Abstract
The underlying mechanisms explaining the differential course of SARS-CoV-2 infection and the potential clinical consequences after COVID-19 resolution have not been fully elucidated. As a dysregulated mitochondrial activity could impair the immune response, we explored long-lasting changes in mitochondrial functionality, circulating cytokine levels, and metabolomic profiles of infected individuals after symptoms resolution, to evaluate whether a complete recovery could be achieved. Results of this pilot study evidenced that different parameters of aerobic respiration in lymphocytes of individuals recuperated from a severe course lagged behind those shown upon mild COVID-19 recovery, in basal conditions and after simulated reinfection, and they also showed altered glycolytic capacity. The severe groups showed trends to enhanced superoxide production in parallel to lower OPA1-S levels. Unbalance of pivotal mitochondrial fusion (MFN2, OPA1) and fission (DRP1, FIS1) proteins was detected, suggesting a disruption in mitochondrial dynamics, as well as a lack of structural integrity in the electron transport chain. In serum, altered cytokine levels of IL-1β, IFN-α2, and IL-27 persisted long after clinical recovery, and growing amounts of the latter after severe infection correlated with lower basal and maximal respiration, ATP production, and glycolytic capacity. Finally, a trend for higher circulating levels of 3-hydroxybutyrate was found in individuals recovered after severe compared to mild course. In summary, long after acute infection, mitochondrial and metabolic changes seem to differ in a situation of full recovery after mild infection versus the one evolving from severe infection.
Supplementary information
The online version contains supplementary material available at 10.1007/s10565-024-09976-0.
Keywords: COVID-19, SARS-CoV-2, Disease severity, Immune system, Cytokines, Mitochondria, Metabolomics
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) triggered the known disease outbreak in late 2019 (COVID-19) which caused a vast number of deaths worldwide. Clinically, COVID-19 patients presented a wide spectrum of symptoms ranging from asymptomatic or flu-like symptoms to severe pneumonia, respiratory failure, and death. While the symptoms of COVID-19 are usually mild, severe disease results in approximately 10% of the cases, and previous conditions such as diabetes, obesity, chronic lung diseases, organ transplant, immunodeficiency, and cardiovascular comorbidities, as well as older age and male sex, are identified risk factors increasing mortality rates (Rosenthal et al. 2020). Nonetheless, these risk factors do not always explain COVID-19 severity, and the mechanisms underlying this clinical variation have not been fully elucidated. Different, often interconnected processes have been associated with severity, such as an aberrant energy metabolism in peripheral immune cells and an exaggerated release of proinflammatory cytokines (Gibellini et al. 2020).
T cells play a key role in the immune response against viral infections, and functionally exhausted T cells and lymphopenia have been observed specifically during SARS-CoV-2 infection (Delshad et al. 2021). Emerging evidence describes the impairment of viral clearance, performed by T cells and natural killer cells, due to a dysregulated mitochondrial activity (Burtscher et al. 2020; Srinivasan et al. 2021). Mitochondria have been shown to suppress the production of type I interferons, which are critical for antiviral defense (Khan et al. 2015; Soria-Castro et al. 2021). Indeed, a compromised mitochondrial function of T cells during COVID-19 infection contributes to the enhanced inflammatory response observed in patients (Gibellini et al. 2020; Romão et al. 2022). Several mechanisms explain how SARS-CoV-2 affects mitochondrial activity. Among them, the viral disruption of the electron transport chain (ETC) leads to an increased production of reactive oxygen species (ROS). Even though a certain level of ROS regulates immune responses and promotes virus clearance, ROS excess causes cellular damage (Ajaz et al. 2021; De La Cruz-Enriquez et al. 2021). As in other viral infections, COVID-19 is associated with increased production of free radicals owing to mitochondrial dysfunction. This mitochondrial release of ROS activates the inflammasome and promotes the production of proinflammatory cytokines, such as IL-1β and IL-18 (Nakahira et al. 2011; Saleh et al. 2020). Moreover, SARS-CoV-2 infection has been described to hijack the host mitochondria, disturbing the membrane potential of these organelles, which leads to reduced ATP production and increased oxidative stress, inflammation, and tissue damage, all of them implicated in the etiology of the disease (Gibellini et al. 2020). In addition, SARS-CoV-2 infection interferes with mitochondrial dynamics (Díaz-Resendiz et al. 2022), generating morphological changes in mitochondria which affect the innate immune response (Giacomello et al. 2020). Changes in mitochondrial morphology and gene expression have been reported in lung tissues from COVID-19 patients (Duan et al. 2022). A dampened mitochondrial respiration related to immune dysfunction and inflammation has been observed in peripheral blood mononuclear cells (PBMCs) from acute COVID-19 patients (Ajaz et al. 2021).
As shown, the mitochondial involvement during acute infection seems sufficiently supported. We wondered whether an altered mitochondrial response of immune cells might be associated with COVID-19 severity and if detected, how this alteration might change during post-COVID-19 recovery. Our previous data with patients who recovered from an acute condition, specifically heart failure with preserved ejection fraction, showed long-term metabolic changes (Anguita et al. 2022) and some evidence supports a similar scenario after acute COVID-19 (Montefusco et al. 2021). Therefore, further investigation was required to clarify the need for a clinical intervention, follow-up, and/or the potential treatment of these individuals.
Numerous studies describe increased serum levels of pro-inflammatory cytokines in the acute phase of the disease. The cytokine storm in COVID-19 has been related to increased severity and mortality rate, and specific therapeutic strategies target proinflammatory cytokines trying to promote recovery and reduce mortality (Ramasamy and Subbian 2021). In addition, distinct serum protein signatures in long COVID-19 (LC) have been identified (Talla et al. 2023). However, little is known in individuals who recovered after COVID-19 and, to the best of our knowledge, only one study has evaluated their serum cytokine profiles showing similar concentrations to those found in healthy controls (Chi et al. 2020). These results warrant further investigation, as authors did not identify whether the included subjects had suffered severe or mild COVID-19 disease.
The host metabolism shaped under evolutionary pressure is able to respond to endemic viruses, but may be dysfunctional in emerging infections. Recent findings identified glucose restriction as a physiological mechanism to bring the body into a heightened state of responsiveness to viral pathogens, an immune–endocrine circuit disrupted in hyperglycemia (Šestan et al. 2024). The present pilot study seeks to elucidate whether a metabolic recovery runs in parallel to the resolution of COVID-19 symptomatology or else persistent damage caused by SARS-CoV-2 could transcend long after remission of the acute infection.
In summary, the host response to the virus is a major factor directing disease course and we aimed to evaluate long-lasting changes in mitochondrial functionality, circulating cytokine levels, and metabolomic profiles. To this purpose, we analyzed these parameters through comparison in PBMCs of subjects that seemingly recovered from either severe (with Intensive Care Unit, ICU stay) or mild COVID-19. Given the extreme clinical reaction to the virus observed in individuals who suffered a severe COVID-19, we tried to unveil whether differences in the specific response of lymphocytes stimulated by SARS-CoV-2 proteins would still be detected when comparing cells from individuals with past severe and mild courses.
Methods
Study population
The study included a total of 24 subjects (33.3% females) who recovered from COVID-19: 12 severe cases (CSC) and another 12 cases that had experienced mild COVID-19 (CMC), recruited after clinical release and resolution of the symptoms and matched by age, sex, and ethnicity. Known comorbidities reported to increase the risk of severe COVID-19, such as hypertension, cardiovascular disease, or hypothyroidism, precluded recruitment. All CSC and CMC participants were included after informed consent and analyzed between June and October 2022. The study was approved by the Ethics Committee of Hospital Clínico San Carlos.
CSCs admitted to the ICU underwent mechanical respiratory support during their hospital stay, and did not have persistent symptoms of the disease or a diagnosis of long COVID-19 (LC) at the moment of sample collection. Participants in this group were recruited between 5 and 14 months after ICU admission from March to December 2020, with a mean age at sample collection of 54.42 ± 10.7 years. The time to clinical recovery ranged from 4 and 5 months in two participants to the longest recovery time of 13 months for other two cases.
CMCs, with a mean age at recruitment of 54.18 ± 11.8 years, presented common cold symptoms or no symptoms during infection that resolved at home from April 2020 to May 2021, within the year before sample collection when they did not report any symptoms.
Demographic, clinical, and laboratory data of each individual were extracted from electronic medical records (Table 1). Laboratory data included complete blood count, coagulation profile, and different biochemical parameters.
Table 1.
