Abstract
We have investigated a herpes simplex virus mutant that expresses low levels of thymidine kinase (TK), a phenotype associated with drug resistance and pathogenicity, despite a single-base deletion in the gene. Using a dual-reporter system, a 39-nt sequence including the mutation was shown to direct expression of the downstream reporter gene in reticulocyte lysate. Translation of the downstream reporter was not impaired when the mRNA lacked a 5′ cap or had a stable stem loop 5′ of the upstream reporter and was relatively resistant to edeine, an antibiotic that prevents AUG codon recognition by the 40S-eIF2-GTP/Met-tRNAi complex. Twelve nucleotides were as active as the original sequence for translation of the downstream reporter. Surprisingly, this sequence lacks an AUG codon. Analysis of point mutations showed that a CUG codon in the sequence was important. However, many single-base changes had only limited effects, and introduction of AUG codons did not increase translation. A mutant virus containing both the single-base deletion and a mutation that reduced downstream translation in vitro had significantly less TK activity than a virus with the single-base deletion alone. Thus, a remarkably short internal ribosome entry site (IRES) that lacks an AUG codon resides in the viral tk gene. The IRES appears to be responsible for TK expression from a drug-resistant mutant that would otherwise express no TK, which may contribute to pathogenicity. Because we found numerous short sequences with IRES activity, there might be many hitherto unrecognized polypeptides expressed at low levels from eukaryotic mRNAs.
Keywords: drug resistance, pathogenesis, proteomics, translation, acyclovir
On most eukaryotic mRNAs, translational initiation is a complex process whereby protein factors recognize the cap structure at the 5′ end of an mRNA and then, through a mechanism known as scanning, position the 40S ribosome subunit at the 5′-most AUG codon that is in an appropriate nucleotide context. At this point, the 60S ribosome subunit is joined to the 40S subunit, and translation ensues (1). However, on a minority of eukaryotic mRNAs, translation initiates by means of a cap-independent mechanism, as first observed with picornaviruses (2, 3). In picornaviruses, a sequence element known as an internal ribosome entry site (IRES), which is a long structured region of RNA, recruits the translational machinery to the AUG codon, dispensing with the need for the 5′ cap (reviewed in ref. 4). Since their initial discovery, there have been numerous reports of IRES elements in a variety of viral and cellular mRNAs, which in most cases appear to employ mechanisms similar to those of picornavirus IRESs.
The herpes simplex virus (HSV) gene that encodes thymidine kinase (TK) has long served as a model for studies of eukaryotic gene expression (5, 6). The viral TK is also important for the treatment of HSV infections because this enzyme activates the antiviral drug acyclovir (ACV). Although ACV is an effective antiviral agent, drug-resistant virus that causes severe herpetic disease is sometimes observed in the immunocompromised (7). The most common mutations observed in these ACV-resistant (ACVr) clinical isolates are frameshift lesions in the tk gene that would be expected to abolish TK activity (8). This observation has raised the question of how these ACVr mutants can cause severe disease, because TK-negative (TK-) mutants are highly compromised for pathogenicity in animal models of HSV infection (9, 10).
Our laboratory has previously investigated one common mutation observed in ACVr clinical isolates, a single guanine insertion into the tk gene in a run of seven Gs (G7+1G). We found that viruses carrying this frameshift mutation synthesize low levels (≈1%) of active TK, despite the insertion, and that this level of TK expression can restore at least some pathogenicity to the virus (11, 12). This low level of TK, however, still results in substantial ACV resistance (13). The mechanism that permits TK expression in this case is an atypical net +1 ribosomal frameshift (11, 14).
In this study, we have examined another frameshift mutation in the tk gene frequently associated with drug-resistant clinical disease: a deletion in a run of six cytosines known as the C-chord (C6-1C) (8). We found that a virus containing this mutation synthesizes low levels of active TK. Analysis of the mechanism responsible revealed the existence of an unusual IRES in the tk gene.
Materials and Methods
Cells and Viruses. African green monkey kidney (Vero) and TK- human osteosarcoma (143B) cells (American Type Culture Collection) were maintained in DMEM supplemented with 10% FBS at 37°C and 5% CO2. The viruses used in this study were HSV-1 strain KOS and a series of mutant viruses that express various levels of active TK: LS-111/-101//-56/-46 (2% of WT activity), 615.9 (1%), LS-29/-18 (0.5%), and tkLTRZ1 (0%) (6, 9, 11, 15–19).
Plasmids. Plasmid pAG5 (20) contains the BamHI P fragment of strain KOS cloned into pBluescript SK (+) (Promega). Plasmid pAG6.TKC6-1C was made by introducing the C6-1C mutation into pAG5 by using the QuikChange mutagenesis kit (Stratagene), following the manufacturer's instructions, using two complementary oligonucleotides (Integrated DNA Technologies, Coralville, IA) (the sequences for these oligonucleotides and all others are provided in Table 1, which is published as supporting information on the PNAS web site). Plasmid pTKC6-1C.IFS, which contains a stop codon in the TK ORF in addition to the C6-1C mutation, was made similarly, except that pAG6.TKC6-1C was used as the template. Plasmid pTKC6-1C.CUC contains a CUG-to-CUC (both encoding leucine) change.
