Abstract
Although many redox signaling molecules are present at low concentrations, typically ranging from micromolar to sub-micromolar levels, they often play essential roles in a wide range of biological pathways and disease mechanisms. However, accurately measuring low abundant analytes has been a significant challenge due to the lack of sensitivity and quantitative capability of existing measurement methods. In this study, we introduced a novel chemically induced amplifiable system for quantifying low-abundance redox signaling molecules in living cells. We utilized H2O2 as a proof-of-concept analyte and developed a probe that quantifies cellular peroxide levels by combining the NanoBiT system with androgen receptor (AR) dimerization as a reporting mechanism. Our system demonstrated a highly sensitive response to cellular peroxide changes induced both endogenously and exogenously. Furthermore, the system can be adapted for the quantification of other signaling molecules if provided with suitable probing chemistry.
Graphical Abstract
Introduction
Fluorescent probes enable the studies of signaling molecules in live cells and have become indispensable tools in cell biology research. The development of the calcium probes by the Tsien group completely revolutionized the field of calcium signaling and took our knowledge about Ca2+ biological functions to a new level.1 These calcium probes reversibly bind Ca2+ to shift their absorption maxima, thus producing ratiometric readouts that can quantify calcium concentrations in cells. Following the same concept, our group developed the first reversible reaction-based ratiometric glutathione (GSH) probe (ThiolQuant Green) that can quantify GSH concentrations in live cells,2 along with many improved GSH probes with faster reaction kinetics and organelle specificities reported by our and other groups.3-9 However, the success of GSH probes relies heavily on the high concentrations of GSH (1-10 mM) in cells, enabling fast reactions between GSH and the probes.
Redox signaling molecules, such as hydrogen peroxide (H2O2), hydrogen sulfide (H2S) and nitric oxide (NO), have estimated concentrations in the micromolar or sub-micromolar range.10-15 The pioneering work by the groups of Chang, Xian, Pluth, Nagano, Ai, Lippert, and many others has led to a myriad of reaction-based fluorescent probes for these redox signaling molecules.16-22 Based on careful examination of the literature and our in-house experiments, we found that micromolar concentrations of probes are usually necessary to treat cells in the presence of exogenously added excessive redox signaling molecules to obtain meaningful qualitative fluorescence signals using conventional confocal microscopes within a reasonable time frame. The high concentrations of probes can significantly consume the analytes and disturb the biological system. Additionally, these probes are usually qualitative and cannot respond to the endogenous levels of the redox signaling molecules.
We set three criteria to design probes for low concentrations of analytes. (i) The probe concentration used should be less than 1% of the analyte concentration to minimize perturbation of the biological system. (ii) The probe should be able to quantitatively measure the analyte concentrations in live cells. (iii) The assay time should be within a convenient experimental time frame, ideally 1-2 hours.
With these criteria in mind, we initially attempted to follow our work on GSH probes2 to identify reversible reactions with redox signaling molecules and realized the technical challenges for this route. For most of the reaction-based probes, one equivalent of probes produces an equal amount of either fluorescent species, or enzyme substrates, or photons from chemiluminescence upon reacting with the analytes. 22-27 Considering the analytes are in the micromolar range, we would need to keep the probe concentration in the range of low nanomolar. For equal molar conversion from the nanomolar probe to signals, it would be very challenging to detect with either conventional confocal fluorescence microscope or luminescence readouts, which could explain why micromolar concentrations of probes were applied in previous studies.
To address these technical issues, strategies to amplify the output signals are needed. The groups of Renslo and Wells reported a seminal study to develop amplifiable ferrous iron (Fe2+) probes.28 In this work, a Fe2+ responsive group caged puromycin reacts with intracellular Fe2+ to regenerate puromycin, which can be incorporated into the C-terminus of elongating nascent peptide chains to terminate translation and detected using horseradish peroxidase (HRP)-based immunofluorescence (IHC) after fixing the cells. Essentially, one equivalent of released puromycin can be converted to an equal amount of HRP, an enzyme that produces amplified signals. The Chang group applied this strategy to develop peroxymycin-1, a H2O2 responsive group caged puromycin.29
The caveats of the puromycin system are two-folds. First, micromolar concentrations of the caged puromycin probes are still needed, which could consume significant amount of analytes in a similar concentration range. Considering the Michaelis-Menten constant (Km) or dissociation equilibrium constant (Kd) for puromycin as a substrate for ribosomes is in the μM range,30-33 it is not surprising that micromolar of the caged puromycin probes are necessary for this strategy. Ideally, we need to identify small molecules that have Km or Kd in the low nM or sub-nM range. Second, this strategy can only be applied in fixed cells. However, the fixation process may change the level of these redox analytes.
Hormones are Nature’s chemical biology tools to regulate gene expression through hormone receptors.34 For example, estrogens bind estrogen receptors (ER) with nM binding affinities to activate gene expression.35 This progress can be hijacked as an ER activity reporter assay by expressing a plasmid that has the estrogen responsive element (ERE) with downstream reporter genes, such as fluorescent proteins or luciferases.36 Our initial attempt to develop an amplifiable system for low concentrations of analytes is to develop a caged estrogen (E2) that can be released in the presence of H2O2 to subsequently drive firefly luciferase expression. We chose H2O2 as the proof-of-concept study because the H2O2 responsive chemistry has been well established and the reaction rates between H2O2 and boronic esters are relatively fast.27,37 Unfortunately, our initial design has two major drawbacks. In the absence of estrogens, ER has leaky background activities, which can be as high as 10% of the maximum activity in the presence of saturating concentration of estrogens, thus limiting the dynamic range of the assay (Figure S1A). To avoid the high background noise issue, the AR/ARE (androgen receptor/androgen response element) system was evaluated as well (Figure S1B).38 However, one notable drawback of such systems is that the transcription-based assays take 12-24 hours to accumulate enough reporter proteins for detection. Oxidative stress may change the rate of transcription and translation of the reporter proteins, which renders it difficult to dissect the levels of reporter proteins change is due to the uncaging of pro-estrogens/pro-androgens or secondary effects.39,40
With the setbacks in mind and inspired by our work in proteolysis targeting chimeras (PROTACs),41,42 we turned to the chemically induced dimerization (CID) systems,43 in which proteins are expressed before perturbation to the biological system is introduced. Among the CID systems, we chose androgen receptor (AR) as our platform because dihydrotestosterone (DHT) triggers rapid homo-dimerization of AR with a tight intermolecular interaction.44-46 For reporters, we chose luciferase-based bioluminescence instead of fluorescent proteins due to their high sensitivities. Promega developed nano-luciferase (nLuc), a bioluminescent enzyme with an extremely high catalytic efficiency (kcat/KM = 183 μM−1s−1), allowing detection of the enzyme in the picomolar range.47,48 Additionally, Promega also developed a split version of nLuc, namely NanoBiT.49 The complementary fragments of nLuc are called LgBit and SmBit and can be fused to proteins to study protein-protein interactions. AR-LgBit and AR-SmBit fusions have been developed as a reporter assay for androgens, such as DHT (Figure 1A). 50,51 With these tools in hands, we developed a Chemically Induced Dimerization-based Amplifiable Probe (CIDAP) to quantify H2O2 levels in living cells.
Figure 1.