Clinical and demographic data of individuals who recovered after mild (CMC) and severe (CSC) COVID-19
| Normal Range | CMC | CSC | p Value | ||
|---|---|---|---|---|---|
| (n = 11) | (n = 12) | ||||
| Age, mean (± SD), yrs | 54.18 (± 11.8) | 54.42 (± 10.7) | p = 0.961 | ||
| Male, n (%) | 8 (73) | 8 (67) | p = 0.752 | ||
| Ethnicity, n (%) | p = 0.538 | ||||
| • Europe | 8 (73) | 10 (83) | |||
| • South American | 3 (27) | 2 (17) | |||
| Biochemistry [mean/median (± SD)] | |||||
| CRP (mg/dL) | 0.1- 0.5 | 0.3 (± 0.05) | 0.3 (± 0.16) | p = 0.013 | |
| IL-6 | 0.1- 7 | 2.04 (± 1.32) | 2.52 (± 2.09) | p = 0.621 | |
| Ferritin (ng/mL) | 10- 350 | 102.7 (± 118.54) | 78.3 (± 147.46) | p = 0.818 | |
| Cholesterol (mg/dL) | 140- 200 | 212.18 (± 36.50) | 217.33 (± 54.39) | p = 0.794 | |
| Triglycerides (mg/dL) | 30- 150 | 131.75 (± 61.15) | 118.86 (± 37.19) | p = 0.669 | |
| ALT (U/L) | 5- 40 | 25.00 (± 8.10) | 20.00 (± 12.72) | p = 0.254 | |
| AST (U/L) | 5- 40 | 27.91 (± 7.29) | 20.36 (± 5.64) | p = 0.013 | |
| Glucose (mg/dL) | 60- 100 | 101.7 (± 13.86) | 106.75 (± 6,50) | p = 0.274 | |
| Creatinine (mg/dL) | 0.51- 1.17 | 1.02 (± 0.16) | 0.74 (± 0.25) | p = 0.157 | |
| Haemogram and Coagulation [mean/median (± SD)] | |||||
| Hematocrit (%) | 35- 47 | 43.14 (± 3.45) | 43.82 (± 2.14) | p = 0.572 | |
| Erythrocytes (× 1012/L) | 3.8- 5.3 | 4.78 (± 0.33) | 4.89 (± 0.31) | p = 0.414 | |
| Hemoglobin (g/dL) | 11.7- 16 | 14.56 (± 1.40) | 14.82 (± 0.68) | p = 0.569 | |
| Platelet (× 109/L) | 150- 400 | 196.10 (± 25.97) | 240.92 (± 53.61) | p = 0.026 | |
| Prothrombin Time (s) | 9.2- 15.1 | 12.12 (± 0.57) | 11.57 (± 0.50) | p = 0.024 | |
| INR | 0.8- 1.2 | 0.96 (± 0.05) | 0.91 (± 0.04) | p = 0.029 | |
| Thrombin Time (s) | 12- 19 | 16.34 (± 0.87) | 16.14 (± 1.79) | p = 0.736 | |
ALT Alanine transaminase, AST Aspartate transaminase, CRP C reactive protein, INR Standarized prothrombin time ratio, yrs Years. Statistical significance was assessed by two-tailed unpaired Student’s t-test, except for CRP, IL-6, ferritin, ALT and creatinine, which were evaluated by non-parametric Mann–Whitney U tests. Non-parametric data were represented by median and parametric data by mean. Qualitative parameters (sex and ethnicity) were analysed by Pearson’s chi-squared test
Sample collection
For cellular studies, peripheral blood samples were collected and 107 PBMCs cryopreserved in freezing medium {fetal bovine serum (F9665, Sigma Aldrich, St. Louis, MO, US) with 10% dimethylsulfoxide (DMSO, LM-0102-M010.0–005, Biotech, Berlin, Germany)} until further analysis. Serum aliquots were frozen at –80 °C until usage.
Cell metabolism
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were simultaneously evaluated in a Seahorse XFp extracellular flux analyzer (Agilent Technologies, Santa Clara, CA, US). First, PBMCs were counted after thawing to seed 1.5 × 106 cells per mL in RPMI 1640 (15–041-CV, Corning, Corning, NY, US). The next day, another cell counting was carried out to perform the stimulation, as previously described (Stanczak et al. 2021). Specifically, for each participant, cells were stimulated with 2 μg/mL of Spike Peptide Mix (SPM) (PepMix™ of SARS-CoV-2 Spike glycoprotein, PM-WCPV-S-1, JPT Peptide Technologies, Berlin, Germany), 50 U/mL of IL-2 human protein (DA3559, OriGene Technologies, Rockville, MD, US), and 2 μg/mL of CD28 (#16–0289-81, Thermo Fisher Scientific, Waltham, MA, US) for 48 h, to assess the metabolism of stimulated PBMCs, and with DMSO (used to dilute the viral peptide) to test basal metabolism. The Spike PepMix includes a total of 315 14-mer peptides covering the whole spike protein. Then, after 48 h of SPM-stimulation, a rigorous final cell count (6 times per experimental group) was performed, and split in triplicates with 2.5 × 105 PBMCs per well seeded onto poly-D-lysine coated XFp plates (103,022–100, Agilent Technologies, Santa Clara, CA, US), which allowed us to certify reproducibility. Upon a non-carbonated incubation for 30 min at 37ºC, both conditions, basal and SPM-stimulated, were assayed in a Seahorse run for 45 min in XF DMEM medium, pH 7.4 (103,575–100, Agilent Technologies, Santa Clara, CA, US), supplemented with glucose (10 mM), glutamine (2 mM), and pyruvate (1 mM). Mitochondrial and glycolytic functions were then assessed with the Cell MitoStress Test kit (103,010–100, Agilent Technologies, Santa Clara, CA, US), following the manufacturer’s instructions. Briefly, cells were sequentially treated with mitochondrial oxidative phosphorylation (OXPHOS) selective inhibitors: 1.5 μM oligomycin to inhibit ATP synthase and evaluate the ATP production efficiency of the cells; 0.5 μM FCCP (carbonylcyanide-p-trifluoromethoxyphenylhydrazone) which uncoupled mitochondrial respiration to determine the maximal substrate oxidation capacity; and a cocktail of rotenone and antimycin A at 0.5 μM to inhibit mitochondrial respiration, and evaluate non-mitochondrial OCR. Agilent Seahorse Wave allowed to analyze these data, with each parameter calculated as follows: Non-mitochondrial Oxygen Consumption: Minimum OCR measured after rotenone/antimycin A injection. Basal Respiration: (Last OCR measurement before oligomycin injection) – (Non-Mitochondrial Respiration). Maximal Respiration: (Maximum OCR measurement after FCCP injection) – (Non-Mitochondrial Respiration). Proton Leak: (Minimum OCR measurement after oligomycin injection) – (Non-Mitochondrial Respiration). ATP Production: (Last OCR measurement before Oligomycin injection) – (Minimum OCR measurement after Oligomycin injection). Spare Respiratory Capacity: (Maximal Respiration) – (Basal Respiration). Basal Glycolytic Capacity: Last ECAR measurement before Oligomycin injection. Maximal Glycolytic Capacity: Maximum ECAR measurement after FCCP injection.
Flow cytometry
Gating strategies (CytoFLEX cytometer, Beckman Coulter, Brea, CA, US) with antibodies used according to the manufacturer’s instructions (Suppl. Table S1) are presented in Figs. S1 and S2 (Supplementary Material). To detect lymphocyte subpopulations {CD3+CD19− (T lymphocytes), CD3−CD19+ (B lymphocytes), and CD3−CD19− (mostly NK cells)}, samples of 2 × 105 PBMCs were stained with anti-CD3-PE and anti-CD19-APC. In parallel, unstimulated and stimulated PBMCs were stained with anti-CD3-APC-FIRE and anti-CD69-FITC to evaluate CD3+ lymphocyte activation in response to PepMix™ of SARS-CoV-2. Mitochondrial mass (MitoTracker™ Green FM probe, M46750, Invitrogen, Waltham, MA, US) and mitochondrial superoxide (MitoSOX™ Red reagent, M36008, Invitrogen, Waltham, MA, US) of PBMCs were measured following manufacturer’s instructions. Specifically, MitoTracker™ Green FM probe was added to PBMCs incubated in RPMI 1640 (15–041-CV, Corning, Corning, NY, US) to a final concentration of 1 mM for 30 min at 37ºC; alike, MitoSOX™ Red reagent in a final concentration of 3.8 µg/µL was incubated during 15 min. Upon incubation of each probe, we performed a centrifugation in order to remove RPMI medium. Samples from both MitoTracker™ Green FM and cell activation assays were also stained with 7-AAD to exclude non-viable cells (A07704, Beckman Coulter, Brea, CA, US). Data were analyzed with Kaluza v2.1. (Beckman Coulter, Brea, CA, US).
Western blot
Western blot analyses of PBMCs were performed at a concentration of 5 × 105 cells in 14 μl of 2X SDS-PAGE sample buffer (100 mM Tris/HCl, pH 6.8, 3% SDS, 1 mM EDTA, 2% 2-β-mercaptoethanol, 5% glycerol). The protein separation were accomplished by using 4–20% Mini-PROTEAN® TGX™ Precast Protein gels (4,561,096, Bio-Rad®, Hercules, CA, US) under the following conditions: 80 V/15 min and 120 V/75 min. Molecular weight marker (Precision Plus Protein Dual Color Standards, #1,610,374, Bio-Rad®, Hercules, CA, US) was also loaded in every gel. Then, proteins were transferred to PVDF membranes (Immobilon®-FL PVDF, IPFL85R, Merck, Darmstadt, Germany) previously activated with methanol. After transferring, the membranes were blocked 1 h with 5% milk powder diluted in Tween Tris-buffered saline (TTBS). PVDF membranes were incubated overnight at 4ºC with primary antibodies (Suppl. Table S2) diluted in TTBS with 5% BSA and 0.02% sodium azide. Primary antibodies were immunodetected using IRDye®800CW Donkey anti-rabbit (green fluorescence, 1:10.000 in TTBS with 0.01% SDS; 925–32,213, LI-COR Biosciences, Lincoln, NE, US) or IRDye®680RD Goat anti-mouse (red fluorescence, 1:10.000 in TTBS with 0.01% SDS; 925–68,070, LI-COR Biosciences, Lincoln, NE, US), as appropriate, incubated during an hour, and quantified using Image Studio™ Lite v5.2. (LI-COR Biosciences, Lincoln, NE, US). Loading was normalized by anti-α-tubulin (Suppl. Table S2).
Circulating cytokines
To quantify serum cytokines, we used the LEGENDplex™ Human Cytokine Panel 2 kit (740,102, Biolegend, San Diego, CA, US) which includes: granulocyte–macrophage colony-stimulating factor (GM-CSF), interferon alpha-2 (IFN-α2), thymic stromal lymphopoietin (TSLP), and selected interleukins (IL): IL-1α, IL-1β, IL-11, IL-12p40, IL-12p70, IL-15, IL-18, IL-23, IL-27, IL-33, according to the manufacturer’s instructions. Fluorescent signals (CytoFLEX cytometer, Beckman Coulter, Brea, CA, US) were analyzed with LEGENDplex™ data analysis v8.0. Cytokines concentrations derived from the median fluorescence intensity (MFI) interpolated from 8 duplicated standard controls. Serum levels of those cytokines reaching detection thresholds in at least 25% of the samples were compared between CSC and CMC groups (under this condition, seven cytokines were excluded from the analyses: IL-1α, IL-11, IL-12p70, IL-15, IL-23, IL-33, TSLP).
