Abstract
Background
The jellyfish green fluorescent protein (GFP) can be inserted into the middle of another protein to produce a functional, fluorescent fusion protein. Finding permissive sites for insertion, however, can be difficult. Here we describe a transposon-based approach for rapidly creating libraries of GFP fusion proteins.
Results
We tested our approach on the glutamate receptor subunit, GluR1, and the G protein subunit, αs. All of the in-frame GFP insertions produced a fluorescent protein, consistent with the idea that GFP will fold and form a fluorophore when inserted into virtually any domain of another protein. Some of the proteins retained their signaling function, and the random nature of the transposition process revealed permissive sites for insertion that would not have been predicted on the basis of structural or functional models of how that protein works.
Conclusion
This technique should greatly speed the discovery of functional fusion proteins, genetically encodable sensors, and optimized fluorescence resonance energy transfer pairs.
Background
The discovery that the jellyfish green fluorescent protein (GFP) can form a functional fluorophore without other gene products or co-factors [1] was rapidly followed by reports that GFP can be used to create fluorescent fusion proteins [e.g. [2,3]]. For the first time, it became possible to create a wide variety of genetically encodable fluorescent fusion proteins that could be followed in living systems [reviewed in:[4]]. Most GFP fusion proteins have been built by placing GFP at either the N- or C-terminus of the host protein. This can, however, destroy the function of some host proteins. The alternative is to insert GFP into the middle of the host protein [5-8]. Unfortunately, finding a permissive location for insertion of the GFP can be problematic and time consuming.
One way of speeding the process is to randomly generate libraries of GFP fusion proteins and then screen for clones that encode functional, fluorescent proteins. One group used a combination of nick translation and nuclease S1 treatment to randomly insert GFP into a cAMP-dependent protein kinase regulatory subunit from Dictyostelium[6]. A surprisingly large number of the resulting fusion proteins were fluorescent and retained cAMP binding, demonstrating that this can be a powerful approach. A weakness of this strategy, however, is that it can produce deletions in the host sequence. Another approach is to use the random behavior of a transposon to insert GFP into many different places in a target protein. Two synthetic transposons have been reported that can produce GFP fusion proteins [9,10]. The design of these transposons included additional protein domains or linkers between the GFP and the target protein, however, and little is known about how many of the resulting proteins continued to function. We reasoned that a Tn5 transposon [11,12] could be designed that would generate GFP fusion proteins with relatively short linkers (~7 amino acids) between the GFP and the host protein analogous to GFP fusion proteins that have already been shown to function [6,8]. To test this approach, we targeted the G protein subunit αs and the glutamate receptor subunit GluR1.
Results
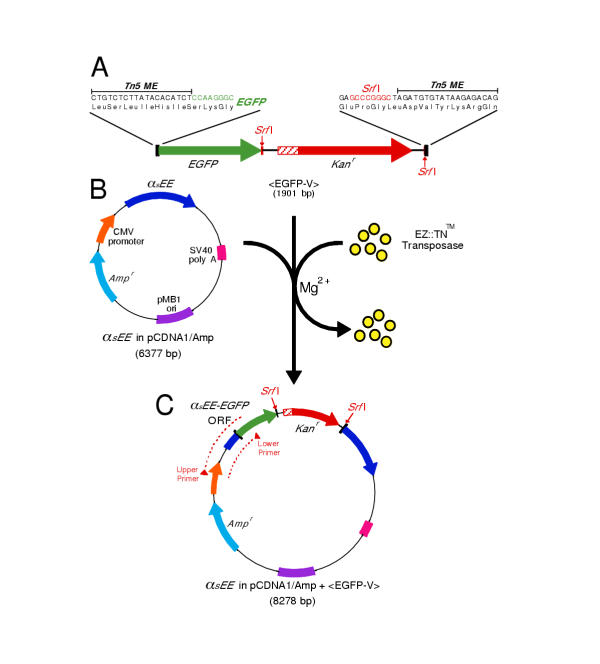
Changes have been made to the Tn5 transposon, and its transposase, that in concert produce a hyperactive transposon capable of a 1% insertion frequency in an in vitro reaction [reviewed in: [12]]. This hyperactive Tn5 transposon is defined as any sequence flanked by the inverted 19 base pair repeats known as mosaic ends (MEs). The recombinant Tn5 transposase binds these ME sequences and, in the presence of Mg2+, catalyzes the random insertion of the transposon into target DNA in a complex process that involves generating a 9 base pair staggered nick in the target. This staggered nick is subsequently repaired to produce a 9 base pair duplication of the target sequence that flanks the inserted transposon. Two possible reading frames extend through the MEs of the Tn5 transposon. Our initial GFP transposon, <EGFP-V>, was created by placing the sequence encoding enhanced green fluorescent protein (EGFP) in one of these frames such that if the transposon landed in another coding sequence, in the correct orientation and frame, it would produce a GFP fusion protein (figure 1A). The low probability of transposition in an in vitro reaction made it necessary to include antibiotic resistance, so Kanr was added to the transposon flanked by Srf I restriction sites that can be used to subsequently remove it.
Figure 1.
