Abstract
Adenosine deaminase (ADA), a protein whose deficit leads to severe combined immunodeficiency, binds to the cell surface by means of either CD26, A1 adenosine receptors, or A2B adenosine receptors. The physiological role of these interactions is not well understood. Our results show that by a 3-fold reduction in the EC50 for the antigen, ADA potentiated T cell proliferation in autologous cocultures with antigen-pulsed immature or mature dendritic cells. Costimulation was not due to the enzymatic activity but to the interaction of ADA–CD26 complexes in T cells with an ADA-anchoring protein in dendritic cells. From colocalization studies, it is deduced that ADA colocalizing with adenosine receptors on dendritic cells interact with CD26 expressed on lymphocytes. This costimulatory signal in the immunological synapse leads to a marked increase (3- to 34-fold) in the production of the T helper 1 and proimmflamatory cytokines IFN-γ, TNF-α, and IL-6.
Keywords: adenosine deaminase, costimulation, immunosynapse
Adenosine deaminase (ADA; EC 3.5.4.4) an enzyme involved in purine metabolism, catalyzes the hydrolytic deamination of adenosine or 2′-deoxyadenosine to inosine or 2′-deoxyinosine and ammonia. Congenital defect of ADA causes severe combined immunodeficiency, which is characterized by the absence of functional T and B lymphocytes in affected individuals (1). For many years, ADA was considered to be cytosolic, but it has been found on the cell surface of many cell types; therefore, it can be considered an ectoenzyme. In addition, ecto-ADA has been proposed to have a catalytic-independent function as a costimulatory molecule in lymphocytes (2).
So far, two types of surface anchoring proteins for ecto-ADA have been described. The first type, with only one member, is CD26, a multifunctional protein of 110 KDa strongly expressed on epithelial cells (kidney proximal tubules, intestine, and bile duct) and on several types of endothelial cells and fibroblasts and on leukocyte subsets (3–5). The second type of ecto-ADA-binding proteins includes the adenosine receptors (AR) A1 (A1R) (6) and A2B (A2BR) (7). The association between ADA and CD26 on the T cell surface has been proposed to have a costimulatory function during T cell antigen receptor–CD3 complex engagement (2). Because CD26 has a short cytoplasmatic tail, it needs partners to transduce the signal. Ishii et al. (8) have described that CD26-mediated signaling occurs through its association with CD45RO. At present, it is not known whether ADA generates a signal when it binds to AR. However, we have previously demonstrated that ADA binding to A1R or A2BR is required for high efficiency affinity binding of the agonist and for efficient agonist-dependent signaling (6, 7).
Dendritic cells (DC) are the most potent antigen-presenting cells (APC) specialized in the initiation of immune responses by directing the activation and differentiation of naïve T lymphocytes (9, 10). Immature DC (iDC) reside in most tissues to uptake antigen; they are engaged when exposed to danger signals produced by microorganisms, inflammatory cytokines, nucleotides, and cell damage (11). Upon exposure to these factors, DC lose their phagocytotic capacity, migrate to secondary lymphoid organs, and undergo a maturation process that involves acquisition of high levels of membrane MHC and costimulatory molecules, such as CD54, CD80, and CD86, as well as CD83. In secondary lymphoid organs, mature DC (mDC) present the captured and processed antigens to T cells and direct the development of immune responses (10). Depending on the context, DC can stimulate the polarized out-growth of distinct T cell subsets, such as T helper 1 (Th1) and T helper 2 (Th2). Th1 or Th2 polarization orchestrates the immune effector mechanism that is more appropriate for the invading pathogen. Th1 cells promote cellular immunity, protecting against intracellular infection and cancer, whereas Th2 cells promote humoral immunity, are highly effective against extracellular pathogens, and are involved in tolerance mechanisms and allergic diseases (12). During their activation, T cells require at least two signals. The first signal is provided by stimulation of T cell antigen receptor–CD3 complex by specific peptide–MHC contacts. The second signal is provided by costimulatory surface molecules that interact with their ligands on the surface of APC and are involved in a series of antigen nonspecific interactions between APC and T cells at the immunological synapse. The critical role of costimulation in the activation of naïve T cells is reflected by the fact that, in the absence of a costimulatory signal, antigen presentation induces T cells to become anergic and tolerant (13, 14). The molecular events underlying the immunological synapse formation during T cell activation are not fully understood and probably depend on the physiological context of the interaction between DC and T cells. Although not required for T cell antigen receptor signal initiation (15), synapse formation has been associated with the induction of T cell proliferation, cytokine production, negative thymocyte selection, and lytic granule release (16–19).
The two types of ecto-ADA anchoring proteins have recently been identified in DC, but their involvement on events taking place in the immunological synapse has not yet been investigated. It should also be noted that CD26 is expressed on a restricted subpopulation of DC from afferent lymph and lymph nodes (20), whereas the AR are expressed in myeloid DC (21–24) as much as in plasmacytoid DC (25), which are the two major known subsets of DC.
In this work, the role of CD26, ADA, and AR on the immunological synapse has been investigated. Our results show that ecto-ADA is expressed on monocyte-derived DC surface selectively colocalizing with A2BR. Ecto-ADA expressed in DC, acting through CD26 expressed in T cells, potentiates T cell activation by increasing proliferation and inducing high production levels of the Th1 cytokine IFN-γ and of proinflammatory cytokines, such as IL-6 and TNF-α, but without effect in Th2 cytokine production.