Metabolomic analysis
Serum samples (300 μL) were thawed and prepared for nuclear magnetic resonance (NMR) analyses (Biosfer Teslab, Reus, Spain). High-resolution 1H-NMR spectroscopy data (Bruker 600 MHz Spectrometer), 1D Nuclear Overhauser Effect Spectroscopy (NOESY), Carr-Purcell-Meiboom-Gill (CPMG) allowed to characterize small molecules such as amino acids and sugars; and LED Diffusion (Diff) to detect larger molecules such as lipoproteins, glycoproteins, and choline compounds. All sequences were analyzed at 37 °C in quantitative conditions. Lipid extracts for NMR measurement were prepared through a biphasic extraction with methanol/ methyl-tert-butyl ether, dried and reconstituted in 0.01% tetramethylsilane solution (0.067 mM TMS) and deuterated solvents.
From intact serum samples, the Liposcale test® (IVD-CE) was used to obtain the composition, mean size, and number of lipoprotein particles of large, medium, and small subtypes of the main lipoprotein types (VLDL, LDL, and HDL), as reported (Mallol et al. 2015). From the same NMR spectra, circulating glycoproteins were derived from the specific region where glycoproteins resonate through deconvolution with analytical functions, to quantify the area, proportional to the concentration of the acetyl groups of N-acetylglucosamine and N-acetyl galactosamine (GlycA), and of N-acetylneuraminic acid (GlycB) (Fuertes-Martín et al. 2019). Complementary, low molecular-weight metabolites (LMWM) were profiled from intact serum 1H-NMR spectra to obtain the concentration of aqueous metabolome, including a set of amino acids and sugars (Ozcariz et al. 2023). Finally, the concentration of the major lipid classes (fatty acids, glycerolipids, phospholipids, and sterols) and some individual species from the 1H-NMR spectra of the serum samples were estimated by using a previous protocol (Gil et al. 2019). Lipid quantification was obtained based on the Lipspin software, a quantification algorithm relying on lineshape fitting analysis of spectral regions (Barrilero et al. 2018). Since the spectral area is equivalent to the molecular abundance, individual signal areas were converted into molar concentrations by normalizing to external measurements. Lipid species obtained by this NMR approach included: cholesterol (free and esterified), unsaturated fatty acids (omega-6, omega-7, omega-9, omega-3), saturated fatty acids, monounsaturated fatty acids, linoleic acid, docosahexaenoic acid, arachidonic and eicosapentaenoic; glycerides and phospholipids (total cholines, triglycerides, phosphoglycerides, phosphatidylcholine, sphingomyelin, and lysophosphatidylcholine). The association between COVID-19 severity and lipoprotein, glycoprotein, low molecular weight metabolites, and lipid profiles were analyzed at two sample collection times separated by 3 months.
Statistical analysis
The normality of the variables was assessed with the Shapiro–Wilk test (less than 30 data). Outliers were detected and discarded with Grubbs’ test (https://www.graphpad.com/quickcalcs/Grubbs1.cfm, GraphPad online tool). Differences were assessed using unpaired two-tailed Student’s t-tests and unpaired non-parametric Mann–Whitney U tests, to compare CMC and CSC groups, as well as paired two-tailed Student’s test and paired non-parametric Wilcoxon tests between non-stimulated and stimulated condition in the same group. For unequal variances after the Student’s t-test, Welch’s t-test was applied. Qualitative parameters (sex and ethnicity) were analyzed by Pearson’s chi-squared test. Correlation analyses were performed using Spearman’s rank correlation tests. A p < 0.05 was considered statistically significant. SPSS v.15.0.1. (SPSS Inc., Chicago, IL, US) and Prism v8.0. (GraphPad Software Inc., San Diego, CA, US) allowed the analyses and graphical representations. For the metabolomic data, quantitative variables were represented by median and interquartile range and categorical variables by frequencies (total and percentage in brackets) and 95% confidence interval. The Wilcoxon-Mann–Whitney U test was used to compare the different groups through graphical representation, considering the comparison of samples between times as paired data. The possible interaction between sample collection time and COVID-19 severity was determined using a Kruskal–Wallis test. R Bioconductor v4.1.1. was used to perform data analysis (https://www.bioconductor.org/).
Results
Description of the studied cohorts
Months after the resolution of the symptoms of COVID-19, 24 individuals were enrolled in this pilot study. Two groups were established including subjects who underwent ICU stay with mechanical respiratory support, defined as cases who recovered after severe COVID-19 (CSC), and those who experienced minor symptoms without hospitalization, defined as cases who recovered after mild COVID-19 (CMC). As mentioned, the described diseases associated with worse prognosis upon SARS-CoV-2 infection precluded enrollment. Demographic and clinical data upon recruitment of these recovered subjects (Table 1) showed that both groups did not differ in terms of age, gender or ethnicity. However, C reactive protein and platelets count displayed significantly higher levels in the CSC group than in the CMC one, while prothrombin time, standardized prothrombin time ratio (INR), and aspartate transaminase (AST), were significantly decreased in CSC.
Decreased mitochondrial activity in individuals who recovered from severe COVID-19
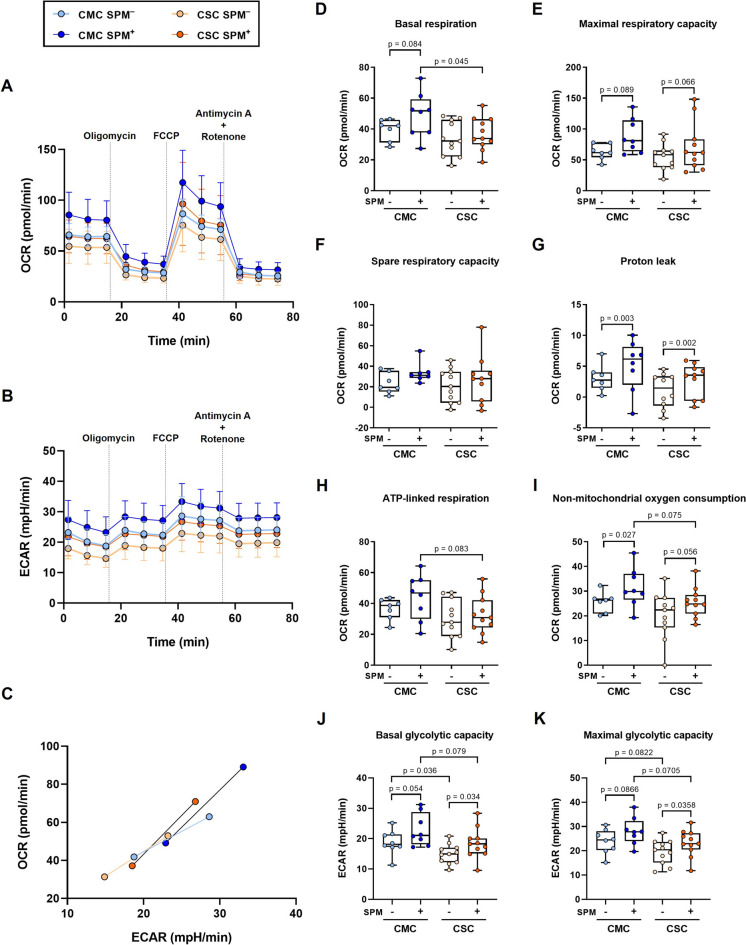
The metabolic integrity in both basal and spike peptide mix (SPM)-stimulated PBMCs was assessed using a mitochondrial stress test based on the sequential addition of oxidative phosphorylation inhibitors. As shown in Figs. 1A and 1B, profiles of aerobic respiration and extracellular acidification of CSC lagged behind those of CMC, in such a way that upon activation with the peptide mix, the SPM-stimulated kinetics of the CSC group overlapped the unstimulated condition of the CMC group. These changes, summarized in the energy phenotypes, uncovered more energetic situations for both unstimulated and SPM-stimulated CMC subjects (Fig. 1C). The energy phenotype represents a simultaneous comparison of mitochondrial respiration and glycolysis under baseline and stressed conditions, to define the metabolic potential in response to an energy demand.
Fig. 1.
Lymphocyte metabolism in individuals who recovered after mild (CMC) and severe (CSC) COVID-19. A Oxygen consumption rate (OCR) and (B) extracellular acidification rate (ECAR) profiles of non-stimulated or spike peptide mix (SPM) stimulated PBMCs from CMC SPM− (n = 8), CMC SPM+ (n = 8), CSC SPM− (n = 11) and CSC SPM+ (n = 11) groups (traces represent means). C Energy phenotype of the human PBMCs of CMC (n = 8) and CSC (n = 11) groups, where minimum and maximum values of OCR and ECAR were confronted to obtain an estimation of the metabolic range of the cells (points represent means of basal respiration and glycolytic capacity and the stressed condition displays the maximal respiration and glycolytic capacity). D-I Mitochondrial parameters of the CMC and CSC groups (n = 6–11). J-K Basal and maximal glycolytic capacities comparing the different groups (n = 6–11). Results are expressed using box and whiskers diagrams, where lines represent median values, boxes the 25 and 75 percentiles, and whiskers mark maximum and minimum values. Statistical significance was assessed by two-tailed unpaired Student’s and Paired t-tests, except for spare respiratory capacity data evaluated by unpaired non-parametric Mann–Whitney U and Wilcoxon tests
A detailed analysis of basal respiration showed a significant difference between CMC and CSC groups in the SPM-stimulated condition (p = 0.045, Fig. 1D), and maximal respiratory capacities followed a similar pattern (Fig. 1E). As a result, the spare respiratory capacities (maximal minus basal respirations) seemed comparable between CMC and CSC groups (Fig. 1F). Upon SPM-stimulation, mild and severe groups showed significant increases in the uncoupling rate, reflecting statistically significant increments of proton leak (Fig. 1G). Again, the ATP-linked respiration evidenced a more advantageous situation for CMC as compared to CSC, rendering a trend for a significant difference between the SPM-stimulated conditions of both groups (Fig. 1H). In parallel, the non-mitochondrial oxygen consumption also exposed a trend for significance between the SPM-stimulated subgroups, with higher values in CMC (Fig. 1I). As shown, the different mitochondrial parameters consistently displayed reduced values for both basal and SPM-stimulated conditions in the CSC subgroups.
