Abstract
Human cells harbor a variety of factors that function to block the proliferation of foreign nucleic acid. The APOBEC3G enzyme inhibits the replication of retroviruses by deaminating nascent retroviral cDNA cytosines to uracils, lesions that can result in lethal levels of hypermutation. Here, we demonstrate that APOBEC3G is capable of deaminating genomic cytosines in Saccharomyces cerevisiae. APOBEC3G expression caused a 20-fold increase in frequency of mutation to canavanine-resistance, which was further elevated in a uracil DNA glycosylase-deficient background. All APOBEC3G-induced base substitution mutations mapped to the nuclear CAN1 gene and were exclusively C/G → T/A transition mutations within a 5′-CC consensus. The APOBEC3G preferred sites were found on both strands of the DNA duplex, but were otherwise located in hotspots nearly identical to those found previously in retroviral cDNA. This unique genetic system further enabled us to show that expression of APOBEC3G or its homolog APOBEC3F was able to inhibit the mobility of the retrotransposon Ty1 by a mechanism that involves the deamination of cDNA cytosines. Thus, these data expand the range of likely APOBEC3 targets to include nuclear DNA and endogenous retroelements, which have pathological and physiological implications, respectively. We postulate that the APOBEC3-dependent innate cellular defense constitutes a tightly regulated arm of a conserved mobile nucleic acid restriction mechanism that is poised to limit internal as well as external assaults.
Keywords: APOBEC3F, endogenous retrotransposon Ty1, hypermutation, cytodeamination, Saccharomyces cerevisiae
The human apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) protein uses cytosine to uracil deamination to inhibit the replication of a variety of retroviruses, including HIV-1 (refs. 1–6; reviewed by in refs. 7–10). APOBEC3G localizes predominantly to the cytoplasm of mammalian cells (4, 11). In a retrovirus-infected cell, this localization may facilitate the incorporation of APOBEC3G into viral particles, which are released from the plasma membrane. APOBEC3G is also specifically incorporated into virions through an association with the viral Gag protein and/or viral genomic RNA (12–18). Once a retrovirus enters a cell, its genomic RNA is reverse transcribed, and during this process, APOBEC3G is capable of deaminating cDNA cytosines to uracils (C → U). These lesions occur at such a high frequency that they ultimately inactivate the virus (causing G → A hypermutation, as read-out on the genomic strand of the virus). APOBEC3G has several human homologs, and at least one, APOBEC3F, restricts HIV-1 infection by a similar mechanism (19–22). However, APOBEC3F and -G deaminate cytosines within different local contexts, preferring 5′-TC and 5′-CC, respectively (19). Both of the resulting mutational signatures are apparent in patient-derived HIV-1 sequences (e.g., ref. 23), suggesting that the APOBEC3-dependent restriction mechanism has a critical role in vivo.
To counteract the APOBEC3-dependent cellular defense, HIV-1 and most other related retroviruses encode a small protein, virion infectivity factor (Vif), which mediates the degradation of APOBEC3F and -G and thereby facilitates retroviral replication (reviewed in refs. 9 and 10). Vif binds APOBEC3G and mediates its polyubiquitination by a cellular E3 ligase complex containing Cullin 5 and Elongins B/C (24–26). Thus, the relative levels of Vif and APOBEC3 proteins appear to be key determinants of retroviral infection.
Although the APOBEC3-Vif interaction is likely important in primates, this relationship does not appear to be ubiquitous. First, APOBEC3 proteins appear to have evolved before Vif-encoding retroviruses (e.g., refs. 8, 27, and 28). All known mammals have APOBEC3G or at least one APOBEC3G-like protein, yet many of their retroviruses appear to lack an effective counterdefense protein such as HIV-1 Vif. For instance, rodents encode a single APOBEC3G homolog and host a variety of retroviruses, none of which appear to encode Vif or a Vif-like counterdefense. Second, many of the APOBEC family member proteins, including APOBEC3F and -G, show an extraordinarily high number of amino acid replacement mutations, indicating that they have been under a strong positive selection (27, 29). Evidence for positive selection predates by millions of years the time that HIV is predicted to have entered the human population. Thus, these observations have fueled speculation that the human APOBEC3 proteins possess another, possibly primary, cellular function (e.g., refs. 8, and 27–30).
Retroelements comprise a large proportion of the murine and human genomes (reviewed in refs. 31 and 32). Although the majority of these elements are apparently nonfunctional, their cis- or trans-mobilization can contribute to a variety of genome destabilizing events including recombination and insertional mutagenesis. There are two major classes of endogenous retroelements. The first and largest is exemplified by long interspersed nucleotide elements (LINEs), which replicate upon integration via an LTR-independent mode. The second major class forms intracellular virus-like particles and replicates by using LTR sequences that flank the coding genes, much like HIV and related retroviruses. Examples of this class include Saccharomyces retrotransposons Ty1 and Ty3, Drosophila copia and gypsy elements and human endogenous retroviruses (HERVs). Although human LINE replication appears uninhibited by APOBEC3G expression (30), a role in controlling the spread of LTR-dependent retroelements remained a distinct possibility.
Here, we test this possibility by using Saccharomyces cerevisiae as a model system. We first ask whether human APOBEC3G is capable of functioning in yeast and, second, we apply this system to assess whether APOBEC3G is capable of inhibiting Ty1 retrotransposition. We show that APOBEC3G expression in yeast caused a mutator phenotype fully attributable to the deamination of genomic cytosines. This surprising functionality enabled us to further show that expression of APOBEC3G or -F inhibited replication of the yeast LTR-dependent retrotransposon Ty1 by a mechanism that involves cDNA cytosine deamination. These data indicate that the APOBEC3-dependent mechanism of retroelement restriction is highly conserved and that the range of APOBEC3 substrates may be far broader than originally anticipated.