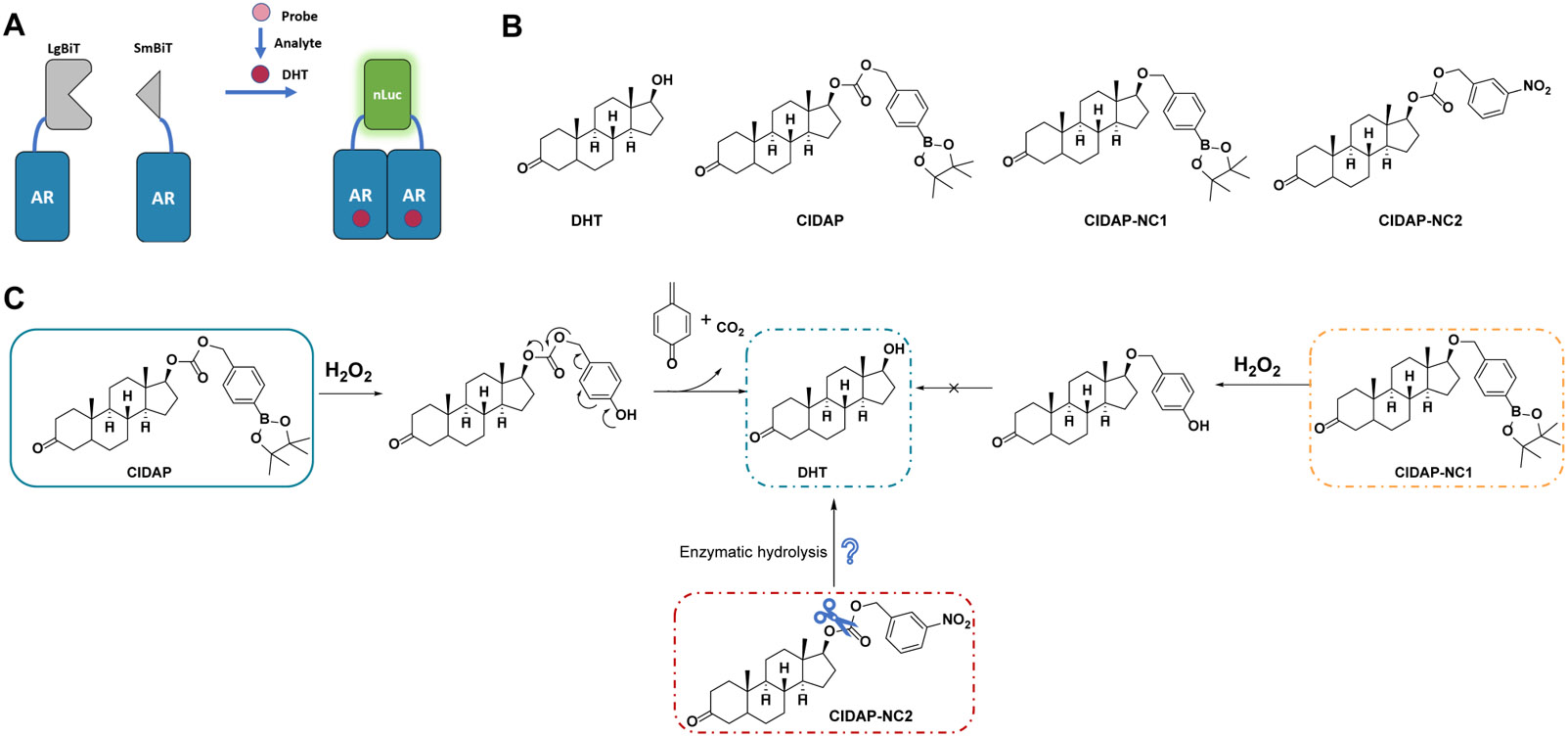
Design of Chemically Induced Dimerization-based Amplifiable Probes (CIDAP). (A) LgBiT or SmBiT are fused to androgen receptor (AR) and transiently co-expressed in mammalian cells. Addition of androgens (such as DHT) triggers AR dimerization and the reconstitution of LgBiT and SmBiT to form nLuc. DHT can be converted to a prodrug (i.e., the probe), which reacts with redox signaling molecules (such as H2O2) to regenerate DHT. (B) The structures of DHT, CIDAP, CIDAP-NC1, and CIDAP-NC2. (C) The reaction mechanism of CIDAP and its cascade to release DHT. CIDAP-NC1 is the negative control that cannot regenerate DHT. CIDAP-NC2 is used as the control to investigate the impact of enzymatic hydrolysis on carbonate.
Results
Design of CIDAP and Evaluation of Its Activity in Cells.
Examining the co-crystal structure of DHT and AR, we found that 17-β-OH in DHT forms a key interaction with AR (PDB: 5JJM).52 We developed a CIDAP platform by caging the 17-β-OH in DHT with a H2O2 responsive boronic ester group to abolish its binding to AR (Figure 1B and 1C).16 We initially developed a probe without the carbonate group (CIDAP-NC1) but failed to release DHT in the presence of H2O2, given that alcohol is a poor leaving group. CIDAP-NC1 serves as a negative control in the following experiments.
To evaluate whether the carbonate group can be directly hydrolyzed by intracellular enzymes to release DHT in the absence of H2O2 (Figure 1C), we developed a CIDAP analog without the boronic ester group, CIDAP-NC2. Density functional theory (DFT) calculations showed that the carbonate carbons in CIDAP and CIDAP-NC2 have similar partial charge densities, suggesting similar hydrolysis rates (Table S1). Moreover, CIDAP was directly incubated with recombinant nLuc protein to confirm that it does not suppress or enhance the luminescence of nano-Luciferase (Figure S4).
We used HeLa cells as a model system to test if CIDAP can respond to endogenous levels of H2O2. AR-LgBit and AR-SmBit were transiently transfected and co-expressed in HeLa cells, followed by incubation with CIDAP, a positive control DHT, and two negative controls CIDAP-NC1 and CIDAP-NC2 at 1.1 nM (Figure 2A). As expected, nM levels of DHT and CIDAP trigger bioluminescence with a high signal-to-background ratio (S/N = ~40). The lower signals from the CIDAP group could be attributed to its incomplete reaction with H2O2 under the experimental conditions. CIDAP-NC1 and CIDAP-NC2 produce minimal bioluminescence compared to CIDAP, indicating that bioluminescence produced by CIDAP is not contributed from CIDAP direct binding to AR without uncaging or uncaging from direct hydrolysis of the carbonate. Additionally, the reactivity of CIDAP towards other reactive oxygen species (ROS) is insignificant, indicating a high degree of selectivity towards H2O2 (Figure S3A), consistent with previous studies.27 We further examined CIDAP's reactivity towards lower concentrations of peroxynitrite and hypochlorite. As anticipated, hypochlorite did not react with CIDAP. In contrast, consistent with previous findings, boronic acid rapidly reacted with peroxynitrite. Interestingly, we observed that, following the oxidation of boronic acid, peroxynitrite proceeded to react with the produced DHT and oxidize the 17-β-OH group (Figure S3B). Despite these results suggesting that peroxynitrite can react with CIDAP and deplete DHT, considering the final product would not induce the reconstitution of nLuc, this side reaction is unlikely to impact the final quantification of peroxide.
Figure 2.