Transposition with <EGFP-V> (A) The transposon, <EGFP-V>, is flanked by 19 bp inverted repeats, the MEs. The EGFP coding region is positioned such that when <EGFP-V> inserts between the codons of a target gene, a fusion protein will be produced. <EGFP-V> also carriesKanr. There is a stop codon in the 5' end of the Kanr cassette in the same frame as the EGFP coding sequence, so if the transposon lands in an open reading frame, in the correct orientation and frame, a truncated, EGFP-tagged, protein will initially be produced. Removal of theKanr cassette by Srf I digestion and re-ligation produces a reading frame that extends across the entire transposon. (B) The target plasmid, αsEE in pcDNA1/Amp, encodes an epitope tagged version of the G protein subunit αs. (C) Transposed plasmids carry Ampr and Kanr. <EGFP-V> insertions within the target gene produce a PCR product when <EGFP-V> is inserted in the correct orientation, and the size of the PCR product reveals which <EGFP-V> insertions are in the coding sequence.
An epitope tagged version of the G-protein subunit αs (αsEE) was chosen as the first target (figure 1B). Previous studies have shown that the N- and C-termini of αs are important for its interactions with receptors, G-protein β and γ subunits, and the plasma membrane [13,14], so placing GFP within internal regions of αs is more likely to generate a functional, fluorescent subunit [8]. Moreover, the structure of αs has been solved [15], making it possible to interpret the results in the context of the three-dimensional structure. After transposition and transformation, colonies expressing dual antibiotic resistance were screened with PCR to identify clones in which <EGFP-V> had landed in the correct orientation within the coding region (figure 1C). Assuming that Tn5 behavior is random, the probability that <EGFP-V> will land in the αsEE coding sequence during transposition should be the ratio of the coding sequence to the size of the total plasmid (18.5%). However, transpositions that disrupt critical elements of the plasmid (the plasmid origin or the Ampr gene) should not be recovered after transformation, so the predicted probability of observed transpositions within the αsEE coding sequence increases to 23.8%, with half of these (11.9%) being in the correct orientation. PCR screening of 384 Ampr + Kanr resistant colonies identified 44 clones with <EGFP-V> insertions within the αsEE coding region in the correct orientation (11.4%).
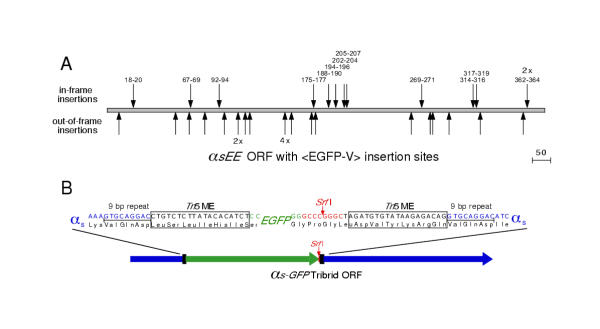
Each clone containing an in-frame insertion should encode a truncated αs protein with GFP at the carboxy-terminus due to a stop codon in the Kanr. Thirty-five of the PCR-positive clones were transiently expressed in HEK 293 cells, and 13 were fluorescent. Sequencing confirmed that the 13 fluorescent constructs were truncated αs-GFP fusion proteins (with 12 being unique insertions) and that the remaining 22 <EGFP-V> insertions were out of frame (figure 2A). The 12 clones encoding unique αs-GFP fusion proteins were digested with Srf I and re-ligated to create full-length fusion proteins (figure 2B). Transient expression of each of the 12 αs-GFP fusion proteins in HEK 293 cells produced a fluorescent signal. This is surprising because several insertions appear to be in internal and/or rigid secondary protein structures (figure 3). It appears that the folding of GFP to form a fluorophore is thermodynamically favorable at most insertion sites.
Figure 2.
<EGFP-V> Insertions in αsEE. (A) Location of 35 <EGFP-V> insertions in the αsEE coding region. The 13 labeled insertions marked above the coding region are in the correct reading frame and encode fluorescent fusion proteins (12 unique insertions, 1 duplication). The insertions are named for the 3 amino acids of αs duplicated during transposition. The 22 unlabeled insertions (18 unique) marked below the coding region are those in which <EGFP-V> landed out-of-frame with respect to αsEE. Redundant insertions are indicated by the number of clones recovered at that site (e.g. 2X, 4X). (B) Srf I digestion of the transposed clone, followed by religation, removes theKanr selection cassette and produces the full-length fusion protein. In the final fusion protein, the EGFP domain is bordered by amino acid linkers encoded by the Tn5 MEs as well as the 9 bp duplication of the target sequence that is generated during the transposition process.
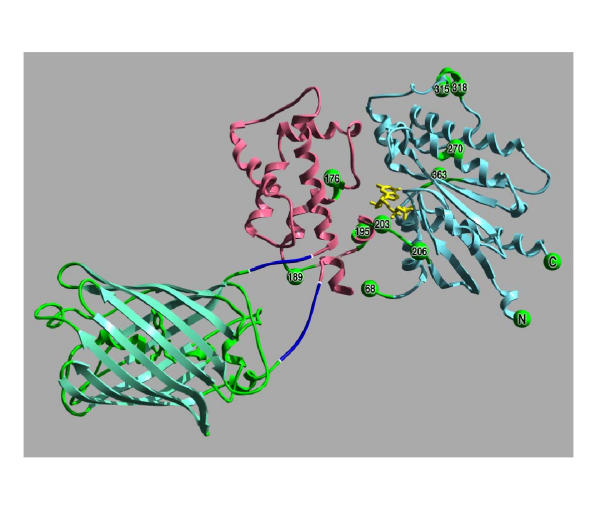
Figure 3.