Materials and Methods
Yeast Strains and Plasmids. See Supporting Text, which is published as supporting information on the PNAS web site.
Yeast Mutation Assays. pHybLex-Zeo, pJG4–5, pYES3-CT, and their derivatives were transformed into L40 and selected by using a synthetic complete medium containing zeocin and lacking tryptophan (SC+ZEO-TRP) (33). Several thousand viable cells from independent colonies were used to inoculate 2.5 ml SC+GAL+RAF+ZEO-TRP. Cultures were grown at 30°C for 3–4 days and concentrated 5-fold, and a fraction was plated to SC+CAN-ARG to obtain CanR mutants. Viable cell counts were obtained by plating a dilution to rich medium (33). Viable cells were counted after 2 days, and CanR colonies were counted after 3–4 days of incubation at 30°C. The CAN1 gene of CanR colonies was amplified by PCR and sequenced as reported (34). Accurate values for the mutation frequencies were obtained by using multiple independent cultures (n = 6–8) for each strain in each experiment and by repeating each experiment at least twice and as many as seven times. Sequencher (GenesCodes) was used for mutational analyses.
Immunoblotting. Cell pellets from a 10-ml log phase culture were washed with 1 ml of 20% trichloroacetic acid (TCA), resuspended in 50 μl of 20% TCA, and then lysed by vortexing with an equal volume of glass beads at 4°C. The supernatant was centrifuged to pellet the proteins. Pelleted proteins were resuspended in 100 μlof SDS-gel loading buffer, separated by SDS/PAGE, transferred to a poly(vinylidene difluoride) membrane, and probed with antibodies to APOBEC3G (35), LexA (Invitrogen), or Vif (36–38).
Ty1 Retrotransposition Assays. Ty-his3AI, TyHRT-his3AI, Ty-lucAI, or TyHRT-lucAI plasmids were cotransformed with pJG4–5, pJG4–5-APOBEC3G, or pJG4–5-APOBEC3F into DG1251 or GRY1990 (39) and selected by using SC-URA-TRP+GLC (33). his3AI transformants were grown in SC-URA-TRP+GLC to saturation. 106 cells were subcultured in 1 ml of SC-URA-TRP+GAL for 12 h, and an aliquot was plated to SC-HIS. Cell viability was determined by plating a dilution to rich medium. Retrotransposition was quantified by determining the frequency of His+ colonies. lucAI transformants were grown 1 day in SC-URA-TRP+GLC. Cells were transferred to SC-URA-TRP+GAL and grown for an additional 2 days at 30°C to induce retroelement expression and reverse transcription. Retrotransposition was quantified by measuring the relative active levels of luciferase to β-galactosidase (D.V.N. and J. Strathern, unpublished data). All incubations for plasmid-based Ty1 assays were at 30°C.
For endogenous retrotransposition assays, DG1141 was transformed with pJG4-5, pJG4-5-APOBEC3G, or pJG4-5-APOBEC3F. Single colonies were resuspended in water, and 10,000–50,000 cells were transferred to 2 ml of SC-TRP+GAL and grown at 20°C for 7–10 days until the cultures reached saturation. Dilutions of the starting and ending cultures were plated to rich media to determine the number of viable cells, and the equivalent of 1 ml of the saturated culture was plated to SC-HIS to score retrotransposition events.
Ty1 DNA Sequencing. Retrotransposed Ty1 and TyHRT cDNAs were isolated by growing His+ colonies overnight in 10 ml of SC-HIS at 30°C and preparing DNA with a standard glass bead/phenol extraction method. The resulting DNA was used to amplify a 1,026-bp (Ty) or 971-bp (TyHRT) region spanning the RT gene and HIS3 by using 5′-TTC ATG TGG GAC ACT AGA GAT (TyRT) or 5′-CCT GAG TGG GAG TTG TTA (TyHRT) and 5′-TAT GAT ACA TGC TCT GGC CAA (HIS3). PCR products were purified (Qiagen) and sequenced with 5′-GT CTG CGA GGC AAG AAT GAT. GFP-negative retrotransposition events were obtained from pools of His+ colony genomic DNA by transformation into Escherichia coli. GFP-negative colonies were identified with fluorescent light, and the resident plasmid DNA was amplified (as above except the product was 2.1 kb for Ty RT) and sequenced by using 5′-C GTT ATC CGG ATC ATA TGA and 5′-G TAG TTC CCG TCA TCT TGA.
Results
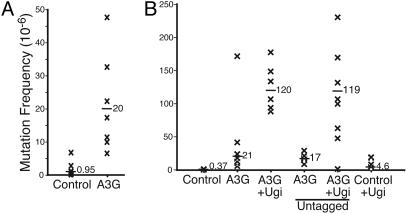
APOBEC3G Stimulates Mutation in S. cerevisiae via the Uracil Excision Pathway. To test whether human APOBEC3G could elicit its hallmark mutator activity in yeast, we expressed a LexA-APOBEC3G fusion protein in the haploid strain L40 and monitored the accumulation of mutations that conferred resistance to the toxic amino acid canavanine. Liquid cultures were grown from individual colonies expressing APOBEC3G or a control vector and then plated onto a solid medium containing canavanine. The numbers of canavanine-resistant (CanR) colonies were determined after 3- to 4-days growth. In contrast to cells expressing a control vector, those expressing LexA-APOBEC3G showed a 20-fold increase in the median frequency of CanR mutation, suggesting that APOBEC3G was capable of deaminating cytosines within yeast genomic DNA (Fig. 1A and Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
APOBEC3G stimulates mutation in S. cerevisiae by the uracil excision pathway. (A) APOBEC3G expression causes an increase in the median frequency of mutation to CanR. Each X represents the frequency derived from an independent culture and the median is indicated. The y axis reports the observed number of CanR colonies per million viable yeast cells. Yeast expressing the control vector showed a frequency of spontaneous mutation to CanR similar to that reported previously (42, 43). The data are representative of seven independent experiments (Fig. 6 reports the mean of the median values for these experiments). (B) APOBEC3G and Ugi coexpression triggers a synergistic increase in the frequency of mutation to CanR. APOBEC3G is tagged with the DNA binding domain of LexA unless noted. The parameters are identical to A.
