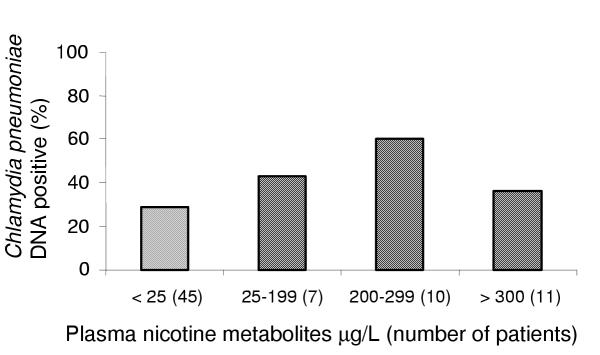Abstract
Background
The prevalence and role of Chlamydia pneumoniae in chronic obstructive pulmonary disease (COPD) remain unclear.
Methods
Peripheral blood mononuclear cells were obtained from 100 outpatients with smoking-related, clinically stable COPD, and induced sputum was obtained in 62 patients.
Results
Patients had mean age (standard deviation) of 65.8 (10.7) years, mean forced expiratory volume in one second of 1.34 (0.61) L, and 61 (61.0%) were male. C. pneumoniae nucleic acids were detected by nested polymerase chain reaction in 27 (27.0%). Current smoking (odds ratio {OR} = 2.6, 95% confidence interval {CI}: 1.1, 6.6, P = 0.04), season (November to April) (OR = 3.6, 95% CI: 1.4, 9.2, P = 0.007), and chronic sputum production (OR = 6.4, 95% CI: 1.8, 23.2, P = 0.005) were associated with detection of C. pneumoniae DNA.
Conclusions
Prospective studies are needed to examine the role of C. pneumoniae nucleic acid detection in COPD disease symptoms and progression.
Background
Chronic obstructive pulmonary disease (COPD) is a major cause of morbidity and mortality throughout the world [1]. By 2020, the World Health Organization predicts that COPD will become the 5th (currently 12th) most prevalent disease worldwide, and the 3rd (currently 6th) most common cause of death [2]. Smoking is the major identified risk factor, and approximately 15% of smokers develop COPD [1]. Smoking cessation reduces the rate of decline in forced expiratory volumes, but no pharmacological intervention has been shown to modify the progression of disease. Bacterial and viral infections of the respiratory tract trigger acute exacerbations of chronic bronchitis, and may contribute to disease progression [3].
A role for Chlamydia pneumoniae in patients with COPD remains unclear. C. pneumoniae infection, diagnosed serologically, caused 5 to 10% of acute exacerbations in patients with chronic bronchitis [4,5]. The seroprevalence and geometric mean titres of C. pneumoniae IgG [5,6]. and IgA [7] antibodies were higher in COPD patients than controls, irrespective of exacerbation status. Von Hertzen et al measured serum IgG and IgA, salivary secretory IgA, circulating immune complexes, and nucleic acid, and found evidence of "chronic" C. pneumoniae infection in 65% of COPD patients [8], and recently Lieberman et al showed an association between C. pneumoniae serology, determined by a commercial immunoassay, and COPD exacerbation [9]. However, the reproducibility and validity of serologic testing remain uncertain [10].
Given these limitations of serology, the direct detection of C. pneumoniae nucleic acids may facilitate studies of its role in chronic disease etiology. The presence of C. pneumoniae has been directly demonstrated in lung tissue [11], and correlated with detection of C. pneumoniae DNA in peripheral blood mononuclear cells (PBMC) by PCR (Blasi et al, Proceedings of the Fourth Meeting of the European Society for Chlamydia Research, Helsinki, Finland, 2000). However, there are limited data regarding the prevalence of C. pneumoniae DNA in PBMC of COPD patients.
In a previous study, we found that PBMC C. pneumoniae DNA detection among cardiac patients [12] was associated with current smoking and winter/spring season. The primary objective of this study was to examine the prevalence and clinical determinants of C. pneumoniae DNA in COPD patients, and in particular to validate the observed association with current smoking and season. Secondary objectives were to explore whether C. pneumoniae detection was associated with unique symptoms, pulmonary function, or sputum cell counts, and to contrast the prevalence among respirology and cardiology patients.
