Abstract
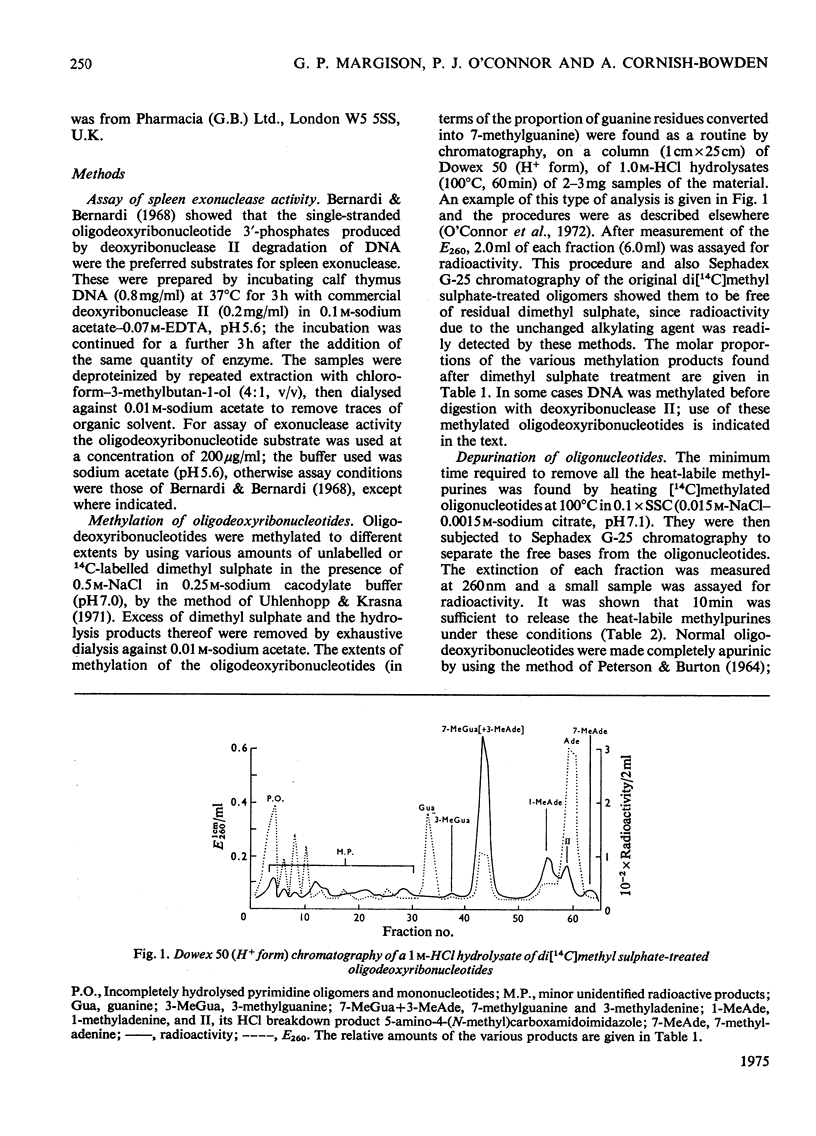
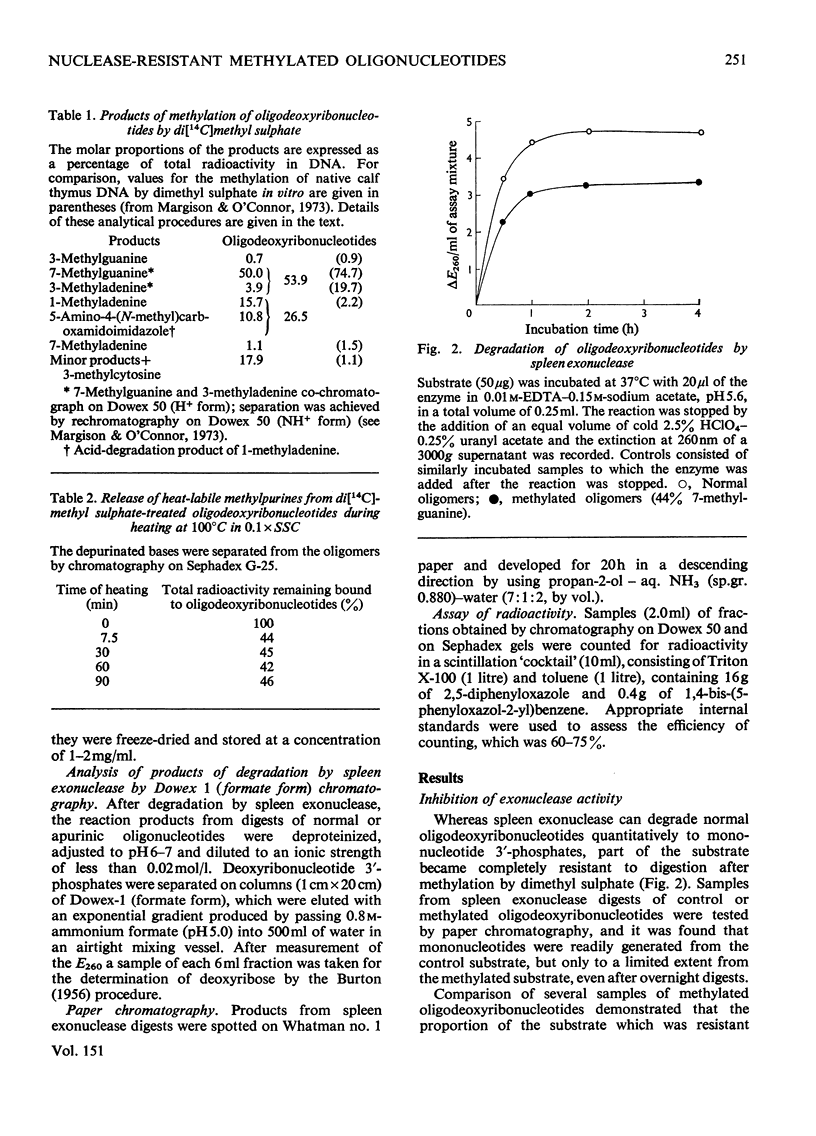
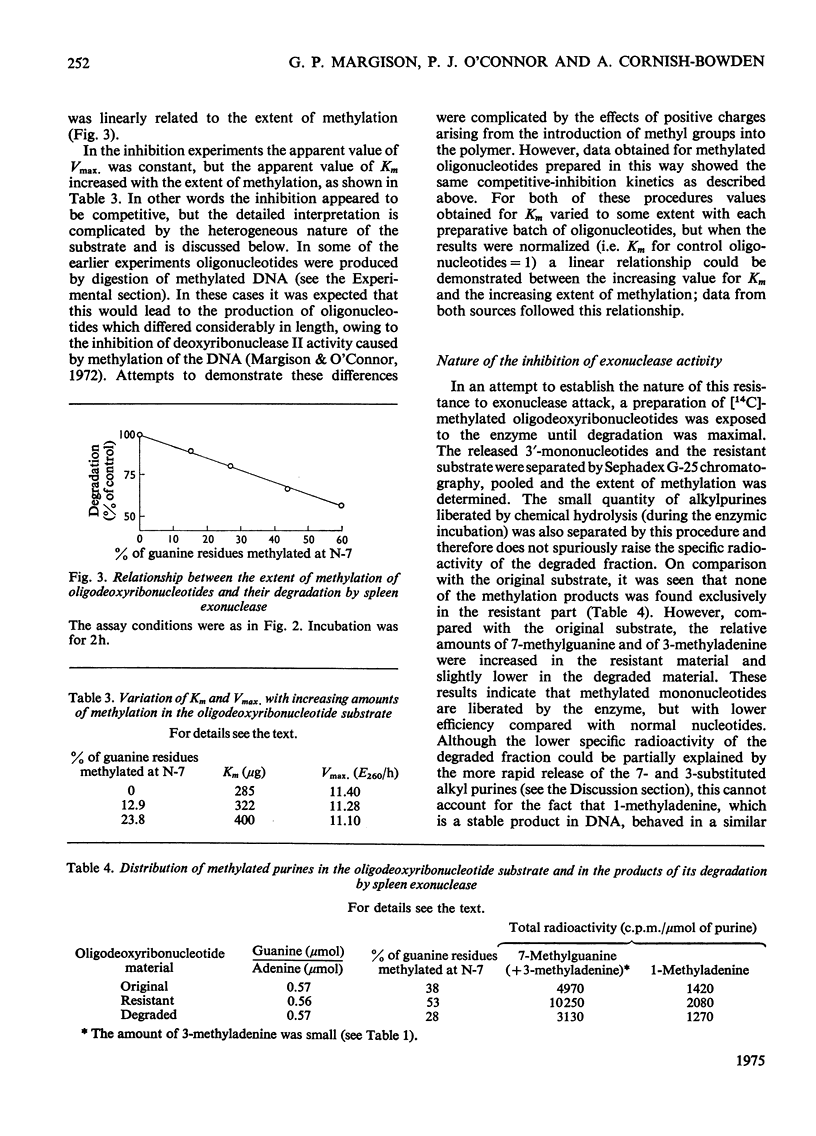
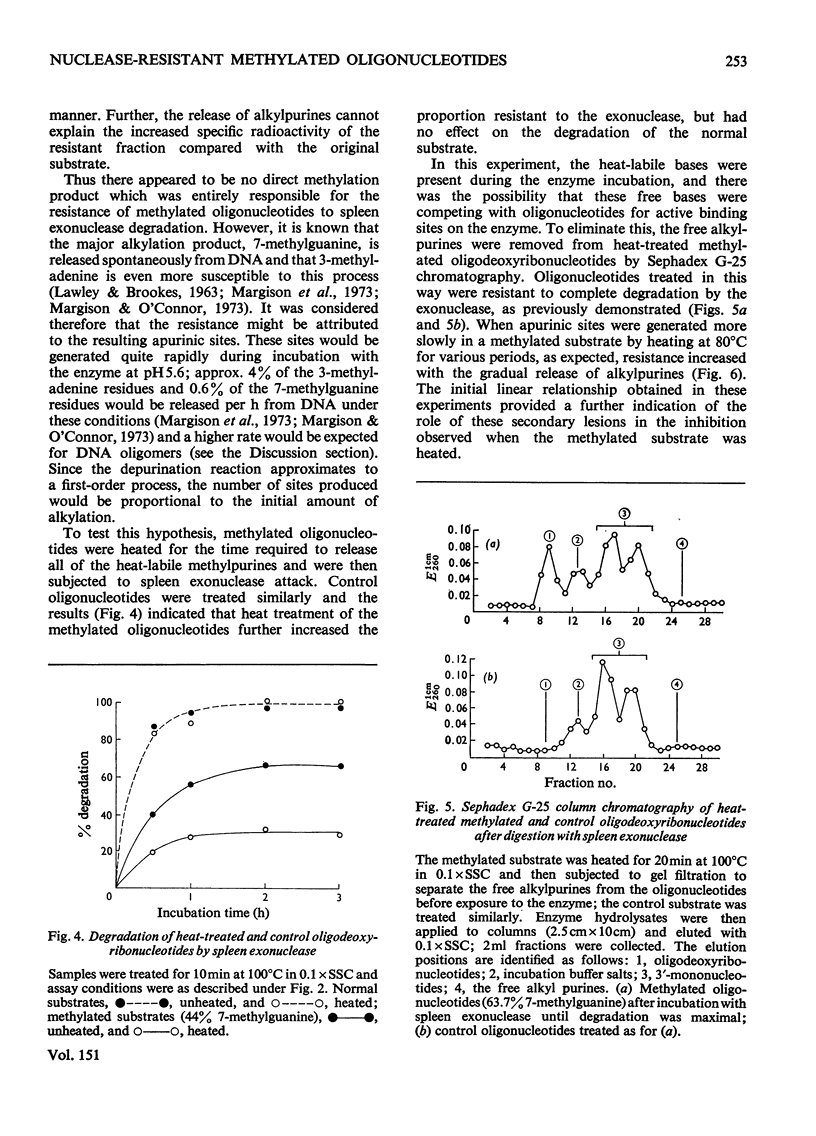
The effect of introducing methyl groups into DNA substrates was studied by using the spleen exonuclease (EC 3.1.4.1), an enzyme which hydrolyses oligonucleotides in a sequential manner by splitting off 3'-phosphomononucleotides starting from the 5'-hydroxyl terminus. Analyses of oligodeoxyribonucleotide 3'-phosphate substrates after reaction in vitro with dimethyl sulphate demonstrated that the resultant methylation pattern differed from the previously found for native DNA, particularly with respect to the relative amounts of 1- and 3-methyladenine produced. Although after treatment with increasing amounts of dimethyl sulphate the substrate became progressively resistant to degradation by the exonuclease, the methylation products themselves were only partially responsible for the observed inhibition of enzyme activity. The incomplete degradation encountered was apparently due to the presence of apurinic sites, which arose as secondary lesions after the spontaneous release of the labile alkyl purines from the methylated substrate. Inhibition of enzyme activity appeared to be competitive, being characterized by constant values for apparent Vmax, and increased values for apparent Km. the interpretation of this, however, is complicated by the complex nature of the substrate, and these aspects are considered in some detail.