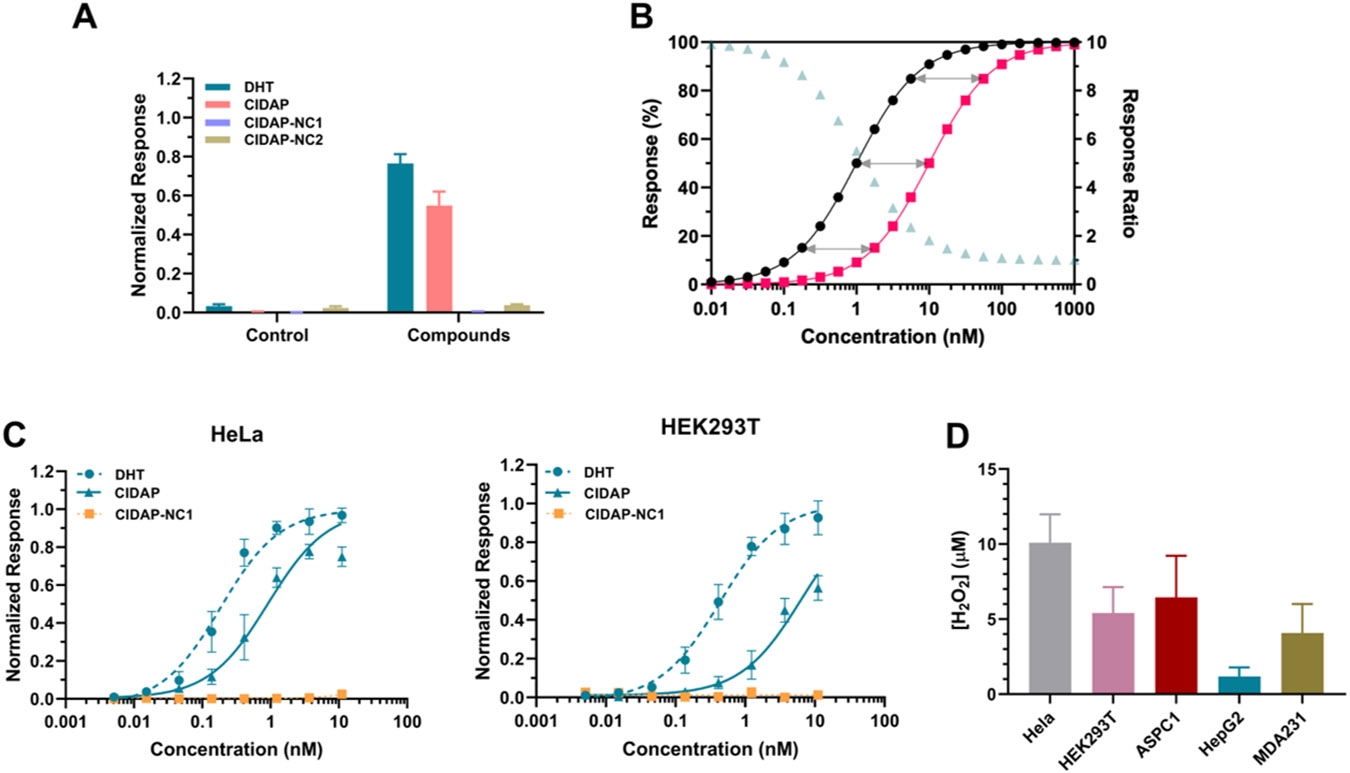
Refinement and assessment of CIDAP system as a quantifying method for cellular peroxide level. (A) The bioluminescence response of DHT, CIDAP, CIDAP-NC1 and CIDAP-NC2. AR-LgBit and AR-SmBit are co-expressed in HeLa cells for 24 h, followed by the incubation with control (ethanol) and each compound at 1.1 nM, separately. The readouts were obtained after 2 h of incubation. Data are mean ± SEM of experimental replicates (n = 4). (B) Simulated Data for DHT and Probe responses. Black trace: DHT response; red trace: Probe response; pale green trace: the ratio of DHT and Probe responses at the same concentrations. (C) The dose-response curves of DHT, CIDAP, and CIDAP-NC1 in HeLa and HEK293T cells. Data are mean ± SEM of experimental replicates (n = 4). Both cell lines were incubated with different concentrations of DHT, CIDAP and CIDAP-NC1 (from 11 nM to 5 pM with 3-fold dilutions) for 2 h. Bioluminescence was measured in the presence of furimazine. (D) H2O2 levels in different cell lines. The SEM on the H2O2 level calculated using the global fitting program provided by GraphPad Prism (see Supporting Information for details).
Quantitative Method to Convert CIDAP Signals to Peroxide Concentrations.
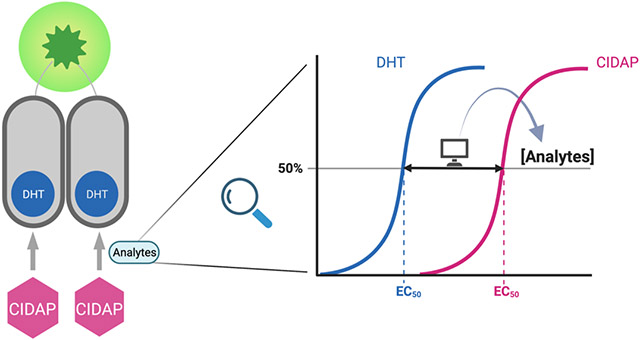
The next question is how we can quantitatively convert CIDAP signals to H2O2 concentrations. We adopt the concept of dose-response shift in pharmacology as the solution. In cells co-expressing AR-LgBiT and AR-SmBiT, addition of DHT triggers nLuc formation and bioluminescence with a classical sigmoidal dose-response curve (black trace in Figure 2B). Addition of the probe with the same concentration series as DHT to cells would also generate a dose response curve (red trace in Figure 2B). When the concentrations of the probe used are significantly lower than those of H2O2, the peroxide concentration remains constant throughout the assay. Consequently, the reaction between the probe and peroxide follows the pseudo first-order reaction kinetics. In a first-order reaction, the percentage of conversion remains constant, regardless of the initial concentration of the reactant (grey arrows in Figure 2B). During the measurement, different probe concentrations can be introduced to the cells to initiate partial reactions with peroxide. The constant percentage of CIDAP-to-DHT conversion results in a right shift of the dose-response curve. According to the pseudo first-order reaction law, the relationship between the peroxide concentration and the percentage of probe conversion can be expressed by , where is the secondary reaction rate constant between H2O2 and CIDAP, is the percentage of probe conversion, and is the reaction time. The secondary reaction rate constant is determined as 196.6 M−1min−1 (Figures S2A and S2B). The percentage of CIDPA conversion α can be calculated using the ratio of the EC50 values of DHT and CIDAP (EC50, DHT/EC50, CIDAP) from the dose-response curves.
Our dose-response shift approach offers a theoretical framework to quantify low concentration analytes using amplifiable systems. In Spangler et al.’s puromycin-based system, a series of concentrations of puromycin and the probe indeed generated dose-response shift curves (Figure 2 in reference 28), consistent with our analysis. However, in the following applications, they only employed a single concentration of puromycin and the probe (Figures 3 and 4 in reference 28). It is worth noting that the response ratios (pale green triangles in Figure 2B) are dependent on the initial concentration used for probe and benchmark (e.g., DHT or puromycin). A single concentration of probe and benchmark can only provide a qualitative measurement.
Quantification of Endogenous Peroxide Levels.
HeLa and HEK293T cells were employed as model systems to refine the experimental protocols. The optimal incubation time was determined to be 2 h to balance signal outputs and prevent complete CIDAP conversion (Figure S5). Following the optimized protocol, cells expressing AR-LgBit and AR-SmBit were incubated with DHT, CIDAP, and CIDAP-NC1 (concentrations ranging from 11 nM to 5 pM with 3-fold dilutions) to generate dose-response curves (Figure 2C). Based on the equation, we determined the concentrations of peroxide in HeLa and HEK293T cells to be 10.09 ± 1.89 and 5.4 ± 1.73 μM, respectively. Moreover, we applied this approach to three additional cell lines, namely AsPC1, HepG2, and MDA-MB-231, and obtained H2O2 concentrations ranging from 1.8 to 10 μM (Figure 2D, Figure S6). It should be noted that the fact that the CIDAP concentration applied is <1% of measured H2O2 levels supports our pseudo-first-order reaction assumption.
Quantification of Peroxide Level Changes upon Exogeneous H2O2 Treatment.
In order to expand the scope of application for this system and evaluate its responsiveness to changes in the cellular environment, an exogenous H2O2 concentration of 50 μM was introduced to elevate the changes of H2O2 levels in cells. This resulted in an increase in cellular peroxide levels from 5.4 ± 1.73 μM to 21.9 ± 9.3 μM (Figure 3A). Additionally, a range of H2O2 concentrations between 0 and 50 μM were introduced, with the calculated peroxide levels demonstrating an increasing trend that was consistent with the increasing amounts of exogenous H2O2 added (Figure S7). These findings confirmed the sensitivity of the method for monitoring changes in H2O2 levels within cells. However, it is important to note that the detected changes in overall peroxide concentration in the cellular environment may not necessarily reflect accurate changes in intracellular peroxide levels, as both peroxide and the probe can permeate membranes.53,54
Figure 3.