To begin to determine whether the LexA-APOBEC3G-induced mutator phenotype occurred by a C → U deamination mechanism, we asked whether a uracil DNA glycosylase deficiency would exacerbate this phenotype. Because most DNA-based organisms use uracil DNA glycosylase to rid their genomes of uracil (40), it was likely that, were this the mechanism, many of the APOBEC3G-induced uracil lesions would have been repaired and that the observed mutation frequency would be an underestimate of APOBEC3G activity. Indeed, yeast expressing both APOBEC3G and a uracil DNA glycosylase inhibitor (Ugi) protein showed a 320-fold increase in the median frequency of mutation to CanR (Fig. 1B). This stimulation was ≈6- and 26-fold higher than that observed in LexA-APOBEC3G-expressing and in Ugi-expressing yeast cells, respectively, indicating that many of the APOBEC3G-dependent uracils were repaired by a uracil excision mechanism.
In yeast, the major uracil DNA glycosylase is Ung1p (uracil DNA N-glycosylase 1 protein). Ung1p and most other Ung proteins from bacteria to humans are strongly inhibited by Ugi (41). However, Ugi-resistant uracil excision activities occur in mammalian cells, such as those elicited by the SMUG1 and TDG1 proteins (40). To eliminate the possibility the some of the APOBEC3G-induced uracils might be repaired by auxiliary systems in yeast, we used homologous recombination to construct an Ung1p deletion strain, L40 ung1::kanMX4. This strain showed levels of CanR mutation virtually indistinguishable from Ugi-expressing cells in the presence or absence of APOBEC3G (Fig. 7, which is published as supporting information on the PNAS web site). Thus, the majority of APOBEC3G-induced lesions in yeast were repaired by an Ung1pdependent mechanism. Together with the exquisite specificity that Ung1p has for uracil, these data indicated that the APOBEC3G-dependent mutator phenotype was attributable to a DNA cytosine deamination mechanism.
APOBEC3G is localized predominantly to the cytoplasm of mammalian cells (4, 11). Therefore, we were surprised by the high mutation frequencies caused by its expression in yeast and concerned that this might have been attributable to the DNA-binding properties of the LexA tag. Therefore, we monitored the CAN1 mutation frequency of cells expressing either LexA-APOBEC3G or untagged APOBEC3G. Little difference in the overall median frequencies of CanR mutation was observed demonstrating that the DNA binding domain of LexA was not responsible for the APOBEC3G-dependent mutator phenotype (Fig. 1B). The broader implications of human APOBEC3G mutating a genomic target will be considered below and in Discussion.
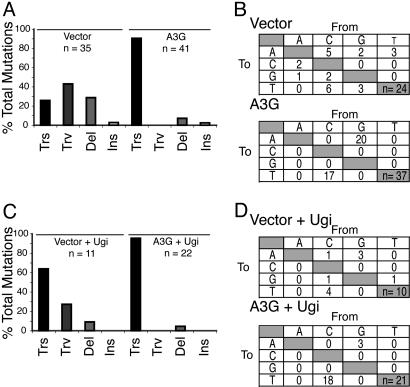
APOBEC3G Triggers Predominantly C/G → T/A Transition Mutations in Yeast. CAN1 encodes a membrane-spanning arginine transporter that must be inactivated for growth to occur in the presence of the toxic arginine analog canavanine. A wide variety of base substitution, insertion, deletion and more complex mutations can confer CanR (e.g., refs. 42 and 43). To further investigate the mechanism of the APOBEC3G-induced mutator phenotype, the CAN1 gene of a large number of CanR colonies was sequenced. In agreement with previous studies, cells containing a control vector displayed a wide range of CAN1 mutations including transitions (26%), transversions (43%), insertions (3%), and deletions (28%) (Fig. 2 and Fig. 8, which is published as supporting information on the PNAS web site). In contrast, the vast majority (90%) of the CAN1 mutations in APOBEC3G expressing cells were C/G → T/A transitions. APOBEC3G-induced transitions occurred at the expense of other types of mutations, accounting for the elevated CanR mutation frequency.
Fig. 2.
APOBEC3G triggers C/G → T/A transition mutations in S. cerevisiae. (A) Histograms summarizing the types of mutations found in the CAN1 gene of S. cerevisiae expressing a control vector or APOBEC3G. Data from Lex-APOBEC3G and untagged APOBEC3G-expressing cells were nearly identical and were pooled for these analyses. Mutations were categorized as transitions (Trs), transversions (Trv), deletions (Del), or insertions (Ins). (B) Summary of the base substitution mutations found in the CAN1 gene of S. cerevisiae expressing a control vector or APOBEC3G. (C) Histograms summarizing the types of mutations found in the CAN1 gene of S. cerevisiae expressing Ugi and a control vector or APOBEC3G. Labels are as in A. (D) Summary of the base substitution mutations found in the CAN1 gene of S. cerevisiae expressing Ugi and a control vector or APOBEC3G.
Yeast lacking Ung1p because of Ugi expression also displayed an increased level of C/G → T/A transition mutations (64%), as would be expected of cells lacking uracil excision repair (Figs. 2 and 8). However, five of seven of these transitions occurred at positions that were not mutated in APOBEC3G expressing cells. Coexpression of Ugi and APOBEC3G resulted in an even stronger C/G → T/A transition bias (95%), and 19 of 21 of these mutations occurred at sites that were also mutated in APOBEC3G expressing (Ugi negative) yeast cells. These data further demonstrated that APOBEC3G is capable of triggering genomic hypermutation in yeast by a C → U deamination mechanism.