Finally, the estimation of basal and maximal glycolytic capacities (Figs. 1J and K, respectively) evidenced comparable changes upon SPM-stimulation in CMC and CSC groups, but with significantly lower values (or trends) of basal and maximal glycolytic capacities for the severe course when compared to the mild one, in both unstimulated and SPM-stimulated conditions.
Severity of COVID-19 does not affect lymphocyte activation upon SPM-stimulation
We checked whether the SPM-stimulation was associated with lymphocyte activation by analyzing CD69 expression. This membrane receptor, one of the most studied activation cell markers, is rapidly induced after lymphocyte stimulation. Upon SPM-stimulation of PBMCs, CD69 surface expression significantly increased in CD3+CD19− cells, indicative of activation in the T subset of lymphocytes from CMC and CSC groups (Suppl. Fig. S3).
SPM-stimulation in CSC enhances superoxide production without changes in mitochondrial mass
To quantify the increase in mitochondrial mass after cell activation, the ratio between SPM-stimulated and unstimulated sample pairs was calculated. No significant changes were observed between CMC and CSC groups, neither in overall PBMCs nor in the cell subpopulations studied (Suppl. Fig. S4).
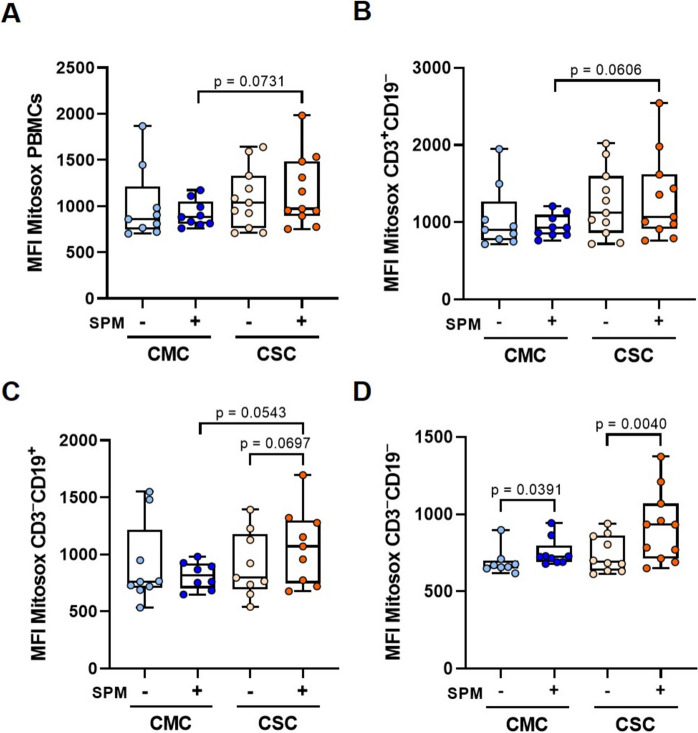
In addition, we evaluated mitochondrial ROS upon SPM-stimulation. PBMCs from CSC samples showed an increased ROS content compared to the CMC ones (Fig. 2A). Similar trends were also observed in either CD3+CD19− (Fig. 2B) or CD3−CD19+ cells (Fig. 2C), but statistical significant difference was not reached (p = 0.111) when considering CD3−CD19− (mostly NK cells, Fig. 2D), although a statistically significant SPM-stimulation was reached in both CMC and CSC groups.
Fig. 2.
Mitochondrial superoxide detection and comparison between individuals who recovered from mild (CMC) and severe (CSC) COVID-19. Diagrams present the mitochondrial superoxide anion levels produced before and after spike peptide mix (SPM) stimulation, measured by flow cytometry with MitoSOX™ Red reagent for (A) PBMCs, and (B) CD3+CD19−, C CD3−CD19+, D CD3−CD19− lymphocyte subpopulations from CMC (n = 8–9) and CSC (n = 9–11) groups. Results are depicted using box and whiskers diagrams, where lines represent median values, boxes the 25 and 75 percentiles, and whiskers mark maximum and minimum values. Statistical significance was assessed by two-tailed unpaired Student’s and Paired t-tests. The comparison between the stimulated groups in PBMCs, CD3+CD19− and CD3−CD19+ showed unequal variances after Student’s t-test, therefore Welch’s t-test was applied
Unbalance in mitochondrial fusion and fission protein levels according to past COVID-19 severity
Different proteins implicated in mitochondrial fusion and fission processes, as well as the oxidative phosphorylation complexes (Figs. 3 and 4, respectively), were analyzed to elucidate expression changes putatively associated with COVID-19 course.
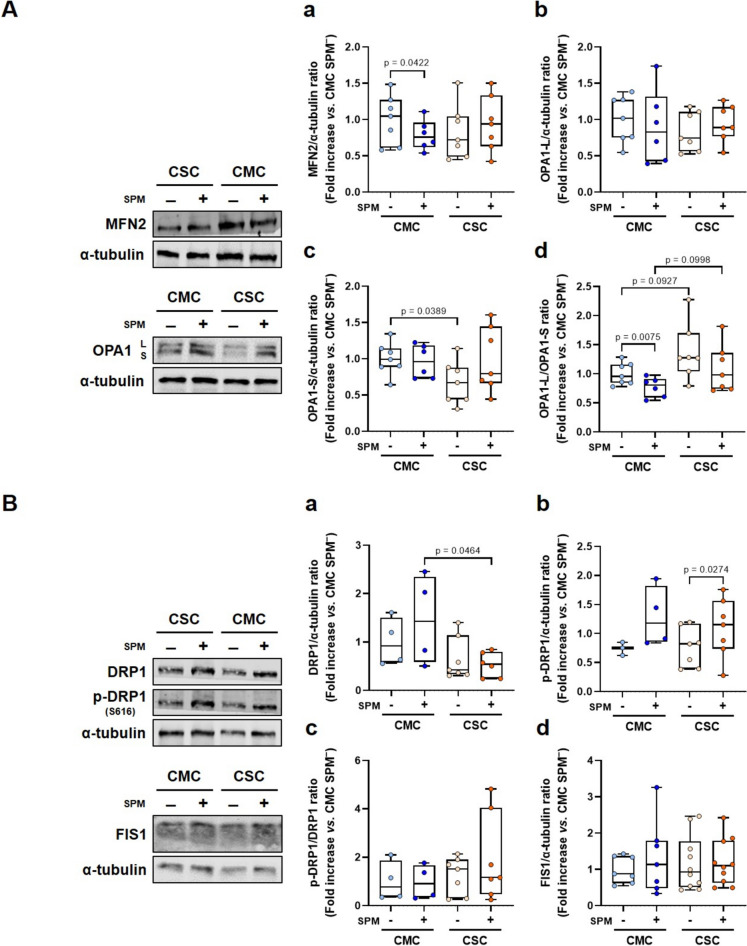
Fig. 3.
Expression of pivotal proteins involved in mitochondrial fusion and fission in individuals who recovered after mild (CMC) and severe (CSC) COVID-19. A Left, representative Western blots of mitochondrial fusion proteins, OPA1 and MFN2 (α-tubulin was used as loading control), in non-stimulated and stimulated PBMCs of CMC (n = 8) and CSC (n = 10). Right, histograms presenting the protein/ α-tubulin and the OPA1-L/ OPA1-S ratio quantifications of band intensities in CMC SPM− (n = 7), CMC SPM+ (n = 6), CSC SPM− (n = 7) and CSC SPM+ (n = 7). B Left, representative Western blot analyses of mitochondrial fission proteins, DRP1, phospho-DRP1 (p-DRP1; Ser616) and FIS1 (α-tubulin was used as loading control), in non-stimulated and stimulated PBMCs of CMC (n = 7) and CSC (n = 10). Right, histograms displaying the protein/ α-tubulin and p-DRP1/ DRP1 ratio quantifications of band intensities in CMC SPM− (n = 3–7), CMC SPM+ (n = 4–7), CSC SPM− (n = 7–10) and CSC SPM+ (n = 6–10). Results are expressed using box and whiskers diagrams, where lines represent the median value, the box the 25 and 75 percentiles, and whiskers mark maximum and minimum values. Statistical significance was analyzed by two-tailed unpaired Student’s and Paired t-tests, except for the CSC SPM– group in the DRP1 data, which were evaluated by unpaired non-parametric Mann–Whitney U and Wilcoxon tests. Additionally, the comparison between the non-stimulated groups in the OPA1-L/ OPA1-S ratio quantification, and between the stimulated and non-stimulated CMC groups in FIS1 protein levels presented unequal variances after Student’s t-test, therefore Welch’s t-test was applied
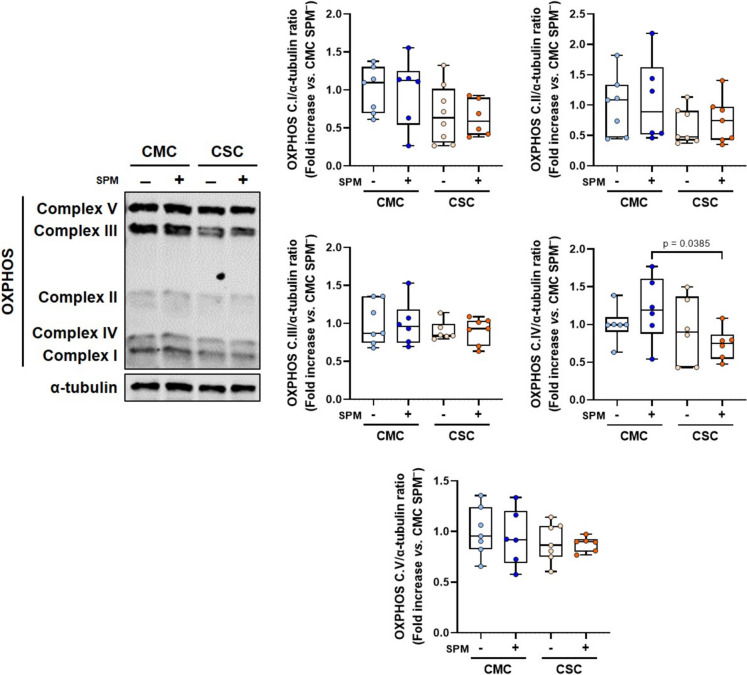
Fig. 4.