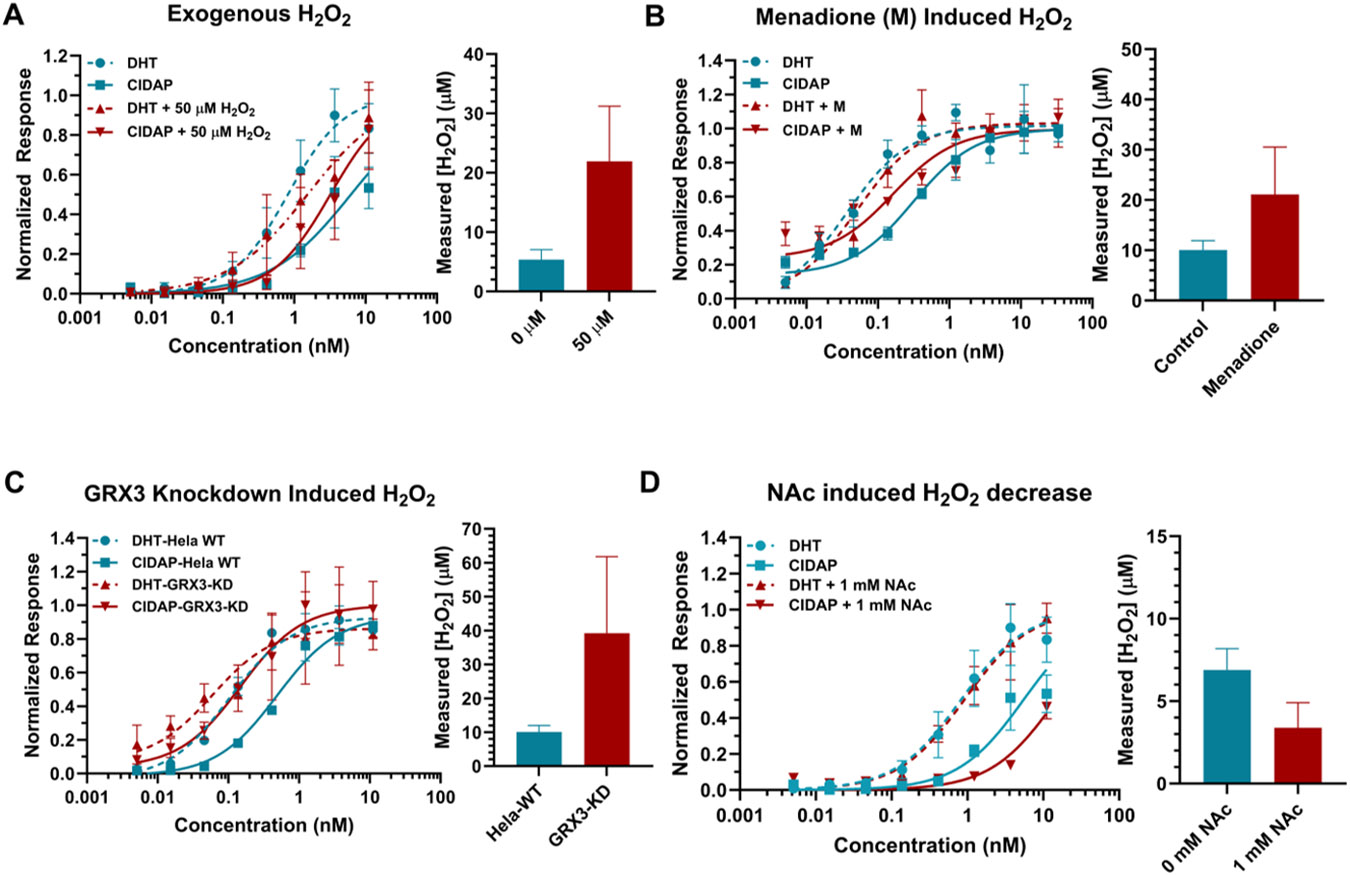
Quantification of H2O2 levels in cells under various conditions. The response of CIDAP is accurately reflecting the fluctuation of cellular peroxide level. (A) H2O2 levels in HEK293T cells upon exogenous 50 μM H2O2 treatment. Data on the left are mean ± SEM of experimental replicates (n = 4). (B) H2O2 levels in HeLa cells pretreated with 10 μM menadione. Data on the left are mean ± SEM of experimental replicates (n = 4). (C) H2O2 levels in HeLa wild type and Grx-3 knockdown cell. Data on the left are mean ± SEM of experimental replicates (n = 3). (D) H2O2 levels in 293 cells pretreated with 1 mM NAc. The H2O2 level was measured after 1 hour of incubation with DHT/CIDAP. Data on the left are mean ± SEM of experimental replicates (n = 3). The SEM on the H2O2 level in (A) – (D) were calculated using the global fitting program provided by GraphPad Prism.
Quantification of Peroxide Level Changes under Biological Perturbations.
First, we assessed the responsiveness of the CIDAP system to fluctuations in endogenous peroxide levels induced by small molecule treatment. Menadione has been reported to induce H2O2 through intracellular redox cycling.55,56 DHT and CIDAP were added to HeLa cells pretreated with menadione. Our findings revealed a significant increase in H2O2 level from 10 μM to 21 μM upon menadione treatment (Figure 3B).
Furthermore, we aim to explore the utility of CIDAP to evaluate peroxide level changes due to genetic perturbations. Previous research has demonstrated that mammalian glutaredoxin 3 (Grx3) plays a critical role in maintaining cellular redox homeostasis, and its downregulation results in high oxidative stress.57 To assess the potential applicability of our system in such biological contexts, we applied it to both wild-type and Grx3 knockdown HeLa cells. Our results showed a significant increase in peroxide levels from 10 μM in the parental cells to approximately 39 μM in the Grx3 knockdown cells (Figure 3C), thereby demonstrating the feasibility of this system in quantifying peroxide levels upon genetic perturbations.
Moreover, the downregulation of peroxide level was also assessed with CIDAP system. N-acetylcysteine (NAc), a well-established antioxidant molecule, was utilized to scavenger ROS within the cells.58,59 Pre-treatment of cells within 1 mM NAc prior to the addition of DHT and CIDAP resulted in a proximate one-fold reduction in peroxide levels as determined by our CIDAP system. This demonstrates the versatility of the CIDAP system, which is capapble of quantifying both increases and decreases in peroxide concentration effectively. (Figure 3D)
A Simulation Model for CIDAP to Validate Experimental Measurements.
A previous study using protein-based peroxide sensor, such as HyPer, estimated the endogenous basal level of H2O2 is in the low nM range for K562 cells,60 which is in disagreement with our measurements of micromolar peroxide levels in typical cancer cell lines. One possible explanation for the discrepancy can be that HyPer is mainly localized in the cytoplasm with no access to peroxide inside organelles like mitochondria or peroxisomes, which generate the majority of H2O2 inside cells. As a small molecule, CIDAP can diffuse freely through membranes. The results from CIDAP are considered an overall resting status of peroxide in the whole cellular environment instead of one particular compartment. To corroborate our experimental measurements, we developed Matlab Simbiology models to simulate the entire process, incorporating all possible steps and the corresponding kinetic parameters from the treatment of the compounds to the final reconstitution of nLuc led by AR dimerization (Figures S8 and S9, Tables S2 and S3). The kinetic parameters used in this simulation are based on our measurements and values reported in the literature. If the peroxide level were set to 10 nM (i.e., the pseudo-first-order reaction law is no longer applicable), it would take more than 8 hours to achieve signal saturation at 10 nM of CIDAP concentration, (Figure S10), which contradicted to the experimental results, in which 2 h is sufficient to achieve signal saturation with high concentrations of probes. In another word, if the intracellular H2O2 concentrations were in the low nM range under our experimental conditions, we would not observe the results in Figures 2 and 3. The simulation results also revealed that higher peroxide levels in the μM range resulted in a leftward shift of the simulated CIDAP curves at 2 h, which agrees with our experimental data.
