Abstract
Interleukin (IL)-6 signaling through its soluble receptor (IL-6 transsignaling) directs transition between innate and acquired immune responses by orchestrating the chemokine-directed attraction and apoptotic clearance of leukocytes. Through analysis of mononuclear cell infiltration in WT and IL-6-deficient mice during peritoneal inflammation, we now report that IL-6 selectively governs T cell infiltration by regulating chemokine secretion (CXCL10, CCL4, CCL5, CCL11, and CCL17) and chemokine receptor (CCR3, CCR4, CCR5, and CXCR3) expression on the CD3+ infiltrate. Although blockade of IL-6 trans-signaling prevented chemokine release, chemokine receptor expression remained unaltered suggesting that this response is regulated by IL-6 itself. To dissect the signaling events promoting T cell migration, inflammation was established in knock-in mice expressing mutated forms of the universal signal-transducing element for IL-6-related cytokines gp130. In mice (gp130Y757F/Y757F) deficient in SHP2 and SOCS3 binding, but presenting hyperactivation of STAT1/3, T cell recruitment and CCL5 expression was enhanced. Conversely, both of these parameters were suppressed in mice with ablated gp130-mediated STAT1/3 activation (gp130ΔSTAT/ΔSTAT). T cell migration was related to STAT3 activity, because monoallelic deletion of Stat3 in gp130Y757F/Y757F mice (gp130Y757F/Y757F:Stat3+/-) corrected the exaggerated responses observed in gp130Y757F/Y757F mice. Consequently, STAT3 plays a defining role in IL-6-mediated T cell migration.
Keywords: chemokines, cytokines, gp130
Interleukin (IL)-6 is traditionally considered a regulator of acute-phase responses and a lymphocyte stimulatory factor (1). However recent advances have highlighted a pivotal role for this cytokine in directing leukocyte trafficking and facilitating transition between innate and acquired immune responses (2, 3). Identification of these IL-6 activities has largely been achieved through an increased understanding of the regulatory properties of its soluble receptor, whereas better appreciation of IL-6 signaling has led researchers to consider the interplay between IL-6 and other cytokines (4–7). These studies collectively underline a role for IL-6 in governing inflammation and have emphasized the therapeutic potential of targeting IL-6 as a strategy for the management of infectious and inflammatory diseases (8, 9).
IL-6 responses are transmitted through gp130, which serves as the universal signal-transducing receptor subunit for all IL-6-related cytokines (10, 11). Although this classically occurs through IL-6 binding to its membrane-bound receptor (IL-6R), it is clear that a soluble form of the cognate IL-6 receptor (sIL-6R) affords IL-6 with an alternative mechanism of gp130 activation. This additional mode of cell activation is termed IL-6 trans-signaling and results from formation of a sIL-6R/IL-6 complex, which can directly bind cellular gp130 (2). Because gp130 is ubiquitously expressed within tissue, trans-signaling provides IL-6 with the capacity to activate cells that would not intrinsically respond to IL-6 itself (2).
Studies have documented inherent roles for IL-6 trans-signaling in a number of biological processes, including its involvement in leukocyte trafficking and activation (2, 3). Although initial observations in IL-6-deficient (IL-6-/-) mice noted that IL-6 suppressed neutrophil accumulation at sites of infection or inflammation (12, 13), it is evident that neutrophil clearance and their subsequent replacement by a more sustained population of mononuclear leukocytes is governed by the sIL-6R (5, 14). In particular, IL-6 trans-signaling defines the nature of the inflammatory infiltrate by controlling leukocyte apoptosis and the expression of inflammatory chemokines and adhesion molecules (5, 13–18). This event is a critical step in the successful resolution of any inflammatory response and defines an immunological switch from innate to acquired immunity at sites of inflammation.
In terms of mononuclear leukocytes, IL-6-mediated signaling is associated with the activation of a number of cellular events important in host defense and chronic disease progression. With respect to monocytes/macrophages, IL-6 affects cellular differentiation (19–21). However, the role of IL-6 in lymphocyte activation may have more profound consequences, where it controls T cell polarization (22–24), IL-2-dependent cell proliferation (25, 26), L-selectin (CD62L) adhesion (18), B cell activation, and antibody production (25). Furthermore, the ability of IL-6 to rescue T cells from entering apoptosis (17, 27–30) may have a considerable bearing in the progression of chronic inflammation. Indeed, sIL-6R-mediated trans-signaling promotes T cell expression of antiapoptotic regulators (17, 30), whereas blockade of sIL-6R signaling in experimental models suppresses the pathogenesis of Crohn's disease and rheumatoid arthritis (17, 31). Through in vivo analysis of experimental peritoneal inflammation, studies presented herein now show that IL-6 activation of STAT3 promotes T cell recruitment.