Materials and methods
Subjects
One hundred consecutive consenting outpatients were recruited from the Firestone Institute for Respiratory Health at St. Joseph's Healthcare between March 2000 and October 2001. The study protocol was approved by the hospital's Research Ethics Board, and all subjects gave written consent. Patient characteristics are summarized in Table 1. Patients were current or former smokers who had a previous diagnosis of smoking-related chronic airflow limitation, with best forced expiratory volume in one second (FEV1) of < 70% predicted and FEV1/Vital Capacity < 70%, were clinically stable, and had not received antibiotics in the prior month. Spirometry was obtained before and 10 minutes after inhalation of 200 μg inhaled salbutamol. Detailed smoking history, symptoms, use of corticosteroids, and past history of pneumonia, diabetes mellitus, angina pectoris, and myocardial infarction were obtained. Clinical and laboratory data were collected independently by blinded study personnel. Seven patients with no history of prior smoking were enrolled, and their spirometry data and sputum cell counts were excluded from the analyses.
Table 1.
Patient characteristics by Chlamydia pneumoniae DNA detection status.
| Entire Study Group (%) | DNAa Present (%) | DNA Absent (%) | Unadjusted ORb (95% CI) | |
| Number | 100 | 27 (27.0) | 73 (73.0) | |
| Male | 61 / 100 (61.0) | 20 / 27 (74.1) | 41 / 73 (56.2) | 2.2 (0.8, 5.9) |
| Current smoking | 32 / 100 (32.0) | 13 / 27 (48.1) | 19 / 73 (26.0) | 2.6 (1.1, 6.6) |
| Season (Nov. to April) | 44 / 100 (44.0) | 18 / 27 (66.7) | 26 / 73 (35.6) | 3.6 (1.4, 9.2) |
| Smoker or Season | 63 / 100 (63.0) | 25 / 27 (92.6) | 38 / 73 (52.1) | 11.5 (2.5, 52.2) |
| Chronic cough | 50 / 98 (51.0) | 16 / 26 (61.5) | 34 / 72 (47.2) | 1.8 (0.7, 4.5) |
| Chronic sputum | 64 / 99 (64.6) | 24 / 27 (88.9) | 40 / 72 (55.6) | 6.4 (1.8, 23.2) |
| Pneumonia in 1 year | 9 / 98 (9.2) | 2 / 26 (7.7) | 7 / 72 (9.7) | 0.8 (0.2, 4.0) |
| Prednisone | 18 / 98 (18.4) | 3 / 27 (11.1) | 15 / 71 (21.1) | 0.5 (0.1, 1.8) |
| Inhaled corticosteroids | 67 / 98 (68.4) | 19 / 27 (70.4) | 48 / 71 (67.6) | 1.1 (0.4, 3.0) |
| Coronary artery disease | 10 / 92 (10.9) | 2 / 27 (7.4) | 8 / 65 (12.3) | 0.6 (0.1, 2.9) |
| Diabetes mellitus | 5 / 92 (5.4) | 2 / 27 (7.4) | 3 / 65 (4.6) | 1.7 (0.3, 10.5) |
aC. pneumoniae DNA detected by nested polymerase chain reaction [14]. b Unadjusted odds ratios (OR) with 95% confidence intervals (SPSS).
Sample size
A sample size of 100 produces a 95% confidence interval equal to the sample proportion ± 0.08 when the estimated proportion is 0.20. For an estimated 50% prevalence of each of current smoking and season, assuming independence, the study had 64% power to detect an odds ratio (OR) of 5.0 or higher, and 40% power to detect an odds ratio of 3.0 or higher, for each of these factors as predictors of C. pneumonia DNA detection, using X2 test with continuity correction and with a significance level of 0.05, two-tailed (Power Analysis and Sample Size for Windows 2000, NCSS, Kaysville, UT, USA).
Sputum induction and examination
Sputum induction and processing was performed as described by Pizzichini et al [13] for the first 73 patients, and no sputum was collected for remaining patients. Sputum induction was discontinued based on low prevalence of C. pneumoniae DNA and to increase enrollment. A differential count of 400 non-squamous cells was performed by experienced laboratory technologists, and a sample of sputum was frozen at -70°C for PCR testing in 62 patients. Insufficient sputum was available on the remaining 11 patients after performance of sputum cytology.