Plasmid pAG3 was constructed by using the Renilla luciferase (Rluc) and firefly luciferase (Fluc) genes from pRL-null and pGL-basic, respectively (Promega). pRL-null-link was made by removing the polylinker from pRL-null by digesting with PstI and BglII. After removing sticky ends with T4 DNA polymerase, a new polylinker was inserted downstream of the Rluc gene to make pRL-link by the insertion of a duplex of synthetic oligonucleotides containing BglII, MluI, XhoI, and SmaI recognition sites into the XbaI site of pRL-null-link, destroying the ensuing 5′ XbaI site while retaining the 3′ site. The Fluc sequence was removed from pGL3-basic by digesting with NcoI and XbaI [the NcoI overhang was removed with mung bean nuclease (NEB, Beverly, MA), which removes the initiating methionine codon] and then cloned into SmaI- and XbaI-digested pRL-link to give pAG1. To remove an in-frame stop codon from the 5′ end of the Rluc gene in pAG1, a fragment containing the gene was generated by PCR and cloned into NheI- and XhoI-digested pAG1 to give pAG3. Subsequent pAG3 series plasmids were constructed by cloning complementary nucleotides into the BglII and XhoI sites of pAG3. These constructs were designed such that the WT tk triplets are in the same register as Fluc and the mutant tk triplets are in the same register as Rluc. To reduce background, stop codons were placed upstream of the test sequence in the Fluc frame and downstream of the test sequence in the other two frames.
pAG3.103.SL, which is based on pAG3.103 and has a stable stem loop (ΔG = -39.2 kcal/mol) inserted upstream of Rluc, was constructed by inserting an annealed duplex of oligonucleotides into the NheI site of pAG3.103.
Correct introduction of all engineered mutations, in the absence of additional mutations, was confirmed by sequencing.
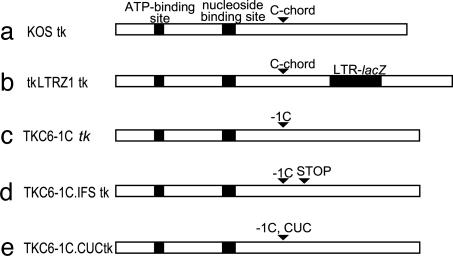
Construction of Recombinant Viruses. Viruses TKC6-1C, TKC6-1C.IFS, and TKC6-1C.CUC (Fig. 1) were made after cotransfection of the respective plasmid and infectious DNA from mutant virus tkLTRZ1 by using Effectene (Qiagen, Valencia, CA) according to the manufacturer's instructions. tkLTRZ1 lacks TK activity because of an insertion of LacZ driven by a strong promoter in tk (18). Screening for recombinant viruses is described in refs. 20 and 21 and exploits a blue (nonrecombinant)/white (recombinant) screen. The three mutants formed plaques with WT size and morphology. The correct introduction of mutations and the absence of unwanted tk mutations were confirmed by sequencing.
Fig. 1.
Structure of the tk coding regions of viruses used in this study. (a) KOS (WT) TK, the ATP-binding site, and nucleoside-binding sites are marked with black boxes, and the C-chord is indicated by an arrow. (b) tkLTRZ1 (tk with LTR-lacZ, indicated by a black box, inserted into the PstI site; the C-chord is indicated by an arrow). (c) TKC6-1C (tk with a single C, indicated by an arrow, deleted from the C-chord of KOS tk). (d) TKC6-1C.IFS (tk with a stop codon in the tk ORF, indicated by an arrow, in addition to the C6-1C mutation). (e) TKC6-1C.CUC (tk with a CUG-to-CUC change, indicated by an arrow, in addition to the C6-1C mutation).
In Situ Measurement of TK Activity. Plaque autoradiography of infected 143B cells lacking TK was performed as described in refs. 19 and 20. Viruses that had previously been shown to have a range of TK activities between 0.5% and 2% were used to calibrate the assay.
In Vitro Transcription and Translation Assays. pAG3 series plasmids were linearized with BamHI. Capped transcripts were synthesized by using T7 mMessage mMachine (Ambion, Austin, TX), and uncapped transcripts were synthesized by using Megascript T7 (Ambion), both according to manufacturer's instructions, except for a 5-min 70% ethanol wash after isopropanol precipitation. These RNAs were translated by using rabbit reticulocyte lysate (Amersham Pharmacia Biotech) according to the manufacturer's instructions (final potassium ion concentration was 130 mM). Fluc and Rluc activities were measured by using dual-luciferase assay reagents (Promega) and measuring luminescence on a Victor 2 reader (Wallac, Gaithersburg, MD), which was generously made available by the Institute of Chemistry and Cell Biology, Harvard Medical School. Translations that were performed in the presence of edeine (a gift from the National Cancer Institute, Bethesda) were performed as described in ref. 22, preincubating the lysate with the drug for 5 min at 30°C before the addition of mRNA.
Polypeptides were also analyzed by using SDS/PAGE from [35S]methionine-labeled translations followed by autoradiography.