Methods
Mouse Strains. Experiments were performed in weight-matched 7- to 12-week-old IL-6-/- mice (32), various knock-in gp130 mutant strains, and genetically matched wild-type (WT) controls. Engineering of homozygous gp130ΔSTAT/ΔSTAT and gp130Y757F/Y757F mice, and the compound gp130Y757F/Y757F:Stat3+/- mice has been described in refs. 11, 19, 33, and 34. Procedures were performed in accordance with Home Office approved project license PPL-40/2131.
Staphylococcus epidermidis (SES)-Induced Peritoneal Inflammation. Peritoneal inflammation was induced in mice through i.p. administration of a cell-free supernatant prepared from a clinical isolate of S. epidermidis (SES) (5, 14). At defined intervals, the peritoneal cavity was lavaged, and the leukocyte infiltrate assessed by direct counting (Coulter Z2, Beckman Coulter), differential cell staining, and flow cytometric analysis. Lavage fluids were rendered cell-free by centrifugation for analysis of inflammatory mediators. Soluble gp130 was purchased from R & D Systems and was added as indicated.
Murine Leukocyte Isolation. Unfractionated hematopoietic cells from peripheral blood, spleens, and femoral bone marrow of mice were rendered free from red blood cell contamination by using an ammonium chloride lysis buffer (155 mM NH4Cl/7 mM K2CO3/0.1 mM EDTA) and washed in PBS before antibody labeling for flow cytometry.
Antibodies. Fluorochrome-conjugated antibodies were purchased from the following sources: rat anti-mouse monoclonal antibodies to CD3 (17A2), CD4 (GK1.5), CD8a (53–6.7), CD69 (H1.2F3), CD25 (PC61), CD28 (37.51), CD62L (MEL-14), CCR5 (C34–3448), CXCR5 (2G8), B220 (RA3–6B2), and IL-6R (D7715A7) were from BD Pharmingen. Phytoerythrin-conjugated rat anti-mouse monoclonal antibodies to CXCR3 (220803) and CCR3 (83101.111) were from R & D Systems. Murine polyclonal anti-CCR4 (sc-7936) (Santa Cruz Biotechnology) was used in combination with Alex Fluor488 (Fab' fragment) goat anti-rabbit IgG from Molecular Probes. Antibodies against STAT1, STAT3, and extracellular signal-regulated kinase 1/2 were from Santa Cruz Biotechnology, and the phospho-specific STAT1 and STAT3 antibodies were obtained from Cell Signaling Technology (Beverly, MA).
Flow Cytometric Analysis. Leukocytes were incubated with mouse Fc block (BD Pharmingen) before immunolabeling for 30 min at 4°C with primary fluorochrome-conjugated or nonconjugated antibodies. Where necessary, cells were incubated for a further 30 min at 4°C with appropriate fluorochrome-conjugated secondary antibodies. Cells were analyzed by using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) based on previously defined murine leukocyte settings. Data were acquired from 10,000 gated events and staining was compared with fluorochrome-conjugated isotype control antibodies.
ELISA Determinations. Murine CCL3 (MIP-1α), CCL5 (RANTES), CCL11 (eotaxin), CCL17 (TARC), CXCL10 (IP-10), and CXCL13 (BCA-1) were quantified by using commercially available ELISA kits (R & D Systems).
Western Blot Analysis of Peritoneal Lining. Protein lysates were prepared from frozen sections of parietal peritoneal membrane. Samples were cleared of cellular debris and separated by SDS/PAGE for Western blot analysis with specific primary antibodies (19). Immunolabeled proteins were detected by using appropriate HRP-conjugated secondary antibodies, followed by visualization with enhanced chemiluminescence (Amersham Pharmacia Biotech).
Statistical Analysis. Data are expressed as mean ± SEM, and statistical analysis were performed by using a Student's unpaired t test (statview se + graphics 1.03 software, Abacus Concepts, Berkeley, CA). P < 0.05 was considered significantly different.