Blood collection
Peripheral blood mononuclear cells (PBMC) were obtained by venipuncture into an 8 mL cell preparation tube (CPT, BD Vacutainer Systems, Franklin Lakes, NJ, USA). Within 2 hours of venipuncture, specimens were mixed by inversion and centrifuged at 1500 × g for 30 minutes. The mononuclear cells layer was aspirated and frozen at -70°C. Samples were thawed in batches, 200–500 μL aliquots were pelleted and extracted using QIAamp DNA mini-kits (Qiagen, Mississauga ON) into 50–100 μL of elution buffer.
Detection of chlamydia pneumoniae DNA
A 5–10 μL aliquot was amplified in triplicate by a nested polymerase chain reaction (PCR) targeting the ompA gene [14], consisting of 40 amplification cycles for a 333 base pair product and 30 cycles for a 207 bp product, followed by separation on a 2.0% (weight/volume) TAE agarose gel containing ethidium bromide, with ultraviolet light visualization. The 207 base pair product was confirmed as C. pneumoniae by hybridization with a specific fluorescein-labeled oligonucleotide probe (ECL, Amersham). A probe-confirmed positive in any of the three reactions was considered a positive, as previously described [12,15]. Eight controls, consisting of one low strength positive control and one intermediate strength positive control, six negative controls without DNA, and one additional tube with master mix open to the air throughout specimen addition, were run for every 13 patient specimens. PCR extraction and amplification were performed in separate rooms.
Nicotine metabolites assay
Using the plasma fraction from the citrated CPT tube, nicotine metabolites were assayed in 73 patients by enzyme immunoassay (Immulite Nicotine Metabolite detection kit, Diagnostic Products Corporation, Los Angeles CA, USA). The Immulite nicotine metabolite assay has a calibration range of 10 to 500 μg/L, and non-smokers are defined by a nicotine metabolite level of < 25 μg/mL.
Statistical analysis
Continuous variables were tested by unpaired t-test or Mann Whitney U test, and proportions were compared using X2 or Fisher exact tests (unmatched data) or the McNemar test (matched data). Multiple logistic regression modeling (SPSS for Windows 10.0, SPSS Inc., Chicago IL) was undertaken using DNA status (in blood or sputum) as the response variable. A priori, an association with current smoking and with winter/spring season was expected. All associations with clinical history, spirometry and cell counts were considered hypothesis-generating. To examine the effect of missing data, smoking and season were analyzed after imputation of C. pneumoniae DNA status for PBMC negative subjects in whom sputum data was missing. Imputation was performed based on proportion sputum DNA positive among subjects who were DNA negative in PBMC (SPSS).
Prevalence of PBMC C. pneumoniae DNA in COPD patients was compared with our coronary angiography study[12] using gender, smoking and season-stratified Maentel-Haentzel X2. Predictors of FEV1 were modeled by linear regression, for the entire study group and separately by age ≤ 65 years or >65 years. A P-value of < 0.05, two-tailed, was considered statistically significant for the primary hypothesis of an association with current smoking or season, and a P-value of < 0.01, two-tailed, was considered significant for all secondary analyses.
Results
C. pneumoniae DNA prevalence was 24 of 100 (24.0%) in PBMC and 7 of 62 (11.3%) in induced sputum, for a total of 27 of 100 (27.0%) patients positive in at least one specimen type. Among 62 subjects from whom both PBMC and sputum were obtained, PBMC detected 21 of 24 (87.5%) positives, whereas sputum detected 7 of 24 (29.2%) positives (P = 0.003, McNemar test).
C. pneumoniae DNA detection in PBMC or sputum by categorical clinical characteristics is tabulated in Table 1, with unadjusted odds ratios, and by continuous clinical and laboratory characteristics in Table 2.
Table 2.