Model of αs-GFP(92–94). The GFP insertions into αs can be interpreted in the context of the structures of GFP (PDB file: 1EMA) and αs-GTPγ S (PDB file: 1AZT). In this image, the structure of GFP [42] is green, while the helical domain of the αsubunit [15] is pink, and the GTPase domain is blue. GTPγ S is yellow. The GFP insertion αs-GFP(92–94) that produced a functional G protein subunit is illustrated by the short linkers (encoded by the Tn5 MEs) between GFP and αs (dark blue). The other sites of <EGFP-V> insertion are shown as green spheres. The numbers on the spheres indicate the second of the three duplicated residues that flank the transposon insertions (the numbers are based on the long form of αs).
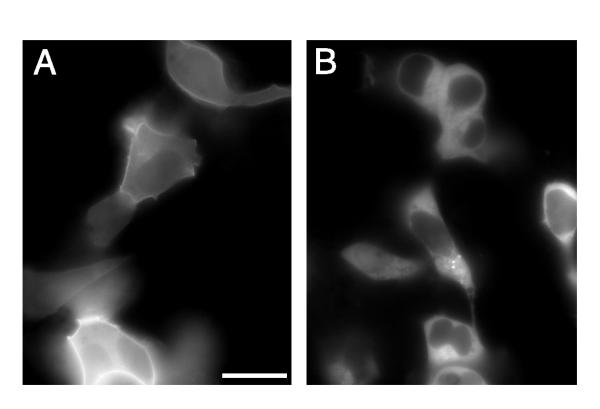
To determine what effect the GFP insertions had on αs localization, the full-length fusion proteins were transiently co-expressed in HEK 293 cells with G protein subunits β1 and γ7, which have been shown to mediate signaling between the β-adrenergic receptor and Gs[16]. Amino- and carboxy-terminus GFP fusions, αs-GFP(N) and αs-GFP(C), respectively, were also co-expressed with β1 and γ7 for comparison. The end-labeled GFP fusions and two of the transposon insertions, αs-GFP(18–20) and αs-GFP(92–94), showed clear localization to the plasma membrane (figure 4A). The remaining 10 fusion proteins displayed a uniform fluorescence signal throughout the cytoplasm (figure 4B).
Figure 4.
Localization of αs-GFP Fusion Proteins in Living Cells. (A) Membrane localization of tribrid fusion, αs-GFP(92–94), in HEK 293 cells ~24 hr after co-transfection with β1 and γ7. Similar localization patterns were observed when αs-GFP(18–20), αs-GFP(N), or αs-GFP(C) fusions were co-expressed with β1 and γ7 (A non-linear representation of the image brightness was used to illustrate both the dimly fluorescent cells in the upper right corner and the very bright ones at the bottom, scale bar = 20 μm). (B) The remaining 10 tribrid fusion proteins were evenly distributed throughout the cytosol (with little fluorescence in the nucleus) as seen here in HEK 293 cells transiently expressing αs-GFP(362–364), β1 and γ7.
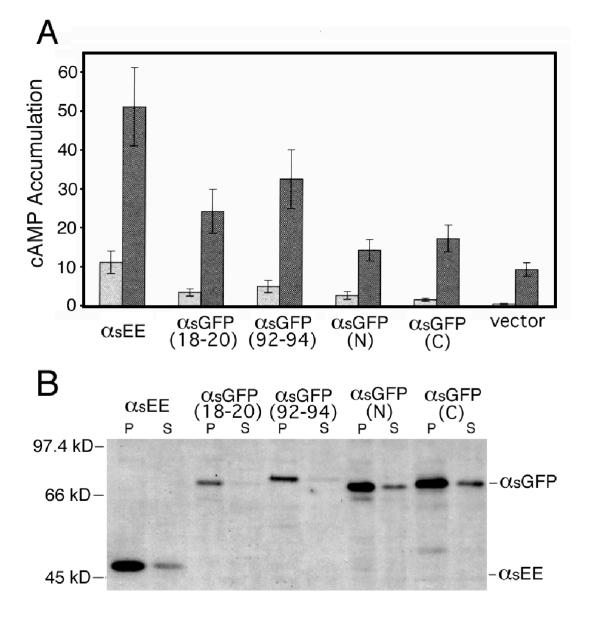
The fusion proteins were tested for function by assaying their abilities to stimulate adenylyl cyclase in response to receptor stimulation. They were co-expressed with the luteinizing hormone (LH) receptor in HEK 293 cells and cAMP accumulation was measured in both the presence and absence of the LH receptor agonist, human chorionic gonadotropin (hCG). Basal and stimulated cAMP accumulation in cells expressing αs-GFP(18–20), αs-GFP(92–94), αs-GFP(N), or αs-GFP(C) were higher than in cells expressing vector alone (figure 5A). However, only in cells expressing αs-GFP(92–94) were these differences statistically significant (p < 0.05). The basal and stimulated activities of αs-GFP(92–94) were less than those of αs, although these differences were not statistically significant (p < 0.05). The remaining 10 of the12 fusion proteins exhibited no detectable activity. One possible explanation for the decreased activities of the αs-GFP fusion proteins relative to αsEE would be a decrease in protein expression level. Cell fractionation and immunoblotting with an anti-EE monoclonal antibody showed that both αs-GFP(92–94) and αs-GFP(18–20) were expressed at lower levels than αsEE, in contrast to αs-GFP(N) and αs-GFP(C) (figure 5B).
Figure 5.