Results
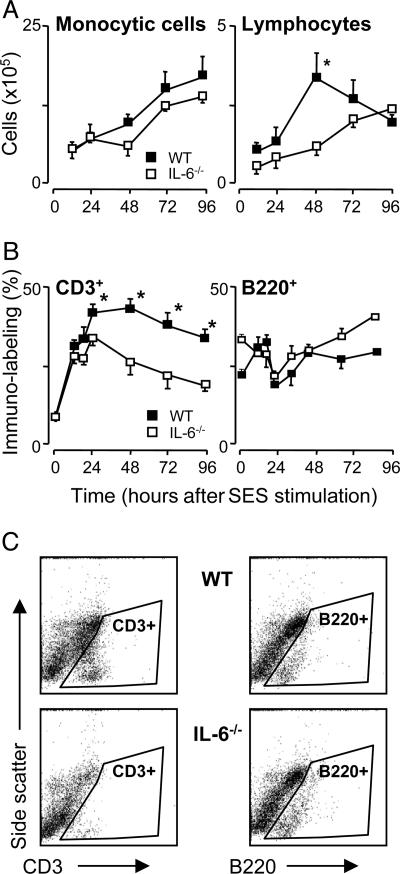
IL-6-Deficient Mice Exhibit Impaired T Cell Recruitment in Vivo. After induction of peritoneal inflammation in WT and IL-6-/- mice by using a cell-free supernatant (termed SES) derived from a clinical isolate of S. epidermidis, it was evident that IL-6 deficiency results in an impaired recruitment of lymphocytes but not monocytic cells (Fig. 1). This reduction in lymphocyte migration was confined to the CD3+ population, with no significant difference observed in the trafficking of B220+ B-cells (Fig. 1). Specific phenotypic analysis of the T cell population from WT and IL-6-/- mice showed that the composition of the CD3+ subpopulation was largely unaffected by the absence of IL-6, suggesting that the impaired infiltration was not attributable to a global T cell defect (see Tables 1 and 2, which are published as supporting information on the PNAS web site).
Fig. 1.
Affect of IL-6 deficiency on mononuclear cell infiltration in vivo. SES-induced peritoneal inflammation was established in WT and IL-6-/- mice. (A) At defined time points, the peritoneal cavity was lavaged, and total lymphocyte (T and B cells) and monocytic cell numbers were determined by differential cell counting. (B) Infiltrating lymphocytes were phenotyped according to CD3+ or B220+ staining by flow cytometry. (C) Representative scatter plots for CD3+ or B220+ staining at 48 h are presented. Isotype antibody control staining is shown in Fig. 7, which is published as supporting information on the PNAS web site. Values represent the mean ± SEM (n = 10 mice per time point; *, P < 0.05).
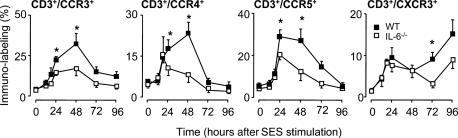
T Cell Chemokine Receptor Expression and Accompanying Chemokine Production. Given the involvement of IL-6 trans-signaling in the regulation of inflammatory chemokines (13–16), experiments next determined whether the impaired T cell migration presented in Fig. 1 was attributable to a defective chemokine response. Peritoneal inflammation was induced in WT and IL-6-/- mice, and the profile of the chemokine receptors CCR3, CCR4, CCR5, and CXCR3 was defined on the T cell infiltrate (Fig. 2). Although activation with SES resulted in a temporal increase in CD3+ T cells expressing CCR3, CCR4, CCR5, and CXCR3, the proportion of cells bearing these receptors was noticeably lower in the absence of IL-6. Furthermore, the profile of expression for each chemokine receptor was distinct and does not parallel the recruitment of CD3+ cells presented in Fig. 1. This finding suggests that T cell chemokine receptors are differentially and temporally regulated during the course of an inflammatory response.
Fig. 2.
T cell chemokine receptor expression after peritoneal inflammation. SES-induced peritoneal inflammation was established in WT and IL-6-/- mice. At defined intervals, the peritoneal cavity was lavaged and the CD3+ infiltrate analyzed by flow cytometry for changes in CCR3, CCR4, CCR5, and CXCR3. Values represent the mean ± SEM (n = 10 mice per group; *, P < 0.05).