Materials and Methods
Abs. FITC-conjugated mAbs against IgG-γ1 isotype-matched control, HLA-DR, CCR5, CD4, CD14, CD19, and CD45RA; phycoerythrin (PE)-conjugated mAbs against IgG-γ1 isotype-matched control, HLA-DR, CXCR4, CD1a, CD11c, CD14, CD19, CD40, CD45, CD45RO, and CD56; and peridinin chlorophyll protein-conjugated mAbs against IgG-γ1 isotype-matched control and CD3 were purchased from BD Biosciences (Erembodegem-Aalst, Belgium). Nonconjugated mAb against CD26 (Ba5); nonconjugated mouse IgG2a isotype control; and PE-conjugated mAbs against CD80, CD83, and CD86 were from Coulter–Immunotech Diagnostic (Marseille, France), and PE-conjugated mAb against CD209 was from eBioscience (San Diego). The polyclonal Abs (pAbs) used were PC21, an affinity chromatography-purified pAb against the second extracellular loop of A1R (26); MPE, an affinity chromatography-purified pAb against the third extracellular loop of A2BR (27); nonconjugated or FITC-conjugated pAb against purified calf ADA (26); and nonconjugated irrelevant rabbit IgG (Sigma–Aldrich). Secondary Abs Alexa Fluor 488-conjugated goat anti-mouse IgG and Texas red-conjugated goat anti-rabbit IgG were purchased from Molecular Probes, and PE-conjugated goat anti-rabbit from Sigma–Aldrich.
Generation of Monocyte-Derived DC and Isolation of Lymphocytes. Human peripheral blood mononuclear cells (PBMC) were obtained immediately after extraction from the heparinized blood of healthy donors by using the standard Ficoll gradient method. To obtain monocytes, PBMC (3 × 106 cells per ml) were incubated in serum free XVIVO-15 medium (BioWhittaker) supplemented with 1% autologous serum, 50 μg/ml gentamycin (Braun, Melsungen, Germany), and 2.5 μg/ml fungizone (Bristol-Myers Squibb) (DC medium) for 2 h at 37°C in a humid atmosphere of 5% CO2. Adherent cells were washed four times with prewarmed, serum-free XVIVO-10 medium (BioWhittaker) and then cultured in DC medium at 37°C in a humid atmosphere of 5% CO2. Cells were differentiated for 7 days to iDC by adding 1,000 units/ml IL-4 (Prospec-Tany Technogene LTD, Rehovot, Israel) and 1,000 units/ml granulocyte–macrophage colony-stimulating factor (Prospec-Tany Technogene, Rehovot, Israel) at days 0, 3, and 5. To obtain mDC, 1,000 units/ml TNF-α (Sigma–Aldrich) or 500 ng/ml lipopolysaccharide (Sigma–Aldrich) were added for the last 48 h. DC immunophenotypes were confirmed by flow cytometry using commercially labeled mAbs against surface markers. iDC obtained were CD3-, CD4-, CD8-, CD14-, CD19-, CD56-, HLA-DR+, CD80+, CD83+, CD86+, CD1a+, CD11c+, CD40+, CD45RO+, CD45RA-, CD209+, CXCR4+, and CCR5-. In mDC, the expression of HLA-DR, CD80, CD83, and CD86 was highly increased compared with iDC. As a source of a T cell-enriched population for cocultures with autologous DC, after the 2-h period of plastic adherence, nonadherent PBMC were collected and washed three times with XVIVO-10 medium.
ADA and Hg2+-Treated ADA Preparations. ADA from calf intestine (Roche Diagnostics) was desalted by passage through a PD-10 column (Amersham Pharmacia), and its enzymatic activity was tested. Enzymatic activity of ADA was evaluated by measuring the decrease of absorbance at 265 nm (28). To obtain ADA without enzymatic activity, desalted ADA was treated with HgCl2 as described in ref. 6 and passed again through a PD-10 column to eliminate remaining HgCl2. The enzymatic activity of Hg2+-treated ADA (ADA-Hg2+) was completely abolished.
Cocultures. iDC or mDC were harvested at day 7, and fresh autologous T cells were prepared. DC were resuspended in DC medium (4 × 105 cells per ml) and pulsed with the indicated concentration of staphylococcal enterotoxin A (SEA) (Sigma–Aldrich) for 4 h at 37°Cina humid atmosphere of 5% CO2. Pulsed DC were washed four times and resuspended at 4 × 105 cells per ml with XVIVO-10 medium. When T cell proliferation was measured, pulsed DC were γ-irradiated before coculture. Autologous cocultures were performed in 96-well plates containing T cells (2 × 105 cells per well), effectors (when indicated; see Figs. 1, 2, 3, 4), and DC (2 × 104 cells per well) in XVIVO-10 medium at a final volume of 200 μl/well. Cocultures were incubated for the times indicated (see Figs. 1, 2, 3, 4) at 37°C in a humid atmosphere of 5% CO2.
Fig. 1.