Spirometrya and sputum cell counts by Chlamydia pneumoniae DNA detection status among 92 patients with chronic obstructive pulmonary disease.
| Number of Subjects | Entire Study Group | DNAb Present | DNA Absent | P-value (t-test) | |
| Age in years (SD) | 93 | 65.8 (10.7) | 66.9 (11.9) | 65.3 (10.3) | 0.53 |
| Smoking pack-years (SD) | 93 | 43.1 (25.1) | 48.9 (29.3) | 41.8 (24.1) | 0.22 |
| FEV1c (SD), L | 93 | 1.35 (0.61) | 1.34 (0.64) | 1.36 (0.61) | 0.88 |
| FEV1 % predicted (SD) | 93 | 46.9 (16.8) | 44.3 (15.1) | 48.0 (17.5) | 0.33 |
| FEV1/VCd % (SD) | 93 | 45.7 (15.4) | 42.6 (15.8) | 47.0 (15.2) | 0.22 |
| Total cell count, 106/g (SD) | 73 | 10.9 (15.4) | 9.9 (12.5) | 11.4 (16.8) | 0.70 |
| Viability, % (SD) | 73 | 61.5 (24.2) | 60.4 (22.7) | 62.0 (25.0) | 0.79 |
| Squamous Epithelial, % (SD) | 73 | 3.8 (8.6) | 3.7 (5.3) | 3.9 (10.0) | 0.92 |
| Neutrophils, % (SD) | 73 | 63.6 (25.3) | 60.6 (27.2) | 65.2 (24.5) | 0.46 |
| Macrophages, % (SD) | 73 | 25.6 (19.2) | 26.4 (21.2) | 25.2 (18.3) | 0.78 |
| Eosinophils, median % (Inter-quartile range)e | 73 | 1.2 (3.0) | 1.2 (3.7) | 1.0 (2.9) | 0.71e |
aAll spirometry after inhalation of 200 μg salbutamol. bC. pneumoniae DNA detected in peripheral blood mononuclear cells or induced sputum by nested polymerase chain reaction [14]. cFEV1: forced expiratory volume in 1 second. dVC: vital capacity. eComparison by Mann Whitney U test (SPSS).
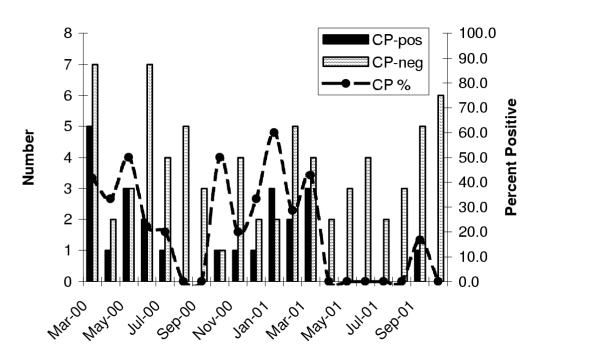
Current smoking (OR = 3.5, 95% CI: 1.2, 9.9, P = 0.02) and season (November to April, OR = 3.9, 95% CI: 1.4, 10.9, P = 0.008) were associated with DNA detection, independent of age and gender, whereas male gender was not (OR = 2.2, 95% CI: 0.8, 6.3, P = 0.14). A month-by-month breakdown for C. pneumoniae DNA positive PBMC is illustrated in Figure 1. The composite of current smoking or season identified 25 of 27 (92.6%) C. pneumoniae positives, and was strongly associated with DNA detection (age and gender adjusted OR = 11.1, 95% CI: 2.4, 50.5, P = 0.002). The composite of smoking or season was also associated with DNA detection in PBMC alone (adjusted OR = 9.0, 95% CI: 2.0, 41.4, P = 0.005).
Figure 1.
Relation of Chlamydia pneumoniae (CP) DNA prevalence in blood peripheral mononuclear cells versus month of study visit among 100 patients presenting to a respirology referral clinic. CP-DNA positive (solid), CP-DNA negative (thatched) or proportion of all monthly specimens which were CP-DNA positive (line).
To further explore the relationship between current smoking status and the detection of C. pneumoniae DNA, we measured plasma nicotine metabolites in 73 subjects. Levels = 25 μg/L indicate current smoking status. At this threshold, seven former smokers were reclassified as current smokers, and one current smoker as a former smoker. Nicotine metabolites = 25 μg/L were associated with C. pneumoniae DNA with OR of 2.1 (95% CI: 0.8, 5.7, P = 0.13). There was no relationship between higher plasma nicotine levels and the probability of detecting Chlamydia pneumoniae DNA (see Figure 2.)