Activity and Expression Levels of αs-GFP Fusion Proteins. (A) HEK 293 cells were transfected with 2 μg/106 cells of the indicated αs-GFP constructs or vector alone (pcDNA1/Amp) and 0.2 μg/106 cells of plasmid encoding the LH receptor. cAMP accumulation was measured in the presence (dark gray bars) or absence (light gray bars) of hCG, an LH receptor agonist. Cells expressing each of the αs-GFP fusion proteins exhibited increased basal and stimulated cAMP accumulation relative to cells expressing vector alone, but only the increases in cells expressing αs-GFP(92–94) were significantly greater (p < .05). Values represent the mean ± S.E. of 5 independent experiments. (B) Immunoblots of the membrane pellets (P) and supernatant (S) fractions from transiently transfected HEK 293 cells. Expression levels of tribrid fusion proteins αs-GFP(18–20) and αs-GFP(92–94), but not of amino- and carboxy-terminus fusions, αs-GFP(N) and αs-GFP(C), respectively, were decreased in both fractions relative to that of unlabeled αsEE. Similar results were obtained in an additional experiment.
Interpreting these results in the context of the structure of αs leads to a surprising result. A rational approach to designing a fluorescent, functional αs-GFP fusion protein would have most likely targeted the exposed loops [e.g. [8]], yet these insertions were not functional. The most functional protein was produced by the insertion of GFP into an α-helix that one would have avoided (figure 3).
The discovery that all of the in-frame insertions in αs produced truncated fluorescent fusion proteins suggested that we could identify in frame insertions by transiently expressing all of the transposed clones and visually screening them for fluorescence. This alternative screening strategy could be particularly useful for large coding regions where a PCR-based screen might fail. To reduce the number of transient transfections required, a second transposon was created with enhanced cyan fluorescent protein (ECFP). Two separate transpositions with the different colored transposons, followed by co-transfections in the visual screen (one potential green clone and one potential cyan clone per well), can identify twice as many in-frame insertions in a given number of transfections. This approach could be expanded to encompass many different fluorophores.
In the experiments with αs, several clones were recovered with identical transposon insertions. This is consistent with previous reports of Tn5 preferentially inserting into particular locations in the target sequence [17,18]. Since these "hotspots" could become a limiting factor in the number of unique insertions recovered within a target sequence, the second reading frame through the Tn5 MEs was used for the ECFP transposon. This doubles the number of potentially useful insertion sites within a given target sequence.
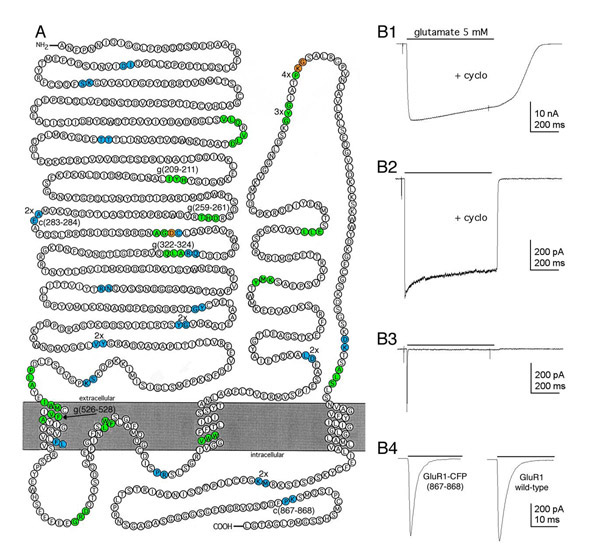
The glutamate receptor subunit GluR1 [19] was used to test the new transposons and the visual screening process. Independent transpositions of the GluR1 plasmid were performed with the EGFP and ECFP transposons (<TgPT-0> and <TcPT-1>, respectively). In 288 co-transfections, there were 20 wells with EGFP fluorescence, 21 wells with ECFP fluorescence, and 2 wells with both EGFP and ECFP fluorescence. Sequencing revealed 35 unique insertions (17 <TgPT-0> and 18 <TcPT-1>) and 10 repetitive insertions (figure 6A). The recovery of 45 fluorescent clones from 576 colonies (7.8%) agrees with the predicted frequency of transpositions resulting in GluR1-EGFP/ECFP fusions (7.7%), which is consistent with the interpretation that all in-frame insertions produce a fluorescent protein. Clones representing unique fluorescent fusion proteins were digested with Srf I to remove the Kanr selection cassette and re-ligated to generate full-length GluR1-EGFP/ECFP fusions. These fusion proteins were screened, in transiently transfected HEK 293 cells, for glutamate-gated ion channel function. Of the 29 unique tribrid fusion constructs tested, all produce detectable fluorescence and 6 were functional (figure 6B).
Figure 6.
Insertion Sites and Functional Screening of GluR1-GFP Fusion Proteins (A) A model of GluR1 topology showing the locations of the GFP insertion in 45 fluorescent fusion proteins. In-frame insertions of <TgPT-0> result in a 3 amino acid duplication (green) flanking the insertion site. In-frame insertions of <TcPT-1> generate only a 2 amino acid duplication (cyan) in the target because of the different reading frame. The orange amino acids are overlapping insertion sites of the two transposons (See supplemental diagram for the two reading frames). Multiple clones with identical transpositions are identified as 2x, 3x, etc. The six insertions resulting in functional, fluorescent GluR1-GFP/CFP tribrid fusion proteins are identified by the duplicated target amino acids (e.g. g209–211, c867–868). This figure was adapted from [43]. (B) AMPA receptor-mediated current from GluR1-CFP(867–868). (B1) Large whole-cell current elicited by the rapid sustained application of 5 mM glutamate (bar) in a cell transiently expressing GluR1-CFP(867–868) after reducing desensitization with cyclothiazide (100 μM). (B2) Current elicited by 5 mM glutamate in an outside-out patch pulled from a cell transiently expressing GluR1-CFP(867–868). (B3) Current elicited in the same patch as B3, but in the absence of cyclothiazide. Note the rapid and nearly complete desensitization of the current. (B4) The trace on the left is an expanded time scale of B3, the trace on the right is from an outside-out patch pulled from a cell transiently expressing wild-type GluR1 in the presence of 5 mM glutamate without cyclothiazide.