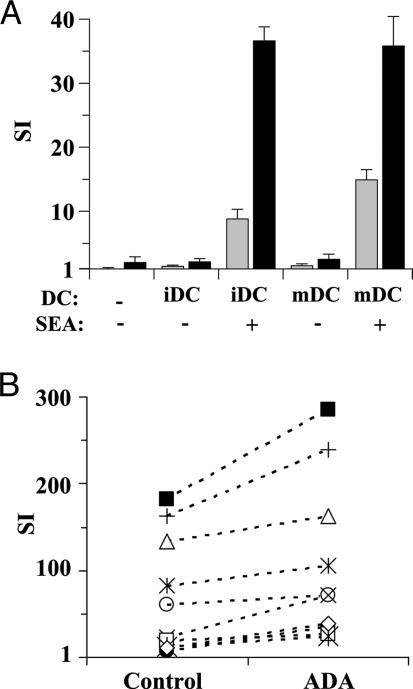
ADA potentiates T cell proliferation induced by superantigen-pulsed DC. (A) iDC or mDC were pulsed or not with 100 pg/ml SEA, irradiated, and then preincubated without (gray bars) or with (black bars) 4 μM ADA for 1 h. DC were cocultured with autologous T cells for 7 days, and proliferation was determined as [3H]thymidine incorporation. Values are expressed as SI, determined as the ratio of [3H]thymidine incorporated in cocultured T cells versus noncocultured T cells. Data are the mean ± SD of triplicates. Representative data of one of 10 independent experiments is shown. (B) Data of SEA-pulsed iDC without (control) or with ADA from 10 healthy donors is shown (each symbol represents a different donor). P < 0.005 ADA versus control by a paired Student t test.
Fig. 2.
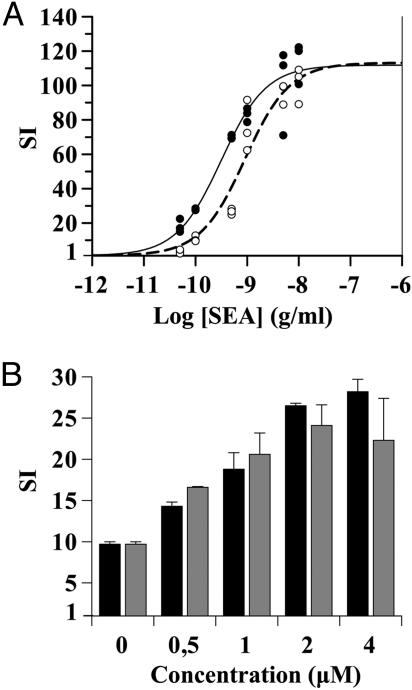
Dose–response curves of SEA stimulation and ADA costimulation in T cell proliferation induced by DC. (A) iDC were pulsed with increasing SEA concentrations, irradiated, and then preincubated without (open circles, dotted line) or with (filled circles, solid line) 4 μM ADA for 1 h. DC were cocultured with autologous T cells. (B) iDC were pulsed with 100 pg/ml SEA, irradiated, and then preincubated with increasing concentrations of ADA (black bars) or ADA-Hg2+ (gray bars) for 1 h. Values are mean ± SD of triplicates. Subsequently in A and B, DC were cocultured with autologous T cells for 7 days, and proliferation was determined as [3H]thymidine incorporation. Values are expressed as SI, determined as indicated in the legend for Fig. 1. Experiments were performed in triplicates. Representative data of one of three independent experiments is shown.
Fig. 3.
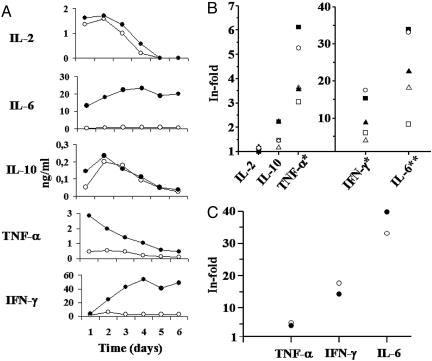
ADA increases the secretion of IL-6, TNF-α, and IFN-γ in cocultures of T cells with SEA-pulsed DC. iDC were pulsed with 100 pg/ml SEA and then preincubated with or without 4 μM ADA for 1 h. DC were cocultured with autologous T cells for different days, and cytokine levels were determined in the supernatants as indicated in Materials and Methods.(A) Cytokine levels versus days of coculture in the absence (open circles) or presence (filled circles) of ADA. One representative experiment is shown. (B) Cytokine levels of coculture supernatants with or without ADA from five healthy donors determined at day 2. Each symbol represents the value of a cytokine level from a given healthy donor. (C) Cytokine levels of coculture supernatants with ADA (open circles) or ADA-Hg2+ (filled circles) from a healthy donor determined at day 2. For each donor, control experiments were performed in the absence of T cells (cytokine levels < 20 pg/ml). In the absence of DC, cytokine levels produced by T cells were <20 pg/ml, except for IL-6, which was between 60 and 130 pg/ml in the absence of ADA and between 2 and 6 ng/ml in the presence of ADA (data not shown). (B and C) Increase of cytokine levels promoted by ADA or ADA-Hg2+ are expressed as the ratio of cytokine levels in the presence of ADA versus cytokine levels in the absence of ADA. The value of 1 indicates no changes in cytokine production in treated versus untreated cells. *, P < 0.05 for TNF-α and IFN-γ; **, P < 0.01 for IL-6 (treated versus untreated), paired Student's t test.
Fig. 4.