Results
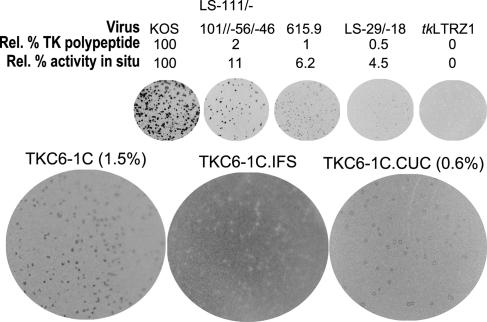
Active TK Is Synthesized in Cells Infected with Virus TKC6-1C. A single-base deletion in the C-chord of the tk gene has previously been identified as a common mutation in ACVr HSV from patients who suffer herpetic disease despite ACV therapy (7, 8, 23). Gaudreau et al. (8) described two HSV-1 isolates with the C-chord deletion; by plaque autoradiography, one had a TK-low phenotype, and the other had a TK- phenotype. To define the phenotypes resulting from this mutation while avoiding problems that may be encountered due to using clinical isolates with poorly defined genotypes, we elected to engineer this mutation into HSV-1 KOS, a well characterized laboratory strain. To this end, we recombined a plasmid-borne tk gene carrying this mutation with virus tkLTRZ1, which is derived from KOS and has lacZ inserted into tk. This method permits recombinant viruses to be isolated by using a blue/white screen, which obviates the need to introduce a selection pressure (ACV is frequently used to select TK mutants) to help enrich recombinant viruses, because the probability of the recombinant virus acquiring unwanted mutations is increased. Additionally, this approach eliminates a potential source of contaminating TK+ virus (12, 20). We named the virus containing the frameshift mutation TKC6-1C. Plaques from TKC6-1C had ≈1.5% of the TK activity of strain KOS, as measured by quantitative plaque autoradiography (Fig. 2).
Fig. 2.
Quantitative plaque autoradiography of viruses. The top line shows the names of the viruses that were used to calibrate the assay. The second line shows the levels of active TK polypeptide expressed by each virus relative to WT strain KOS. The third line shows the average amounts of radioactivity measured per plaque as a percentage relative to that measured for KOS. The next line shows the radiographic images from the plates. Below, the three larger plates present plaque autoradiography of the viruses generated for this study, with the names of the viruses and their activities relative to that of KOS indicated above the plates. The TK activity levels for TKC6-1C (1.5%) and TKC6-1C.CUC (0.6%) were estimated from a graph (not shown) of the relative percentage of active TK polypeptide plotted against the relative percentage of TK activity in situ for the viruses that have a range of TK activities (LS-111/-101//-56/-46, 615.9, LS-29/-18, and tkLTRZ1).
Although the frameshift mutation in TKC6-1C is in the middle of the gene, the polypeptide that is generated should contain the nucleoside and ATP binding sites (Fig. 1). We therefore considered the possibility that this largely out-of-frame polypeptide was ≈1.5% as active as WT TK. To address this question, we constructed virus TKC6-1C.IFS (Fig. 1), which would encode the identical non-TK frame polypeptide as TKC6-1C but with a stop codon introduced into the WT TK frame a short distance downstream of the frameshift mutation. No detectable TK activity was observed with TKC6-1C.IFS (Fig. 2). We therefore concluded that expression of active TK by TKC6-1C requires synthesis of WT TK amino acids downstream of the frameshift mutation, raising the question of how this synthesis occurs.
Sequences from the Mutant tk Gene Permit Expression of Downstream Sequences. We hypothesized that a translational event permitted the synthesis of active TK. To investigate this possibility, we used a dual-luciferase reporter system, pAG3 (Fig. 3a), similar to one previously reported (24). In this system, the test sequence is placed between a downstream reporter gene (Fluc) and an upstream reporter (Rluc) that permits an internal control for translation efficiency. The plasmid is transcribed in vitro and then translated in rabbit reticulocyte lysate, a standard system for studies of translational mechanisms, and Rluc and Fluc activities measured separately. In our studies, the test sequence was inserted so that the two reporter genes were out of frame with each other. Importantly, the initiating AUG codon sequence was removed from the Fluc gene. The test sequence was surrounded by stop codons, upstream in the Fluc frame and downstream in the other two frames, to reduce the potential effects of recoding events outside of the test sequence resulting in expression of Fluc. An in-frame Fluc/Rluc fusion serves as a normalization control in which the ratio of Fluc to Rluc expression is set to 100%. The background level for plasmids that did not promote Fluc expression was ≈0.2% relative to the in-frame fusion.
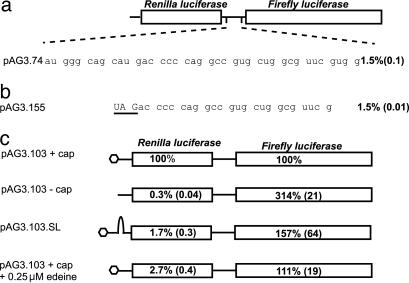
Fig. 3.
