T., Bondulich M.K., Danby E.C.E., Phillips J., Taxy B.A., Kordasiewicz H.B., and Bates G.P. (2022). Alternative processing of human HTT mRNA with implications for Huntington’s disease therapeutics. Brain 145, 4409–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franich N.R., Hickey M.A., Zhu C., Osborne G.F., Ali N., Chu T., Bove N.H., Lemesre V., Lerner R.P., Zeitlin S.O., Howland D., Neueder A., Landles C., Bates G.P., and Chesselet M.F. (2019). Phenotype onset in Huntington’s disease knock-in mice is correlated with the incomplete splicing of the mutant huntingtin gene. J Neurosci Res 97, 1590–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker R.E., Kashin S., Reed N.M., Steven Tan S., Lee W.-S., Mcdonald T.M., Morris K., Kamitaki N., Mullally C.D., Morakabati N., Goldman M., Lind G., Kohli R., Lawton E., Hogan M., Ichihara K., Berretta S., and Mccarroll S.A. (2024). Long somatic DNA-repeat expansion drives neurodegeneration in Huntington disease. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey M.A., Zhu C., Medvedeva V., Lerner R.P., Patassini S., Franich N.R., Maiti P., Frautschy S.A., Zeitlin S., Levine M.S., and Chesselet M.F. (2012). Improvement of neuropathology and transcriptional deficits in CAG 140 knock-in mice supports a beneficial effect of dietary curcumin in Huntington’s disease. Mol Neurodegener 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoschek F., Natan J., Wagner M., Sathasivam K., Abdelmoez A., Von Einem B., Bates G.P., Landwehrmeyer G.B., and Neueder A. (2024). Huntingtin HTT1a is generated in a CAG repeat-length-dependent manner in human tissues. Mol Med 30, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliano M., Seeley C., Sapp E., Jones E.L., Martin C., Li X., Difiglia M., and Kegel-Gleason K.B. (2021). Disposition of Proteins and Lipids in Synaptic Membrane Compartments Is Altered in Q175/Q7 Huntington’s Disease Mouse Striatum. Front Synaptic Neurosci 13, 618391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel K.B., Kim M., Sapp E., Mcintyre C., Castano J.G., Aronin N., and Difiglia M. (2000). Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci 20, 7268–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landles C., Milton R.E., Ali N., Flomen R., Flower M., Schindler F., Gomez-Paredes C., Bondulich M.K., Osborne G.F., Goodwin D., Salsbury G., Benn C.L., Sathasivam K., Smith E.J., Tabrizi S.J., Wanker E.E., and Bates G.P. (2020). Subcellular Localization And Formation Of Huntingtin Aggregates Correlates With Symptom Onset And Progression In A Huntington’S Disease Model. Brain Commun 2, fcaa066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landles C., Osborne G.F., Phillips J., Canibano-Pico M., Nita I.M., Ali N., Bobkov K., Greene J.R., Sathasivam K., and Bates G.P. (2024). Mutant huntingtin protein decreases with CAG repeat expansion: implications for therapeutics and bioassays. Brain Commun 6, fcae410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L.M., Trottier Y., Richards W.G., Osmand A., Paganetti P., and Bates G.P. (2010). Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem 285, 8808–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., and Bates G.P. (1996). Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506. [DOI] [PubMed] [Google Scholar]
- Missineo A., Tomei L., Alaimo N., Martufi P., Zavattieri M., Ferrari F., Piai A., Seguin J., Esquina C., Huang N., Wu H.-Y., Nicotra E., Pace J., and Doherty E.M. (Year). “Highly selective monoclonal antibodies targeting the HTT exon1 neo-epitope”, in: 19th Annual Huntington’s Disease Therapeutics Conference). [Google Scholar]