Figure 2.
Relation of Chlamydia pneumoniae DNA prevalence in blood or sputum versus plasma nicotine metabolite levels among 73 patients with stable chronic obstructive pulmonary disease. Nicotine metabolites of < 25 μg/L indicate no recent exposure to smoking. For nicotine metabolites = 25 μg/L, odds ratio = 2.1 (95% CI: 0.8, 5.7, P = 0.13) for detection of C. pneumoniae DNA (SPSS).
To assess the influence of missing C. pneumoniae DNA status of sputum, imputation of one, three or seven additional sputum positives was performed, corresponding to the proportion of observed sputum DNA positives with 95% confidence interval among subjects who were PBMC DNA negative. Imputation was performed without regard to smoking or season. The OR associated with season was 3.7 in the non-imputed model, and 3.4, 3.1, and 2.4 respectively for one, three or seven imputed positives, in a model adjusted for smoking, age and gender. The ORs for smoking in these models were 3.7, 3.4, 2.6, and 2.0, for the zero, one, three or seven imputed positives, respectively.
Self-reported chronic sputum production was associated with C. pneumoniae DNA detection (age, gender, smoking and season-adjusted OR = 4.2, 95% CI: 1.1, 16.0, P = 0.04), whereas self-reported chronic cough, use of corticosteroids, or a history of coronary artery disease or diabetes mellitus were not (see Table 1). Chronic sputum production identified 24 of 27 (88.9%) of patients with C. pneumoniae DNA detected. In a forward selection model including sputum production, smoking, season, gender and age, only sputum production (OR = 5.3, 95% CI: 1.4, 19.6, P = 0.01) and season (OR = 2.8, 95% CI: 1.1, 7.5, P = 0.04) were independent predictors. Two-way interactions were not statistically significant.
We examined the association between C. pneumoniae DNA and measures of pulmonary function (Table 2), and sputum cell counts. Pulmonary function was moderately to severely impaired, and no relationship was found with C. pneumoniae DNA detection. Sputum neutrophil, macrophage, and eosinophil counts were not associated with C. pneumoniae detection.
We also examined the association between C. pneumoniae DNA and spirometry by linear regression modeling of FEV1. Age, gender, current smoking, C. pneumoniae DNA status, and season were included in the model (Table 3). In the entire study group, age and male gender were predictors of FEV1. However, if we used the per cent predicted FEV1 as the outcome variable, no variable was significant at the P < 0.05 level (data not shown), and only smoking approached this level of significance (P = 0.09). We then examined subjects aged 65 years or younger, and those older than 65 years, separately. In the younger age group, age predicted FEV1, whereas current smoking, male gender and C. pneumoniae DNA status were associated at P < 0.10. In the older age group, male gender was associated with FEV1, but age, current smoking, season or DNA status were not.
Table 3.
Clinical predictors of forced expiratory volume in one second among 93 patients with chronic obstructive pulmonary disease.