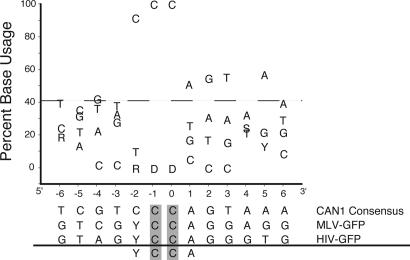
The Local APOBEC3G Mutation Preference in Yeast Is Nearly Identical to That Observed in Model Retroviral Substrates. A closer examination of the C/G → T/A transitions triggered by APOBEC3G expression revealed that 37 of 37 occurred within the dinucleotide 5′-CC, which could be found on either strand of the DNA duplex (Figs. 2, 3, and 8). APOBEC3G expression alone triggered C/G → T/A transition mutations at 14 distinct sites within the CAN1 gene. Coexpression of APOBEC3G and Ugi caused C/G → T/A transition mutations at six identical and two additional sites. The three most frequently APOBEC3G mutated 5′-CC dinucleotide sites, C356, C656, and C1195, accounted for 48% of the total combined APOBEC3G- and APOBEC3G-plus-Ugi-dependent base substitution mutations. The extended sequence preference of APOBEC3G in the yeast system was compared to that defined previously in model HIV and murine leukemia virus retroviral systems as 5′-YCCA (Y = C or T, ref. 19). Interestingly, APOBEC3G exhibited a strikingly similar 5′-CCCA preference in yeast (Fig. 3), indicating that its preference, as observed in other systems, was intact.
Fig. 3.
The local sequence preference of APOBEC3G-induced hypermutation. A graph illustrating the percentage that each base was found at the indicated position relative to the C/G → T/A transition mutation site in APOBEC3G expressing cells (n = 37). The base found most frequently is indicated below. APOBEC3G consensus sites observed in model retroviral substrates, HIV-GFP or MLV-GFP, are shown (19). Multiple bases had the same percentage and are indicated by the one letter code where D = A/G/T, R = A/G, S = G/C, and Y = C/T. The G/C content of the CAN1 gene is indicated by the dashed line.
It is further notable that in addition to a large number of C/G → T/A transition mutations, four deletions and a single insertion were detected in the CAN1 gene of APOBEC3G-expressing yeast cells (Figs. 2 and 8; combined data including the Ugi experiments). Three of five of these alterations occurred either in or immediately adjacent to a preferred or potential APOBEC3G hotspot, 5′-CCC. In contrast, only 1 of 12 of the deletions and insertions found in control vector containing cells occurred at similar sites. The remainder (11 of 12) were distributed throughout the CAN1 gene and were presumably caused by a variety of mechanisms. The presence of deletions and insertions associated with APOBEC3G hotspots suggested that C → U deamination events are able to precipitate gross genomic instability. This finding is further supported by our observation that a small (≈5%) proportion of CanR mutants failed to yield a CAN1 gene-specific PCR product, potentially representing larger-scale lesions.
Affect of HIV-1 Vif on APOBE3G-Induced Yeast Hypermutation. In primates such as humans and chimpanzees, Vif counteracts the antiretroviral activity of APOBEC3G by targeting it for proteasomal degradation (reviewed in refs. 9 and 10). Vif accomplishes this by binding to APOBEC3G. Some data suggest that this association alone may directly impair APOBEC3G function (44). Therefore, we wished to assess whether the interaction between Vif and APOBEC3G could be detected with this yeast assay system.
HIV-1 Vif, derived from the YU2 or the IIIB provirus, was expressed alongside APOBEC3G by using yeast two-hybrid bait or prey vectors. All possible pairwise combinations were tested for the ability to drive the yeast two-hybrid reporter genes lacZ or HIS3.No significant β-galactosidase activity or histidine prototrophy was observed despite repeated attempts (data not shown). This result was not attributable to an expression failure, as both proteins could be detected in cell lysates by immunoblotting (Fig. 9, which is published as supporting information on the PNAS web site).
However, because some weak or transient interactions may escape detection by the yeast two-hybrid assay, we reasoned that the sensitive CAN1 mutation assay might provide a more robust method for monitoring this interaction. To examine whether HIV-1 Vif could affect APOBEC3G-mediated hypermutation in yeast, we compared the CanR mutation frequencies of cells coexpressing Vif and APOBEC3G with those of cells expressing either protein alone. The robust hypermutability of APOBEC3G was not significantly affected by HIV-1 Vif coexpression (Fig. 9). Therefore, we have been unable to detect a Vif-APOBEC3G interaction in yeast.
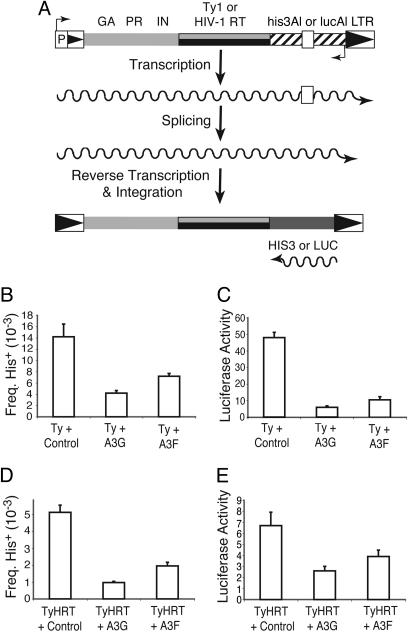
APOBEC3F and APOBEC3G Inhibit Ty1 Retrotransposition. To explore the possibility that APOBEC3 proteins function to impede the mobility of endogenous retroelements that replicate by using LTR sequences, we assayed the ability of the yeast retrotransposon Ty1 to replicate in the presence of APOBEC3G or its homolog APOBEC3F. Ty1 activity was monitored by using an intron-disrupted retrotransposition indicator gene (Fig. 4A). Ty1 RNA expression, splicing, reverse transcription, and integration yield functional reporter gene cDNA copies, encoding either histidine prototrophy (45, 46) or luciferase activity.