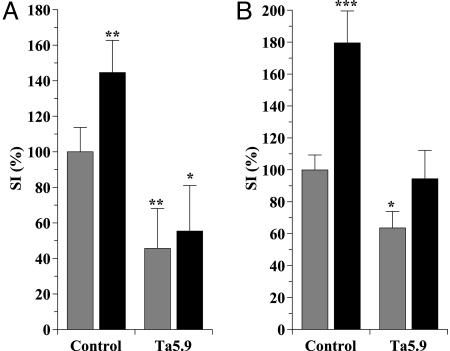
Ta5.9 inhibits the costimulatory ADA effect in T cell proliferation induced by superantigen-pulsed DC or by anti-CD3 stimulation. (A) Before coculture, iDC were pulsed with 100 pg/ml SEA, irradiated, and then incubated without (gray bars) or with (black bars) 4 μM ADA for 1 h. Autologous T cells were preincubated without (control) or with 33 μg/ml of mAb anti-CD26 Ta5.9 (Ta5.9) for 15 min and then added to the DC prepared as indicated above. Proliferation was determined as [3H]thymidine incorporation after 7 days in coculture. (B) T cells were preincubated without (control) or with 33 μg/ml mAb anti-CD26 Ta5.9 for 15 min and then incubated without (gray bars) or with (black bars) 4 μM ADA for 1 h. T cell activation was induced by 1 ng/ml OKT3 for 4 days, and proliferation was determined as [3H]thymidine incorporation. Values are expressed as percentages of SI (see Fig. 1 legend) with respect to the SI value obtained in untreated control cells. Data from four independent experiments using blood from four different donors performed in triplicates are shown. Data are the mean ± SD. *, P < 0.005; **, P < 0.001; ***, P < 0.0001 versus control without ADA by Student's t test.
Proliferation Assays and Cytokine Determination. In anti-CD3-induced proliferation experiments, T cells (2 × 105 cells per well) were incubated in XVIVO-10 medium with the indicated effectors and with 1 ng/ml of mouse mAb anti-CD3 (OKT3) for 4 days at 37°C in a humid atmosphere of 5% CO2 (29, 30). In control experiments, 1 μg/ml OKT3 was immobilized on the plates for 20 h at 4°C and washed twice with PBS before addition of cells (2 × 105 cells per well) and effectors. When T cell proliferation was measured in cocultures, DC and T cells were incubated for 7 days at 37°C in a humid atmosphere of 5% CO2. In both experimental conditions proliferation was measured as [3H]thymidine incorporation. Cells were pulsed for the last 18 h with 1 μCi (1 Ci = 37 GBq) thymidine per well ([3H]methyl, 2 Ci/mmol; Moravek Bioquemicals, Brea, CA) and fixed in 3.7% formaldehyde for 30 min, harvested onto filters and extensively washed with an LKB 1295-001 cell harvester (Wallac, Gaithersburg, MD). Tritium incorporation was determined by a liquid scintillation counter (1205 Betaplate, Wallac). IL-2, IL-4, IL-6, IL-10, TNF-α, and IFN-γ levels were determined by using a BD Cytometric Bead Array human Th1/Th2 cytokine kit (Pharmingen) according to the manufacturer's protocol, with 50 μl of conveniently diluted coculture supernatants.
RT-PCR. Total RNA was isolated from cells with a QuickPrep Total RNA extraction kit (Amersham Pharmacia). Aliquots of 1 μg of total RNA and 1 μg of random hexamers (Invitrogen) as primers for retrotranscription dissolved in 14 μl of RNase-free water were heated to 70°C for 5 min and then cooled to 4°C. Reverse transcriptase buffer 5× (5 μl; 250 mM Tris·HCl, pH 8.3/375 mM KCl/15 mM MgCl2/50 mM DTT) (Promega), 1.5 μl of dNTPs (mix, 10 mM each; Sigma), 1 μl of reverse transcriptase enzyme [Moloney murine leukemia virus reverse transcriptase (H-); Promega], 1 μl of RNase inhibitor (RNasa OUT, Invitrogen), and 2.5 μl of RNase-free water were added to a final volume of 25 μl. The incubation was continued at 25°C for 10 min and 42°C for 52 min and terminated at 70°C for 15 min. PCR was carried out for 50 cycles (1 min at 95°C, 45 sec for annealing, and 2 min at 72°C) in a final volume of 50 μl containing 25 μl of 2× PCR Master Mix (Promega), 5 μl of cDNA, and 0.6 μM (each) primers. RT-PCR negative control was performed by loading diethylpyrocarbonate water instead of cDNA. Specific primers were selected according to the GeneBank database resource, and annealing temperatures were as follows. A1R: annealing at 60.5°C, 646-bp amplimer, 5′-GCG GTG AGG CAG GGG AGT CT-3′ (forward), and 5′-GGG GCG CCT ACA GTC AAC AC-3′ (reverse); A2BR: annealing at 59.4°C, 542-bp amplimer, 5′-CCT GGG CTT CTG CAC TGA CTT CTA-3′ (forward), and 5′-GTA ACC AGC ACA GGG CAA AAA TCC-3′ (reverse); CD26: annealing at 53.6°C, 507-bp amplimer; 5′-CGT GCC CGT GGT TCT GCT-3′ (forward), 5′-TCT TTC CCC GTC CAT GTG ATT CT-3′ (reverse); GAPDH: annealing at 59.9°C; 425-bp amplimer; 5′-GCG GGG CTC TCC AGA ACA TCA T-3′ (forward); 5′-GGT GGT CCA GGG GTC TTA CTC C-3′ (reverse).