An IRES in the tk gene. (a) The Rluc and Fluc genes (Upper) separated by the test sequence (Lower). To the left of the test sequence is the name of the plasmid. To the right of the test sequence is the normalized percentage of Fluc activity observed after in vitro translation in rabbit reticulocyte lysate, with the standard deviation in parentheses. The result in b is presented similarly. (b) pAG3.155 contains a stop codon (UAG, underlined) in the Fluc reading frame upstream of the test sequence. (c) The Rluc and Fluc genes are represented by the boxes, and all of these constructs had identical test sequences to pAG3.103 (shown in Fig. 4). Inside the boxes are the levels of each enzyme activity expressed as a percentage of the levels in the capped control construct (observed Fluc activities were in the thousands of raw light units). The standard deviations are noted in parentheses. The first line represents the mRNA that has standard levels of Fluc activity and was used as a control. The hexagon represents the cap. The second line represents the mRNA synthesized from the same template but without the cap. The third line represents an mRNA that has a cap but has a stable stem-loop structure placed upstream of the Rluc gene. The fourth line represents the same mRNA as the first line, but 0.25 μM edeine is added to the translation mix.
Thirty-nine nucleotides including the C-chord of TKC6-1C were inserted as a test sequence into this system so that the TK reading frame downstream of the C-chord was out of frame with Rluc but in-frame with Fluc (pAG3.74). This plasmid directed expression of the downstream reporter at ≈1.5% efficiency relative to the in-frame fusion control (Fig. 3a), a value remarkably similar to the efficiency of TK expression observed from the TKC6-1C virus. This efficiency of Fluc expression will be referred to from here on as “standard levels.” We have also tested reporter constructs containing the tk sequence in transfected Vero cells. In this system, we observe ≈0.4% Fluc activity (a negative control construct expressed levels <0.08%; unpublished data).
The tk Sequence Is an IRES. We considered two possible mechanisms to account for Fluc expression downstream of the frameshift mutation: ribosomal frameshifting on the test sequence, as we had previously observed on a run of eight Gs of tk sequence (11, 14), or translational initiation on the test sequence. To distinguish between these two mechanisms, we constructed a plasmid (pAG3.155) in which a stop codon was placed in the Rluc frame upstream of the test sequence, which would be expected to prevent Fluc expression as a result of ribosomal frameshifting but not initiation. Fig. 3b shows that standard levels of Fluc expression were observed with such a dicistronic construct, consistent with initiation but not frameshifting. Interestingly, increased potassium in the translation reactions resulted in increased Fluc expression (unpublished data), which has been observed in IRESs (e.g., 25).
One possible mechanism for initiation on the test sequence is that translational initiation complexes assemble by means of interactions with the m7G cap on the 5′ end of mRNA, and a small fraction of these complexes “skip” upstream AUG codons and continue scanning until they initiate on the test sequence, or complexes that did initiate translation on upstream AUG codons then reinitiate after translation termination, or both. A second potential cap-dependent mechanism is ribosomal shunting, in which signals in the mRNA direct the scanning complex to bypass regions of mRNA (1). Alternatively, initiation could occur by means of an IRES-mediated event, independently of a 5′ cap and ribosomal scanning or shunting (reviewed in ref. 26). To ask whether a 5′ cap was important for the expression of Fluc, uncapped mRNA synthesized from pAG3.103 was translated, and Rluc and Fluc activities were compared with those from capped pAG3.103 message (Fig. 3c). Despite an ≈300-fold reduction in Rluc activity from uncapped mRNA, Fluc was translated at least as efficiently from uncapped mRNA as from capped mRNA. As an alternative technique, we generated an mRNA with a stable stem loop (ΔG = -39.2 kcal/mol) inserted upstream of the Rluc AUG to occlude scanning ribosomes. Despite an ≈60-fold reduction in Rluc activity, Fluc was synthesized at least as efficiently as from constructs lacking the stem loop (Fig. 3c), suggesting that Fluc was synthesized independently of scanning ribosomes. Interestingly, in both the uncapped and stem-loop constructs, the Fluc activity was reproducibly greater than that of the control construct.
A third method by which to eliminate cap-dependent translation is to translate mRNAs in the presence of the antibiotic edeine. At low concentrations, edeine interferes with the ability of the 40S-eIF2-GTP/Met-tRNAi complex to recognize the AUG codon (27, 28). Under these conditions, despite a drastic reduction in Rluc translation, Fluc expression was similar to that of the control construct (Fig. 3c).
A fourth possibility is that the message becomes altered in such a way as the 5′ end of a portion of the mRNAs are brought closer to the 5′ end of Fluc (e.g., through a cryptic promoter or a break in the mRNA). To test for a sequence-specific event that results in translation initiating somewhere in Fluc, we made a construct in which the WT tk triplets were not in the same register as Fluc (pAG3.88). Only background levels of Fluc activity were observed with this construct (Fig. 4f). We also purified mRNAs from a denaturing urea gel that were then translated, followed with analysis by Northern blot hybridization. Fluc activity was observed with RNA from pAG3.155 but not from a control construct, and no difference in RNA size distribution could be seen between these two transcripts (see Fig. 6 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site).