Evaluation of oxidative phosphorylation complexes in non-stimulated and stimulated PBMCs from individuals who recovered from mild (CMC) and severe (CSC) COVID-19. Left, representative Western-blot analysis of mitochondrial respiratory chain complexes, from complex I to complex V, in CMC (n = 7–8) and CSC (n = 10) (α-tubulin used as loading control). Right, histograms displaying the protein/ α-tubulin ratio upon quantification of band intensities from CMC SPM− (n = 6–7), CMC SPM+ (n = 6), CSC SPM− (n = 6–8) and CSC SPM+ (n = 6–7). Results are expressed using box and whiskers diagrams, where lines represent the median value, boxes the 25 and 75 percentiles, and whiskers mark maximum and minimum values. Statistical significance was assessed by two-tailed unpaired Student’s and Paired t-tests
The mitochondrial fusion protein mitofusin-2 (MFN2) showed a statistically significant decrease upon SPM-stimulation in PBMCs of CMC and, in contrast, a non-significant increase was observed in CSC (Fig. 3Aa). Furthermore, statistically significant lower levels of the short isoform of the optic atrophy type 1 (OPA1-S) were evidenced by comparison of basal conditions of the CSC and CMC groups (Fig. 3Ac), while a parallel change did not reach significance in the long isoform (Fig. 3Ab). The OPA1-L/ OPA1-S ratio showed non-significant increases both in basal and stimulated conditions in the CSC groups compared to the CMC ones (Fig. 3Ad). Upon SPM-stimulation, a statistically significant decrease in this ratio was observed in the samples from CMC (p = 0.0075, Fig. 3Ad).
Regarding mitochondrial fission processes, the dynamin-related protein (DRP1) levels in the SPM-stimulated groups were significantly lower in CSC compared to CMC (Fig. 3Ba). The activated form, p-DRP1, showed significantly higher levels in the SPM+ groups in comparison with the non-stimulated PBMCs (Fig. 3Bb). No significant differences were observed in the p-DRP1/DRP1 ratio in the studied groups (Fig. 3Bc), and they also presented comparable values in mitochondrial fission 1 protein (FIS1) levels (Fig. 3Bd).
The expression of the five complexes that integrate the ETC and are involved in oxidative phosphorylation were analyzed (Fig. 4). The complex IV levels were significantly decreased in the SPM-stimulated CSC group compared to the homologous CMC, while no differences were detected for the unstimulated groups. In terms of complex I and II, although non-stimulated and SPM-stimulated PBMCs from CSC groups seemed to present median expression levels lower than those of CMC, no statistically significant changes were reached. No additional differences were observed in levels of complex III and V (Fig. 4).
Circulating cytokines levels differ between CMC and CSC groups
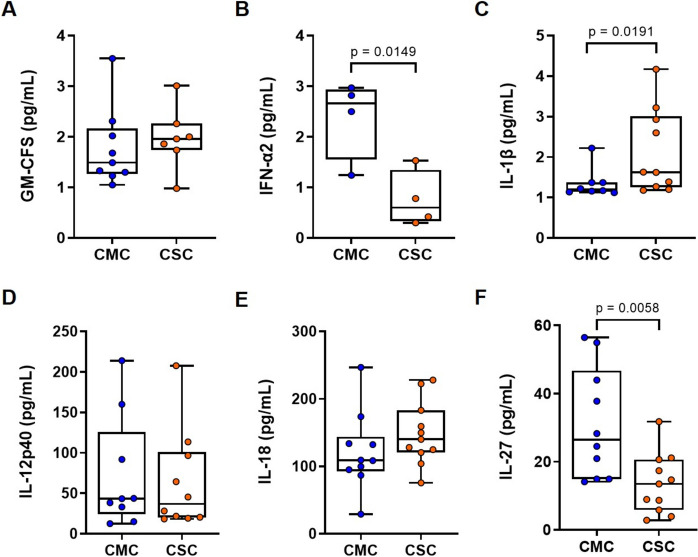
Our results revealed that IL-1β levels were significantly higher in CSC compared with CMC samples (Fig. 5C) and the same trend was observed in IL-18 levels, although the increase did not reach statistical significance (Fig. 5E). On the contrary, IFN-α2 (Fig. 5B) and IL-27 (Fig. 5F) were significantly lower in the CSC group. No significant differences in GM-CFS and IL-12p40 levels were detected between the two groups (Figs. 5A and 5D, respectively).
Fig. 5.
Circulating cytokines levels in subjects who recovered from mild and severe COVID-19. Serum cytokines levels were measured by flow cytometry and were compared between CMC (n = 12) and CSC (n = 10). Specifically, the assessed cytokines were: A GM-CFS, B IFN-α2, C IL-1β, D IL-12p40, E IL-18, and (F) IL-27. Results are expressed using box and whiskers diagrams, where lines represent median values, boxes the 25 and 75 percentiles, and whiskers mark maximum and minimum values. Statistical significance was assessed by two-tailed unpaired Student’s t-test in IFN-α2, IL-18, and IL-27 data, while IL-1β, GM-CFS, and IL-12p40 data were evaluated by unpaired non-parametric Mann–Whitney U tests
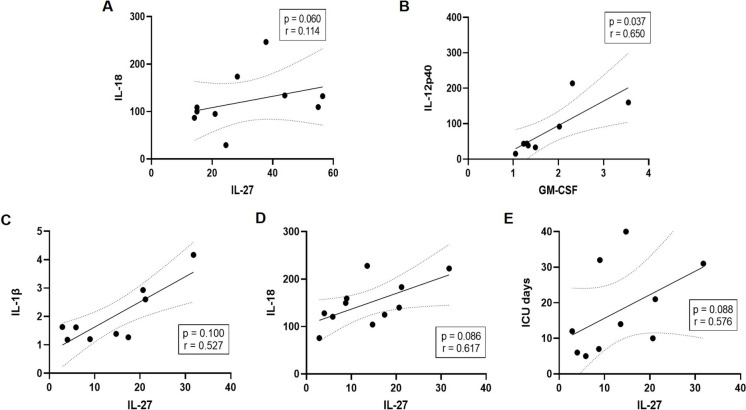
Correlations between the different cytokines, as well as between cytokines and ICU days in CSC were assessed. Using Spearman’s correlation analysis, IL-27 showed a weak positive correlation with IL-18 in the CMC group (Fig. 6A), and a stronger positive correlation in the CSC group (Fig. 6D) with some evidence of correlation with ICU days (Fig. 6E). Then, correlations between IL-18 and IL-27 with mitochondrial respiration, mitochondrial mass, and superoxide anion levels, as well as protein expression levels (OPA1-S and OPA1-L/OPA1-S ratio), and ICU days were evaluated (Suppl. Figs. S5-S7). In the CSC group, inverse correlations of IL-18 with basal and maximal respiration were detected, in contrast with the lack of correlation observed in the CMC group (Suppl. Fig. S5). Parallel inverse correlations of IL-27 with maximal respiratory (p = 0.024, r = −0.669) and glycolytic capacities (p = 0.015, r = −0.709) were evidenced in the CSC but not in the CMC group (Suppl. Fig. S6), as well as a direct correlation with the OPA-L/OPA-S ratio (p = 0.038, r = 0.835; Suppl. Fig. S7B). Conversely, a significant correlation of IL-27 with mitochondrial mass in T cells (p = 0.041, r = 0.687; Suppl. Fig. S7A) and similar trends in the other studied subpopulations of PBMCs were detected only in the CMC group, not in CSC.
Fig. 6.
Correlation between cytokines levels and Intensive Care Unit (ICU) days. Scatter plots showing the correlation between A IL-27 and IL-18 and B GM-CSF and IL-12p40 in CMC; and the correlation between IL-27, and C IL-1β, D IL-18, and E ICU days, in the CSC group. Spearman’s rank correlation coefficient test was used for assessing the degree of linear correlation
Metabolomic fingerprint reveals a trend for elevated levels of circulating 3-hydroxybutyrate in CSC
To investigate the specificity of cellular metabolic alterations and the potential implications for overall metabolism, we conducted a comprehensive comparison of lipoprotein, glycoprotein, low molecular weight metabolites, and lipid profiles using nuclear magnetic resonance (1H-NMR). Interestingly, no significant differences between CSC and CMC groups were observed in lipoprotein and glycoprotein profiles, or in the lipid profile (Suppl. Tables S3-S5). However, we identified higher circulating levels of two metabolites, 3-hydroxybutyrate and glutamate, in the CSC group compared to the CMC group (Table 2). Kruskal–Wallis test of those two metabolites considering the values obtained in both draw blood samples from each individual separated by three months, confirmed a trend for significance (p = 0.083) between CSC (median: 36 μM [31.4; 36.0] and 27 μM [26.8; 27.6]) and CMC (median: 27.6 μM [20.4; 34.9] and 24 μM [17.6; 36]) groups in 3-hydroxybutyrate, but not in glutamate (p = 0.240). An inverse correlation of 3-hydroxybutyrate with ICU stay (p = 0.059; r = −0.613) was found.
Table 2.