| Beta Coefficient (95% Confidence Interval)a | |||
| Risk Factor | Entire Study Group | Age ≤ 65 years (n = 34) | Age > 65 years (n = 59) |
| Age (L/year) | -0.03 (-0.04, -0.01)b | -0.05 (-0.08, -0.02)b | -0.01 (-0.04, 0.02) |
| Male | 0.43 (0.21, 0.66)b | 0.35 (-0.03, 0.74)d | 0.41 (0.12, 0.70)c |
| Current Smoker | -0.17 (-0.41, 0.08) | -0.40 (-0.80, 0.00)d | -0.05 (-0.36, 0.27) |
| Chlamydia pneumoniae DNAe | -0.16 (-0.41, 0.10) | -0.40 (-0.86, 0.06)d | -0.10 (-0.43, 0.23) |
| November to April | 0.19 (-0.04, 0.41) | 0.19 (-0.20, 0.57) | 0.17 (-0.12, 0.47) |
a Multiple linear regression beta coefficient (litres) associated with the presence of clinical variables for prediction of FEV1 (SPSS). Age in years, all other variables coded as present (1) or absent (0). b P < 0.001 c P < 0.05 d P < 0.10 eC. pneumoniae DNA detected in peripheral blood mononuclear cells or induced sputum by nested polymerase chain reaction [14]
PBMC results from the COPD patients in this study were compared with patients from the coronary angiography study reported previously [12]. COPD patients were more likely to have C. pneumoniae DNA in PBMC than patients undergoing coronary angiography (24 of 100 versus 24 of 208, adjusted OR = 2.9, 95% CI: 2.4, 9.9, P < 0.001), independent of current smoking status, gender, or season. The highest monthly prevalence was found in March and April, irrespective of year or study group. Among cardiology patients, the C. pneumoniae prevalence in March and April 1999 was 16% (20 of 122), compared with a prevalence of 5% (3 of 60) in the months of May to October. Similarly, in COPD patients studied in 2000 and 2001, March and April prevalence was 50% (9 of 18), compared with 15% (6 of 41) in the months of May to October. In the combined dataset, season (November to April OR = 4.3, 95% CI: 2.0, 9.2, P < 0.001) and smoking (OR = 2.7, 95% CI: 1.3, 5.4, P = 0.005) predicted C. pneumoniae DNA detection in PBMC, whereas male gender did not (OR = 1.5, 95% CI: 0.7, 3.2, P = 0.28). The combined measure of current smoking or season was associated with DNA detection (OR = 10.1, 95% CI: 3.0, 33.8, P < 0.001).
Discussion
Among clinically stable patients with smoking-related COPD and moderate to severe pulmonary function impairment, we detected a relatively high prevalence of C. pneumoniae DNA. Peripheral blood (PBMC) specimens were superior to induced sputa, and an association between C. pneumoniae DNA detection, current smoking and winter/spring months was demonstrated. Chronic sputum production was associated with C. pneumoniae, but we found no clear association with other clinical symptoms, measures of lung function, or sputum cell counts.
Data on the prevalence of C. pneumoniae DNA in PMBC of COPD patients have been published solely in abstract form. Black et al found a prevalence of 14 of 20 (70.0%, 95% CI: 48.1, 85.5) Australian patients, using whole blood or PBMC (Black et al, Proceedings of the Fourth Meeting of the European Society for Chlamydia Research, Helsinki, Finland, 2000). Blasi et al found a prevalence of 10 among 21 (47.6%, 95% CI: 28.3, 67.6) Italian patients, compared with 11 of 21 (52.4%) who had C. pneumoniae DNA in trans-bronchial lung biopsy specimens (Blasi et al, Proceedings of the Fourth Meeting of the European Society for Chlamydia Research, Helsinki, Finland, 2000). Our prevalence of 24.0% in PBMC of Canadian COPD patients is slightly lower than these previous estimates, although within the 95% confidence interval for Blasi et al's estimate. Although this estimate was higher for patients with chronic respiratory disease compared with patients undergoing coronary angiography, we have recently found a high prevalence (31/103, 29.8%) among family practice controls without acute cardiorespiratory symptoms (Smieja et al, unpublished data).
We found a low prevalence (11%) of C. pneumoniae DNA in induced sputum among patients with chronic stable COPD. By contrast, Von Hertzen et al detected DNA in spontaneous sputum of 59% of patients with severe COPD [8], and Black et al detected DNA in 94% of patients hospitalized for COPD exacerbation (Black et al, Proceedings of the Fourth Meeting of the European Society for Chlamydia Research, Helsinki, Finland, 2000). We obtained induced sputum to measure sputum cell counts by a validated methodology, but spontaneous sputum may be superior for routine C. pneumoniae detection. Conversely, sputum positivity may be higher during acute exacerbations, and our study intentionally excluded such patients. Our data support the utility of PBMC in preference to sputum for C. pneumoniae DNA detection.
A clearer relationship between C. pneumoniae and smoking is emerging. Hahn first proposed that smoking was a potential confounder of the association between C. pneumoniae antibody levels and cardiovascular disease [16]. Von Hertzen et al showed that smoking was associated with higher C. pneumoniae IgA levels in smoking-discordant twins [17], and Karvonen et al showed that C. pneumoniae IgG seropositivity to be higher in smokers in a population-based study [18]. This study has validated our previous observation that current smoking was associated with a higher prevalence of C. pneumoniae nucleic acid detection [12]. Whether C. pneumoniae associations with chronic diseases such as COPD or coronary artery disease are confounded by inaccurate ascertainment of smoking status, or whether infection is directly involved in the causal pathway for smoking-associated diseases, will require careful investigation.