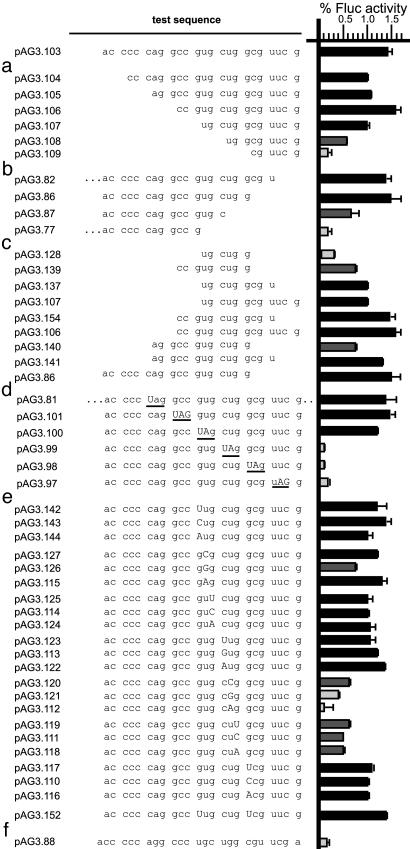
Fig. 4.
Sequence requirements for the tk IRES. The indicated test sequence was cloned into the dual-luciferase vector, and mRNAs from these constructs were translated in rabbit reticulocyte lysate. The relative Fluc activities were then measured. To the left of the test sequence is the plasmid name. To the right of test sequence is a bar graph in which the length of the bar indicates the level of Fluc activity, and the error bars indicate standard deviations. For sequences that support “standard” levels of Fluc expression (≥1%), the bars are black. For sequences that support “intermediate” levels of Fluc expression (≥0.5% but <1%), the bars are dark gray. For sequences that have low or background levels of recoding (<0.5%), the bars are light gray. The sequence (pAG3.103) is a tk sequence expressing standard levels of Fluc expression. (a) A series of 5′ three-base deletions. (b) 3′ three-base deletions. (c) Rebuilding the constructs to regain Fluc activity. (d) Introduction of stop codons (underlined, and in capital letters when different from the tk sequence) into the Fluc reading frame. “...” and “..” denote that the construct has an additional 12 nt of tk sequence upstream or 3 nt of the tk sequence downstream, respectively, in b and d.(e) Point mutations (capital letters). (f) Changing the register of the tk sequence with respect to the Fluc ORF. A base was deleted immediately upstream of the tk sequence, and a base was added immediately downstream.
Taken together, these observations argue strongly that the tk test sequence does not support ribosomal frameshifting or internal initiation after ribosomal scanning, shunting, transcriptional initiation, or RNA breakage; rather, they suggest that the sequence is an IRES.
A 12-nt Segment Suffices for IRES Activity. To define a minimal sequence sufficient for Fluc expression, a series of deletions in the test sequence, three bases at a time from the 5′ end, was generated and analyzed (Fig. 4a). Constructs were scored as expressing standard levels, intermediate levels, or low/background levels of Fluc activity. Interestingly, removal of part of the C-chord reduced expression to intermediate levels, but deletion of the entire C-chord resulted in standard levels. Test sequences of 12 and 9 bases exhibited intermediate levels of expression, and Fluc activity was only abolished when the test sequence was 6 bases long (pAG3.109). A similar series of 3′ deletions (Fig. 4b) demonstrated that bases near the 3′ end of the test sequence were important for Fluc activity (pAG3.77). These data suggested that sequences important for activity include CC GUG CUG G (triplets are in the Fluc and tk frames). However, this sequence alone supported only intermediate levels of Fluc expression (pAG3.139). Therefore, we took a short sequence (UG CUG G) that did not support Fluc expression above background (pAG3.128) and added back bases to the 5′ or 3′ end and measured Fluc activity (Fig. 4c). These data indicated that the minimal sequence sufficient for standard levels of expression was CC GUG CUG GCG U (pAG3.154).
Ribosomes Enter the Fluc Frame at or 3′ of CUG. To ask at which codon ribosomes enter the Fluc frame, we replaced test sequence codons in that frame with stop codons (Fig. 4d). Expression of Fluc was not reduced after the addition of stop codons in a sequential 5′-to-3′ fashion until the CUG was changed to UAG. The introduction of stop codons downstream of the CUG also prevented expression of Fluc. These data are consistent with the ribosome entering the Fluc frame at the CUG codon. Alternatively, replacement of the CUG with UAG could have altered a crucial regulatory sequence.
Point Mutagenesis. The deletion and stop codon analyses suggested that the sequence GUGCUGG was most important. We therefore mutagenized each of these bases (Fig. 4e). Changing most of the nucleotides in the sequence had little or no effect. However, changes to the U or G of the CUG codon (in the tk and Fluc frames) had more substantial effects on Fluc expression. Expression of Fluc was reduced to background levels when the U was changed to an A. The other changes to the U or the G had intermediate effects.
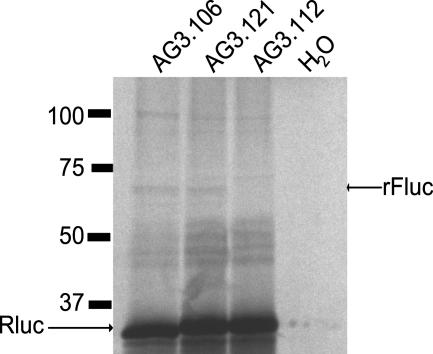
Polypeptides Expressed from the Reporter Construct Are Consistent with Internal Initiation. If Fluc expression is due to internal translational initiation, then one would expect that constructs that expressed Fluc activity would direct the synthesis of a product roughly the size of Fluc. We therefore radiolabeled the products of cell-free translations from all of our constructs and analyzed them by SDS/PAGE. Examples from constructs that had high, intermediate, or low Fluc activities are shown in Fig. 5. An ≈60-kDa band that comigrated with authentic Fluc was expressed from all constructs that expressed Fluc activity (e.g., AG3.106 and AG3.121; Fig. 5) and not from those that exhibited background levels of activity (e.g., AG3.112; Fig. 5). A band of ≈100 kDa, which is slightly smaller than one would predict for the size of an Rluc–Fluc fusion protein, was also observed, but its presence did not correlate with Fluc enzyme activity. These data bolster the case for internal initiation of Fluc.
