Abstract
Transcription initiation involves the coordination of multiple events, starting with activators binding specific DNA target sequences, which recruit transcription coactivators to open chromatin and enable binding of general transcription factors and RNA polymerase II to promoters. Two key human transcriptional coactivator complexes, ATAC (ADA-two-A-containing) and SAGA (Spt-Ada-Gcn5 acetyltransferase), containing histone acetyltransferase (HAT) activity, target genomic loci to increase promoter accessibility. To better understand the function of ATAC and SAGA HAT complexes, we used in vitro biochemical and biophysical assays to characterize human ATAC and SAGA HAT module interactions with nucleosomes and how a transcription factor (TF) coordinates these interactions. We found that ATAC and SAGA HAT modules bind nucleosomes with high affinity, independent of their HAT activity and the tested TF. ATAC and SAGA HAT modules directly interact with the VP16 activator domain and this domain enhances acetylation activity of both HAT modules. Surprisingly, ATAC and SAGA HAT modules increase TF binding to its DNA target site within the nucleosome by an order of magnitude independent of histone acetylation. Altogether, our results reveal synergistic coordination between HAT modules and a TF, where ATAC and SAGA HAT modules (i) acetylate histones to open chromatin and (ii) facilitate TF targeting within nucleosomes independently of their acetylation activity.
Graphical Abstract
Graphical Abstract.
Introduction
The eukaryotic genome is packaged in the nucleus as a highly condensed structure called chromatin. The basic repeating unit of chromatin is the nucleosome, which contains ∼147 base pairs (bp) of DNA wrapped ∼1.65 times around two copies of each histone protein (H2A, H2B, H3 and H4) (1–3). Human genomic DNA is wrapped into ∼20 million nucleosomes, which compact into dynamic higher-order structures that regulate DNA accessibility and control genome functions, including transcription and DNA repair/replication (4,5). Access to DNA is critical for DNA-mediated events (6). For example, transcription factors (TFs) must access specific DNA target sequences within promoters and enhancers to initiate the activation of genes by facilitating the recruitment of coactivators, including chromatin-modifying complexes (7,8). However, the mechanisms by which TFs and chromatin-modifying complexes coordinate their gene-activating functions are not yet fully understood.
Chromatin-modifying complexes recognize (readers), deposit (writers) and remove (erasers) post-translational modifications (PTMs) of histones in a dynamic manner. Two highly conserved large multi-subunit transcriptional regulatory coactivator complexes that can acetylate histones at distinct residues are ATAC (ADA-two-A-containing) and SAGA (Spt-Ada-Gcn5 acetyltransferase) (9,10).
The metazoan ATAC and SAGA coactivator complexes contain 10 and 18–20 well-characterized subunits, respectively, which are organized in functional modules (9–11). SAGA is conserved from yeast to humans and organized into the histone acetyltransferase (HAT), histone H2Bub1 deubiquitinase, activator binding (AM), splicing and core modules (9–15). ATAC, which has been described in metazoans but is not found in yeast, also contains a HAT module (16) and a core module (17); however, the structural organization of ATAC is not yet known.
Importantly, ATAC and SAGA HAT modules (hereafter ATACHAT and SAGAHAT) are highly similar in that three of their subunits KAT2A/KAT2B, TADA3 and SGF29 are shared between them. The fourth and distinctive subunit is either TADA2A or TADA2B for ATAC or SAGA HAT modules, respectively (Figure 1A) (9,18,19). The similar HAT modules of these two endogenous coactivator complexes seem to have different histone acetylation targets in vivo, where SAGA acetylates H3K9 and H3K14 (16,20,21), while it has been suggested that ATAC acetylates both histones H3 and H4 (15,22–26). In addition, ATAC and SAGA play different regulatory roles in transcription regulation and/or cellular homeostasis (24,26–33). How these two coactivator complexes containing very similar HAT modules target histone acetylation differentially remains an open question.
Figure 1.