Low molecular weight metabolite profile of individuals who recovered from mild (CMC) and severe (CSC) COVID-19
| CMC (n = 12) | CSC (n = 12) | p Value | |
|---|---|---|---|
| Low molecular weight metabolites (μM) | |||
| 3-Hydroxybutyrate | 27.6 [20.4;34.9] | 36.0 [31.4;36.0] | 0.032 |
| Acetone | 12.6 [9.75;16.2] | 12.0 [9.58;15.4] | 0.908 |
| Alanine | 308 [246;379] | 288 [274;309] | 0.603 |
| Creatine | 46.7 [40.1;56.1] | 35.1 [25.9;49.3] | 0.166 |
| Creatinine | 45.6 [36.8;55.0] | 57.5 [47.5;72.4] | 0.065 |
| Glucose | 3947 [3812;4130] | 4106 [3969;4257] | 0.356 |
| Glutamate | 103 [90.7;118] | 133 [108;150] | 0.043 |
| Glutamine | 316 [296;335] | 305 [287;333] | 0.686 |
| Glycine | 146 [136;153] | 160 [135;172] | 0.356 |
| Histidine | 53.9 [44.0;64.1] | 56.0 [50.8;61.1] | 0.773 |
| Lactate | 232 [212;281] | 315 [246;349] | 0.149 |
| Tyrosine | 39.2 [33.6;47.7] | 41.9 [28.3;50.3] | 1.000 |
| Valine | 185 [149;206] | 193 [168;207] | 0.686 |
| Isoleucine | 30.9 [24.3;36.6] | 33.0 [23.5;34.9] | 0.773 |
| Leucine | 84.4 [74.5;100] | 85.8 [66.4;97.2] | 0.908 |
Statistical significance was evaluated by using the Mann–Whitney U test
Discussion
Exploring the metabolism of immune cells in previously infected individuals provides interesting information regarding COVID-19 recovery, as shown in this pilot study. Our results could suggest potential biomarkers of COVID-19 sequelae even in the absence of symptoms, and could explain the observed vulnerability of some patients (Sharma and Bayry 2023). Biochemical and coagulation parameters such as INR or CRP levels had been found increased in acute severe COVID-19 patients compared to mild cases (Chen et al. 2020; Klok et al. 2020), mirroring our clinical data in recovered subjects (Table 1). Reportedly, a mitochondrial dysfunction along with an inflammatory state led to platelet damage in COVID-19 patients, especially in those who suffered worse symptomatology (Tang et al. 2020). In the present work, although significant differences in platelet number and AST levels were observed between CMC and CSC groups, both groups showed values within normal ranges and the ALT/AST ratio was higher in CSC. In line with our results, Cao and cols. (Cao X et al. 2024) described higher AST levels in asymptomatic than in mild COVID-19 patients (mean: 27.93 vs. 25.62 U/L, respectively). At the moment, the prognostic role of liver enzymes is not exempt from controversy (Aloisio and Panteghini 2021; Yip et al. 2021). A recent report identified longer recovery in participants with previous suboptimal health and, once adjusted for all prepandemic conditions, only clinical cardiovascular disease was associated with recovery. Even though acute infection severity partially mediated this association, the authors emphasized the need of further investigation on alternative mechanisms (Oelsner et al. 2024). In this sense, the physiological changes that we found in diverse parameters could still mark a subclinical condition, which could gain full meaning in situations of cellular stress.
Mitochondria is a key factor in the immune response to combat viral infections. Our findings show that peripheral cells from the CSC group exhibited reduced metabolic activity when compared with those of the CMC group, suggesting a possible compromised mitochondrial function (Fig. 1). Concordantly, during acute infection, decreased basal and maximal respiration have been reported in PBMCs of COVID-19 patients admitted to the ICU compared to patients with a chest infection and healthy controls (Ajaz et al. 2021). In PBMCs of acute COVID-19 patients, cellular stress was found to alter mitochondrial function leading to reduced immune responses and increased inflammatory responses associated with disease severity. In our pilot study, the bioenergetics of immune cells from the CSC group remained reduced long after symptoms resolution when compared to the CMC group, with lower glycolysis and mitochondrial respiration in both basal and stimulated conditions (Fig. 1C). Many viruses, including SARS-CoV-2, enhance replication by metabolically reprogramming cells towards glycolysis (Karagiannis et al. 2022). Concordantly, we found increased rates of either basal or maximal glycolytic capacities in the SPM-stimulated conditions (Figs. 1J-K). These metabolic changes driven by mitochondria persist long after symptoms onset, as shown by our energy phenotype findings comparing both groups with different acute-infection severity in either non-stimulated or SPM-stimulated conditions (Fig. 1C). As observed, in both CMC and CSC groups, the sham infection with SPM induced statistical trends for higher basal and maximal glycolytic capacities (Figs. 1J-K), and evidenced lower values in basal conditions in the CSC compared to the CMC group, in parallel to non-mitochondrial oxygen consumptions (Fig. 1I).
The CSC group presented higher levels of ROS, both in basal and stimulated conditions. Even though the determinations of these flow cytometry probes rely on mitochondrial membrane potential and morphology, upon SPM-stimulation, we consistently found trends for significantly higher levels in the T- and B-lymphocyte subpopulations of the CSC group (Fig. 2), which might reflect an altered ETC and higher uncoupling (and therefore lower proton leak) in CSC individuals, as already shown (Fig. 1G). Moreover, ROS levels fit with the observed changes in OPA1-S, as discussed below.
In patients with acute SARS-CoV-2 infection, Liu and cols (Liu et al. 2021) found elevated mitochondrial mass correlated to T-cell apoptosis, which could drive clinical lymphocytopenia. In contrast, and concordantly to our results, Castro-Sepulveda and cols (Castro-Sepulveda et al. 2022) did not detect mass differences (Suppl. Fig. S4). Viruses perturb the equilibrium in mitochondrial dynamics to foster their survival. The mediator of mitochondrial fusion, MFN2, acts as a direct inhibitor of mitochondrial antiviral signaling, thereby reducing antiviral immunity (Yasukawa et al. 2009). In terms of MFN2, our findings show how the activation of PBMCs with SPM generated opposed tendencies in CMC and CSC groups (Fig. 3Aa). In PBMCs of the CMC group, the response to viral proteins showed significantly lower MFN2 levels warranting an adequate antiviral response, while in the CSC group, the reinfection-like stimulus still drives an increase in MFN2 levels that would prevent the adequate antiviral response. Furthermore, OPA1-L showed parallel effects to those observed with MFN2 (Figs. 3Ab and 3Aa, respectively). The OPA1-S isoform showed a different behavior in CMC and CSC groups (Figs. 3Ac and 3Ad), with statistical trends to increased OPA1-L/ OPA1-S ratios found in the basal and stimulated CSC groups (Fig. 3Ad). The stress-induced cleavage of OPA1 and generation of OPA1-S promote survival in cells facing adverse conditions, as recently postulated (Lee H et al. 2020). Herein, higher ROS levels in CSC subjects (Fig. 2A) correlated with lower concentrations of OPA1-S (Fig. 3Ac) in either basal or SPM-stimulated conditions. The ratios of OPA1-L/ OPA1-S displayed higher values in the CSC group, with similar statistical trends when basal and SPM-stimulated groups were compared to the CMC groups (Fig. 3Ad), although most probably due to limited statistical power, those differences did not reach significance. Moreover, OPA1’s role for cristae maintenance is critical for the OXPHOS activity of cells, as it directs the proper functional assembly of respiratory complexes. The pro-fission factor DRP1 is frequently targeted by viruses: the human immunodeficiency virus reduces overall DRP1 and p-DRP1 levels and impairs mitochondrial fission (Fields et al. 2016). Conversely, the hepatitis B virus induces mitochondrial fission, which facilitates mitophagy and enhances viral survival (Kim et al. 2013). As previously reported, SARS-CoV-1 modulates mitochondrial dynamics by inducing DRP1 degradation (Shi et al. 2014). Given the genome similarities between SARS-CoV-1 and SARS-CoV-2, the significant decrease in DRP1 levels observed between SPM-stimulated CMC and CSC groups suggests an analogous disruption in mitochondrial dynamics (Fig. 3Ba). Similar generation of p-DRP1 was detected in both CMC and CSC groups upon SPM-stimulation (Fig. 3Bb), and no changes were observed regarding FIS1 (Fig. 3Bd). In summary, the correct balance of mitochondrial dynamics is a key factor in antiviral defense, and our data seem to suggest some long-lasting perturbations.
Mitochondrial integrity is fundamental for an adequate antiviral response, and therefore we aimed to study the status of oxidative phosphorylation complexes. As shown (Fig. 4), even though complexes I and II displayed lower levels in the CSC group, only complex IV reached a significant difference between the SPM-stimulated CMC and CSC groups. Concordantly, Soria-Castro et al. also found a lack of mitochondrial structural integrity and ETC disruption in postmortem samples from patients who died from COVID-19 (Soria-Castro et al. 2021).
The cross-talk between acute inflammatory processes induced by SARS-CoV-2 and the underlying oxidative stress (Forcados et al. 2021; Shenoy 2020) could contribute to COVID-19 severity and might explain the observed differences between our mild and severe groups. In our study IL-1β, IFN-α2, and IL-27 showed significant changes between CSC and CMC groups (Fig. 5). In agreement with our findings, higher levels of one of the hallmarks of COVID-19, IL-1β, were described in severe patients (Kalinina et al. 2022; Ling et al. 2021; Saleh et al. 2020) and synergistically with IFNγ promote a physiological response of hyperinsulinemia, responsible for the systemic glucose restriction following severe infection (Šestan et al. 2024). We also identified a reduction in IL-27 and IFN-α2 levels in the CSC group (Fig. 5). In line with our results, previous studies pointed to lower levels of IL-27 at admission in severe COVID-19 patients, acting as a biomarker of poor clinical outcome or disease severity (Tamayo-Velasco et al. 2021; Zamani et al. 2022). We detected, specifically in the CSC group, inverse correlations between IL-27 and maximal respiratory and glycolytic capacities (p = 0.024, r = −0.669 and p = 0.015, r = −0.709, respectively; Suppl. Fig. S6) and a direct correlation with the OPA1-L/OPA1-S ratio (p = 0.038; r = 0.835; Suppl. Fig. S7). Once again, our data would show that altered cytokine levels persist long after clinical recovery. Concordantly, a recent study reported improper adaptive immune responses in LC (Yin et al. 2024). Shedding light on this scenario, another work established serum protein signatures which identified heterogeneity with a subset of LC patients showing persistent inflammation, and only those with higher IFN-γ presented higher IL-27, IL-18 and NF-κB (Talla et al. 2023).