- Neueder A., Landles C., Ghosh R., Howland D., Myers R.H., Faull R.L.M., Tabrizi S.J., and Bates G.P. (2017). The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington’s disease patients. Sci Rep 7, 1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’reilly D., Belgrad J., Ferguson C., Summers A., Sapp E., Mchugh C., Mathews E., Boudi A., Buchwald J., Ly S., Moreno D., Furgal R., Luu E., Kennedy Z., Hariharan V., Monopoli K., Yang X.W., Carroll J., Difiglia M., Aronin N., and Khvorova A. (2023). Di-valent siRNA-mediated silencing of MSH3 blocks somatic repeat expansion in mouse models of Huntington’s disease. Mol Ther 31, 1661–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressl C., Matlik K., Kus L., Darnell P., Luo J.D., Paul M.R., Weiss A.R., Liguore W., Carroll T.S., Davis D.A., Mcbride J., and Heintz N. (2024). Selective vulnerability of layer 5a corticostriatal neurons in Huntington’s disease. Neuron 112, 924–941 e910. [DOI] [PubMed] [Google Scholar]
- Sapp E., Seeley C., Iuliano M., Weisman E., Vodicka P., Difiglia M., and Kegel-Gleason K.B. (2020). Protein changes in synaptosomes of Huntington’s disease knock-in mice are dependent on age and brain region. Neurobiol Dis 141, 104950. [DOI] [PubMed] [Google Scholar]
- Sapp E., Valencia A., Li X., Aronin N., Kegel K.B., Vonsattel J.P., Young A.B., Wexler N., and Difiglia M. (2012). Native mutant huntingtin in human brain: evidence for prevalence of full-length monomer. J Biol Chem 287, 13487–13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Bondulich M.K., Smith D.L., Faull R.L., Roos R.A., Howland D., Detloff P.J., Housman D.E., and Bates G.P. (2013). Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A 110, 2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G.P., Davies S.W., Lehrach H., and Wanker E.E. (1997). Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549–558. [DOI] [PubMed] [Google Scholar]
- Shelbourne P.F., Keller-Mcgandy C., Bi W.L., Yoon S.R., Dubeau L., Veitch N.J., Vonsattel J.P., Wexler N.S., Group U.S.-V.C.R., Arnheim N., and Augood S.J. (2007). Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum Mol Genet 16, 1133–1142. [DOI] [PubMed] [Google Scholar]
- Smith E.J., Sathasivam K., Landles C., Osborne G.F., Mason M.A., Gomez-Paredes C., Taxy B.A., Milton R.E., Ast A., Schindler F., Zhang C., Duan W., Wanker E.E., and Bates G.P. (2023). Early detection of exon 1 huntingtin aggregation in zQ175 brains by molecular and histological approaches. Brain Commun 5, fcad010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk J.M., Murphy Z., Slow E.J., Leavitt B.R., and Hayden M.R. (2005). Selective degeneration and nuclear localization of mutant huntingtin in the YAC128 mouse model of Huntington disease. Hum Mol Genet 14, 3823–3835. [DOI] [PubMed] [Google Scholar]
- Vonsattel J.P., and Difiglia M. (1998). Huntington disease. J Neuropathol Exp Neurol 57, 369–384. [DOI] [PubMed] [Google Scholar]
- Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., and Richardson E.P. Jr. (1985). Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol 44, 559–577. [DOI] [PubMed] [Google Scholar]
- Wheeler V.C., Gutekunst C.A., Vrbanac V., Lebel L.A., Schilling G., Hersch S., Friedlander R.M., Gusella J.F., Vonsattel J.P., Borchelt D.R., and Macdonald M.E. (2002). Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum Mol Genet 11, 633–640. [DOI] [PubMed] [Google Scholar]
- Wheeler V.C., White J.K., Gutekunst C.A., Vrbanac V., Weaver M., Li X.J., Li S.H., Yi H., Vonsattel J.P., Gusella J.F., Hersch S., Auerbach W., Joyner A.L., and Macdonald M.E. (2000). Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum Mol Genet 9, 503–513. [DOI] [PubMed] [Google Scholar]
- Yamada K., Hariharan V.N., Caiazzi J., Miller R., Ferguson C.M., Sapp E., Fakih H.H., Tang Q., Yamada N., Furgal R.C., Paquette J.D., Biscans A., Bramato B.M., Mchugh N., Summers A., Lochmann C., Godinho B., Hildebrand S., Jackson S.O., Echeverria D., Hassler M.R., Alterman J.F., Difiglia M., Aronin N., and Khvorova A. (2024). Enhancing siRNA efficacy in vivo with extended nucleic acid backbones. Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.