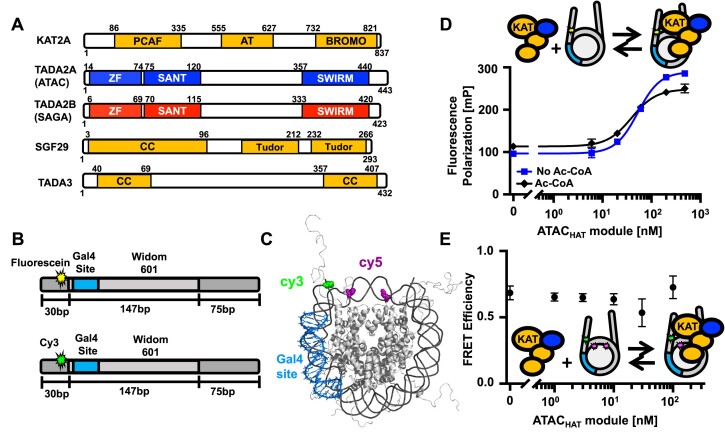
ATAC and SAGA HAT module organization and ATAC HAT module interactions with nucleosomes. (A) Schematic representations of the shared subunits of the ATAC and SAGA HAT modules (KAT2A, SGF29 and TADA3), together with TADA2A or TADA2B, which are either ATAC or SAGA subunits, respectively (10). PCAF: PCAF homology domain; AT: acetyltransferase domain; BROMO: bromo domain; ZF: zinc-finger domain; SANT: Swi3, ADA2, N-Cor and TFIIIB domain; Tudor: Tudor domain; CC: coiled coil domain. (B) (Upper part) Schematic of the DNA used in fluorescence polarization experiments. DNA is 252 bp long and consists of the 147-bp Widom 601 sequence (light grey, center) flanked by a 30-bp (dark grey, left) and a 75-bp linker DNA (dark grey, right). Within the 601 sequence is a 19-bp Gal4 binding site at 8–26 bp in the 601 nucleosome positioning sequence (NPS). Fluorescein (star) is attached to a modified thymine 4 bp into the 30-bp linker DNA away from the 601 NPS. (Lower part) Schematic of the DNA used in Förster resonance energy transfer (FRET) experiments. Same sequence as above but with Cy3 (star) attached to a modified thymine 4 bp into the 30-bp linker DNA away from the 601 NPS. (C) Nucleosome structure (PDB 1KX5) indicating the estimated distance between Cy3 on the DNA (green) and Cy5 on H2A (magenta). The histone octamer is gray, DNA is dark gray and Gal4 binding site is blue. (D) Fluorescence polarization in mP of nucleosomes with increasing amounts of the ATACHAT module. The titrations were fit to binding isotherms to determine the S1/2 values. The ATACHAT module binds with an S1/2 of 52 ± 2 nM (blue square), reaching saturating conditions around 100 nM. The ATACHAT module with acetyl-CoA (Ac-CoA) binds at an S1/2 of 40 ± 1 nM (black diamond). (E) FRET efficiency of the Cy3–Cy5-labeled nucleosome with increasing concentrations of ATACHAT. Each FRET and fluorescence polarization value was determined in triplicate and the uncertainty is determined from the standard error of the three measurements.
Recent cryo-electron microscopy (cryo-EM) studies (12,14,15) have revealed the structure of most of the SAGA complex at high resolution. However, SAGAHAT could not be resolved in the structures, indicating that the module has a high degree of flexibility within the SAGA complex. This flexibility is further enhanced by nucleosome binding, which appears to result in the displacement of the HAT module (15). In addition, a metazoan SAGAHAT has been reported to function on its own as the ADA complex (34) and with the addition of the protein Chiffon (35).