Discussion
Creating functional, fluorescent fusion proteins involves finding a permissive site for the insertion of GFP, a process that in most cases still involves some guesswork. The results of both the αs and GluR1 transpositions illustrate this point. Based on previous studies with the G protein subunit αq [8] we anticipated that an insertion within an exterior flexible loop region of αs would be most likely to produce a functional fusion protein. Surprisingly, the αs fusion protein that was the most functional, αs-GFP (92–94), resulted from an insertion into an α helix (figure 3), while the insertions in exposed loops, αs-GFP (67–69) and αs-GFP (188–190), were not functional. Similarly, in the case of GluR1, one of the insertions that produced a functional channel, GluR1-GFP(526–528), was within the hydrophobic region thought to be the first transmembrane domain (see Additional File: Figure 7). Additionally, within a given region of GluR1, one insertion will produce a functional channel while another nearby insertion does not (e.g. the intracellular carboxy-terminus region or the amino terminus between amino acids 210 and 330). The reasons for these discrepancies are not obvious.
The discovery that GFP will still fold and form a fluorophore when placed virtually anywhere in another coding region suggests that the limiting step in the process is whether the target protein it is inserted into folds and functions correctly. Indeed, GFP fusion constructs have been used to assay, and improve upon, the folding of a variety of proteins in a bacterial expression system [20]. The relatively random nature of the transposition events we recovered in this study suggests that it might be possible to insert GFP at nearly every position in a given protein, but there are two potential limits. First, the laws of probability predict that there will be rapidly diminishing returns in the search for unique Tn5 insertions as one recovers each additional clone. Second, the behavior of the Tn5 transposon is not entirely random. Goryshin and colleagues [18] have shown that there is a weak consensus site for Tn5 insertion which is consistent with our results. It appears that the resolution limit will be an insertion each three amino acids on average in a target protein.
Inserting a reporter domain such as GFP into another protein always has the potential of perturbing the target and destroying it's ability to function. In this study 16% of the tribrid fusion proteins were still functional. One explanation for why GFP can be used for internal insertion is that the N- and C-termini of GFP exit the structure quite close to one another and are unlikely to displace the surrounding domains of the target protein a great deal. This is analogous to the use of the bovine pancreatic trypsin inhibitor for internal insertions [21]. The transposons described here could potentially be improved upon by optimizing the length and flexibility of the linkers between the target and the GFP. Another potential improvement to the process would be to use bacterial expression to screen for transposon insertions that produce a fluorescent protein. This could, however, be problematic with proteins from the mammalian nervous sytem, such as ion channels, that are difficult to express in bacteria.
The approach described here should speed the discovery of genetically encodable fluorescent sensors. The pioneering work of Siegel and Isacoff showed that GFP placed within a portion of the Shaker K+ channel C-terminus produced a fluorophore that responded to changes in membrane voltage [5], but they built a number of different constructs before finding one that worked. Similarly, Ataka and Pieribone created an EGFP-Na+ channel fusion protein that changes fluorescence in response to membrane depolarizations on a time-scale that would be sufficent to image action potentials. This discovery, however, was the result of designing, building, and testing eight different tribrid fusion proteins [22]. Little is known about the mechanism whereby changes in channel conformation are converted to changes in the fluorophore, so it remains to be determined whether GFP can signal conformational changes in other kinds of proteins. Nevertheless, the use of the transposons described here should shift the work from building the constructs to devising high throughput assays for function.
Finally, random GFP tagging will facilitate the creation of potential fluorescence resonance energy transfer (FRET) reagents to study protein interactions in living systems. To date, a few studies have demonstrated the potential power of GFP-FRET by labeling different proteins [6,23-26] or by fusing two different fluorophores to the same protein [27-32]. Creating efficent donor and acceptor fusion proteins is difficult, however, because FRET only occurs when the two fluorophores are attached to surfaces that are very close to one another. The approach described here makes it possible to rapidly generate libraries of potential donor and acceptor tribrid fusion proteins that can be screened, in pairwise combinations, for function and FRET signals.
Conclusions
The transposons described here make it possible to rapidly generate large numbers of different GFP fusion proteins. The results show that GFP can be inserted into a wide variety of other protein domains and it will continue to fold and form a fluorophore. The rapid and random nature of the transposition process makes it possible to generate and screen many different fusion constructs to identify those that continue to function. In the case of the two proteins tested here, roughly 1 in 6 of the fusion proteins retained their signaling function, and the random nature of the transposition process revealed permissive sites for insertion that would not have been predicted on the basis of structural or functional models of how that protein works. This simple tool should speed the search for a wide variety of new biological probes for the study of nervous system.