Fig. 5.
Synthesis of Fluc-sized polypeptides from the IRES. An autoradiogram of the translation products of the constructs indicated at the top of the gel were radiolabeled and analyzed by SDS/PAGE and autoradiography. The positions of Rluc polypeptide (Rluc), authentic recombinant Fluc polypeptide (rFluc), and molecular weight markers are indicated.
A Mutation That Affects IRES Function in Vitro also Affects TK Activity of the C6-1C Mutant. If the IRES that we identified by using in vitro translation is important for expression of TK from the ACVr C6-1C virus, then a mutation that reduces IRES activity in vitro without changing the tk ORF should also reduce TK expression from the mutant. We therefore constructed a recombinant virus that carried such a mutation (CUG-to-CUC mutation) that resulted in an ≈3-fold decrease in IRES activity (pAG3.111; Fig. 4; both CUG and CUC encode leucine). Virus C6-1C.CUC was engineered to have both the C6-1C and CUC mutations (Fig. 1). This virus had ≈0.6% tk activity (Fig. 2). A Northern blot showed that the tk mRNA from this virus migrated similarly to those from KOS and TKC6-1C (Fig. 7, which is published as supporting information on the PNAS web site). These data are consistent with the TK activity from the TKC6-1C virus being dependent on the IRES.
Discussion
In this study, we have found that a virus containing a frameshift mutation in tk, which is frequently found in clinical ACVr isolates (7, 8), expresses low levels of TK activity. Analyses using cell-free translation lead us to conclude that the mutant sequence is adjacent to an unusual IRES. We discuss below the evidence that this sequence is an IRES, what makes it unusual, how it may function, how it may contribute to TK activity and pathogenesis by the virus, and its implications for genomic and proteomic analyses.
The tk Sequence Is an IRES. To conclude that a sequence functions as an IRES, it is important to rule out other mechanisms that have been suggested to explain some reports of IRESs and, in particular, to rule out ribosome scanning mechanisms (29). Several results exclude the possibility that a scanning mechanism was responsible for expression of Fluc in our assays. First, the downstream gene in the reporter construct lacked an initiating AUG codon. Second, in experiments where scanning-dependent translation was perturbed by making uncapped transcripts, by introducing a stable stem loop, or by translating the mRNA in the presence of edeine (Fig. 3d), Fluc was expressed as efficiently as from controls. Indeed, Fluc expression was reproducibly greater than the controls in these experiments. This finding would be difficult to explain on the basis of a scanning-dependent or ribosomal shunting mechanism but could be explained if ribosomes that have translated the Rluc gene and are entering the test sequence impede IRES-dependent translation of Fluc. Third, (Fig. 4e) two constructs introduced AUG codons into the test sequence (pAG3.144 and pAG3.122). If ribosomes were scanning the test sequence, an increase in expression of Fluc would be expected (particularly with pAG3.122, because it has a good Kozak consensus sequence). Such an increase was not observed. Fourth, although the crucial CUG codon that we identified is in a reasonably good Kozak consensus, changing to an unfavorable context did not affect Fluc expression (pAG3.152; Fig. 4d). Fifth, we observed no evidence for short transcripts that contained a 5′ end near the Fluc ORF that could account for Fluc expression (Fig. 6); but even if we had, the results summarized above would not be expected to occur by means of expression of Fluc from such short transcripts.
The tk IRES Is Unusual. IRESs are typically long, highly structured RNA sequences that serve to recruit elements of the translational machinery to the message (reviewed in ref. 30). The tk IRES is only 12 bases long. RNA folding programs such as mfold (31) predict that this sequence exhibits little, if any, structure. There have been reports of short sequences of between 9 and 22 nt that can serve as IRESs (32–35). Unlike the tk IRES, these sequences were analyzed in reporter constructs that placed the test sequence in close proximity to the AUG of the downstream reporter gene.
Although the IRES contains two codons, a GUG and the crucial CUG codon, that can substitute in cap-dependent initiation for AUG using Met-tRNAi (36), several of the lines of evidence outlined above suggest that the tk IRES does not use Met-tRNAi-dependent initiation. There have been reports of methionine-independent initiation in IRESs (37, 38), but these IRESs are much longer and more structured than the tk IRES.
A third unusual feature of the tk IRES is that it occurs in a DNA virus. Thus far, the only DNA viruses that have been shown to contain IRESs are gammaherpesviruses (39–42). In those cases, unlike the tk sequence, the IRES lies in an intergenic region rather than in the middle of a coding sequence.