Targeting of the SAGA complex by TFs occurs at least in part through interactions with the AM module, which in humans is TRRAP (34,36). In addition, Myc has been described to interact directly with TRRAP and KAT2A (37–40). Interestingly, the SAGAHAT module has a number of nucleosome binding domains indicating that it can target nucleosomes directly (10). This in combination with the observation that the AM module is not included in the ATAC complex, nor in the ADA complex, raises questions about how TFs target these endogenous coactivator complexes to chromatin.
Uncovering how these human HAT modules engage with TFs to target, acetylate and open chromatin is important for understanding mechanisms by which these coactivator complexes function to facilitate transcription initiation. Here we report systematic biochemical and biophysical studies of the interactions of SAGAHAT and ATACHAT with the activator Gal4–VP16 and nucleosomes that contain the Gal4 target sequence. We find that both HAT modules directly interact strongly with nucleosomes and the VP16 activator. We then asked how the chimeric TF, Gal4–VP16, functions with each HAT module to open and acetylate nucleosomes. We focused on the Gal4 target site located in the nucleosome entry–exit region, a common position for TF binding sites (41–43). We determined that Gal4–VP16 significantly enhances histone acetylation of both ATACHAT and SAGAHAT, with a larger impact on ATACHAT, and that this depends on the VP16 activator domain (AD). We then investigated the impact of each HAT module on Gal4–VP16 binding to its target site within nucleosomes. Interestingly, we found that both ATACHAT and SAGAHAT increase Gal4–VP16 binding by an order of magnitude, but these increases did not depend on the histone acetylation activity of either complex. These results indicate that both ATACHAT and SAGAHAT function with TFs to not only acetylate nucleosomes, but also facilitate TF binding within nucleosomes independent of their histone acetylation activity.
Materials and methods
DNA preparation and purification
DNA constructs for reconstituting nucleosomes for FRET and acetylation assay experiments were prepared using polymerase chain reaction (PCR) with Cy3-labeled oligonucleotides and a plasmid containing the Widom 601 NPS with the Gal4 binding site (5′-CCGGAGGGCTGCCCTCCGG-3′) at bases 8–26 (44–46). Oligonucleotides were labeled with Cy3 NHS ester (GE Healthcare) at an amine-modified internal thymine and HPLC purified on a 218TP C18 reverse-phase column (Grace/Vydac) as previously described (47). DNA constructs for reconstituting nucleosomes for fluorescence anisotropy (FA) were prepared using PCR with fluorescein-labeled oligonucleotides and a plasmid containing the Widom 601 NPS. Note that we refer to the 147-bp Widom 601 sequence, while structural studies have shown that the central 145 bp of the Widom 601 sequence is wrapped within the nucleosome (48). Following PCR amplification, DNA was purified using a MonoQ column (GE Healthcare).
Core histone expression, purification and histone octamer preparation
The histone octamer was prepared as previously described (47). Human histones H2A(K119C), H2B, H3 and H4 were purchased from Histone Source. Lyophilized histones were resuspended in unfolding buffer [20 mM Tris–HCl, pH 7.5, 7 M guanidinium and 10 mM dithiothreitol (DTT)] at 5 mg/ml for 1 h and then spun to remove aggregates. To determine concentration, absorption at 276 nm was measured for each unfolded histone. Histone octamers were refolded by combining (H2A and H2B):(H3 and H4) together at a ratio of 1.2:1. Double dialysis was performed to form the histone octamer into refolding buffer [10 mM Tris–HCl, pH 7.5, 1 mM ethylenediaminetetraacetic acid (EDTA), 2 M NaCl and 5 mM β-mercaptoethanol (BME)]. The octamer was removed from dialysis and labeled with Cy5 maleimide (GE Healthcare) at H2A(K119C) as previously described (49,50). Labeled octamer was purified over a Superdex 200 (GE Healthcare) gel filtration column to remove excess dimer, tetramer and dye. Labeling efficiency was determined by ultraviolet–visible absorption, and purity of each octamer was confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) (Supplementary Figure S1).