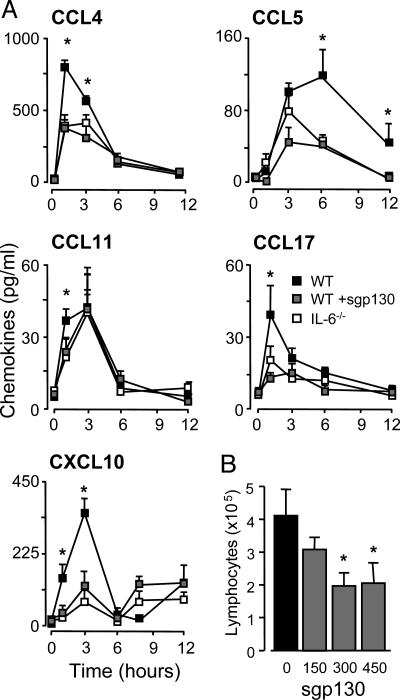
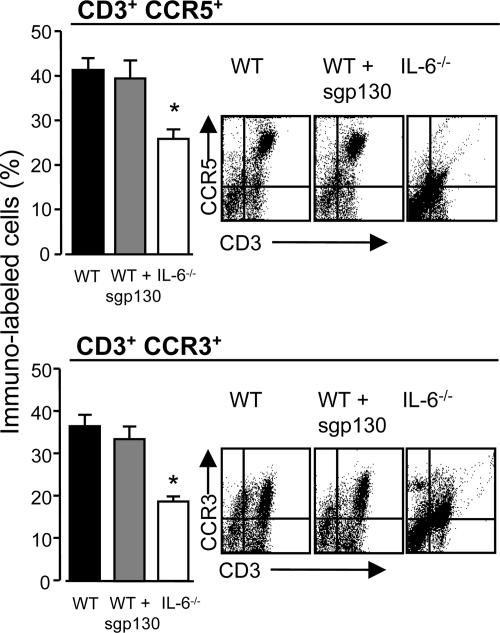
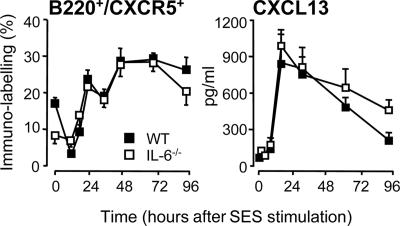
Dysregulation of chemokine-mediated T cell trafficking was substantiated by quantification of chemokine levels in peritoneal lavage fluid. These analyses showed that IL-6 deficiency is associated with defective expression of CCL4, CCL5, CCL17, and CXCL10, whereas a marginal difference was observed for CCL11 (Fig. 3A). Administration of WT mice with sgp130 (the natural antagonist for sIL-6R-mediated signaling) reverted the pattern of chemokine expression to that observed in IL-6-/- mice (Fig. 3A), and dose-dependently suppressed lymphocyte migration into the peritoneal cavity (Fig. 3B). Although sgp130 blocked peritoneal chemokine expression, analysis of CCR5 and CCR3 expression on the CD3+ infiltrate showed that blockade of IL-6 trans-signaling did not affect the T cell expression of these chemokine receptors (Fig. 4). Similar data for CCR4 and CXCR3 were also obtained (data not shown). To test whether the defect in chemokine and cellular chemokine receptor expression was a universal feature of IL-6-/- mice, CXCR5 levels on the B220+ population was monitored in conjunction with the peritoneal expression of its ligand CXCL13. As shown in Fig. 5, IL-6 deficiency did not affect the proportion of B220+CXCR5+ cells or CXCL13 expression.
Fig. 3.
Peritoneal chemokine levels after SES-induced inflammation. (A) SES-induced peritoneal inflammation was established in WT and IL-6-/- mice, and at defined intervals, the peritoneal cavity was lavaged and chemokine levels quantified by ELISA. To block IL-6 trans-signaling, WT mice were administered with 150 ng per mouse sgp130. (B) Dose-dependent blockade of SES-induced lymphocyte recruitment (48 h) by sgp130 administration of WT mice. Values represent the mean ± SEM (n = 5 mice per group; *, P < 0.05).
Fig. 4.
Affect of sgp130 on T cell chemokine receptor expression. SES-induced peritoneal inflammation was established in IL-6-/-, WT, and WT mice treated with sgp130. After 36 h, the peritoneal cavity was lavaged and the proportion of CD3+CCR3+ and CD3+CCR5+ cells determined. Representative scatter plots for each condition are also shown. Values represent the mean ± SEM (n = 5 mice per group; *, P < 0.05).
Fig. 5.
CXCR5 and CXCL13 expression after SES-induced inflammation. SES-induced peritoneal inflammation was established in WT and IL-6-/- mice. At defined intervals, the peritoneal cavity was lavaged, and the proportion of B220+CXCR5+ (Left) cells was analyzed by flow cytometry. (Right) CXCL13 was quantified by using ELISA. Values represent the mean ± SEM (n = 10 mice per group; *, P < 0.05).