Fig. 4.
APOBEC3F and -G inhibit Ty1 retrotransposition. (A) A schematic depicting retrotransposition by Ty1 or TyHRT yielding histidine prototrophy or luciferase activity. GA, PR, IN, RT, and LTR represent gag, protease, integrase, reverse transcriptase, and long-terminal repeat, respectively. (B and D) APOBEC3F or -G expression diminishes Ty1 or TyHRT retrotranspostion as monitored by the number of His+ colonies. For each condition, at least eight independent cultures were analyzed, and the error bars depict one standard error of the mean. (C and E) APOBEC3F or -G expression diminishes Ty1 or TyHRT retrotranspostion as monitored by luciferase activity. The conditions were identical to those described above.
We first measured the ability of Ty1-his3AI to retrotranspose in the presence of human APOBEC3F or -G (Fig. 4B). In comparison to cells containing a control vector, an average of 51% or 70% fewer His+ colonies were detected in the presence of APOBEC3F or -G, respectively. Slightly larger APOBEC3-dependent declines in Ty1-lucAI retrotransposition were observed, as monitored by the relative levels of luciferase present in liquid cultures (Fig. 4C). However, an almost total inhibition (94–98%) was observed when retrotransposition of a genomic Ty1-his3AI element was assayed in the presence of APOBEC3F or -G, suggesting that the ratio of APOBEC3 protein to retrotransposition intermediate (and/or Ty host factors) is a key determinant of this inhibitory mechanism (Fig. 10, which is published as supporting information on the PNAS web site). Together, these data clearly demonstrated that APOBEC3F or -G can inhibit Ty1 retrotransposition.
To assess whether the APOBEC3-dependent inhibition of retrotransposition in yeast could be influenced by the reverse transcriptase or the integration pathway, similar assays were performed with Ty1 constructs in which the normal reverse transcriptase was replaced with that from HIV-1 (TyHRT; refs. 45 and 46). TyHRT integration occurs predominantly by homologous recombination, whereas Ty1 integration mostly uses its own integrase. Retrotransposition of both TyHRT-hisAI and TyHRT-lucAI (i.e., the accumulation of HIV-1 reverse transcriptase products) was also inhibited by APOBEC3F or APOBEC3G expression (Fig. 4 D and E). Levels of inhibition were roughly similar to those observed with Ty1 reverse transcriptase, indicating that neither the reverse transcriptase nor the integration pathway were key effectors of the APOBEC3-imposed retrotransposition block. These data further highlight the utility of the yeast Ty1 system for studying aspects of both APOBEC3 and HIV-1 biology.
Ty1 Restriction by APOBEC3F and APOBEC3G Involves a cDNA Cytosine Deamination Mechanism. As cDNA C → U deamination is a hallmark antiretroviral activity of APOBEC3F and -G, we asked whether this could account for the observed Ty1 retrotransposition block. If so, we expected to find an inordinate number of retrotransposon minus strand C → T transition mutations amongst the His+ integrants (equivalent to plus strand G → A transitions). We sequenced >26 and 47 kbp of TyRT-HIS3 template generated in the presence of APOBEC3F and -G, respectively, and found only two C → T transitions among the APOBEC3G-exposed templates. One transition occurred within a dinucleotide consensus 5′-GC that is rarely preferred by this protein, and it therefore likely represents a reverse transcription or PCR error. The second transition occurred within the trinucleotide 5′-CCC, which is the most common APOBEC3G-preferred site. However, this meager number of base substitutions may have been in part due the fact that functional His+ (and not His–) integrants were analyzed. It is further possible that uracil residues within the retrotransposon cDNA triggered its degradation, as hypothesized originally for retroviruses (2).
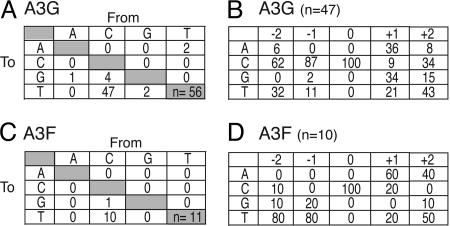
Therefore, to address the former possibility and to enrich for mutations, we used a modified version of the Ty-his3AI system in which a GFP cassette was placed upstream of his3AI (Fig. 11, which is published as supporting information on the PNAS web site). This system enabled the selection of His+ integrants and a subsequent screen for unselected GFP-negative variants. Twenty independent GFP mutants were recovered from retrotransposition experiments in which APOBEC3G was expressed. Each sequence contained at least one mutation and as many as 15 mutations. In total, 56 base substitution mutations were identified, and 47 of these were minus strand C → T transitions (Figs. 5A and 11). Almost all of the APOBEC3G-dependent transitions occurred within the consensus 5′-YCC, very similar to the preferred cytosine deamination consensus site in the CAN1 gene and in a variety of other systems (e.g., compare Figs. 5B and 3). Moreover, many of the C → T transitions occurred at positions that were identical to those observed previously in GFP-encoding HIV or MLV (2, 19). A similar strand-specific transition bias and sequences with multiple transitions were found in GFP-negative templates produced in the presence of APOBEC3F (Figs. 5C and 11). However, in contrast to APOBEC3G, the APOBEC3F-dependent mutations occurred within a distinct 5′-TTC consensus (Fig. 5D; observed previously with an HIV substrate; ref. 19). We conclude that Ty1 retrotransposition can be inhibited by APOBEC3F and -G and that much (and possibly all) of this effect can be attributed to a cDNA cytosine deamination mechanism.
Fig. 5.