Nucleosome preparation
Nucleosomes were reconstituted and purified as previously described (47,51). DNA and histone octamers were combined at a ratio of 1.25:1 in high-salt refolding buffer (0.5× TE, pH 8.0, 2 M NaCl, 0.5 mM EDTA and 1 mM benzamidine–HCl) and reconstituted using double dialysis with 4 L of reconstitution buffer at 4°C for 5–6 h and then changed into a new 4 L bucket of buffer at 4°C overnight (0.5× TE, pH 8.0, 0.5 mM EDTA, 1 mM benzamidine–HCl). Dialyzed nucleosomes were purified in a 5–30% (w/v) sucrose gradient with an SW41 Ti (Beckman Coulter) rotor in an Optima L-90K ultracentrifuge (Beckman Coulter) spinning at 41 000 rpm for 22 h at 4°C. Sucrose gradients were fractionated into 0.4 mL fractions and analyzed by native PAGE. Fractions containing center positioned nucleosomes were concentrated and buffer exchanged into 0.5× TE (pH 8) with a 30-kDa centrifugal filter. To verify purity, nucleosomes were analyzed by a native 5% acrylamide gel and 0.3× TBE (Supplementary Figure S1). The slow mobility bands are the center positioned nucleosomes, which were purified to at least 80%.
Recombinant Gal4-DBD and Gal4–VP16 expression and purification
The Gal4 DNA binding domain from amino acids 1 to 147 (hereafter called Gal4-DBD) (46,52) and Gal4 DNA binding domain fused to the VP16 AD (hereafter called Gal4–VP16) (53) were expressed in Rosetta™ BL21(DE3) pLysS cells. Cells were grown in 2xYT and induced at OD600 of 0.5 with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 100 mM zinc acetate. Cells were expressed for 3 h and then spun at 4000 × g for 15 min. The supernatant was removed, and cell pellets were frozen with liquid nitrogen.
Cells were thawed and resuspended in 30 mL of Gal4 buffer A [50 mM Tris–HCl, pH 8, 200 mM NaCl, 10 mM imidazole, 10 mM BME, 20 μM zinc acetate, 1 mM DTT and 1 mM phenylmethylsulfonyl fluoride (PMSF)] with leupeptin and pepstatin added at a final concentration of 20 μg/ml. Resuspended cells were sonicated and spun at 23 000 × g for 15 min. Lysate containing the protein was collected. The lysate was added to a Ni-NTA column and washed with Gal4 buffer A. The protein was eluted with Gal4 buffer B (50 mM Tris–HCl, pH 7.5, 200 mM NaCl, 200 mM imidazole, 20 μM zinc acetate, 1 mM DTT, 1 mM PMSF and 0.2% Tween 20). Fractions were run on a 12% acrylamide SDS gel, pooled and dialyzed into Gal4 buffer C with 200 mM NaCl (25 mM Tris–HCl, pH 7.5, 200 mM NaCl, 20 μM zinc acetate, 1 mM DTT and 1 mM PMSF). The protein was further purified after dialysis using cation exchange chromatography with a gradient from 200 to 600 mM NaCl using Gal4 buffer C with appropriate salt concentrations. Fractions containing the protein were pooled, concentrated, exchanged into Gal4 buffer D (10 mM HEPES, pH 7.5, 200 mM NaCl, 10% glycerol, 20 μM zinc acetate, 1 mM DTT and 1 mM PMSF) and then flash frozen for storage at −80°C. The final yield was ∼5 mg/l of culture grown. Purity was confirmed by SDS–PAGE (Supplementary Figure S2).