Immunostaining, Flow Cytometry, and Confocal Laser Microscopy. When directly labeled Abs were used, cells were washed with PBS, resuspended at 2 × 106 cells per ml (50 μl per tube), and incubated with FITC-, PE-, and peridinin chlorophyll protein-conjugated Abs for 30 min at 4°C. Cells were washed with PBS, fixed with 1% formaldehyde in PBS, and analyzed by flow cytometry. FITC-, PE-, and peridinin chlorophyll protein-conjugated isotype-matched Abs were used for negative controls. When unlabeled primary Abs were used, cells were washed with PBS, fixed in PBS containing 2% paraformaldehyde for 15 min at room temperature, and washed twice with PBS containing 20 mM glycine to quench the aldehyde groups. Cells were incubated with PBS containing 1% BSA, 20 mM glycine, and 0.1% NaN3 (blocking buffer) for 15 min and labeled with 10 μg/ml PC21, 10 μg/ml MPE, 20 μg/ml anti-ADA, 20 μg/ml irrelevant rabbit IgG (used as a negative control of pAbs), a 1:10 ratio of Ba5, or a 1:10 ratio of mouse IgG2a isotype control (used as a negative control of Ba5) for 1 h at room temperature. Cells were washed three times with blocking buffer and stained for flow cytometry analysis with the secondary Ab PE-conjugated goat anti-rabbit IgG (1:20) or Alexa Fluor 488-conjugated goat anti-mouse IgG (1:1,000) or for confocal microscopy analysis with Texas red-conjugated goat anti-rabbit IgG (1:2,000) or Alexa Fluor 488-conjugated goat anti-mouse IgG (1:1,000) for 1 h at room temperature. When FITC-conjugated rabbit anti-ADA Ab was used, cells were washed three times with blocking buffer after secondary Ab of the first labeling and then incubated with FITC-conjugated anti-ADA for 1 h at room temperature. FITC-conjugated irrelevant rabbit IgG was used as negative control of FITC-conjugated anti-ADA Ab. Coverslips were mounted with ImmunoFluore mounting medium (ICN). Confocal microscope observations were made with a LeicaTCS-SP (Leica) confocal scanning laser microscope adapted to an inverted Leitz DMIRBE microscope. Flow cytometry analysis was done with an EPICS Profile flow cytometer (Coulter). Cell populations were selected by forward and side light scatter parameters.
Results and Discussion
To test whether ADA has a costimulatory role during antigen presentation, we assessed the effect of ADA in the T cell activation induced by superantigen-pulsed DC. SEA-pulsed and unpulsed iDC and mDC were preincubated with or without ADA and then cocultured with autologous T cells. ADA increased T cell proliferation in cocultures with pulsed iDC and mDC (Fig. 1A), and the effect was observed for every donor tested (Fig. 1B). The increase in the stimulation index (SI) value obtained in the presence of ADA was statistically significant for iDC (P < 0.005) (Fig. 1B) and mDC (P < 0.05) (data not shown). When T cells were cultured in the presence or absence of ADA without DC or with DC not pulsed with the superantigen, T lymphocyte proliferation was negligible. When pulsing iDC with different doses of SEA, a sigmoid curve was apparent for T cell proliferation with an EC50 of 900 ± 200 pg/ml (Fig. 2A). A sigmoid curve was also observed with DC preincubated with ADA, reaching the same maximal effect as without ADA but displaced to the left, with an EC50 of 310 ± 60 pg/ml. The decrease in the EC50 value obtained in the presence of ADA is statistically significant (64 ± 4%, n = 3). Similar results were observed with mDC (data not shown). Therefore, ADA costimulation requires much less an amount of antigen to attain a certain level of T cell activation. Because physiological antigen levels are very low and ADA action seems prominent at very low antigen levels, we decided to characterize the phenomenon with a suboptimal superantigen concentration (SEA = 100 pg/ml). The ADA-induced costimulatory effect was dose-dependent with a maximal effect at 2 μM ADA (Fig. 2B). According to the catalytically independent function of ecto-ADA as a costimulatory molecule in lymphocytes (2), the Hg2+-treated ADA (ADA-Hg2+), which lacks enzymatic activity, showed a dose-dependent costimulatory effect similar to the one observed with catalytically active ADA (Fig. 2B). Comparable results were obtained with mDC (data not shown). Therefore, the costimulatory effect of ADA was due to its extra-enzymatic properties. However, it is known that adenosine, through A2AR, promotes a down-regulation of the immune response acting in T cells as much as in DC (21, 22, 24, 31–33). Taking this into account, it is predictable that ADA, by degradating adenosine, is capable of enhancing T cell and DC functions. Therefore, the enzymatic and extra-enzymatic properties of ADA seem to contribute to the regulation of the immune function.
Because SEA binds to MHC class II, the observed T cell response is CD4+-mediated (34). Two major CD4+ subsets are known, Th1 and Th2. Th1 or Th2 polarization orchestrates the immune effector mechanism that is more appropriate for the invading pathogen, each one having a particular cytokine pattern. Whereas Th1 response is mainly characterized by secretion of IFN-γ and IL-2, the main cytokines of Th2 response are IL-4 and IL-10 (12). To investigate the role of ADA in Th1 or Th2 polarization, serial measurements of Th1/Th2 and proinflammatory cytokines in the coculture supernatants were performed at different times. Our results show that without ADA, there was a moderate secretion of the Th1 cytokines IL-2 and IFN-γ and of the proinflammatory cytokine TNF-α. When ADA was present in cocultures, an increase of IFN-γ, TNF-α, and IL-6 was observed, whereas IL-2 and IL-10 were not significantly modified (Fig. 3). IL-4 was also tested, but it was undetectable in the absence or presence of ADA. By using blood from a donor and ADA-Hg2+, it was confirmed that the ADA-mediated increase in IFN-γ, TNF-α, and IL-6 production does not depend on the enzymatic activity (Fig. 3C). All these results indicate that the costimulatory effect of ADA is due to the up-regulation of the Th1 cytokine IFN-γ and the proinflammatory cytokines TNF-α and IL-6, having no effect on IL-2, IL-10, and IL-4 levels. This finding suggests that ADA induces a proinflammatory profile of the Th1 T cell subset.