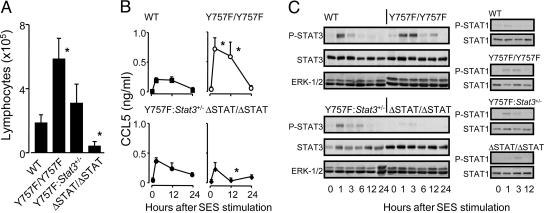
IL-6 Control of T Cell Recruitment Requires gp130-Dependent STAT1/3 Activation. To dissect the signaling mechanisms associated with the IL-6 regulation of T cell infiltration, SES-induced peritoneal inflammation was established in knock-in mice that express mutated forms of gp130 (11, 19, 33). Mice unable to elicit gp130-dependent STAT1/3 activation (gp130ΔSTAT/ΔSTAT) showed a poor capacity to promote T cell migration (Fig. 6A). In contrast, T cell recruitment to the peritoneal cavity was dramatically elevated in gp130Y757F/Y757F mice deficient in SHP-2 and SOCS3 binding to gp130, but displaying enhanced STAT1/3 activity (Fig. 6A). This defect in migration was directly attributable to gp130-mediated events after inflammatory activation, because both basal CCL5 levels (Figs. 3 and 6B) and the basal expression of CCR3 and CCR5 on circulating CD3+ T cells remained unaffected by the genetic composition of the various knock-in strains (see Table 3, which is published as supporting information on the PNAS web site). Regulation of this process does however appear to involve a chemokine component, and analysis of peritoneal CCL5 levels showed that alterations in the activation of STAT1/3 through gp130 signaling significantly modified the profile of CCL5 release (Fig. 6B). To delineate the potential involvement of STAT3 signaling over that of STAT1, SES-induced inflammation was established in gp130Y757F/Y757F mice that had been crossed onto a Stat3 heterozygous (gp130Y757F/Y757F:Stat3+/-) background (11). To verify the specific reduction in peritoneal STAT3 expression within gp130Y757F/Y757F:Stat3+/- mice, STAT3 levels and its activation after SES stimulation was monitored by Western blot analysis in peritoneal membranes from the various mouse strains tested (Fig. 6C). Consistent with previous results (34), cellular STAT1 levels and its initial activation remained unaffected by the monoallelic deletion of STAT3 (Fig. 6C). Quantification of T cell infiltration in gp130Y757F/Y757F:Stat3+/- mice showed that reduction of gp130-mediated STAT3 signaling reduced the exaggerated T cell migration seen in gp130Y757F/Y757F mice (Fig. 6A) and suppressed CCL5 expression back to that observed in WT mice (Fig. 6B). The specificity of these STAT3-mediated responses is further suggested by experiments that demonstrate that gp130 activation of peritoneal mesothelial cells selectively activates STAT3 but not STAT1 (35). Thus, the defect in T cell trafficking seen in IL-6-deficiency appears to require gp130-mediated STAT3 activation.
Fig. 6.
Genetic modification of gp130-mediated STAT3 signals in vivo affects T cell recruitment after SES-induced inflammation. SES-induced peritoneal inflammation was established in the indicated strains, and at appropriate time intervals, the peritoneal cavity was lavaged and lymphocytes assessed by differential counting (A) and intraperitoneal CCL5 levels quantified by ELISA (B). Results expressed as mean ± SEM (n = 5 mice per group, *, P < 0.05). (C) Temporal changes in STAT1 and STAT3 phosphorylation were monitored by Western blot analysis of the peritoneal lining of WT, gp130Y757F/757F, gp130ΔSTAT/ΔSTAT, and gp130Y757F/Y757F:Stat3+/- mice after SES activation. Levels of extracellular signal-regulated kinase (ERK)-1/2 were monitored as an internal control, and densitometry of the banding intensity is presented as Fig. 8, which is published as supporting information on the PNAS web site.
Discussion
Transition from innate to acquired immunity represents a pivotal event in the control of any inflammatory response. Regulation of this immunological switch relies on the coordinated expression of chemotactic agents and a precise regulation of apoptotic events. Through differential control of these processes, recent studies have highlighted that gp130 signaling has the capacity to suppress innate immune responses (12, 14, 23) while concurrently promoting development of acquired immunity (18, 23, 25–27). In this study, we have extended these observations by demonstrating that IL-6 deficiency specifically disrupts chemokine control of T cell trafficking, yet leaves monocytic and B cell recruitment unaffected. This response is somewhat surprising given that many of the inflammatory chemokines shown to be dysregulated in IL-6-/- mice also attract monocytic cells. Because IL-6 also modulates the balance between macrophage and dendritic cell differentiation (19–21), it remains to be established whether IL-6 deficiency has specific affects on the phenotype of a recruited monocytic cell population. Consequently, the nature of the monocytic cells may be as important as the total cell numbers. Dysregulated T cell migration is however consistent with the role IL-6 performs in more chronic inflammatory conditions, where the ability of IL-6 to rescue T cells from entering apoptosis has been linked with the pathology of several models of chronic disease (4, 17, 36).