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon P., Verly W. Alkylation of phosphates and stability of phosphate triesters in DNA. Eur J Biochem. 1972 Nov 21;31(1):103–111. doi: 10.1111/j.1432-1033.1972.tb02506.x. [DOI] [PubMed] [Google Scholar]
- Bernardi A., Bernardi G. Studies on acid hydrolases. IV. Isolation and characterization of spleen exonuclease. Biochim Biophys Acta. 1968 Feb 26;155(2):360–370. [PubMed] [Google Scholar]
- Burnotte J., Verly W. G. A kinetic approach to the mechanism of deoxyribonucleic acid cross-linking by HNO 2 . J Biol Chem. 1971 Oct 10;246(19):5914–5918. [PubMed] [Google Scholar]
- Cordonnier C., Bernardi G. A comparative study of acid deoxyribonucleases extracted from different tissues and species. Can J Biochem. 1968 Aug;46(8):989–995. doi: 10.1139/o68-148. [DOI] [PubMed] [Google Scholar]
- Hadi S. M., Goldthwait D. A. Endonuclease II of Escherichia coli. Degradation of partially depurinated deoxyribonucleic acid. Biochemistry. 1971 Dec 21;10(26):4986–4993. doi: 10.1021/bi00802a024. [DOI] [PubMed] [Google Scholar]
- Hendler S., Fürer E., Srinivasan P. R. Synthesis and chemical properties of monomers and polymers containing 7-methylguanine and an investigation of their substrate or template properties for bacterial deoxyribonucleic acid or ribonucleic acid polymerases. Biochemistry. 1970 Oct 13;9(21):4141–4153. doi: 10.1021/bi00823a017. [DOI] [PubMed] [Google Scholar]