The mutational preferences of APOBEC3G and -F in Ty1 cDNA. (A and C) Summaries of the GFP gene (and surrounding region) base substitution mutations observed in pools of His+ retrotranspositions, which had occurred in the presence of APOBEC3G or -F, respectively. (B and D) Base preferences surrounding the Ty1 cDNA C → T transition sites attributable to expression of APOBEC3G or -F, respectively. APOBEC3G shows a clear preference for 5′-YCC, whereas APOBEC3F prefers 5′-TTC (Y = C or T; the mutated cytosine is underlined). Fig. 11 for additional information.
Discussion
The studies reported here demonstrate that APOBEC3G expression in S. cerevisiae results in a genomic mutator phenotype entirely attributable to a DNA cytosine deamination mechanism. The 5′-CC → CT transitions occurred within a broader local preference that could be found on either strand of the DNA duplex. In addition, the fact that APOBEC3G was functionally expressed in this system enabled us to show that both it and its homolog APOBEC3F could inhibit the mobility of the yeast retrotransposon Ty1 by a mechanism also involving DNA cytosine deamination. However, all of the Ty1 C → T transitions occurred preferentially on the cDNA (minus) strand of the retroelement. Thus, strand specificity distinguishes the mechanisms of genomic hypermutation and retroelement restriction.
A second distinguishing feature of these mechanisms is the fact that they manifest at dramatically different frequencies. CAN1 mutation occurred at a low frequency, with ≈1 of 50,000 of the APOBEC3G-expressing cells eliciting CanR. Because CAN1 is 1,771 bp, this frequency translates to ≈0.0000112 mutations per kb. In contrast, Ty1 restriction by APOBEC3G was highly efficient, because the vast majority of potential retrotransposition events could be inhibited [70–98% or more depending on whether Ty1 transcripts were present in relatively high copy (plasmid-based, GAL promoter) or low copy (chromosome-based, Ty promoter), respectively (Figs. 4 and 10)]. The frequency of mutation in this system can be gauged by calculating the number of additional (hitch-hiking) mutations associated with those that inactivate the Ty1 GFP cassette. For APOBEC3G, 36 additional minus strand base substitutions (including 33 C → T transitions) were found among 20 independent 1,488-bp Ty1 cDNA sequences, translating to ≈1.2 mutations per kb. This 5-log potency differential suggested that the APOBEC3G-dependent genomic lesions are occurring nonspecifically (possibly genome-wide), whereas Ty1 restriction may be due to the specific targeting of APOBEC3G (or -F) to the replicating retroelement. This observation is supported by the recent finding that APOBEC3G could associate with Ty1 Gag in an RNA-dependent manner (47). A similar specific association appears to be required for the restriction of HIV-1 by APOBEC3G (reviewed in refs. 7 and 8). Thus, taken together with our studies here, which uniquely revealed a major role for cDNA deamination in Ty1 restriction, it is likely that APOBEC3G and -F exploit an ancient and perhaps a structurally conserved pathway to accomplish the destruction of replicating retroelements.
Schwartz and colleagues (48) recently reported that APOBEC3G was able to block the transposition of two types of murine retroelements in HeLa cells. As for Ty1 and several retroviruses, much of the inhibitory effect was attributable to the deamination of cDNA cytosines. However, because the APOBEC3 proteins do not exist outside of mammals (e.g., refs. 8 and 28), our data showing that APOBEC3F or -G could inhibit yeast Ty1 retrotransposition were unexpected. Therefore, the Ty1 data not only demonstrate the remarkable conservation of this mechanism but, importantly, they also show that mammalian factors (in addition to APOBEC3F or -G) are not required for retroelement restriction.
This is the first report that a two-domain DNA cytosine deaminase such as APOBEC3G can mutate a nuclear gene within a eukaryotic cell. This was particularly surprising because APOBEC3G had shown a clear cytoplasmic localization in a variety of mammalian cell lines (4, 11). Therefore, the simple presence of a nuclear membrane or chromatinized DNA in eukaryotic cells is not sufficient to prevent APOBEC3G from deaminating genomic cytosines. Thus, our data imply the existence of regulatory mechanisms that serve to ensure the proper cytoplasmic compartmentalization of APOBEC3G in mammalian cells. A further implication is that the loss of such regulatory mechanisms might lead to genomic hypermutation, larger-scale lesions, and eventual carcinogenesis. The likelihood of such a pathological role is supported by data indicating that APOBEC3G is overexpressed in some breast cancer tissues (49).
The APOBEC3G-dependent yeast CAN1 mutations were found on both DNA strands, indicating for the first time that both the nontranscribed (displaced) and transcribed DNA strands can be substrates for its deamination activity. Previously, APOBEC3G had shown only a marked preference for single-strand DNA or for DNA that may have been displaced by transcription (e.g., refs. 2 and 49–51). Such an association with transcription was supported by our data, where a 6-fold bias in displaced strand C → T mutation was observed in APOBEC3G-expressing cells lacking Ung1p (Fig. 2). However, the fact that APOBEC3G-dependent C → T transitions were distributed equally over both DNA strands in the repair proficient parent strain indicated that the preferential repair of uracils in the displaced strand might also be occurring. Additional experiments will be required to further delineate this possibility.