To determine whether the costimulatory effect of ADA is due to the association between ADA and CD26 on the T cell surface in the immunological synapse, cocultures were performed in conditions at which the ADA–CD26 interaction was interfered by using the anti-CD26 mAb Ta5.9, which is directed against the ADA binding domain of CD26 (35, 36). When T cells were preincubated with Ta5.9 before the coculture with ADA-treated DC, the costimulatory effect of ADA in T cell activation was abrogated (Fig. 4A), showing that ADA binding to T cell CD26 is the first step in the ADA-induced costimulatory effect. According to this finding, the ADA effect also occurs in OKT3-induced T cell proliferation, and the costimulation is also inhibited by Ta5.9 (Fig. 4B). In absence of ADA, T cell proliferation induced by OKT3 or triggered by T cell–DC contacts at the immunological synapse is slightly reduced by Ta5.9 (Fig. 4). This effect is probably due to the shedding of endogenous ADA in isolated T cells produced by the Ab–CD26 interaction. As a negative control it was tested that Ta5.9 did not affect the stimulation induced by immobilized OKT3 (data not shown). Therefore, the costimulatory effect of ADA observed in T cell activation is due to the ADA–CD26 interaction at the T cell surface.
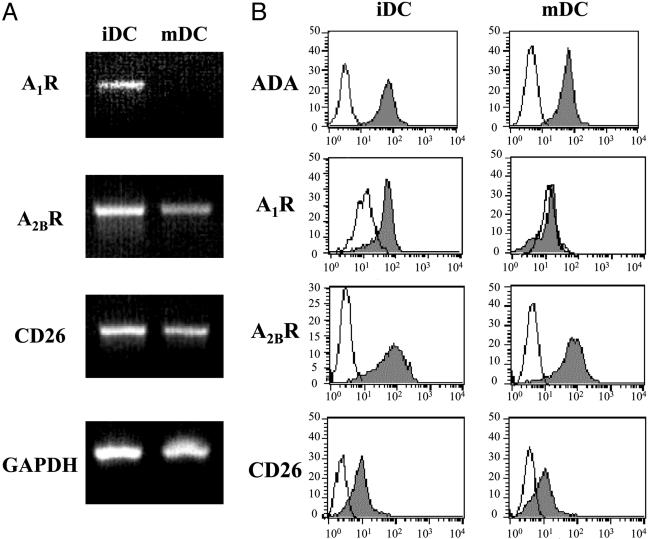
To know whether there are ADA-anchoring proteins in DC presenting the protein to T cells in the immunological synapse, CD26 and AR expression and the degree of colocalization with ADA were investigated. It has been described that both types of ADA-anchoring proteins are expressed in DC (20–25). The monocyte-derived DC used in this work express ADA-anchoring proteins at the mRNA and protein levels. By RT-PCR it was shown that iDC express all ADA-anchoring proteins mRNA (A1R, A2BR, and CD26 mRNA), whereas mDC express mRNAs for A2BR and CD26 but not for A1R (Fig. 5A). At the protein level, the expression of ecto-ADA and ADA-anchoring proteins were tested by flow cytometry with nonpermeabilized cells and specific Abs (Fig. 5B). The results were consistent with data on A1R, A2BR, and CD26 mRNA expression and also confirm the presence of endogenously expressed ecto-ADA in iDC and in mDC (Fig. 5). Our results concerning A2BR expression disagree with those of Panther et al. (21) and Fossetta et al. (23). It should be noted that, in these papers, monocytes were isolated from buffy coats by a process that involves strong and long manipulation of cells and then differentiated to DC by using RPMI medium 1640 containing FCS. In contrast, monocytes used in our study were from fresh blood and differentiated to DC by using autologous serum and a XVIVO-15 medium, which can be included in investigations into medicinal protocols/products for human use (37). This circumstance combined with the Good Manufacturing Practice standards of our laboratory leads to the obtention of physiologically relevant DC, which, according to our results, express the A2BR. The expression of the receptor in DC did not change upon exposure of cocultures to SEA (data not shown).
Fig. 5.
ADA and ADA-anchoring protein expression in DC. (A) mRNA from iDC and mDC was isolated, and RT-PCR analysis of A1R (646 bp), A2BR (542 bp), and CD26 (507 bp) was performed as described in Materials and Methods. GAPDH mRNA (425 bp) was analyzed as positive control. (B) Indirect immunostaining of iDC and mDC was performed as indicated in Materials and Methods. A PE-conjugated goat anti-rabbit secondary Ab was used to detect the primary Abs [rabbit pAb anti-ADA, rabbit pAb anti-A1R (PC21), or rabbit pAb anti-A2BR (MPE)]. An Alexa Fluor 488-conjugated goat anti-mouse secondary Ab was used to detect the mouse mAb anti-CD26 (Ba5) primary Ab. Immunolabeling was detected by flow cytometry (gray histogram). Negative controls (white histogram) were obtained by using an irrelevant IgG as primary Ab. Representative data of one of five independent experiments are shown.
To identify the protein anchoring ADA to the DC surface, cells were stained with specific Abs directed against ADA and ADA-anchoring proteins, and then colocalization was assessed by confocal microscopy. Only the A2BR is able to colocalize with ADA in iDC (Fig. 6) and mDC (data not shown). In DC, unlike in lymphocytes, ADA is very poorly colocalized with CD26. The crystal structure of the CD26–ADA complex (38) does not show in ADA two but just one motive capable of interacting with CD26. Then it is unlikely that ADA would bind two CD26 molecules, one expressed on the T cell surface and another on the DC surface. Also A1R were very poorly colocalized with ADA in iDC (Fig. 6). Therefore, the endogenously expressed ADA present in DC seems to be predominantly anchored to the cellular surface via the A2BR.