- Kirtikar D. M., Goldthwait D. A. The enzymatic release of O6-methylguanine and 3-methyladenine from DNA reacted with the carcinogen N-methyl-N-nitrosourea. Proc Natl Acad Sci U S A. 1974 May;71(5):2022–2026. doi: 10.1073/pnas.71.5.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAWLEY P. D., BROOKES P. FURTHER STUDIES ON THE ALKYLATION OF NUCLEIC ACIDS AND THEIR CONSTITUENT NUCLEOTIDES. Biochem J. 1963 Oct;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUDLUM D. B. ALKYLATION OF POLYNUCLEOTIDE COMPLEXES. Biochim Biophys Acta. 1965 Apr 19;95:674–676. doi: 10.1016/0005-2787(65)90525-3. [DOI] [PubMed] [Google Scholar]
- Lawley P. D. Some chemical aspects of dose-response relationships in alkylation mutagenesis. Mutat Res. 1974 Jun;23(3):283–295. doi: 10.1016/0027-5107(74)90102-x. [DOI] [PubMed] [Google Scholar]
- Lesca P. Age variations of acid deoxyribonuclease activity in mouse liver nuclei. Nature. 1968 Oct 5;220(5162):76–77. doi: 10.1038/220076a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Ljungquist S., Lindahl T. A mammalian endonuclease specific for apurinic sites in double-stranded deoxyribonucleic acid. I. Purification and general properties. J Biol Chem. 1974 Mar 10;249(5):1530–1535. [PubMed] [Google Scholar]