APOBEC3F and -G are related to activation-induced deaminase (AID), a single-domain DNA cytosine deaminase with a prominent nuclear function in promoting the diversification of Ig genes (reviewed in ref. 8). Although the APOBEC3s and AID are related and likely to share some key properties (DNA cytosine deamination), they are quite distinct. To deaminate antibody gene DNA, AID must transit from the cytoplasm to the nucleus. This is facilitated by its small size (24 kDa) and bona fide nucleocytoplasmic shuttling signals (52). These properties may also contribute to its ability to mutate the yeast CAN1 gene, as reported recently by Poltoratsky et al. (53). In contrast, to accomplish retroelement restriction APOBEC3F and -G may never have to leave the cytoplasm, where they can access assembling retroelements (exogenous or endogenous). These proteins are around twice the size of AID, and they do not appear to possess nuclear localization signals. Nevertheless, both studies underscore the tremendous potential of the yeast system for studying these molecules. For instance, when our data are considered with the fact that a Cullin 5–Elongin B/C ubiquition ligation complex is required for APOBEC3G degradation in human cells (24, 54), it becomes reasonable to use yeast to reconstitute the minimal components required for this interaction and thus open the door to alternative therapeutic screening strategies.
We propose that limiting the mobility of endogenous retroelements is the predominant physiological function of some of the mammalian APOBEC proteins, including the human APOBEC3F and -G DNA cytosine deaminases. A balance between limited mobility and maintained genome integrity is likely to be an important determinant of speciation, and it would certainly account for the fact that many of the APOBEC proteins are positively selected (27, 29). The advantage of having a potent retroelement restrictor clearly outweighs the disadvantage of encoding and regulating potential cellular genome mutators such as APOBEC3F and -G.
Supplementary Material
Acknowledgments
We thank A. Bielinsky, W. Brown, D. Kirkpatrick, D. Livingston, D. MacDuff, A. Rattray, and R. Ricke for helpful comments; K. Christensen, J. Doebler, and E. Rajendra for technical assistance; D. Garefinkel, J. Lingappa, M. Malim, and R. Wright for reagents; J. Strathern for unpublished reagents; and the University of Minnesota AGAC for sequencing. The HIV-1 Vif antibody (no. 319) was obtained from M. Malim through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was supported by the University of Minnesota, a Searle Scholarship, and a Burroughs–Wellcome Fund Hitchings–Elion Fellowship (to R.S.H.), and it was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract NO1-CO-12400 (to D.V.N.).
Author contributions: A.J.S., D.V.N., and R.S.H. designed research; A.J.S. and D.V.N. performed research; D.V.N. contributed new reagents/analytic tools; A.J.S., D.V.N., and R.S.H. analyzed data; and A.J.S., D.V.N., and R.S.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: Vif, virion infectivity factor.
References
- 1.Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. (2002) Nature 418, 646–650. [DOI] [PubMed] [Google Scholar]
- 2.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S. & Malim, M. H. (2003) Cell 113, 803–809. [DOI] [PubMed] [Google Scholar]
- 3.Lecossier, D., Bouchonnet, F., Clavel, F. & Hance, A. J. (2003) Science 300, 1112. [DOI] [PubMed] [Google Scholar]
- 4.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L. & Trono, D. (2003) Nature 424, 99–103. [DOI] [PubMed] [Google Scholar]
- 5.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C. & Gao, L. (2003) Nature 424, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mariani, R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H. & Landau, N. R. (2003) Cell 114, 21–31. [DOI] [PubMed] [Google Scholar]
- 7.Bieniasz, P. D. (2004) Nat. Immunol. 5, 1109–1115. [DOI] [PubMed] [Google Scholar]
- 8.Harris, R. S. & Liddament, M. T. (2004) Nat. Rev. Immunol. 4, 868–877. [DOI] [PubMed] [Google Scholar]
- 9.Navarro, F. & Landau, N. R. (2004) Curr. Opin. Immunol. 16, 477–482. [DOI] [PubMed] [Google Scholar]
- 10.Rose, K. M., Marin, M., Kozak, S. L. & Kabat, D. (2004) Trends Mol. Med. 10, 291–297. [DOI] [PubMed] [Google Scholar]
- 11.Marin, M., Rose, K. M., Kozak, S. L. & Kabat, D. (2003) Nat. Med. 9, 1398–1403. [DOI] [PubMed] [Google Scholar]
- 12.Alce, T. M. & Popik, W. (2004) J. Biol. Chem. 279, 34083–34086. [DOI] [PubMed] [Google Scholar]
- 13.Cen, S., Guo, F., Niu, M., Saadatmand, J., Deflassieux, J. & Kleiman, L. (2004) J. Biol. Chem. 279, 33177–33184. [DOI] [PubMed] [Google Scholar]
- 14.Svarovskaia, E. S., Xu, H., Mbisa, J. L., Barr, R., Gorelick, R. J., Ono, A., Freed, E. O., Hu, W. S. & Pathak, V. K. (2004) J. Biol. Chem. 279, 35822–35828. [DOI] [PubMed] [Google Scholar]
- 15.Douaisi, M., Dussart, S., Courcoul, M., Bessou, G., Vigne, R. & Decroly, E. (2004) Biochem. Biophys. Res. Commun. 321, 566–573. [DOI] [PubMed] [Google Scholar]
- 16.Zennou, V., Perez-Caballero, D., Gottlinger, H. & Bieniasz, P. D. (2004) J. Virol. 78, 12058–12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, K., Liu, B., Xiao, Z., Yu, Y., Yu, X., Gorelick, R. & Yu, X. F. (2004) J. Virol. 78, 11841–11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schafer, A., Bogerd, H. P. & Cullen, B. R. (2004) Virology 328, 163–168. [DOI] [PubMed] [Google Scholar]
- 19.Liddament, M. T., Brown, W. L., Schumacher, A. J. & Harris, R. S. (2004) Curr. Biol. 14, 1385–1391. [DOI] [PubMed] [Google Scholar]
- 20.Bishop, K. N., Holmes, R. K., Sheehy, A. M., Davidson, N. O., Cho, S. J. & Malim, M. H. (2004) Curr. Biol. 14, 1392–1396. [DOI] [PubMed] [Google Scholar]
- 21.Zheng, Y. H., Irwin, D., Kurosu, T., Tokunaga, K., Sata, T. & Peterlin, B. M. (2004) J. Virol. 78, 6073–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiegand, H. L., Doehle, B. P., Bogerd, H. P. & Cullen, B. R. (2004) EMBO J. 23, 2451–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janini, M., Rogers, M., Birx, D. R. & McCutchan, F. E. (2001) J. Virol. 75, 7973–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P. & Yu, X. F. (2003) Science 302, 1056–1060. [DOI] [PubMed] [Google Scholar]
- 25.Yu, Y., Xiao, Z., Ehrlich, E. S., Yu, X. & Yu, X. F. (2004) Genes Dev. 18, 2867–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehle, A., Goncalves, J., Santa-Marta, M., McPike, M. & Gabuzda, D. (2004) Genes Dev. 18, 2861–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawyer, S. L., Emerman, M. & Malik, H. S. (2004) PLoS Biol. 2, E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conticello, S. G., Thomas, C. J., Petersen-Mahrt, S. & Neuberger, M. S. (2005) Mol. Biol. Evol. 22, 367–377. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, J. & Webb, D. M. (2004) Hum. Mol. Genet. 13, 1785–1791. [DOI] [PubMed] [Google Scholar]
- 30.Turelli, P., Vianin, S. & Trono, D. (2004) J. Biol. Chem. 279, 43371–43373. [DOI] [PubMed] [Google Scholar]
- 31.Bannert, N. & Kurth, R. (2004) Proc. Natl. Acad. Sci. USA 101, Suppl. 2, 14572–14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deininger, P. L., Moran, J. V., Batzer, M. A. & Kazazian, H. H., Jr. (2003) Curr. Opin. Genet. Dev. 13, 651–658. [DOI] [PubMed] [Google Scholar]
- 33.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (2002) Short Protocols in Molecular Biology (Wiley, New York), 5th Ed.