Additionally, we tested the effect of compound permeability rate crossing the cell membrane by varying the permeability half-life of the compound from 1 minute to 20 minutes and found that it had a minimal impact on the simulation results (Figure S11). This was also experimentally validated by treating cells with 0.05% digitonin to permeabilize cells (Figure S12). Notably, no significant difference in luminescence signals were observed for DHT induced AR dimerization with or without digitonin treatment, suggesting rapid transmembrane permeability for DHT.
Overall, our simulation model corroborates our experimental results. We tentatively attribute the different H2O2 levels measured in our and previous studies to the difference in experimental conditions and cell lines used. Additionally, two key cysteine residues in HyPer form a disulfide bond upon reacting with H2O2. And the oxidized form of HyPer can be reduced by thioredoxins in an NADPH dependent manner.61 So Hyper could potentially serve as a catalyst for H2O2 decomposition at the expense of NADPH. Therefore, we are uncertain whether the low nM H2O2 levels in cells are indeed perturbed by HyPer. In contrast, CIDAP reacts with H2O2 irreversibly and does not catalyze H2O2 decomposition. CIDAP is not expected to significantly change the H2O2 levels due to the low concentrations of CIDAP used.
Discussion
The Lippert group reported an elegant kinetics-based quantification method for peroxynitrite in solution.62 Overall, the fluorescent probe field for reactive species still remains at a qualitative stage. Our work provides a proof-of-concept strategy to quantify these low abundance reactive species.
It should be noted that in all our plots, we assume the DHT and CIDAP concentrations are the same inside and outside of cells. However, these hydrophobic molecules may preferentially accumulate inside cells. If assuming the enrichment ratios for DHT and CIDAP are the same and the probe concentration in cells is less than 10% of H2O2 concentration to satisfy the pseudo-first-order assumption, our calculated H2O2 levels will still hold. If the enrichment ratios for DHT and CIDAP are not the same, this will introduce a systematic error for the computed H2O2 concentrations.
Although we have successfully demonstrated the capability of quantitatively measuring peroxide using the CIDAP system, there is still room for improvement. Primarily, the entire quantification process relies on the rate constant measured in PBS (pH 7.4) at 37°C. However, this rate may vary within a cellular environment due to factors such as salt concentrations and localized pH variations, introducing potential errors. However, measuring the actual rate constant of CIDAP within cells is challenging due to our inability to precisely control intracellular H2O2 levels.
The system can only measure the average concentration of peroxide and cannot capture rapid and transient changes in live cells. The main factor underlying this limitation is the slow reaction kinetics between the boronic ester in CIDAP and H2O2, with a bimolecular reaction rate constant of only ~200 M−1min−1 (3.33 M−1s−1). It requires 2 hours of incubation to achieve sufficient signal-to-noise ratios. Therefore, the measured H2O2 levels are indeed the averaged levels during the experimental time frame. Thus, when applying this system to determine the variation in peroxide levels among different cellular conditions, the fold variations between conditions are more accurate and relevant, as compared to the absolute numerical output produced by the CIDAP system. Ideally, a probe featuring faster kinetics and enabling completion of the assay within 5 minutes, such as Hyper, would provide a more precise measurement of peroxide level changes in real time.
A potential issue for the CIDAP system pertains to the impact on intrinsic peroxide levels due to androgen and the overexpression of AR, especially in hormone-associated systems like prostate cancer and breast cancer.63-66 In such scenarios, the peroxide levels obtained from the CIDAP system may not accurately represent the natural peroxide levels in these systems. To overcome this limitation, other non-hormone based chemically induced dimerization systems can be considered.
When discussing the measurement and quantification of peroxide, it is impossible to overlook the significance of HyPer and roGFP2-Tsa2ΔCR fluorescent proteins.67-71 These genetically encoded, oxidant-sensitive fluorescent probes have revolutionized the field due to their unique features. Hyper reacts rapidly and allows real-time monitoring of H2O2 level in vivo, while also enabling compartmental resolution through genetic engineering. The latest version, HyPer7, has surpassed its predecessors by addressing the issue of pH sensitivity and is highly sensitive to small changes in peroxide levels.70 With meticulous calibration, HyPer can quantify peroxide levels at the nanomolar level using flow cytometry in K562 cells.60 This result is significantly lower than the concentration detected using the CIDAP system. One possible explanation is that HyPer primarily localizes in the cytoplasm, thus potentially reflecting the overall concentration of cytosolic peroxide. In contrast, CIDAP, as a small molecule, can freely distribute between the cytoplasm and subcellular organelles. Therefore, it represents the overall peroxide level in the entire cellular environment, including organelles such as mitochondria and peroxisomes, which have high peroxide levels. To demonstrate CIDAP's capability of penetrating subcellular organelles, CIDAP was incubated with freshly prepared mitochondria from mouse livers for 2 hours. Results showed that CIDAP is permeable to mitochondrial membranes and detected inside the mitochondria (Figure S13). Furthermore, DHT was also detected within the mitochondria, indicating that portion of CIDAP had already reacted with mitochondrial peroxide (Figure S13).
Additionally, HyPer7 can be easily saturated for moderate to high concentrations of H2O2, limiting its application in pathological conditions. In a comparative study wherein cells expressing either Hyper7 or AR-NanoBit were exposed to varying peroxide concentrations ranging from 0-50 μM, the results from the Hyper7 group aligned consistently with prior reported findings. The observed signal was easily detectable and dosage-dependent within the 0-10 μM peroxide range (Figure S7). However, at peroxide levels surpassing 20 μM, the signal reached saturation and overlapped with the signal observed at 10 μM. In contrast, the CIDAP system demonstrated unique performance characteristics. At lower peroxide levels, signal distinctions were less discernible, yet they became increasingly marked as the peroxide concentration exceeded 10 μM.
This is where the CIDAP system presents itself as an invaluable tool that can adapt seamlessly across a wide range of peroxide levels – particularly within systems with elevated oxidative stress. It should be noted though that whilst the CIDAP system serves as an exceptional alternative for conditions involving heightened peroxide levels surpassing the detection limits of HyPer, it does not replace them entirely. Furthermore, the main purpose of the CIDAP system is to serve as a platform to demonstrate the potential applications of this system on low-abundance analytes in general, rather than being specifically fine-tuned for H2O2 measurement.
Conclusions
In summary, we presented a novel amplifiable system for the quantitative measurement of peroxide. Through rigorous testing and evaluation, our innovative approach effectively met all the criteria we established for developing probes designated to analytes in low abundance. The system demonstrated sufficient flexibility and sensitivity across multiple cell lines, which further emphasizes its adaptability to diverse biological systems. Besides, the low concentration of probe required for the assay introduces minimal disturbance and toxicity in the cellular environment, which suggests potential applications to a broad range of biological systems to investigate H2O2 in redox signaling processes and disease development. Our findings provide proof-of-concept evidence for the feasibility of this approach and shed light on the further improvement of probes for low-abundance analytes. With our strategy being extendable to measure other redox such as H2S and NO, this work serves as a testament to the significance of creating sensitive yet flexible tools capable of investigating essential but elusive analytes involved in intricate and dynamic processes like redox signaling.