ATAC and SAGA HAT module expression and purification
Recombinant ATAC and SAGA HAT modules were produced using the MultiBac system (54) as previously described (16). In short, a construct containing human KAT2A with an N-terminal histidine tag followed by a tobacco etch virus protease cleavage site, human TADA2A (for the ATAC HAT module) or TADA2B (for the SAGA HAT module), mouse TADA3 and human SGF29 was integrated in the EMBacY bacmid via TN7 transposition (55). The resulting recombinant baculoviruses were then used to produce the corresponding HAT modules by infecting SF21 cells. Cells were harvested, flash frozen in liquid nitrogen and stored at −80°C. For the purification, cell pellets of the respective HAT module were thawed and resuspended in HAT TALON buffer A (50 mM Trizma® base, 400 mM NaCl, 10 mM imidazole, pH 8) supplemented with cOmplete™ EDTA-free protease inhibitor tablets (Roche) and Benzonase® nuclease. Resuspended cells were lysed using sonication and spun down at 40 000 × g for 45 min. The supernatant was additionally filtered through a folding filter and then incubated with HisPur™ Cobalt resin or loaded on a prepacked HiFliQ Cobalt-NTA FPLC column with a peristaltic pump. The protein was eluted via a step gradient of 7%, 30%, 50% and 100% HAT TALON buffer B (50 mM Trizma® base, 400 mM NaCl, 250 mM imidazole, pH 8). The elutions were applied on an acrylamide SDS gel and fractions containing the complex were pooled accordingly. Tris(2-carboxyethyl)phosphine (TCEP) was added to a final concentration of 0.25 mM and the imidazole content diluted with HAT TALON buffer A where appropriate, to facilitate removal of the His-tag by tobacco etch virus protease cleavage of the His-tag overnight. Samples were concentrated and loaded onto a Superdex 200 Increase size exclusion chromatography (SEC) column equilibrated with HAT SEC buffer (25 mM Trizma® base, 400 mM NaCl, 0.25 mM TCEP, pH 8). Fractions were evaluated by SDS–PAGE (Supplementary Figure S3), pooled and concentrated. Aliquots were stored frozen in liquid nitrogen.
FRET efficiency measurements
For nucleosome–TF binding measurements, nucleosomes and TF were incubated with either the SAGA or ATAC HAT module without or with 300 μM Ac-CoA in T75 buffer (10 mM Tris–HCl, pH 8, 75 mM NaCl, 10% glycerol and 0.25% Tween 20) in a 20 μl volume at room temperature for at least 10 min and then analyzed using Horiba Scientific Fluoromax 4. Nucleosomes were at 1 nM in all experiments. For nucleosome–HAT module binding measurements, nucleosomes and either the ATAC or SAGA HAT module were incubated in T75 buffer in a 20 μl volume at room temperature for at least 10 min as above. Fluorescence spectra were measured with Fluoromax 4 by exciting at 510 and 610 nm and measuring emission from 530 to 750 nm and 630 to 750 nm for donor and acceptor excitations, respectively. FRET efficiency was then calculated using the ratioA method (56). FRET efficiency values were normalized against the FRET efficiency value of nucleosomes in the absence of titrant. The titrations were carried out in triplicate. The average FRET efficiency and standard error were determined for each TF concentration. The S1/2 and its standard error were then determined from a weighted fit to a non-cooperative binding isotherm using Prism 9 software.
Fluorescence polarization measurements of HAT module binding to nucleosomes
Fluorescent polarization measurements were carried out by mixing increasing amounts of either ATACHAT or SAGAHAT with 5 nM NCPFA in 75 mM NaCl, 25 mM Tris–HCl (pH 7.5) and 0.25% Tween 20 in a 30 μl reaction volume. Ac-CoA (300 μM) was added to buffer for measurements taken with Ac-CoA. Samples were loaded and incubated in a Corning round bottom polystyrene plate. Polarization measurements were acquired using a Tecan infinite M1000Pro plate reader by exciting at 470 nm. Polarized emission was measured at 519 nm with 5 nm excitation and emission bandwidths. Fluorescence polarization was calculated from the emission polarized parallel and perpendicular to the polarized excitation light as previously described (57,58). The S1/2 was determined by fitting the data to a non-cooperative binding isotherm. S1/2 values were averaged over three separate experiments with error calculated as the standard error between the runs.