- 34.Marsischky, G. T., Filosi, N., Kane, M. F. & Kolodner, R. (1996) Genes Dev. 10, 407–420. [DOI] [PubMed] [Google Scholar]
- 35.Newman, E. N., Holmes, R. K., Craig, H. M., Klein, K. C., Lingappa, J. R., Malim, M. H. & Sheehy, A. M. (2005) Curr. Biol. 15, 166–170. [DOI] [PubMed] [Google Scholar]
- 36.Fouchier, R. A., Simon, J. H., Jaffe, A. B. & Malim, M. H. (1996) J. Virol. 70, 8263–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon, J. H., Southerling, T. E., Peterson, J. C., Meyer, B. E. & Malim, M. H. (1995) J. Virol. 69, 4166–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, J. H., Fouchier, R. A., Southerling, T. E., Guerra, C. B., Grant, C. K. & Malim, M. H. (1997) J. Virol. 71, 5259–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gietz, R. D., Schiestl, R. H., Willems, A. R. & Woods, R. A. (1995) Yeast 11, 355–360. [DOI] [PubMed] [Google Scholar]
- 40.Barnes, D. E. & Lindahl, T. (2004) Annu. Rev. Genet. 38, 445–476. [DOI] [PubMed] [Google Scholar]
- 41.Mol, C. D., Arvai, A. S., Sanderson, R. J., Slupphaug, G., Kavli, B., Krokan, H. E., Mosbaugh, D. W. & Tainer, J. A. (1995) Cell 82, 701–708. [DOI] [PubMed] [Google Scholar]
- 42.Huang, M. E., Rio, A. G., Nicolas, A. & Kolodner, R. D. (2003) Proc. Natl. Acad. Sci. USA 100, 11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rattray, A. J., Shafer, B. K., McGill, C. B. & Strathern, J. N. (2002) Genetics 162, 1063–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stopak, K., de Noronha, C., Yonemoto, W. & Greene, W. C. (2003) Mol. Cell 12, 591–601. [DOI] [PubMed] [Google Scholar]
- 45.Nissley, D. V., Garfinkel, D. J. & Strathern, J. N. (1996) Nature 380, 30. [DOI] [PubMed] [Google Scholar]
- 46.Nissley, D. V., Boyer, P. L., Garfinkel, D. J., Hughes, S. H. & Strathern, J. N. (1998) Proc. Natl. Acad. Sci. USA 95, 13905–13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dutko, J. A., Schafer, A., Kenny, A. E., Cullen, B. R. & Curcio, M. J. (2005) Curr. Biol. 15, 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esnault, C., Heidmann, O., Delebecque, F., Dewannieux, M., Ribet, D., Hance, A. J., Heidmann, T. & Schwartz, O. (2005) Nature 433, 430–433. [DOI] [PubMed] [Google Scholar]
- 49.Harris, R. S., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Mol. Cell 10, 1247–1253. [DOI] [PubMed] [Google Scholar]
- 50.Beale, R. C., Petersen-Mahrt, S. K., Watt, I. N., Harris, R. S., Rada, C. & Neuberger, M. S. (2004) J. Mol. Biol. 337, 585–596. [DOI] [PubMed] [Google Scholar]
- 51.Yu, Q., Konig, R., Pillai, S., Chiles, K., Kearney, M., Palmer, S., Richman, D., Coffin, J. M. & Landau, N. R. (2004) Nat. Struct. Mol. Biol. 11, 435–442. [DOI] [PubMed] [Google Scholar]
- 52.Ito, S., Nagaoka, H., Shinkura, R., Begum, N., Muramatsu, M., Nakata, M. & Honjo, T. (2004) Proc. Natl. Acad. Sci. USA 101, 1975–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poltoratsky, V. P., Wilson, S. H., Kunkel, T. A. & Pavlov, Y. I. (2004) J. Immunol. 172, 4308–4313. [DOI] [PubMed] [Google Scholar]
- 54.Mehle, A., Strack, B., Ancuta, P., Zhang, C., McPike, M. & Gabuzda, D. (2004) J. Biol. Chem. 279, 7792–7798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.