Although IL-6 has been defined as a T cell activation factor and shown to induce T cell proliferation and survival (27–30), it is now evident that IL-6 bioactivity is also tightly linked to the homing or migratory capacity of T cells. IL-6 has previously been shown to attract CD4+ T cells to the central nervous system, whereas in vitro studies have described IL-6 as a chemotactic factor (37–39). However, the highly restricted expression of the cognate IL-6R observed clinically and experimentally on circulating and recruited T cells (7, 30) suggests that selective inflammatory chemokines may provide a more prominent mechanism for T cell attraction. In this respect, examination of IL-6-/- mice and mice presenting altered gp130-mediated STAT1/3 activity showed that STAT3 signaling is required for chemokine-driven T cell recruitment. Regulation of this response is further supplemented by the dysregulation of T cell chemokine receptor expression in IL-6-/- mice. This result is particularly interesting given the ability of IL-6 to balance Th1-Th2 polarization (22–24). Expression of individual chemokine receptors has been used to define T cell subsets as possessing either Th1 (CXCR3 and CCR5) or Th2 (CCR3 and CCR4) activities (40–43). Although IL-6 deficiency globally affected the expression of all four of these chemokine receptors, it is worth considering that both Th1 and Th2 populations express IL-6R (27). However it is unclear whether IL-6 directly or indirectly influences the profile of chemokine receptors on T cells. For instance, lymph nodes cells from IL-6-/- mice show reduced expression of IL-2 (22), which is known to promote T cell expression of CCR1, CCR2, CCR5, and CXCR3 (44, 45). In this respect, it is important to note that although blockade of IL-6 trans-signaling in WT mice disrupted chemokine secretion, the T cell chemokine receptor expression remained unaltered.
Retention of inflammatory T cells as sites of immunological challenge is a characteristic feature of many chronic inflammatory diseases and is often associated with the protection of the T cell population from apoptotic clearance (46). Significantly, regulation of survival signals are governed by gp130-mediated STAT3 signaling, and knock-in mice presenting enhanced gp130-dependent STAT3 activity develop spontaneous autoimmune arthritis, which is attributable to an inability of T cells to enter apoptosis (36). The data presented herein extends this paradigm by showing that IL-6-driven STAT3 activity is also important for the chemokine regulation of T cell recruitment. Significantly, blockade of sIL-6R-mediated events in experimental models of colitis and arthritis leads to improved disease outcome, with the therapeutic basis for this intervention strategy affecting either the recruitment or apoptotic clearance of mononuclear leukocytes (17, 31). Consequently, IL-6- and sIL-6R-mediated signaling influences the nature and retention of the inflammatory infiltrate within inflamed tissue by controlling inflammatory chemokines and apoptotic regulators. Inappropriate gp130-mediated control of these events may ultimately promote progression to a more chronic inflammatory state.
Supplementary Material
Acknowledgments
This work was supported through The Wellcome Trust and Arthritis Research Campaign project grants.
Author contributions: R.M.M., B.J.J., N.T., and S.A.J. designed research; R.M.M., B.J.J., D.G., A.S.W., C.A.F., and C.R.P. performed research; B.J.J. and M.E. contributed new reagents/analytic tools; R.M.M., B.J.J., N.T., and S.J. analyzed data; and S.A.J. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SES, Staphylococcus epidermidis; sIL-6R, soluble form of the cognate IL-6 receptor.
References
- 1.Kishimoto, T., Akira, S., Narazaki, M. & Taga, T. (1995) Blood 86, 1243-1254. [PubMed] [Google Scholar]
- 2.Jones, S. A. & Rose-John, S. (2002) Biochim. Biophys. Acta 1592, 251-263. [DOI] [PubMed] [Google Scholar]