Supplementary Material
ACKNOWLEDGMENT
The authors appreciated Dr. Elisa Michelini from University of Bologna for providing the AR-NanoBit plasmids, Dr. Chad Johnston from Baylor College of Medicine for the help on high-resolution mass spectrometry, and Dr. Wenshe Liu at Texas A&M University for raising the potential issue of spontaneous carbonate hydrolysis in cells.
Funding Sources
The research was supported in part by National Institutes of Health (R01-GM115622, and R01-CA250503 to J.W.), the Welch Foundation (Q1912 to M.C.W.), Howard Hughes Medical Institute (to M.C.W.), and the Michael E. DeBakey, M.D., Professorship in Pharmacology (to J.W.).
Footnotes
Supporting Information. The Supporting Information is available free of charge via the Internet at http://pubs.acs.org.” Additional experimental details, materials, and methods, including synthesis procedures, in vitro biochemical assays, cell-based assays, density functional theory calculations, MatLab SimBiology simulation, and 1H NMR, 13C NMR and high resolution mass spec spectra for all compounds.
Conflicts of interest
J.W. is the co-founder Chemical Biology Probes LLC. And is a consultant for CoRegen Inc. D.M.L. is the co-founder of CoRegen Inc.
REFERENCES
- (1).Grynkiewicz G; Poenie M; Tsien RY A New Generation of Ca2+ Indicators with Greatly Improved Fluorescence Properties. J Biol Chem 1985, 260 (6), 3440–3450. [PubMed] [Google Scholar]
- (2).Jiang X; Yu Y; Chen J; Zhao M; Chen H; Song X; Matzuk AJ; Carroll SL; Tan X; Sizovs A; Cheng N; Wang MC; Wang J Quantitative Imaging of Glutathione in Live Cells Using a Reversible Reaction-Based Ratiometric Fluorescent Probe. ACS Chem Biol 2015, 10 (3), 864–874. 10.1021/cb500986w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Jiang X; Chen J; Bajic A; Zhang C; Song X; Carroll SL; Cai ZL; Tang M; Xue M; Cheng N; Schaaf CP; Li F; MacKenzie KR; Ferreon ACM; Xia F; Wang MC; Maletic-Savatic M; Wang J Quantitative Real-Time Imaging of Glutathione. Nature communications 2017, 8, 16087. 10.1038/ncomms16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Chen J; Jiang X; Zhang C; MacKenzie KR; Stossi F; Palzkill T; Wang MC; Wang J Reversible Reaction-Based Fluorescent Probe for Real-Time Imaging of Glutathione Dynamics in Mitochondria. ACS sensors 2017, 2 (9), 1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Jiang X; Zhang C; Chen J; Choi S; Zhou Y; Zhao M; Song X; Chen X; Maletic-Savatic M; Palzkill T; Moore D; Wang MC; Wang J Quantitative Real-Time Imaging of Glutathione with Subcellular Resolution. Antioxidants & redox signaling 2019, 30 (16), 1900–1910. 10.1089/ars.2018.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Liu Z; Zhou X; Miao Y; Hu Y; Kwon N; Wu X; Yoon J A Reversible Fluorescent Probe for Real-Time Quantitative Monitoring of Cellular Glutathione. Angewandte Chemie International Edition 2017, 56 (21), 5812–5816. [DOI] [PubMed] [Google Scholar]
- (7).Umezawa K; Yoshida M; Kamiya M; Yamasoba T; Urano Y Rational Design of Reversible Fluorescent Probes for Live-Cell Imaging and Quantification of Fast Glutathione Dynamics. Nature chemistry 2017, 9 (3), 279–286. [DOI] [PubMed] [Google Scholar]
- (8).Jeong EM; Yoon J-H; Lim J; Shin J-W; Cho AY; Heo J; Lee KB; Lee J-H; Lee WJ; Kim H-J; Son YH; Lee S-J; Cho S-Y; Shin D-M; Choi K; Kim I-G Real-Time Monitoring of Glutathione in Living Cells Reveals That High Glutathione Levels Are Required to Maintain Stem Cell Function. Stem Cell Reports 2018, 10 (2), 600–614. 10.1016/j.stemcr.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chen J; Jiang X; Carroll SL; Huang J; Wang J Theoretical and Experimental Investigation of Thermodynamics and Kinetics of Thiol-Michael Addition Reactions: A Case Study of Reversible Fluorescent Probes for Glutathione Imaging in Single Cells. Organic letters 2015, 17 (24), 5978–5981. 10.1021/acs.orglett.5b02910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Olas B; Brodek P; Kontek B The Effect of Hydrogen Sulfide on Different Parameters of Human Plasma in the Presence or Absence of Exogenous Reactive Oxygen Species. Antioxidants 2019, 8 (12), 610. 10.3390/antiox8120610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sies H. Hydrogen Peroxide as a Central Redox Signaling Molecule in Physiological Oxidative Stress: Oxidative Eustress. Redox Biology 2017, 11, 613–619. 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Huang BK; Sikes HD Quantifying Intracellular Hydrogen Peroxide Perturbations in Terms of Concentration. Redox Biology 2014, 2, 955–962. 10.1016/j.redox.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Whiteman M; Moore PK Hydrogen Sulfide and the Vasculature: A Novel Vasculoprotective Entity and Regulator of Nitric Oxide Bioavailability? Journal of Cellular and Molecular Medicine 2009, 13 (3), 488–507. 10.1111/j.1582-4934.2009.00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Furne J; Saeed A; Levitt MD Whole Tissue Hydrogen Sulfide Concentrations Are Orders of Magnitude Lower than Presently Accepted Values. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology 2008, 295 (5), R1479–R1485. 10.1152/ajpregu.90566.2008. [DOI] [PubMed] [Google Scholar]
- (15).Hall CN; Garthwaite J What Is the Real Physiological NO Concentration in Vivo? Nitric Oxide 2009, 21 (2), 92–103. 10.1016/j.niox.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lippert AR; Van de Bittner GC; Chang CJ Boronate Oxidation as a Bioorthogonal Reaction Approach for Studying the Chemistry of Hydrogen Peroxide in Living Systems. Acc. Chem. Res 2011, 44 (9), 793–804. 10.1021/ar200126t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chen W; Pacheco A; Takano Y; Day JJ; Hanaoka K; Xian M A Single Fluorescent Probe to Visualize Hydrogen Sulfide and Hydrogen Polysulfides with Different Fluorescence Signals. Angew. Chem 2016, 128 (34), 10147–10150. 10.1002/ange.201604892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kojima H; Nakatsubo N; Kikuchi K; Kawahara S; Kirino Y; Nagoshi H; Hirata Y; Nagano T Detection and Imaging of Nitric Oxide with Novel Fluorescent Indicators: Diaminofluoresceins. Anal. Chem 1998, 70 (13), 2446–2453. 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- (19).Liu C; Pan J; Li S; Zhao Y; Wu LY; Berkman CE; Whorton AR; Xian M Capture and Visualization of Hydrogen Sulfide by a Fluorescent Probe. Angew. Chem 2011, 123 (44), 10511–10513. 10.1002/ange.201104305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bailey TS; Pluth MD Chemiluminescent Detection of Enzymatically Produced Hydrogen Sulfide: Substrate Hydrogen Bonding Influences Selectivity for H2S over Biological Thiols. J. Am. Chem. Soc 2013, 135 (44), 16697–16704. 10.1021/ja408909h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chen Z; Ai H A Highly Responsive and Selective Fluorescent Probe for Imaging Physiological Hydrogen Sulfide. Biochemistry 2014, 53 (37), 5966–5974. 10.1021/bi500830d. [DOI] [PubMed] [Google Scholar]