Metabolic abnormalities including hyperlipidemia and altered glucose metabolism were reported in SARS-CoV-1 infection over a decade after recovery (Wu et al. 2017). Metabolic programming of lymphocytes is key for modulating inflammatory responses, and thus we aimed to study the lymphocyte metabolism in recovered subjects. The lack of differences in lipoprotein, glycoprotein, and lipid profiles shown in our results suggests that the metabolic alterations found, rather than being influenced by differences in nutritional or metabolic states among individuals, are specific to the cellular level. Recently, an impaired 3-hydroxybutyrate (BHB) production was reported in individuals with acute respiratory distress syndrome induced by SARS-CoV-2 but not by influenza virus (Karagiannis et al. 2022). T cells from those SARS-CoV-2 patients were exhausted and skewed towards glycolysis, but the authors propose they could be metabolically reprogrammed by BHB to perform OXPHOS, thereby increasing their survival, functionality, and production of IFN-γ. BHB provides an alternative carbon source to fuel OXPHOS and the production of amino acids and glutathione to maintain the redox balance. In the present pilot work, a trend for higher plasma levels of BHB in the CSC group was observed (p = 0.083), although always moving within physiological ranges. Further investigation is warranted to clarify whether these subclinical changes might have an impact under new stress conditions, given the main limitation of our pilot study, a reduced statistical power. Moreover, there are currently no validated clinical biomarkers of postacute sequelae of SARS-CoV-2 infection (Erlandson et al. 2024), and if validated IL-27 or BHB could fill this gap.
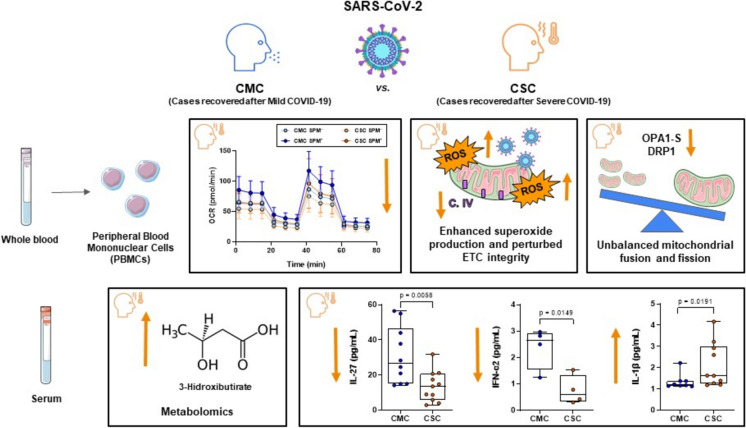
In summary (Fig. 7), the observed enduring changes in metabolism and energy balance upon COVID-19 might suggest important subclinical drivers of future disease under stress conditions. As shown, long after acute infection, mitochondrial changes still differ between a situation of full recovery after mild infection and the potential vulnerability to COVID-derived complications when evolving from a severe infection. The persistence of symptoms long after SARS-CoV-2 infection has been already detected (Notarte et al. 2022) but health consequences might also be present even in seemingly asymptomatic subjects. Ongoing research is critical to ascertain the ultimate cause behind the different course of COVID-19 disease, unveiling the possible influence of a deficient clearance of the virus or the impact of the patients’ genetic architecture, and specifically of genetic markers of mitochondrial function. In this context, mitochondria deserve further interest and, if shown to be causative, offer the potential for therapeutic targets, and to develop effective strategies to achieve full recovery.
Fig. 7.
Mitochondrial and metabolic changes differ between full recovery after mild infection and the evolution from severe infection
Supplementary information
Below is the link to the electronic supplementary material.
(PDF 1.24 MB)
Acknowledgements
The authors wish to thank the “Unidad de citometría de flujo” (UCIF, IdISSC) for their expert technical assistance, Dr. Myriam Calle for the recruitment of recovered patients and the personnel from “Banco de Sangre” of the Hospital Clínico San Carlos (IdISSC) for their support in the recollection of samples. The graphical abstract was partly generated using Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license.
Abbreviations
- 7-AAD
7-Aminoactinomycin D
- ACE2
Angiotensin-converting enzyme 2
- ALT
Alanine transaminase
- AST
Aspartate transaminase
- CMC
Cases who recovered of mild COVID-19
- COVID-19
Coronavirus disease 2019
- CRP
C reactive protein
- CSC
Cases who recovered of severe COVID-19
- DMSO
Dimethylsulfoxide
- DRP1
Dynamin-related protein
- ECAR
Extracellular acidification rate
- ETC
Electron transport chain
- FBS
Fetal bovine serum
- FCCP
Carbonylcyanide-p-trifluoromethoxyphenylhydrazone
- FIS1
Mitochondrial fission 1 protein
- GM-CSF
Granulocyte macrophage colony-stimulating factor
- HBSS
Hank’s balanced salt solution
- ICU
Intensive care unit
- IL
Interleukin
- IFN-α2
Interferon alpha-2
- INR
Standardized prothrombin time ratio
- LC
Long Covid-19
- MFI
Median fluorescence intensity
- MFN2
Mitofusin-2
- NMR
Nuclear magnetic resonance
- OCR
Oxygen consumption rate
- OPA1-L (-S)
Long (short) isoform of optic atrophy type 1
- OXPHOS
Oxidative phosphorylation
- PBMCs
Peripheral blood mononuclear cells
- p-DRP1
Phospho dynamin-related protein
- ROS
Reactive oxygen species
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SPM
Spike Peptide Mix
- TSLP
Thymic stromal lymphopoietin
- TTBS
Tween Tris-buffered saline
Authors’ contributions
IG-D, ARL-P and AG-J performed the Western-blot, flow cytometry and Seahorse extracellular flux analyses; CR-A and YH-G recruited the cases and isolated serum and cells from the blood samples; NM-M and NA completed the metabolomic assays; LE-P supervised the experimental work and assisted in the statistical analyses; and EA and EU designed the study, interpreted the obtained data, and wrote and revised the manuscript.
Funding
Financial support for the study was provided by an intramural Covid-19 program of IdISSC to Drs. Anguita and Urcelay. Adela González-Jiménez holds a “Formación de Profesorado Universitario” contract from Ministerio de Ciencia e Innovación (FPU20/03387), Andrea R. López-Pastor a contract from the REACT-EU INMUNOVACTER-CM and Irene Gómez-Delgado an Investigo contract from Comunidad Autónoma de Madrid (09-PIN1-00011.2/2022).
Data availability
The data that support the findings of this study are available upon request from the corresponding author and the metabolomics data have been deposited in an open access repository: (https://github.com/BiosferTeslab/NMR_Metabolomics_Long_Term_Impairment_After_Severe_Covid).
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Hospital Clínico San Carlos (CE16/211-E and CE20/740-E_BC). Informed consent was obtained from all subjects involved in the study.
Consent for publication
The authors declare that the research was conducted in the absence of any financial or not financial relationships that could be construed as a potential conflict of interest and all authors agreed in the final presentation of the work.
Competing interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Irene Gómez-Delgado and Andrea R. López-Pastor share first authorship of the work. Eduardo Anguita and Elena Urcelay share last authorship of the work.