- 3.Kaplanski, G., Marin, V., Montero-Julian, F., Mantovani, A. & Farnier, C. (2003) Trends Immunol. 24, 25-29. [DOI] [PubMed] [Google Scholar]
- 4.Hong, F., Jaruga, B., Kim, W. H., Radaeva, S., El-Assal, O. N., Tian, Z., Nguyen, V.-A. & Gao, B. (2002) J. Clin. Invest. 110, 1503-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLoughlin, R. M., Witowski, J., Robson, R. L., Wilkinson, T. S., Hurst, S. M., Williams, A. S., Williams, J. D., Rose-John, S., Jones, S. A. & Topley, N. (2003) J. Clin. Invest. 112, 598-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasukawa, H., Ohishi, M., Mori, H., Murakami, M., Chinen, T., Aki, D., Hanada, T., Takeda, K., Akira, S., Hoshijima, M., et al. (2003) Nat. Immunol. 4, 551-556. [DOI] [PubMed] [Google Scholar]
- 7.Becker, C., Fantini, M. C., Schramm, C., Lehr, H. A., Wirtz, S., Nikolaev, A., Burg, J., Strand, S., Kiesslich, R., Huber, S., et al. (2004) Immunity 21, 491-501. [DOI] [PubMed] [Google Scholar]
- 8.Choy, E. H. S., Isenberg, D. A., Garrood, T., Farrow, S., Ioannou, Y., Bird, H., Cheung, N., Williams, B., Hazleman, B., Price, R., et al. (2002) Arthritis Rheum. 41, 2117-2121. [DOI] [PubMed] [Google Scholar]
- 9.Ito, H., Takazoe, M., Fukuda, Y., Hibi, T., Kusugami, K., Andoh, A., Matsumoto, T., Yamamura, T., Azuma, J., Nishimoto, N., et al. (2004) Gastroenterology 126, 989-996. [DOI] [PubMed] [Google Scholar]
- 10.Heinrich, P. C., Behrmann, I., Haan, S., Hermanns, H. M., Müller-Newen, G. & Schaper, F. (2003) Biochem. J. 374, 1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst, M. & Jenkins. B. J. (2004) Trends Genet. 20, 23-32. [DOI] [PubMed] [Google Scholar]
- 12.Xing, Z., Gauldie, J., Cox, G., Baumann, H., Jordana, M., Lei, X.-F. & Achong, M. K. (1998) J. Clin. Invest. 101, 311-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romano, M., Sironi, M., Toniati, C., Polentarutti, N., Fruscella, P., Ghezzi, P., Faggioni, R., Luini, W., van Hinsbergh, V., Sozzani, S., et al. (1997) Immunity 6, 315-325. [DOI] [PubMed] [Google Scholar]
- 14.Hurst, S. M., Wilkinson, T. S., McLoughlin, R. M., Jones, S., Horiuchi, S., Yamamoto, N., Rose-John, S., Fuller, G. M., Topley, N. & Jones, S. A. (2001) Immunity 14, 705-715. [DOI] [PubMed] [Google Scholar]
- 15.Modur, V., Li, Y., Zimmerman, G. A., Prescott, S. M. & McIntyre, T. M. (1997) J. Clin. Invest. 100, 2752-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marin, V., Montero-Julian, F. A., Gres, S., Boulay, V., Bongrand, P., Farnarier, C. & Kaplanski, G. (2001) J. Immunol. 167, 3435-3444. [DOI] [PubMed] [Google Scholar]
- 17.Atreya, R., Mudter, J., Finotto, S., Müllberg, J., Jostock, T., Wirtz, S., Schütz, M., Bartsch, B., Holtmann, M., Becker, C., et al. (2000) Nat. Med. 6, 583-588. [DOI] [PubMed] [Google Scholar]
- 18.Chen, Q., Wang, W.-C., Bruce, R., Li, H., Scleider, D. M., Mulbery, M. J., Bain, M. D., Wallace, P. K., Baumann, H. & Evans. S. S. (2004) Immunity 20, 59-70. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins, B. J., Grail, D., Inglese, M., Quilici, C., Bozinovski, S., Wong, P. & Ernst, M. (2004) Mol. Cell. Biol. 24, 1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chomarat, P., Banchereau, J., Davoust, J. & Palucka, A. K. (2000) Nat. Immunol. 1, 510-514. [DOI] [PubMed] [Google Scholar]
- 21.Bleier, J. I., Pillarisetty, V. G., Shah, A. B. & DeMatteo, R. P. (2004) J. Immunol. 172, 7408-7416. [DOI] [PubMed] [Google Scholar]
- 22.Ohshima, S., Saeki, Y., Mima, T., Sasai, M., Nishioka, K., Nomura, S., Kopf, M., Katada, Y., Tanaka, T., Suemura, M. & Kishimoto, T. (1998) Proc. Natl. Acad. Sci. USA 95, 8222-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romani, L., Mencacci, A., Cenci, E., Spaccapelo, R., Toniatti, C., Puccetti, P., Bistoni, F. & Poli, V. (1996) J. Exp. Med. 183, 1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diehl, S., Anguita, J., Hoffmeyer, A., Zapton, T., Ihle, J. N., Fikrig, E. & Rincon, M. (2000) Immunity 13, 805-815. [DOI] [PubMed] [Google Scholar]