Materials and Methods
PCR and standard subcloning procedures were use to create the initial transposon, <EGFP-V> (full sequence at: http://momotion.med.yale.edu). The Tn5 MEs were added to the 5' and 3' ends of an EGFP coding sequence, with a Srf I restriction site at its 3' end, such that one continuous reading frame extended through both MEs and EGFP (figure 2). To add antibiotic selection, the Kanr gene from pUniV5-His-TOPO™ (Invitrogen, Carlsbad, CA) was flanked with Srf I sites and inserted into the transposon. The improved transposons, <TgPT-0> and <TcPT-1>, were created in the same way as <EGFP-V>, but Asc I sites were added to facilitate changing the fluorescent protein at a later date (supplemental material). In addition, the two different reading frames present in the MEs were used to create the two different transposons, and ECFP was used in place of EGFP in <TcPT-1>. A primer complementary to the19 bp Tn5 ME (5'-CTGTCTCTTATACACATCT-3') was used to amplify the transposons (1 cycle at 95°C for 3:30 min., 24 cycles of 95°C for 30 sec 47°C for 30 sec 72°C for 1 min., 1 cycle at 72°C for 5 min.) with Pfu polymerase (Stratagene, La Jolla, CA). The PCR product was purified and concentrated with the Geneclean II kit (Bio101 Inc., Vista, CA) and eluted in 1X TE buffer. 0.2 fmoles of transposon were incubated with 5.0 μL of EZ::TN™ transposase (Epicentre Technologies) in 25% glycerol at 25°C for 30 min.
Molar equivalents of transposon and target plasmid (0.4 fmoles ea.) were incubated in reaction buffer (50 mM Tris-acetate (pH 7.5), 150 mM potassium acetate, 10 mM magnesium acetate and 4 mM spermidine) at 37°C for 2 hr in a 10 μL reaction. Transposition was stopped by adding 1 μL of 1% SDS and incubating at 70°C for 10 min. Top 10 F' E. coli (Stratagene) were transformed with 1 μL of the transposition reaction and plated on LB agar with either ampicillin (100 μg/mL) and kanamycin (50 μg/mL) to recover transposed clones, or ampicillin (100 μg/mL) alone to establish the transposition efficiency.
The cDNA encoding the rat αs [33], modified to carry the EE epitope [34], was in pcDNA1/Amp (Invitrogen). GFP was added to the N- or C-terminus of αs to create end-labeled constructs for comparison with the transposed GFP tribrid fusion proteins. The amino-labeled clone, GFP-[GGGPSGGGGS]-αsEE, and carboxy-labeled clone, αsEE-[SGGGGSGQH]-GFP, were generated via overlap extension [35]. Linker sequences are in brackets. The flip variant of rat GluR1 was in the CMV expression plasmid pRK5 (a generous gift from Derek Bowie, Emory University, Atlanta, GA).
PCR screening for <EGFP-V> insertions within the αsEE coding region was performed using a protocol described by Cease et al. [36] using an upper primer complimentary to the 5' UTR (5'-GCTCCCGCGGCTCCTGCTCTGCTC-3'), and a lower primer complimentary to EGFP (5'-GCCGTCGCCGATGGGGGTGTTCTG-3'. The clones that produced clear PCR products within the expected size range were then miniprepped (QIAgen, Germantown, MD).
Insertion sites were identified for all PCR-positive <EGFP-V> transposed clones and all fluorescent <TgPT-0>/<TcPT-1> transposed clones by sequencing out of the transposon with a primer complimentary to the EGFP/ECFP coding region (5'-tggccgtttacgtcgccgtcca-3'). Srf I restriction digestion was then used to remove theKanr cassette from the clones carrying in-frame insertions, thereby creating a sequence encoding a full-length fusion protein. After digestion and re-ligation, Top 10 F' E. coli were transformed with 1 μL of the ligation reaction and plated on LB agar containing ampicillin. The colonies were re-plated the following day on ampicillin and kanamycin to verify loss of theKanr.
The fusion proteins were transiently expressed in HEK 293 cells [37]. Transfections were done using Lipofectamine 2000 (Gibco BRL). Images were collected from live cells 20–48 hr later on an inverted Zeiss microscope fitted with computer controlled (IPLabs, Scanalytics) filter wheels (Ludl Electronics) on the excitation and emission paths. EGFP was imaged with an FITC filter set, while ECFP was distinguished from EGFP in co-expression experiments by changing both the excitation and emission filter sets (Exciters: 440AF21 & 500AF25, Dichroic cat# XF 2063, Emitters 480AF & 545AF35; Omega, Brattleboro, VT).
αs-GFP fusion proteins were assayed for the ability to stimulate adenylyl cyclase in response to luteinizing hormone (LH) receptor stimulation [38]. 106 HEK 293 cells/60 mm-dish were co-transfected with 2 μg of plasmid DNA encoding the αs-GFP fusion protein, and 0.2 μg of plasmid DNA encoding the rat LH receptor in pCIS [39], using 10 μL of Lipofectamine 2000. [3H]-adenine-labeled cells were assayed for cAMP accumulation after incubation at 37°C for 40 min. in the presence of 1 mM 3-isobutyl-1-methylxanthine (IBMX) a phosphodiesterase inhibitor, and in the presence or absence of 7.5 ng/mL human chorionic gonadotropin (hCG), as described previously. Conversion of ATP to cAMP was expressed as:
103 × [3H]cAMP/([3H]ATP + [3H]cAMP).
12 × 106 HEK 293 cells were transfected, using DEAE-dextran [40], with 25 μg of plasmid DNA. Forty-eight hours after transfection, cells were lysed and membrane and supernatant fractions harvested as described previously [8]. 10 μg of membrane proteins and normalized volumes of the supernatants were resolved by SDS-polyacrylamide electrophoresis (10%), transferred to nitrocellulose, and probed with a monoclonal antibody to the EE epitope [34]. The antigen-antibody complexes were visualized with ECL chemiluminescence (Amersham Biosciences, Piscataway, NJ).