- Loveless A. Possible relevance of O-6 alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of nitrosamines and nitrosamides. Nature. 1969 Jul 12;223(5202):206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- Margison G. P., Capps M. J., O'Connor P. J., Craig A. W. Loss of 7-methylguanine from rat liver DNA after methylation in vivo with methylmethanesulphonate or dimethylnitrosamine. Chem Biol Interact. 1973 Feb;6(2):119–124. doi: 10.1016/0009-2797(73)90078-1. [DOI] [PubMed] [Google Scholar]
- Margison G. P., O'Connor P. J. Biological implications of the instability of the N-glycosidic bone of 3-methyldeoxyadenosine in DNA. Biochim Biophys Acta. 1973 Dec 21;331(3):349–356. doi: 10.1016/0005-2787(73)90021-x. [DOI] [PubMed] [Google Scholar]
- Müller W. E., Yamazaki Z., Breter H. J., Zahn R. K. Action of bleomycin on DNA and RNA. Eur J Biochem. 1972 Dec 18;31(3):518–525. doi: 10.1111/j.1432-1033.1972.tb02560.x. [DOI] [PubMed] [Google Scholar]
- O'Connor P. J., Capps M. J., Craig A. W., Lawley P. D., Shah S. A. Differences in the patterns of methylation in rat liver ribosomal ribonucleic acid after reaction in vivo with methyl methanesulphonate and NN-dimethylnitrosamine. Biochem J. 1972 Sep;129(3):519–528. doi: 10.1042/bj1290519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor P. J., Marigison G. P., Craig A. W. Phosphotriesters in rat liver deoxyribonucleic acid after the administration of the carcinogen NN-dimethylnitrosamine in vivo. Biochem J. 1975 Mar;145(3):475–482. doi: 10.1042/bj1450475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette Y., Crine P., Verly W. G. Properties of the endonuclease for depurinated DNA from Escherichia coli. Can J Biochem. 