- (22).Cao J; Lopez R; Thacker JM; Moon JY; Jiang C; Morris SNS; Bauer JH; Tao P; Mason RP; Lippert AR Chemiluminescent Probes for Imaging H 2 S in Living Animals. Chem. Sci 2015, 6 (3), 1979–1985. 10.1039/C4SC03516J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kagalwala HN; Lippert AR Energy Transfer Chemiluminescent Spiroadamantane 1,2-Dioxetane Probes for Bioanalyte Detection and Imaging. Angew Chem Int Ed 2022, 61 (42). 10.1002/anie.202210057. [DOI] [PubMed] [Google Scholar]
- (24).Schäferling M; Grögel DBM; Schreml S Luminescent Probes for Detection and Imaging of Hydrogen Peroxide. Microchim Acta 2011, 174 (1–2), 1–18. 10.1007/s00604-011-0606-3. [DOI] [Google Scholar]
- (25).Van de Bittner GC; Dubikovskaya EA; Bertozzi CR; Chang CJ In Vivo Imaging of Hydrogen Peroxide Production in a Murine Tumor Model with a Chemoselective Bioluminescent Reporter. Proc. Natl. Acad. Sci. U.S.A 2010, 107 (50), 21316–21321. 10.1073/pnas.1012864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Van de Bittner GC; Bertozzi CR; Chang CJ Strategy for Dual-Analyte Luciferin Imaging: In Vivo Bioluminescence Detection of Hydrogen Peroxide and Caspase Activity in a Murine Model of Acute Inflammation. J. Am. Chem. Soc 2013, 135 (5), 1783–1795. 10.1021/ja309078t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Miller EW; Albers AE; Pralle A; Isacoff EY; Chang CJ Boronate-Based Fluorescent Probes for Imaging Cellular Hydrogen Peroxide. J. Am. Chem. Soc 2005, 127 (47), 16652–16659. 10.1021/ja054474f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Spangler B; Morgan CW; Fontaine SD; Vander Wal MN; Chang CJ; Wells JA; Renslo AR A Reactivity-Based Probe of the Intracellular Labile Ferrous Iron Pool. Nat Chem Biol 2016, 12 (9), 680–685. 10.1038/nchembio.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yik-Sham Chung C; Timblin GA; Saijo K; Chang CJ Versatile Histochemical Approach to Detection of Hydrogen Peroxide in Cells and Tissues Based on Puromycin Staining. J Am Chem Soc 2018, 140 (19), 6109–6121. 10.1021/jacs.8b02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pestka S. [45] Peptidyl-Puromycin Synthesis on Polyribosomes from Escherichia Coli. In Methods in Enzymology; Elsevier, 1974; Vol. 30, pp 470–479. 10.1016/0076-6879(74)30047-X. [DOI] [PubMed] [Google Scholar]
- (31).Dinos G. Deacylated tRNA Is Released from the E Site upon A Site Occupation but before GTP Is Hydrolyzed by EF-Tu. Nucleic Acids Research 2005, 33 (16), 5291–5296. 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Fahnestock S; Neumann H; Shashoua V; Rich A Ribosome-Catalyzed Ester Formation. Biochemistry 1970, 9 (12), 2477–2483. 10.1021/bi00814a013. [DOI] [PubMed] [Google Scholar]
- (33).Pestka S; Goorha R; Rosenfeld H; Neurath C; Hintikka H Studies on Transfer Ribonucleic Acid-Ribosome Complexes. Journal of Biological Chemistry 1972, 247 (13), 4258–4263. 10.1016/S0021-9258(19)45069-2. [DOI] [PubMed] [Google Scholar]
- (34).Lonard DM; O’Malley BW Nuclear Receptor Coregulators: Modulators of Pathology and Therapeutic Targets. Nat Rev Endocrinol 2012, 8 (10), 598–604. 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Klinge CM Estrogen Receptor Interaction with Estrogen Response Elements. Nucleic Acids Research 2001, 29 (14), 2905–2919. 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Andruska N; Mao C; Cherian M; Zhang C; Shapiro DJ Evaluation of a Luciferase-Based Reporter Assay as a Screen for Inhibitors of Estrogen-ERα-Induced Proliferation of Breast Cancer Cells. SLAS Discovery 2012, 17 (7), 921–932. 10.1177/1087057112442960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Miller EW; Tulyathan O; Isacoff EY; Chang CJ Molecular Imaging of Hydrogen Peroxide Produced for Cell Signaling. Nat Chem Biol 2007, 3 (5), 263–267. 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- (38).Agoulnik IU; Krause WC; Bingman WE; Rahman HT; Amrikachi M; Ayala GE; Weigel NL Repressors of Androgen and Progesterone Receptor Action. Journal of Biological Chemistry 2003, 278 (33), 31136–31148. 10.1074/jbc.M305153200. [DOI] [PubMed] [Google Scholar]
- (39).Allen RG; Tresini M Oxidative Stress and Gene Regulation. Free Radical Biology and Medicine 2000, 28 (3), 463–499. 10.1016/S0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- (40).Morel Y; Barouki R Repression of Gene Expression by Oxidative Stress. Biochem J 1999, 342 (Pt 3), 481–496. [PMC free article] [PubMed] [Google Scholar]
- (41).Guo W-H; Qi X; Yu X; Liu Y; Chung C-I; Bai F; Lin X; Lu D; Wang L; Chen J; Su LH; Nomie KJ; Li F; Wang MC; Shu X; Onuchic JN; Woyach JA; Wang ML; Wang J Enhancing Intracellular Accumulation and Target Engagement of PROTACs with Reversible Covalent Chemistry. Nat Commun 2020, 11 (1), 4268. 10.1038/s41467-020-17997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Yu X; Guo W-H; Lin H; Cheng R; Monroy EY; Jin F; Ding L; Lu D; Qi X; Wang MC; Wang J Discovery of a Potent BTK and IKZF1/3 Triple Degrader through Reversible Covalent BTK PROTAC Development. Curr Res Chem Biol 2022, 2, 100029. 10.1016/j.crchbi.2022.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Gerry CJ; Schreiber SL Unifying Principles of Bifunctional, Proximity-Inducing Small Molecules. Nature Chemical Biology 2020, 16 (4), 369–378. 10.1038/s41589-020-0469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Grino PB; Griffin JE; Wilson JD Testosterone at High Concentrations Interacts with the Human Androgen Receptor Similarly to Dihydrotestosterone*. Endocrinology 1990, 126 (2), 1165–1172. 10.1210/endo-126-2-1165. [DOI] [PubMed] [Google Scholar]
- (45).He B; Kemppainen JA; Voegel JJ; Gronemeyer H; Wilson EM Activation Function 2 in the Human Androgen Receptor Ligand Binding Domain Mediates Interdomain Communication with the NH2-Terminal Domain. Journal of Biological Chemistry 1999, 274 (52), 37219–37225. 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- (46).Langley E; Kemppainen JA; Wilson EM Intermolecular NH2-/Carboxyl-Terminal Interactions in Androgen Receptor Dimerization Revealed by Mutations That Cause Androgen Insensitivity. Journal of Biological Chemistry 1998, 273 (1), 92–101. 10.1074/jbc.273.1.92. [DOI] [PubMed] [Google Scholar]
- (47).Hall MP; Unch J; Binkowski BF; Valley MP; Butler BL; Wood MG; Otto P; Zimmerman K; Vidugiris G; Machleidt T; Robers MB; Benink HA; Eggers CT; Slater MR; Meisenheimer PL; Klaubert DH; Fan F; Encell LP; Wood KV Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem. Biol 2012, 7 (11), 1848–1857. 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Suzuki K; Kimura T; Shinoda H; Bai G; Daniels MJ; Arai Y; Nakano M; Nagai T Five Colour Variants of Bright Luminescent Protein for Real-Time Multicolour Bioimaging. Nat Commun 2016, 7 (1), 13718. 10.1038/ncomms13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Khare-Pandit S; Hartwell K; Markland W Probing Myc and Max Protein-Protein Interactions using NanoBRETTM and NanoBiT® Assays. https://www.promega.com/resources/pubhub/2017/probing-myc-and-max-protein-protein-interaction-using-nanobret-and-nanobit-assays/ (accessed 2023-03-23). [Google Scholar]