Annual C. pneumoniae seasonal cycles have not, to our knowledge, been previously reported. Among both cardiology and respirology patients, we found peak prevalence in the winter/spring months, particularly in March and April, over three consecutive winters. Whether similar seasonal variation is present in other geographic areas is not known.
We identified no clear relationship between FEV1 and C. pneumoniae DNA detection. In the sub-group of patients aged 65 years or younger, current smoking and C. pneumoniae DNA status were each associated with a reduced of FEV1 approximately 400 mL, although neither was nominally significant at a level of P < 0.05. A larger sample size may allow more precise estimation of this relationship, although only a prospective study design will adequately determine whether DNA detection is associated with greater decline in FEV1. Our data suggest that a study enrolling subjects 65 years or younger may have greater power for examining whether C. pneumoniae is associated with lung function decline independently of smoking status.
We identified self-reported chronic sputum production as a strong risk factor for C. pneumoniae DNA detection. However, this association requires validation, and was not significant at P < 0.01 as we pre-specified for secondary analyses. Nevertheless, C. pneumoniae DNA detection in stable COPD patients may be associated with respiratory symptoms.
In conclusion, C. pneumoniae was prevalent in patients with COPD and associated with current smoking and winter/spring season, and may be associated with chronic sputum production. C. pneumoniae DNA detection in PBMC may facilitate the undertaking of large prospective epidemiologic studies examining the role of C. pneumoniae in COPD etiology and progression.
Competing interests
None declared.
Authors' contributions
MS, RL, and AP were responsible for conception, design, analysis and manuscript writing. FEH, MC, and JBM participated in design and critical review of the manuscript. DK enrolled patients and performed spirometry and sputum induction, SC performed blood processing and nucleic acid amplification. CHG provided design, statistical advice, and critical review of the manuscript. MS acts as guarantor for the accuracy and integrity of the research. All authors read and approved the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
Supported by a grant-in-aide from the Father Sean O'Sullivan Research Centre, St. Joseph's Healthcare, Hamilton ON Canada.
Contributor Information
Marek Smieja, Email: smiejam@mcmaster.ca.
Richard Leigh, Email: rleigh@mcmaster.ca.
Astrid Petrich, Email: petricha@mcmaster.ca.
Sylvia Chong, Email: schong@idirect.com.
Dennis Kamada, Email: kamad@mcmaster.ca.
Frederick E Hargreave, Email: hargreav@mcmaster.ca.
Charles H Goldsmith, Email: goldsmit@mcmaster.ca.
Max Chernesky, Email: chernesk@mcmaster.ca.
James B Mahony, Email: mahonyj@mcmaster.ca.