- 25.La Flamme, A. C. & Pearce, E. J. (1999) J. Immunol. 162, 5829-5837. [PubMed] [Google Scholar]
- 26.Baroja, M. L., Ceuppens, J. L., van Damme, J. & Billiau, A. (1998) J. Immunol. 141, 1502-1507. [PubMed] [Google Scholar]
- 27.Teague, T. K., Schaefer, B. C., Hildemen, D., Bender, J., Mitchell, T., Kappler, J. W. & Marrack, P. (2000) J. Exp. Med. 191, 915-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narimatsu, M., Maeda, H., Itoh, S., Atsumi, T., Ohtani, T., Nishida, K., Itoh, M., Kamimura, D., Park, S.-J., Mizuno, K., et al. (2001) Mol. Cell. Biol. 21, 6615-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeda, K., Kaisho, T., Yoshida, N., Takeda, J., Kishimoto, T. & Akira, S. (1998) J. Immunol. 161, 4652-4660. [PubMed] [Google Scholar]
- 30.Curnow, S. J., Scheel-Toellner, D., Jenkinson, W., Raza, K., Durrani, O. M., Faint, J. M., Rauz, S., Wloka, K., Pilling, D., Rose-John, S., et al. (2004) J. Immunol. 173, 5290-5297. [DOI] [PubMed] [Google Scholar]
- 31.Nowell, M. A., Richards, R. J., Horiuchi, S., Yamamoto, N., Rose-John, S., Topley, N., Williams, A. S. & Jones, S. A. (2003) J. Immunol. 171, 3202-3209. [DOI] [PubMed] [Google Scholar]
- 32.Kopf, M., Baumann, H., Freer, G., Freudenberg, M., Lamers, M., Kishimoto, T., Zinkernagel, R., Bluethmann, H. & Kohler, G. (1994) Nature 368, 339-341. [DOI] [PubMed] [Google Scholar]
- 33.Tebbutt, N. C., Giraud, A. S., Inglese, M., Jenkins, B., Waring, P., Clay, F. J., Malki, S., Alderman, B. M., Grail, D., Hollande, F., et al. (2002) Nat. Med. 8, 1089-1097. [DOI] [PubMed] [Google Scholar]
- 34.Jenkins, B. J., Roberts, A. W., Najdovska, M., Grail, D. & Ernst, M. (2005) Blood 105, 3512-3520. [DOI] [PubMed] [Google Scholar]
- 35.McLoughlin, R. M., Hurst, S. M., Nowell, M. A., Harris, D. A., Horiuchi, S., Morgan, L. W., Wilkinson, T. S., Yamamoto, N., Topley, N. & Jones, S. A. (2004) J. Immunol. 172, 5676-5683. [DOI] [PubMed] [Google Scholar]
- 36.Atsumi, T., Ishihara, K., Kamimura, D., Ikushima, H., Ohtani, T., Hirota, S., Kobayashi, H., Park, S.-J., Saeki, Y., Kitamura, Y., et al. (2002) J. Exp. Med. 196, 979-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamm, I., Cardinale, I., Krueger, J., Murphy, J. S., May, L. T. & Sehgal, P. B. (1989) J. Exp. Med. 170, 1649-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kira, M., Sano, S., Takagi, S., Yoshikawa, K., Takeda, J. & Itami, S. (2002) J. Biol. Chem. 277, 12931-12936. [DOI] [PubMed] [Google Scholar]
- 39.Weissenbach, M., Clahsen, T., Weber, C., Spitzer, D., Wirth, D., Vestweber, D., Heinrich, P. C. & Schaper, F. (2004) Eur. J. Immunol. 34, 2895-2906. [DOI] [PubMed] [Google Scholar]
- 40.Sallusto, F., Mackay, C. R. & Lanzavecchia, A. (1997) Science 277, 2005-2007. [DOI] [PubMed] [Google Scholar]
- 41.Gerber, B. O., Zanni, M. P., Uguccioni, M., Loetscher, M., Mackay, C. R., Pichler, W. J., Yawalkar, N., Baggiolini, M. & Moser, B. (1997) Curr. Biol. 7, 836-843. [DOI] [PubMed] [Google Scholar]
- 42.Qin, S., Rottman, J. B., Myers, P., Weinblatt, M., Loetscher, M., Koch, A. E., Moser, B. & Mackay, C. R. (1998) J. Clin. Invest. 101, 746-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sallusto, F., Lenig, D., Mackay, C. R. & Lanzavecchia, A. (1998) J. Exp. Med. 187, 875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loetscher, P., Seitz, M., Baggiolini, M. & Moser, B. (1996) J. Exp. Med. 184, 569-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loetscher, M., Gerber, B., Loetscher, P., Jones, S. A., Piali, L., Clark-Lewis, I., Baggiolini, M. & Moser, B. (1996) J. Exp. Med. 184, 963-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salmon, M., Scheel-Toellner, D., Huissoon, A. P., Pilling, D., Shamsaadeen, N., D'Angeac, A. D., Bacon, P. A., Emery, P. & Akbar, A. (1997) J. Clin. Invest. 99, 439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.