References
- Ajaz S, McPhail MJ, Singh KK, Mujib S, Trovato FM, Napoli S, et al. Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. Am J Physiol. 2021;320(1):C57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloisio E, Panteghini M. Aspartate aminotransferase in COVID-19: a probably overrated marker. Liver Int. 2021;41(11):2809–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguita E, Chaparro A, Candel FJ, Ramos-Acosta C, Martínez-Micaelo N, Amigó N, et al. Biomarkers of stable and decompensated phases of heart failure with preserved ejection fraction. Int J Cardiol. 2022;361:91–100. [DOI] [PubMed] [Google Scholar]
- Barrilero R, Gil M, Amigó N, Dias CB, Wood LG, Garg ML, et al. LipSpin: a new bioinformatics tool for quantitative for quantitative 1H NMR lipid profiling. Anal Chem. 2018;90(3):2031–40. [DOI] [PubMed] [Google Scholar]
- Burtscher J, Cappellano G, Omori A, Koshiba T, Millet GP. Mitochondria: in the cross fire of SARS-CoV-2 and immunity. iScience. 2020;23(10). [DOI] [PMC free article] [PubMed]
- Cao X, Xie YL, Yi JY, Liu ZL, Han M, Duan JH, et al. Altered liver enzyme markers in patients with asymptomatic, and mild omicron infection: a retrospective study. J Inflamm Res. 2024;17:6875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Sepulveda M, Tapia G, Tuñón-Suárez M, Diaz A, Marambio H, Valero-Breton M, et al. Severe COVID-19 correlates with lower mitochondrial cristae density in PBMCs and greater sitting time in humans. Physiol Rep. 2022;10(14): e15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Ge Y, Wu B, Zhang W, Wu T, Wen T, et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis. 2020;222(5):746–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz-Enriquez J, Rojas-Morales E, Ruiz-Garcia MG, Tobon-Velasco J, Jimenez J. SARS-CoV-2 induces mitochondrial dysfunction and cell death by oxidative stress in leukocytes of COVID-19 patients. Free Radic Res. 2021;55(9–10):982–95. [DOI] [PubMed] [Google Scholar]
- Delshad M, Tavakolinia N, Pourbagheri-Sigaroodi A, Safaroghli-Azar A, Bagheri N, Bashash D. The contributory role of lymphocyte subsets, pathophysiology of lymphopenia and its implication as prognostic and therapeutic opportunity in COVID-19. Int Immunopharmacol. 2021;95: 107586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Resendiz KJG, Benitez-Trinidad AB, Covantes-Rosales CE, Toledo-Ibarra GA, Ortiz-Lazareno PC, Girón-Pérez DA, et al. Loss of mitochondrial membrane potential (ΔΨ m ) in leucocytes as post-COVID-19 sequelae. J Leukoc Biol. 2022;112(1):23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C, Ma R, Zeng X, Chen B, Hou D, Liu R, et al. SARS-CoV-2 achieves immune escape by destroying mitochondrial quality: comprehensive analysis of the cellular landscapes of lung and blood specimens from patients with COVID-19. Front Immunol. 2022;1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandson KM, Geng LN, Selvaggi CA, Thaweethai T, Chen P, Erdmann NB, et al. Differentiation of prior SARS-CoV-2 infection and postacute sequelae by standard clinical laboratory measurements in the RECOVER cohort. Ann Intern Med. Published online August 13, 2024. [DOI] [PMC free article] [PubMed]
- Fields JA, Serger E, Campos S, Divakaruni AS, Kim C, Smith K, et al. HIV alters neuronal mitochondrial fission/fusion in the brain during HIV-associated neurocognitive disorders. Neurobiol Dis. 2016;1(86):154–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcados GE, Muhammad A, Oladipo OO, Makama S, Meseko CA. Metabolic implications of oxidative stress and inflammatory process in SARS-CoV-2 pathogenesis: therapeutic potential of natural antioxidants. Front Cell Infect Microbiol. 2021;26(11):457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes-Martín R, Moncayo S, Insenser M, Martínez-García MÁ, Luque-Ramírez M, Grau NA, et al. Glycoprotein A and B height-to-width ratios as obesity-independent novel biomarkers of low-grade chronic inflammation in women with polycystic ovary syndrome (PCOS). J Proteome Res. 2019;18(11):4038–45. [DOI] [PubMed] [Google Scholar]
- Giacomello M, Pyakurel A, Glytsou C, Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol. 2020;21(4):204–24. [DOI] [PubMed] [Google Scholar]
- Gibellini L, De Biasi S, Paolini A, Borella R, Boraldi F, Mattioli M, et al. Altered bioenergetics and mitochondrial dysfunction of monocytes in patients with COVID‐19 pneumonia. EMBO Mol Med. 2020;12(12). [DOI] [PMC free article] [PubMed]
- Gil M, Samino S, Barrilero R, Correig X. Lipid profiling using 1H NMR spectroscopy. Methods Mol Biol. 2019;2037:35–47. [DOI] [PubMed] [Google Scholar]
- Kalinina O, Golovkin A, Zaikova E, Aquino A, Bezrukikh V, Melnik O, et al. Cytokine storm signature in patients with moderate and severe COVID-19. Int J Mol Sci MDPI. 2022;23(16). [DOI] [PMC free article] [PubMed]
- Karagiannis F, Peukert K, Surace L, Michla M, Nikolka F, Fox M, et al. Impaired ketogenesis ties metabolism to T cell dysfunction in COVID-19. Nature. 2022;609(7928):801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Syed GH, Kim S-J, Siddiqui A. Mitochondrial dynamics and viral infections: a close nexus. Biochim. Biophys. Acta - Mol Cell Res. 2015;1853(10):2822–33. [DOI] [PMC free article] [PubMed]
- Kim SJ, Khan M, Quan J, Till A, Subramani S, Siddiqui A. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013;9(12):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok FA, Kruip MJHA, van der Meer NJM, Arbous MS, Gommers DAMPJ, Kant KM, et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–7. [DOI] [PMC free article] [PubMed]
- Lee H, Smith SB, Sheu SS, Yoon Y. The short variant of optic atrophy 1 (OPA1) improves cell survival under oxidative stress. J Biol Chem. 2020;295(19):6543–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L, Chen Z, Lui G, Wong CK, Wong WT, Ng RWY, et al. Longitudinal cytokine profile in patients with mild to critical COVID-19. Front Immunol. 2021;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhao J, Wang H, Wang W, Su X, Liao X, et al. Metabolic defects of peripheral T cells in COVID-19 patients. J Immunol. 2021;206(12):2900–8. [DOI] [PubMed] [Google Scholar]
- Mallol R, Amigó N, Rodríguez MA, Heras M, Vinaixa M, Plana N, et al. Liposcale: a novel advanced lipoprotein test based on 2D diffusion-ordered 1H NMR spectroscopy. J Lipid Res. 2015;56(3):737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefusco L, Ben Nasr M, D’Addio F, Loretelli C, Rossi A, Pastore I, et al. Acute and long-term disruption of glycometabolic control after SARS-CoV-2 infection. Nat Metab. 2021;3(6):774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VAK, Lee S-J, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarte KI, de Oliveira MHS, Peligro PJ, Velasco JV, Macaranas I, Ver AT, et al. Age, sex and previous comorbidities as risk factors not associated with SARS-CoV-2 infection for long COVID-19: a systematic review and meta-analysis. J Clin Med. 2022;11(24). [DOI] [PMC free article] [PubMed]
- Oelsner EC, Sun Y, Balte PP, Allen NB, Andrews H, Carson A, et al. Epidemiologic features of recovery from SARS-CoV-2 infection. JAMA Netw. 2024;7(6): e2417440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcariz E, Guardiola M, Amigó N, Rojo-Martínez G, Valdés S, Rehues P, et al. NMR-based metabolomic profiling identifies inflammation and muscle-related metabolites as predictors of incident type 2 diabetes mellitus beyond glucose: the Di@bet.es study. Diabetes Res Clin Pract. 2023;202:110772. [DOI] [PubMed]
- Ramasamy S, Subbian S. Critical determinants of cytokine storm and type i interferon response in COVID-19 pathogenesis. Clin Microbiol. Rev. 2021;34(3). [DOI] [PMC free article] [PubMed]
- Romão PR, Teixeira PC, Schipper L, da Silva I, Santana Filho P, Júnior LCR, et al. Viral load is associated with mitochondrial dysfunction and altered monocyte phenotype in acute severe SARS-CoV-2 infection. Int Immunopharmacol. 2022;1:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal N, Cao Z, Gundrum J, Sianis J, Safo S. Risk factors associated with in-hospital mortality in a US national sample of patients with COVID-19. JAMA Netw. 2020;3(12): e2029058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh J, Peyssonnaux C, Singh KK, Edeas M. Mitochondria and microbiota dysfunction in COVID-19 pathogenesis. Mitochondrion. 2020;54:1–7. [DOI] [PMC free article] [PubMed]
- Šestan M, Mikašinović S, Benić A, Wueest S, Dimitropoulos C, Mladenić K, et al. An IFNγ-dependent immune-endocrine circuit lowers blood glucose to potentiate the innate antiviral immune response. Nat Immunol. 2024;25(6):981–93. [DOI] [PubMed] [Google Scholar]
- Sharma C, Bayry J. High risk of autoimmune diseases after COVID-19. Nat Rev Rheumatol. 2023;19(7):399–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S. Coronavirus (Covid-19) sepsis: revisiting mitochondrial dysfunction in pathogenesis, aging, inflammation, and mortality. Inflamm Res. 2020;69(11):1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C-S, Qi H-Y, Boularan C, Huang N-N, Abu-Asab M, Shelhamer JH, et al. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol. 2014;193(6):3080–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria-Castro E, Soto ME, Guarner-Lans V, Rojas G, Perezpeña-Diazconti M, Críales-Vera SA, et al. The kidnapping of mitochondrial function associated with the sars-cov-2 infection. Histol Histopathol. 2021;36(9):947–65. [DOI] [PubMed] [Google Scholar]
- Srinivasan K, Pandey AK, Livingston A, Venkatesh S. Roles of host mitochondria in the development of COVID-19 pathology: could mitochondria be a potential therapeutic target? Mol. Biomed. Springer Science and Business Media LLC. 2021;2(1). [DOI] [PMC free article] [PubMed]
- Stanczak MA, Sanin DE, Apostolova P, Nerz G, Lampaki D, Hofmann M, et al. IL-33 expression in response to SARS-CoV-2 correlates with seropositivity in COVID-19 convalescent individuals. Nat Commun. 2021;12(1):2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talla A, Vasaikar SV, Szeto GL, Lemos MP, Czartoski JL, MacMillan H, et al. Persistent serum protein signatures define an inflammatory subcategory of long COVID. Nat Commun. 2023;14(1):3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo-Velasco Á, Martínez-Paz P, Peñarrubia-Ponce MJ, de la Fuente I, Pérez-González S, Fernández I, et al. HGF, IL-1α, and IL-27 are robust biomarkers in early severity stratification of COVID-19 patients. J Clin Med MDPI. 2021;10(9). [DOI] [PMC free article] [PubMed]
- Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhou L, Sun X, Yan Z, Hu C, Wu J, et al. Altered lipid metabolism in recovered SARS patients twelve years after infection. Sci Rep. 2017;7(1). [DOI] [PMC free article] [PubMed]
- Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, et al. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2(84). [DOI] [PubMed]
- Yin K, Peluso MJ, Luo X, Thomas R, Shin M-G, Neidleman J, et al. Long COVID manifests with T cell dysregulation, inflammation and an uncoordinated adaptive immune response to SARS-CoV-2. Nat Immunol. 2024;25(2):218–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip TC-F, Lui GC-Y, Wong VW-S, Chow VC-Y, Ho TH-Y, Li TC-M, et al. Liver injury is independently associated with adverse clinical outcomes in patients with COVID-19. Gut. 2021;70(4):733–42. [DOI] [PubMed]
- Zamani B, Najafizadeh M, Motedayyen H, Arefnezhad R. Predicting roles of IL-27 and IL-32 in determining the severity and outcome of COVID-19. Int J Immunopathol Pharmacol NLM (Medline). 2022;36. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1.24 MB)
Data Availability Statement
The data that support the findings of this study are available upon request from the corresponding author and the metabolomics data have been deposited in an open access repository: (https://github.com/BiosferTeslab/NMR_Metabolomics_Long_Term_Impairment_After_Severe_Covid).