Whole-cell patch clamp recording was used to test the GluR1 fusion proteins for function in transiently transfected HEK 293 cells as previously described [41]. The external solution was (in mM): 150 NaCl, 3 KCl, 2 CaCl2, 1 MgCl2, 5 glucose, 0.002 glycine and 10 HEPES (pH 7.4). Patch pipettes were filled with a solution containing (in mM): 120 CsF, 33 KOH, 2 MgCl2, 1 CaCl2, 0.1 spermine, 10 HEPES, and 11 EGTA (pH 7.4). Cyclothiazide was prepared as a 20 mM stock solution in DMSO and diluted to 100 μM in external solution. All chemicals were purchased from Sigma. Drugs were applied with a rapid superfusion system made from a pulled theta capillary. The open tip responses obtained with this system had 10–90% rise-times of 150 μs to 300 μs.
Authors' contributions
Author 1 D. L. Sheridan carried out the design and construction of the transposons, conducted the biochemical assays for G-protein signaling, imaged the living cells, and drafted the manuscript. Author 2 C. H. Berlot provided critical reagents and advice for all portions of the G-protein work. Author 3 A. Robert screened the Glutamate receptor subunits for function. Authors 4 F. M. Inglis and 5 K. B. Jakobsdottir provided help in the minipreparation of plasmid DNA for each of the constructs. Authors 6 J. R. Howe and 7 T. E. Hughes participated in the study design, coordination, and analysis.
All authors read and approved the final manuscript.
Supplementary Material
Figure 7
Acknowledgments
Acknowledgments
We thank Tom Hynes for creating figure 3, the members of the Friday Afternoon Lab Meeting for their input, Janet Robishaw for the human β1 in pCMV5 and HA-tagged γ7 in pCI-neo plasmids, Derek Bowie for the GluR1flip in pRK5 plasmid, Jim Boulter for his suggestions, and Michael Hollmann for permission to adapt his GluR1 topology figure. This work was supported by: NIH RO1 EY 08362 (to T.E.H.), NIH RO1 GM 50369 (to C.H.B.) and NIH RO1 NS 37904 (to J.R.H.). D. L. Sheridan is an HHMI Predoctoral Fellow.
Contributor Information
Douglas L Sheridan, Email: Douglas.Sheridan@yale.edu.
Catherine H Berlot, Email: Catherine.Berlot@yale.edu.
Antoine Robert, Email: Antoinerob@yahoo.com.
Fiona M Inglis, Email: fiona.inglis@yale.edu.
Klara B Jakobsdottir, Email: Klarajakobsdottir@hotmail.com.
James R Howe, Email: James.Howe@yale.edu.
Thomas E Hughes, Email: thomas.hughes@yale.edu.
References
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–5. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Wang S, Hazelrigg T. Implications for bcd mRNA localization from spatial distribution of exu protein in Drosophila oogenesis. Nature. 1994;369:400–3. doi: 10.1038/369400a0. [DOI] [PubMed] [Google Scholar]
- Marshall J, Molloy R, Moss GW, Howe JR, Hughes TE. The jellyfish green fluorescent protein: a new tool for studying ion channel expression and function. Neuron. 1995;14:211–5. doi: 10.1016/0896-6273(95)90279-1. [DOI] [PubMed] [Google Scholar]
- Chalfie M. Green fluorescent protein. Photochem Photobiol. 1995;62:651–6. doi: 10.1111/j.1751-1097.1995.tb08712.x. [DOI] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–41. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- Biondi RM, Baehler PJ, Reymond CD, Veron M. Random insertion of GFP into the cAMP-dependent protein kinase regulatory subunit from Dictyostelium discoideum. Nucleic Acids Res. 1998;26:4946–52. doi: 10.1093/nar/26.21.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz PA, Bottcher B, Nassal M. Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc Natl Acad Sci U S A. 1999;96:1915–20. doi: 10.1073/pnas.96.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TE, Zhang H, Logothetis DE, Berlot CH. Visualization of a functional Gαq-green fluorescent protein fusion in living cells. Association with the plasma membrane is disrupted by mutational activation and by elimination of palmitoylation sites, but not by activation mediated by receptors or AlF4. J Biol Chem. 2001;276:4227–35. doi: 10.1074/jbc.M007608200. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Sheehan A, Roeder GS, Snyder M. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:190–5. doi: 10.1073/pnas.94.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkulov GV, Boeke JD. Libraries of green fluorescent protein fusions generated by transposition in vitro. Gene. 1998;222:213–22. doi: 10.1016/S0378-1119(98)00503-4. [DOI] [PubMed] [Google Scholar]
- Goryshin IY, Reznikoff WS. Tn 5 in vitro transposition. J Biol Chem. 1998;273:7367–74. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- Reznikoff WS, Bhasin A, Davies DR, Goryshin IY, Mahnke LA, Naumann T, Rayment I, Steiniger-White M, Twining SS. Tn 5: A molecular window on transposition. Biochem Biophys Res Commun. 1999;266:729–34. doi: 10.1006/bbrc.1999.1891. [DOI] [PubMed] [Google Scholar]
- Sullivan KA, Miller RT, Masters SB, Beiderman B, Heideman W, Bourne HR. Identification of receptor contact site involved in receptor-G protein coupling. Nature. 1987;330:758–760. doi: 10.1038/330758a0. [DOI] [PubMed] [Google Scholar]
- Wedegaertner PB, Chu DH, Wilson PT, Levis MJ, Bourne HR. Palmitoylation is required for signaling functions and membrane attachment of Gqα and Gsα. J Biol Chem. 1993;268:25001–8. [PubMed] [Google Scholar]
- Sunahara RK, Tesmer JJ, Gilman AG, Sprang SR. Crystal structure of the adenylyl cyclase activator Gsα. Science. 1997;278:1943–7. doi: 10.1126/science.278.5345.1943. [DOI] [PubMed] [Google Scholar]