- (50).Calabretta MM; Lopreside A; Montali L; Cevenini L; Roda A; Michelini E A Genetically Encoded Bioluminescence Intracellular Nanosensor for Androgen Receptor Activation Monitoring in 3D Cell Models. Sensors 2021, 21 (3), 893. 10.3390/s21030893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Habara M; Sato Y; Goshima T; Sakurai M; Imai H; Shimizu H; Katayama Y; Hanaki S; Masaki T; Morimoto M; Nishikawa S; Toyama T; Shimada M FKBP52 and FKBP51 Differentially Regulate the Stability of Estrogen Receptor in Breast Cancer. Proc. Natl. Acad. Sci. U.S.A 2022, 119 (15), e2110256119. 10.1073/pnas.2110256119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Nadal M; Prekovic S; Gallastegui N; Helsen C; Abella M; Zielinska K; Gay M; Vilaseca M; Taulès M; Houtsmuller AB; van Royen ME; Claessens F; Fuentes-Prior P; Estébanez-Perpiñá E Structure of the Homodimeric Androgen Receptor Ligand-Binding Domain. Nat Commun 2017, 8 (1), 14388. 10.1038/ncomms14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Wang H; Schoebel S; Schmitz F; Dong H; Hedfalk K Characterization of Aquaporin-Driven Hydrogen Peroxide Transport. Biochimica et Biophysica Acta (BBA) - Biomembranes 2020, 1862 (2), 183065. 10.1016/j.bbamem.2019.183065. [DOI] [PubMed] [Google Scholar]
- (54).Yang NJ; Hinner MJ Getting Across the Cell Membrane: An Overview for Small Molecules, Peptides, and Proteins. In Site-Specific Protein Labeling; Gautier A, Hinner MJ, Eds.; Methods in Molecular Biology; Springer New York: New York, NY, 2015; Vol. 1266, pp 29–53. 10.1007/978-1-4939-2272-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Loor G; Kondapalli J; Schriewer JM; Chandel NS; Vanden Hoek TL; Schumacker PT Menadione Triggers Cell Death through ROS-Dependent Mechanisms Involving PARP Activation without Requiring Apoptosis. Free Radical Biology and Medicine 2010, 49 (12), 1925–1936. 10.1016/j.freeradbiomed.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Kelts JL; Cali JJ; Duellman SJ; Shultz J Altered Cytotoxicity of ROS-Inducing Compounds by Sodium Pyruvate in Cell Culture Medium Depends on the Location of ROS Generation. SpringerPlus 2015, 4 (1), 269. 10.1186/s40064-015-1063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Pham K; Pal R; Qu Y; Liu X; Yu H; Shiao SL; Wang X; O’Brian Smith E; Cui X; Rodney GG; Cheng N Nuclear Glutaredoxin 3 Is Critical for Protection against Oxidative Stress-Induced Cell Death. Free Radical Biology and Medicine 2015, 85, 197–206. 10.1016/j.freeradbiomed.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Liu X; Wang L; Cai J; Liu K; Liu M; Wang H; Zhang H N-Acetylcysteine Alleviates H2O2-Induced Damage via Regulating the Redox Status of Intracellular Antioxidants in H9c2 Cells. Int J Mol Med 2018. 10.3892/ijmm.2018.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Zhitkovich A N -Acetylcysteine: Antioxidant, Aldehyde Scavenger, and More. Chem. Res. Toxicol 2019, 32 (7), 1318–1319. 10.1021/acs.chemrestox.9b00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Lyublinskaya O; Antunes F Measuring Intracellular Concentration of Hydrogen Peroxide with the Use of Genetically Encoded H2O2 Biosensor HyPer. Redox Biology 2019, 24, 101200. 10.1016/j.redox.2019.101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Kritsiligkou P; Shen TK; Dick TP A Comparison of Prx- and OxyR-Based H2O2 Probes Expressed in S. Cerevisiae. Journal of Biological Chemistry 2021, 297 (1), 100866. 10.1016/j.jbc.2021.100866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Kim YL; Plank JT; Li B; Lippert AR Kinetics-Based Quantification of Peroxynitrite Using the Oxidative Decarbonylation of Isatin. Anal. Chem 2022, 94 (51), 17803–17809. 10.1021/acs.analchem.2c03474. [DOI] [PubMed] [Google Scholar]
- (63).Lu JP; Monardo L; Bryskin I; Hou ZF; Trachtenberg J; Wilson BC; Pinthus JH Androgens Induce Oxidative Stress and Radiation Resistance in Prostate Cancer Cells Though NADPH Oxidase. Prostate Cancer Prostatic Dis 2010, 13 (1), 39–46. 10.1038/pcan.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Shiota M; Yokomizo A; Naito S Pro-Survival and Anti-Apoptotic Properties of Androgen Receptor Signaling by Oxidative Stress Promote Treatment Resistance in Prostate Cancer. Endocrine-Related Cancer 2012, 19 (6), R243–R253. 10.1530/ERC-12-0232. [DOI] [PubMed] [Google Scholar]
- (65).Feng T; Zhao R; Sun F; Lu Q; Wang X; Hu J; Wang S; Gao L; Zhou Q; Xiong X; Dong X; Wang L; Han B TXNDC9 Regulates Oxidative Stress-Induced Androgen Receptor Signaling to Promote Prostate Cancer Progression. Oncogene 2020, 39 (2), 356–367. 10.1038/s41388-019-0991-3. [DOI] [PubMed] [Google Scholar]
- (66).Michmerhuizen AR; Spratt DE; Pierce LJ; Speers CW ARe We There yet? Understanding Androgen Receptor Signaling in Breast Cancer. NPJ Breast Cancer 2020, 6, 47. 10.1038/s41523-020-00190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Belousov VV; Fradkov AF; Lukyanov KA; Staroverov DB; Shakhbazov KS; Terskikh AV; Lukyanov S Genetically Encoded Fluorescent Indicator for Intracellular Hydrogen Peroxide. Nat Methods 2006, 3 (4), 281–286. 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- (68).Markvicheva KN; Bilan DS; Mishina NM; Gorokhovatsky A. Yu.; Vinokurov LM; Lukyanov S; Belousov VV A Genetically Encoded Sensor for H2O2 with Expanded Dynamic Range. Bioorganic & Medicinal Chemistry 2011, 19 (3), 1079–1084. 10.1016/j.bmc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- (69).Bilan DS; Pase L; Joosen L; Gorokhovatsky A. Yu.; Ermakova YG; Gadella TWJ; Grabher C; Schultz C; Lukyanov S; Belousov VV HyPer-3: A Genetically Encoded H 2 O 2 Probe with Improved Performance for Ratiometric and Fluorescence Lifetime Imaging. ACS Chem. Biol 2013, 8 (3), 535–542. 10.1021/cb300625g. [DOI] [PubMed] [Google Scholar]
- (70).Pak VV; Ezeriņa D; Lyublinskaya OG; Pedre B; Tyurin-Kuzmin PA; Mishina NM; Thauvin M; Young D; Wahni K; Martínez Gache SA; Demidovich AD; Ermakova YG; Maslova YD; Shokhina AG; Eroglu E; Bilan DS; Bogeski I; Michel T; Vriz S; Messens J; Belousov VV Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metabolism 2020, 31 (3), 642–653.e6. 10.1016/j.cmet.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Morgan B; Van Laer K; Owusu TNE; Ezeriņa D; Pastor-Flores D; Amponsah PS; Tursch A; Dick TP Real-Time Monitoring of Basal H2O2 Levels with Peroxiredoxin-Based Probes. Nat Chem Biol 2016, 12 (6), 437–443. 10.1038/nchembio.2067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