How Might the tk IRES Function? The tk IRES bears certain similarities to another unusual IRES, that of cricket paralysis virus (CrPV). Like the tk IRES, it is active under conditions that disrupt the activity of the eIF2-GTP/Met-tRNAi complex (in the presence of low concentrations of the antibiotic edeine) (22). The tk IRES does not contain an AUG; the CrPV IRES does not initiate with an AUG codon, but with a GCU codon. Our data from experiments introducing stop codons suggest that translation initiates from the tk IRES at or just downstream of a CUG codon.
The CrPV IRES binds directly to 40S ribosomes in the absence of any other factors (22). However, the CrPV IRES is much larger and more complex than the tk IRES, serving to position the ribosome on the message in such a way that the P site is not decoded and translation initiates in the A site. It will be important to learn how a sequence as short as the TK IRES can recruit the translational machinery to the message.
HSV Employs Unusual Translational Mechanisms to Achieve Pathogenic Drug Resistance. The C6-1C mutation is the third example of a frameshift mutation found in an ACVr clinical isolate that appears to be compensated by an unusual translational mechanism to permit expression of active TK (refs. 11 and 20 and unpublished work). In the two previous examples, expression of active TK is explained by ribosomal frameshifting that results in synthesis of active, full-length enzyme. However, in this case, synthesis of a C-terminal fragment of TK appears to be involved. To explain how this fragment leads to active TK, we suggest a mechanism akin to α-complementation in Escherichia coli β-galactosidase protein (43), in which an N-terminal deletion is compensated in trans by a corresponding N-terminal fragment (explained at the atomic level in ref. 44). By analogy, the out-of-frame C-terminal portion of the mutant TK may be compensated by the corresponding in-frame C-terminal fragment synthesized by means of the IRES. This compensation may be assisted by the out-of-frame segment serving as a scaffold and by the dimerization of TK (45).
Regardless of the precise mechanism by which active TK is produced, the levels produced (≈1.5%) are more than those that suffice to permit reactivation from latency from mouse ganglia of a strain that requires TK for reactivation (12). We speculate that the IRES contributes to the ability of clinical ACVr isolates carrying the C6-1C mutation to cause disease in humans.
The tk IRES Suggests the Existence of an Expanded Proteome. The data presented in Fig. 4e show that, at least in vitro, many short sequences can support IRES-mediated translation. These results raise the possibility that there may be many hitherto unrecognized polypeptides synthesized from eukaryotic messages, which could have important implications when considering the coding potential of genomic DNA.
Supplementary Material
Acknowledgments
We thank Fred Wang for suggesting the experiment with C6-1C.1FS, the Harvard Institute of Chemistry and Cell Biology for use of the luminometer, and the National Cancer Institute for providing the edeine. We are grateful to Kevin Bryant for useful discussions and Lee Gehrke for helpful comments on the manuscript. This work was supported by National Institutes of Health Grants P01 NS35138, R01 AI26126, and T32 AI07245.
Author contributions: A.G. and D.M.C. designed research; A.G. performed research; A.G. and D.M.C. analyzed data; and A.G. and D.M.C. wrote the paper.
Abbreviations: HSV, herpes simplex virus; TK, thymidine kinase; IRES, internal ribosome entry site; ACV, acyclovir; ACVr, ACV-resistant; Fluc, firefly luciferase; Rluc, Renilla luciferase.
References
- 1.Hershey, J. W. B. & Merrick, W. C. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Woodbury, NY), pp. 33-88.
- 2.Pelletier, J. & Sonenberg, N. (1988) Nature 334, 320-325. [DOI] [PubMed] [Google Scholar]
- 3.Jang, S. K., Krausslich, H. G., Nicklin, M. J., Duke, G. M., Palmenberg, A. C. & Wimmer, E. (1988) J. Virol. 62, 2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider, R. J. & Mohr, I. (2003) Trends Biochem. Sci. 28, 130-136. [DOI] [PubMed] [Google Scholar]
- 5.McKnight, S. L. (1980) Nucleic Acids Res. 8, 5949-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coen, D. M., Weinheimer, S. P. & McKnight, S. L. (1986) Science 234, 53-59. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert, C., Bestman-Smith, J. & Boivin, G. (2002) Drug Resist. Updat. 5, 88-114. [DOI] [PubMed] [Google Scholar]
- 8.Gaudreau, A., Hill, E., Balfour, H. H., Jr., Erice, A. & Boivin, G. (1998) J. Infect. Dis. 178, 297-303. [DOI] [PubMed] [Google Scholar]
- 9.Coen, D. M., Kosz Vnenchak, M., Jacobson, J. G., Leib, D. A., Bogard, C. L., Schaffer, P. A., Tyler, K. L. & Knipe, D. M. (1989) Proc. Natl. Acad. Sci. USA 86, 4736-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efstathiou, S., Kemp, S., Darby, G. & Minson, A. C. (1989) J. Gen. Virol. 70, 869-879. [DOI] [PubMed] [Google Scholar]