- Wang Q, Mullah BK, Robishaw JD. Ribozyme approach identifies a functional association between the G protein β1γ7 subunits in the β-adrenergic receptor signaling pathway. J Biol Chem. 1999;274:17365–71. doi: 10.1074/jbc.274.24.17365. [DOI] [PubMed] [Google Scholar]
- Berg DE, Schmandt MA, Lowe JB. Specificity of transposon Tn 5 insertion. Genetics. 1983;105:813–28. doi: 10.1093/genetics/105.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryshin IY, Miller JA, Kil YV, Lanzov VA, Reznikoff WS. Tn 5/IS50 target recognition. Proc Natl Acad Sci U S A. 1998;95:10716–21. doi: 10.1073/pnas.95.18.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, O'Shea-Greenfield A, Rogers SW, Heinemann S. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–8. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–5. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- Borjigin J, Nathans J. Bovine pancreatic trypsin inhibitor-trypsin complex as a detection system for recombinant proteins. Proc Natl Acad Sci U S A. 1993;90:337–41. doi: 10.1073/pnas.90.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J. 2002;82:509–16. doi: 10.1016/S0006-3495(02)75415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, Racz A, Lin GC, Barsony J. Dimerization with retinoid X receptors promotes nuclear localization and subnuclear targeting of vitamin D receptors. J Biol Chem. 2000;275:41114–23. doi: 10.1074/jbc.M003791200. [DOI] [PubMed] [Google Scholar]
- Chan FK, Siegel RM, Zacharias D, Swofford R, Holmes KL, Tsien RY, Lenardo MJ. Fluorescence resonance energy transfer analysis of cell surface receptor interactions and signaling using spectral variants of the green fluorescent protein. Cytometry. 2001;44:361–8. doi: 10.1002/1097-0320(20010801)44:4<361::AID-CYTO1128>3.3.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–11. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- Xia Z, Zhou Q, Lin J, Liu Y. Stable SNARE complex prior to evoked synaptic vesicle fusion revealed by fluorescence resonance energy transfer. J Biol Chem. 2001;276:1766–71. doi: 10.1074/jbc.M008741200. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent Indicators For Ca2+ Based On Green Fluorescent Proteins and Calmodulin. Nature. 1997;388:882–7. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Griesbeck O, Heim R, Tsien RY. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Natl Acad Sci U S A. 1999;96:2135–40. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LL, Gandley RE, Han W, Wasserloos K, Stitt M, Kanai AJ, McLaughlin MK, Pitt BR, Levitan ES. Role of metallothionein in nitric oxide signaling as revealed by a green fluorescent fusion protein. Proc Natl Acad Sci U S A. 2000;97:477–82. doi: 10.1073/pnas.97.1.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Sawano A, Eli P, Hama H, Miyawaki A. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry. 2001;40:2502–10. doi: 10.1021/bi002263b. [DOI] [PubMed] [Google Scholar]
- Sakai R, Repunte-Canonigo V, Raj CD, Knopfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur J Neurosci. 2001;13:2314–8. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- Truong K, Sawano A, Mizuno H, Hama H, Tong KI, Mal TK, Miyawaki A, Ikura M. FRET-based in vivo Ca2+ imaging by a new calmodulin-GFP fusion molecule. Nat Struct Biol. 2001;8:1069–73. doi: 10.1038/nsb728. [DOI] [PubMed] [Google Scholar]
- Jones DT, Reed RR. Molecular cloning of five GTP-binding protein cDNA species from rat olfactory neuroepithelium. J Biol Chem. 1987;262:14241–9. [PubMed] [Google Scholar]
- Grussenmeyer T, Scheidtmann KH, Hutchinson MA, Eckhart W, Walter G. Complexes of polyoma virus medium T antigen and cellular proteins. Proc Natl Acad Sci USA. 1985;82:7952–7954. doi: 10.1073/pnas.82.23.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–8. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Cease KB, Potcova CA, Lohff CJ, Zeigler ME. Optimized PCR using Vent polymerase. PCR methods and applications. 1994;3:298–300. doi: 10.1101/gr.3.5.298. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from Human Adenovirus type 5. Journal of General Virology. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Grishina G, Berlot CH. Identification of common and distinct residues involved in the interaction of αi2 and αs with adenylyl cyclase. J Biol Chem. 1997;272:20619–26. doi: 10.1074/jbc.272.33.20619. [DOI] [PubMed] [Google Scholar]
- McFarland KC, Sprengel R, Phillips HS, Kohler M, Rosemblit N, Nikolics K, Segaloff DL, Seeburg PH. Lutropin-choriogonadotropin receptor: An unusual member of the G protein-coupled receptor family. Science. 1989;245:494–9. doi: 10.1126/science.2502842. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent RE, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K, eds Current Protocols in Molecular Biology. New York: John Wiley & Sons, Inc. 1987.
- Robert A, Irizarry SN, Hughes TE, Howe JR. Subunit interactions and AMPA receptor desensitization. J Neurosci. 2001;21:5574–86. doi: 10.1523/JNEUROSCI.21-15-05574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria Green Fluorescent Protein. Science. 1996;273:1392–5. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Maron C, Heinemann S. N-glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron. 1994;13:1331–43. doi: 10.1016/0896-6273(94)90419-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 7