1972 Nov;50(11):1199–1209. doi: 10.1139/o72-163. [DOI] [PubMed] [Google Scholar]
- Petersen G. B., Burton K. Sequences of consecutive cytosine deoxynucleotides in deoxyribonucleic acid. Biochem J. 1964 Sep;92(3):666–672. doi: 10.1042/bj0920666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochon F., Michelson A. M. Polynucleotides. IX. Methylation of nucleic acids, homopolynucleotides and complexes. Biochim Biophys Acta. 1967 Nov 21;149(1):99–106. doi: 10.1016/0005-2787(67)90694-6. [DOI] [PubMed] [Google Scholar]
- Slor H., Lev T. Acid deoxyribonuclease activity in purified calf thymus nuclei. Biochem J. 1971 Aug;123(5):993–995. doi: 10.1042/bj1230993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenhopp E. L., Krasna A. I. Alterations in the structure of deoxyribonucleic acid on chemical methylation. Biochemistry. 1971 Aug 17;10(17):3290–3295. doi: 10.1021/bi00793a020. [DOI] [PubMed] [Google Scholar]
- Verly W. G. Commentary. Monofunctional alkylating agents and apurinic sites in DNA. Biochem Pharmacol. 1974 Jan 1;23(1):3–8. doi: 10.1016/0006-2952(74)90307-4. [DOI] [PubMed] [Google Scholar]
- Verly W. G., Paquette Y. An endonuclease for depurinated DNA in rat liver. Can J Biochem. 1973 Jul;51(7):1003–1009. doi: 10.1139/o73-130. [DOI] [PubMed] [Google Scholar]