- 11.Hwang, C. B., Horsburgh, B. C., Pelosi, E., Roberts, S., Digard, P. & Coen, D. M. (1994) Proc. Natl. Acad. Sci. USA 91, 5461-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths, A., Chen, S. H., Horsburgh, B. C. & Coen, D. M. (2003) J. Virol. 77, 4703-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sacks, S. L., Wanklin, R. J., Reece, D. E., Hicks, K. A., Tyler, K. L. & Coen, D. M. (1989) Ann. Intern. Med. 111, 893-899. [DOI] [PubMed] [Google Scholar]
- 14.Horsburgh, B. C., Kollmus, H., Hauser, H. & Coen, D. M. (1996) Cell 86, 949-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Böni, J. & Coen, D. M. (1989) J. Virol. 63, 4088-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coen, D. M., Irmiere, A. F., Jacobson, J. G. & Kerns, K. M. (1989) Virology 168, 221-231. [DOI] [PubMed] [Google Scholar]
- 17.Irmiere, A. F., Manos, M. M., Jacobson, J. G., Gibbs, J. S. & Coen, D. M. (1989) Virology 168, 210-220. [DOI] [PubMed] [Google Scholar]
- 18.Davar, G., Kramer, M. F., Garber, D., Roca, A. L., Andersen, J. K., Bebrin, W., Coen, D. M., Kosz Vnenchak, M., Knipe, D. M., Breakefield, X. O. & Isacson, O. (1994) J. Comp. Neurol. 339, 3-11. [DOI] [PubMed] [Google Scholar]
- 19.Chen, S. H., Cook, W. J., Grove, K. L. & Coen, D. M. (1998) J. Virol. 72, 6710-6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths, A. & Coen, D. M. (2003) J. Virol. 77, 2282-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths, A., Renfrey, S. & Minson, T. (1998) J. Gen. Virol. 79, 807-812. [DOI] [PubMed] [Google Scholar]
- 22.Wilson, J. E., Pestova, T. V., Hellen, C. U. & Sarnow, P. (2000) Cell 102, 511-520. [DOI] [PubMed] [Google Scholar]
- 23.Sasadeusz, J. J., Tufaro, F., Safrin, S., Schubert, K., Hubinette, M. M., Cheung, P. K. & Sacks, S. L. (1997) J. Virol. 71, 3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grentzmann, G., Ingram, J. A., Kelly, P. J., Gesteland, R. F. & Atkins, J. F. (1998) RNA 4, 479-486. [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson, R. J. (1991) Biochim. Biophys. Acta 1088, 345-358. [DOI] [PubMed] [Google Scholar]
- 26.Pestova, T. V., Kolupaeva, V. G., Lomakin, I. B., Pilipenko, E. V., Shatsky, I. N., Agol, V. I. & Hellen, C. U. (2001) Proc. Natl. Acad. Sci. USA 98, 7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Odom, O. W., Kramer, G., Henderson, A. B., Pinphanichakarn, P. & Hardesty, B. (1978) J. Biol. Chem. 253, 1807-1816. [PubMed] [Google Scholar]
- 28.Kozak, M. & Shatkin, A. J. (1978) J. Biol. Chem. 253, 6568-6577. [PubMed] [Google Scholar]
- 29.Kozak, M. (2001) Mol. Cell. Biol. 21, 1899-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellen, C. U. & Sarnow, P. (2001) Genes Dev. 15, 1593-1612. [DOI] [PubMed] [Google Scholar]
- 31.Zucker, M., Mathews, D. H. & Turner, D. H. (1999) in RNA Biochemistry and Biotechnology, eds. Barciszewski, J. & Clark, B. F. C. (Kluwer, Dordrecht, The Netherlands).
- 32.Chappell, S. A., Edelman, G. M. & Mauro, V. P. (2000) Proc. Natl. Acad. Sci. USA 97, 1536-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chappell, S. A. & Mauro, V. P. (2003) J. Biol. Chem. 278, 33793-33800. [DOI] [PubMed] [Google Scholar]
- 34.Owens, G. C., Chappell, S. A., Mauro, V. P. & Edelman, G. M. (2001) Proc. Natl. Acad. Sci. USA 98, 1471-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, W., Edelman, G. M. & Mauro, V. P. (2003) Proc. Natl. Acad. Sci. USA 100, 4457-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak, M. (1997) EMBO J. 16, 2482-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki, J. & Nakashima, N. (2000) Proc. Natl. Acad. Sci. USA 97, 1512-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, J. B., Hayday, A., Courtneidge, S. & Fried, M. (1986) Cell 44, 477-487. [DOI] [PubMed] [Google Scholar]
- 39.Coleman, H. M., Brierley, I. & Stevenson, P. G. (2003) J. Virol. 77, 13093-13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Low, W., Harries, M., Ye, H., Du, M. Q., Boshoff, C. & Collins, M. (2001) J. Virol. 75, 2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isaksson, A., Berggren, M. & Ricksten, A. (2003) Oncogene 22, 572-581. [DOI] [PubMed] [Google Scholar]
- 42.Bieleski, L. & Talbot, S. J. (2001) J. Virol. 75, 1864-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ullmann, A., Perrin, D., Jacob, F. & Monod, J. (1965) J. Mol. Biol. 12, 918-923. [DOI] [PubMed] [Google Scholar]
- 44.Juers, D. H., Jacobson, R. H., Wigley, D., Zhang, X. J., Huber, R. E., Tronrud, D. E. & Matthews, B. W. (2000) Protein Sci. 9, 1685-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldman, A. S., Haeusslein, E. & Milman, G. (1983) J. Biol. Chem. 258, 11571-11575. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.