Fig. 6.
ADA does not colocalize on the DC surface with CD26 or A1R, but ADA does colocalize on the DC surface with A2BR. (Top Left and Top Center) iDC were immunostained with the pAb FITC-conjugated rabbit anti-ADA (green) (Top Left) and the pAb anti-A2BR, followed by a Texas red-conjugated goat anti-rabbit IgG secondary Ab (red) (Top Center) as indicated in Materials and Methods. (Top Right) Merge of ADA label and A2BR label. Colocalization detected by confocal microscopy is shown in yellow. (Middle Left and Middle Center) For ADA/A1R labeling, iDC were immunostained with the pAb FITC-conjugated rabbit anti-ADA (green) (Middle Left) and the pAb anti-A1R, PC21, followed by a Texas red-conjugated goat anti-rabbit IgG secondary Ab (red) (Middle Center) as indicated in Materials and Methods. (Middle Right) Merge of ADA label and A1R label. Colocalization detected by confocal microscopy is shown in yellow. (Bottom Left and Bottom Center) For ADA/CD26 labeling, iDC were immunostained with the mouse mAb anti-CD26 and Ba5, followed by a Alexa Fluor 488-conjugated goat anti-mouse IgG secondary Ab (green) (Bottom Left) and the pAb rabbit anti-ADA, followed by a Texas Red-conjugated anti-rabbit IgG secondary Ab (red) (Bottom Center) as indicated in Materials and Methods.(Bottom Right) Merge of CD26 label and ADA label. Colocalization detected by confocal microscopy is shown in yellow. (Bars, 10 μm.)
In conclusion, our data demonstrates a costimulatory interaction during the immunosynapse. ADA anchored to the DC surface probably by the A2BR binds to the CD26 expressed on the surface of the T cells, triggering costimulation. This costimulatory signal promotes an augmented T cell activation with a Th1 pattern and proinflammatory cytokine production, therefore enabling an enhanced immune response.
Acknowledgments
We thank all of the blood donors and María Carmen Pardo, Vanesa Villegas, and María de los Ángeles Lopez for technical assistance in blood extraction. This study was supported by Ministerio de Ciencia y Tecnología Grants SAF2002-03293 (to R.F.) and SAF2001-2591 (to J.M.G.), Fundació Marató of Catalonian Telethon Grant 02/021010 (to R.F.), and Ministerio de Salud Grants FIS2003-1200 and FIPSE01-36259 (to T.G.).
Author contributions: R.P., T.G., J.M., C.L., and R.F. designed research; R.P., J.M.M.-N., M.L., and H.O. performed research; N.C. and J.M.G. contributed new reagents/analytic tools; R.P., J.M.M.-N., M.L., and N.C. analyzed data; and R.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DC, dendritic cell(s); iDC, immature DC; mDC, mature DC; ADA, adenosine deaminase; AR, adenosine receptor(s); AnR, AR An; SEA, staphylococcal enterotoxin A; Thn, T helper n; PE, phycoerythrin; pAb, polyclonal antibody.
References
- 1.Franco, R., Valenzuela, A., Lluis, C. & Blanco, J. (1998) Immunol. Rev. 161, 27-42. [DOI] [PubMed] [Google Scholar]
- 2.Martin, M., Huguet, J., Centelles, J. J. & Franco, R. (1995) J. Immunol. 155, 4630-4643. [PubMed] [Google Scholar]
- 3.De Meester, I., Korom, S., Van Damme, J. & Scharpe, S. (1999) Immunol. Today 20, 367-375. [DOI] [PubMed] [Google Scholar]
- 4.Gorrell, M. D., Gysbers, V. & McCaughan, G. W. (2001) Scand. J. Immunol. 54, 249-264. [DOI] [PubMed] [Google Scholar]
- 5.Kameoka, J., Tanaka, T., Nojima, Y., Schlossman, S. F. & Morimoto, C. (1993) Science 261, 466-469. [DOI] [PubMed] [Google Scholar]
- 6.Ciruela, F., Saura, C., Canela, E. I., Mallol, J., Lluis, C. & Franco, R. (1996) FEBS Lett. 380, 219-223. [DOI] [PubMed] [Google Scholar]
- 7.Herrera, C., Casado, V., Ciruela, F., Schofield, P., Mallol, J., Lluis, C. & Franco, R. (2001) Mol. Pharmacol. 59, 127-134. [PubMed] [Google Scholar]
- 8.Ishii, T., Ohnuma, K., Murakami, A., Takasawa, N., Kobayashi, S., Dang, N. H., Schlossman, S. F. & Morimoto, C. (2001) Proc. Natl. Acad. Sci. USA 98, 12138-12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banchereau, J. & Steinman, R. M. (1998) Nature 392, 245-252. [DOI] [PubMed] [Google Scholar]