References
- Barnes PJ. Chronic obstructive pulmonary disease. N Engl J Med. 2000;343:269–280. doi: 10.1056/NEJM200007273430407. [DOI] [PubMed] [Google Scholar]
- Murray CJ, Lopez AD. Alternative projections of mortality and disability by cause 1990–2020: Global Burden of Disease Study. Lancet. 1997;349:1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- Sethi JM, Rochester CL. Smoking and chronic obstructive pulmonary disease. Clin Chest Med. 2000;21:67–86. doi: 10.1016/s0272-5231(05)70008-3. [DOI] [PubMed] [Google Scholar]
- Thom DH, Grayston JT, Campbell LA, Kuo CC, Diwan VK, Wang SP. Respiratory infection with Chlamydia pneumoniae in middle-aged and older adult outpatients. Eur J Clin Microbiol Infect Dis. 1994;13:785–792. doi: 10.1007/BF02111337. [DOI] [PubMed] [Google Scholar]
- Blasi F, Legnani D, Lombardo VM, Negretto GG, Magliano E, Pozzoli R, et al. Chlamydia pneumoniae infection in acute exacerbations of COPD. Eur Respir J. 1993;6:19–22. [PubMed] [Google Scholar]
- Miyashita N, Niki Y, Nakajima M, Kawane H, Matsushima T. Chlamydia pneumoniae infection in patients with diffuse panbronchiolitis and COPD. Chest. 1998;114:969–971. doi: 10.1378/chest.114.4.969. [DOI] [PubMed] [Google Scholar]
- Von Hertzen L, Isoaho R, Leinonen M, Koskinen R, Laippala P, Toyryla M, et al. Chlamydia pneumoniae antibodies in chronic obstructive pulmonary disease. Int J Epidemiol. 1996;25:658–664. doi: 10.1093/ije/25.3.658. [DOI] [PubMed] [Google Scholar]
- Von Hertzen L, Alakarppa H, Koskinen R, Liippo K, Surcel HM, Leinonen M, et al. Chlamydia pneumoniae infection in patients with chronic obstructive pulmonary disease. Epidemiol Infect. 1997;118:155–164. doi: 10.1017/S095026889600725X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman D, Ben-Yaakov M, Lazarovich Z, Ohana B, Boldur I. Chlamydia pneumoniae Infection in Acute Exacerbations of Chronic Obstructive Pulmonary Disease: Analysis of 250 Hospitalizations. Eur J Clin Microbiol Infect Dis. 2001;20:698–704. doi: 10.1007/s100960100596. Ref Type: Generic. [DOI] [PubMed] [Google Scholar]
- Tompkins LS, Schachter J, Boman J, Chernesky MA, Dowell S, Gaydos CA, et al. Collaborative multidisciplinary workshop report: detection, culture, serology, and antimicrobial susceptibility testing of Chlamydia pneumoniae. J Infect Dis. 2000;181:S460–S461. doi: 10.1086/315599. [DOI] [PubMed] [Google Scholar]
- Wu L, Skinner SJ, Lambie N, Vuletic JC, Blasi F, Black PN. Immunohistochemical staining for Chlamydia pneumoniae is increased in lung tissue from subjects with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2000;162:1148–1151. doi: 10.1164/ajrccm.162.3.9912134. [DOI] [PubMed] [Google Scholar]
- Smieja M, Chong S, Natarajan M, Petrich A, Rainen L, Mahony JB. Circulating Nucleic Acids of Chlamydia pneumoniae and Cytomegalovirus in Patients Undergoing Coronary Angiography. J Clin Microbiol. 2001;39:596–600. doi: 10.1128/JCM.39.2.596-600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzichini E, Pizzichini MM, Efthimiadis A, Evans S, Morris MM, Squillace D, et al. Indices of airway inflammation in induced sputum: reproducibility and validity of cell and fluid-phase measurements. Am J Respir Crit Care Med. 1996;154:308–317. doi: 10.1164/ajrccm.154.2.8756799. [DOI] [PubMed] [Google Scholar]
- Tong CY, Sillis M. Detection of Chlamydia pneumoniae and Chlamydia psittaci in sputum samples by PCR. J Clin Pathol. 1993;46:313–317. doi: 10.1136/jcp.46.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smieja M, Mahony JB, Goldsmith CH, Chong S, Petrich A, Chernesky M. Replicate PCR Testing and Probit Analysis for Detection and Quantitation of Chlamydia pneumoniae in Clinical Specimens. J Clin Microbiol. 2001;39:1796–1801. doi: 10.1128/JCM.39.5.1796-1801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn DL, Golubjatnikov R. Smoking is a potential confounder of the Chlamydia pneumoniae-coronary artery disease association. Arterioscler Thromb. 1992;12:945–947. doi: 10.1161/01.atv.12.8.945. [DOI] [PubMed] [Google Scholar]
- Von Hertzen L, Kaprio J, Koskenvuo M, Isoaho R, Saikku P. Humoral immune response to Chlamydia pneumoniae in twin discordant for smoking. J Intern Med. 1998;244:227–234. [PubMed] [Google Scholar]
- Karvonen M, Tuomilehto J, Pitkaniemi J, Naukkarinen A, Saikku P. Importance of smoking for Chlamydia pneumoniae seropositivity. Int J Epidemiol. 1994;23:1315–1321. doi: 10.1093/ije/23.6.1315. [DOI] [PubMed] [Google Scholar]