- 10.Lanzavecchia, A. & Sallusto, F. (2000) Science 290, 92-97. [DOI] [PubMed] [Google Scholar]
- 11.Gallucci, S. & Matzinger, P. (2001) Curr. Opin. Immunol. 13, 114-119. [DOI] [PubMed] [Google Scholar]
- 12.Del Prete, G. (1998) Int. Rev. Immunol. 16, 427-455. [DOI] [PubMed] [Google Scholar]
- 13.Chai, J. G., Vendetti, S., Bartok, I., Schoendorf, D., Takacs, K., Elliott, J., Lechler, R. & Dyson, J. (1999) J. Immunol. 163, 1298-1305. [PubMed] [Google Scholar]
- 14.Greenfield, E. A., Nguyen, K. A. & Kuchroo, V. K. (1998) Crit. Rev. Immunol. 18, 389-418. [DOI] [PubMed] [Google Scholar]
- 15.Lee, K. H., Holdorf, A. D., Dustin, M. L., Chan, A. C., Allen, P. M. & Shaw, A. S. (2002) Science 295, 1539-1542. [DOI] [PubMed] [Google Scholar]
- 16.Kupfer, H., Monks, C. R. & Kupfer, A. (1994) J. Exp. Med. 179, 1507-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delon, J., Stoll, S. & Germain, R. N. (2002) Immunol. Rev. 189, 51-63. [DOI] [PubMed] [Google Scholar]
- 18.Richie, L. I., Ebert, P. J., Wu, L. C., Krummel, M. F., Owen, J. J. & Davis, M. M. (2002) Immunity 16, 595-606. [DOI] [PubMed] [Google Scholar]
- 19.Stinchcombe, J. C., Bossi, G., Booth, S. & Griffiths, G. M. (2001) Immunity 15, 751-761. [DOI] [PubMed] [Google Scholar]
- 20.Gliddon, D. R. & Howard, C. J. (2002) Eur. J. Immunol. 32, 1472-1481. [DOI] [PubMed] [Google Scholar]
- 21.Panther, E., Idzko, M., Herouy, Y., Rheinen, H., Gebicke-Haerter, P. J., Mrowietz, U., Dichmann, S. & Norgauer, J. (2001) FASEB J. 15, 1963-1970. [DOI] [PubMed] [Google Scholar]
- 22.Panther, E., Corinti, S., Idzko, M., Herouy, Y., Napp, M., La Sala, A., Girolomoni, G. & Norgauer, J. (2003) Blood 101, 3985-3990. [DOI] [PubMed] [Google Scholar]
- 23.Fossetta, J., Jackson, J., Deno, G., Fan, X., Du, X. K., Bober, L., Soude-Bermejo, A., De Bouteiller, O., Caux, C., Lunn, C., et al. (2003) Mol. Pharmacol. 63, 342-350. [DOI] [PubMed] [Google Scholar]
- 24.Hofer, S., Ivarsson, L., Stoitzner, P., Auffinger, M., Rainer, C., Romani, N. & Heufler, C. (2003) J. Invest. Dermatol. 121, 300-307. [DOI] [PubMed] [Google Scholar]
- 25.Schnurr, M., Toy, T., Shin, A., Hartmann, G., Rothenfusser, S., Soellner, J., Davis, I. D., Cebon, J. & Maraskovsky, E. (2004) Blood 103, 1391-1397. [DOI] [PubMed] [Google Scholar]
- 26.Gines, S., Ciruela, F., Burgueno, J., Casado, V., Canela, E. I., Mallol, J., Lluis, C. & Franco, R. (2001) Mol. Pharmacol. 59, 1314-1323. [PubMed] [Google Scholar]
- 27.Mirabet, M., Herrera, C., Cordero, O. J., Mallol, J., Lluis, C. & Franco, R. (1999) J. Cell Sci. 112, 491-502. [DOI] [PubMed] [Google Scholar]
- 28.Franco, R., Canela, E. I. & Bozal, J. (1986) Neurochem. Res. 11, 423-435. [DOI] [PubMed] [Google Scholar]
- 29.Kung, P., Goldstein, G., Reinhertz, E. L. & Schlossman, S. F. (1979) Science 206, 347-349. [DOI] [PubMed] [Google Scholar]
- 30.Van Wauwe, J. P., De Mey, J. R. & Goznes, J. G. (1980) J. Immunol. 124, 2708-2713. [PubMed] [Google Scholar]
- 31.Huang, S., Aposov, S., Koshiba, M. & Sitkovsky, M. (1997) Blood 90, 1600-1610. [PubMed] [Google Scholar]
- 32.Ohta, A. & Sitkovsky, M. (2001) Nature 414, 916-920. [DOI] [PubMed] [Google Scholar]
- 33.Hershfield, M. S. (2005) Eur. J. Immunol. 35, 25-30. [DOI] [PubMed] [Google Scholar]
- 34.Newton, D. W., Dohlsten, M., Olsson, C., Segren, S., Lundin, K. E. A., Lando, P. A., Kalland, T. & Kotb, M. (1996) J. Immunol. 157, 3988-3994. [PubMed] [Google Scholar]
- 35.Valenzuela, A., Blanco, J., Callebaut, C., Jacotot, E., Lluis, C., Hovanessian, A. G. & Franco, R. (1997) J. Immunol. 158, 3721-3729. [PubMed] [Google Scholar]
- 36.Blanco, J., Valenzuela, A., Herrera, C., Lluis, C., Hovanessian, A. G. & Franco, R. (2000) FEBS Lett. 477, 123-128. [DOI] [PubMed] [Google Scholar]
- 37.Garcia, F., Lejeune, M., Climent, N., Gil, C., Alcami, J., Morente, V., Alos, L., Ruiz, A., Setoain, J., Fumero, E., et al. (2005) J. Infect. Dis. 191, 1680-1685. [DOI] [PubMed] [Google Scholar]
- 38.Weihofen, W. A., Liu, J., Reutter, W., Saenger, W. & Fan, H. (2004) J. Biol. Chem. 279, 43330-43335. [DOI] [PubMed] [Google Scholar]