Expression, purification of glutathione S-transferase fusion proteins and HAT module binding assays
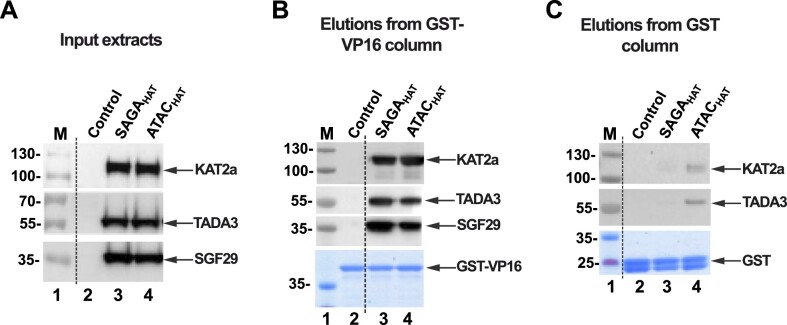
For the binding assays using cell extracts (Figure 2), the expression and binding of glutathione S-transferase (GST) fusion proteins to glutathione Sepharose and HAT binding assay (Supplementary Figure S4) were previously described (59). Briefly, Escherichia coli BL21(DE3) was transformed with either pGEX-2T-GST or pGEX-2T-GST–VP16 vectors. Overnight cultures were diluted 1:100 in LB medium containing ampicillin (100 ug/ml) and incubated at 37°C with shaking. After 1-h incubation, IPTG was added (final concentration 0.1 M) and the cultures were further incubated with shaking for 4 h. Cultures were pelleted by centrifugation at 5000 × g for 5 min at 4°C and pellets were resuspended in 1/10 volume of NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% NP-40); bacteria were lysed by mild sonication and centrifuged at 10 000 × g for 5 min at 4°C. Supernatants were loaded on glutathione Sepharose 4B resin to immobilize either GST or GST–VP16 fusion protein and incubated for 1 h at 4°C. Following several washes of GST- or GST–VP16-bound beads (50 μl each) with NETN buffer, crude SF9 insect whole cell protein extract with no infection (as a negative control) and SF9 cell extract expressing either the four ATACHAT or SAGAHAT module subunits were mixed with the beads (50 μl for each extract) and incubated overnight at 4°C, and then washed with five times NETN buffer containing either 75 or 150 mM NaCl. The resins were boiled in 60 μl of 1× Laemmli buffer and 10% of bound proteins were resolved by SDS–PAGE. Proteins were visualized by western blot analyses with the following antibodies (as indicated in the corresponding figures): anti-GCN5 2GC2C11 mouse monoclonal antibody (mAb) (60), anti-TADA3 #2678 rabbit polyclonal antibody (pAb) (24), anti-SGF29 #2461 pAb (24) and anti-VP16 #5GV2 mAb (61) (Figure 2).
Figure 2.
SAGA and ATAC HAT modules directly interact with VP16 AD. (A) Input extracts (0.2 μl) containing non-infected SF9 cell extract (control, lane 2) and SF9 cell extracts expressing the subunits of either SAGAHAT or ATACHAT (lanes 3 and 4). Fifty microliters of input extracts were then incubated with 50 μl of either GST–VP16 (B) or GST (C) beads, extensively washed and eluted. Ten percent of the total elutions were resolved by SDS–PAGE and analyzed by western blot with the indicated antibodies. Equal loading was tested by Coomassie brilliant blue staining of either GST–VP16 (B) or GST (C) protein containing beads. Each binding measurement was repeated in triplicate.
For the binding assays with purified HAT modules (Supplementary Figure S5), KAT2A in ATACHAT or SAGAHAT expressing cells contains a His-tag, and when KAT2A is expressed alone, it contains a Flag-tag. Extracts from ATACHAT or SAGAHAT expressing cells are loaded on a Ni-NTA agarose matrix and incubated for 2 h at 4°C. After washing the column (20 mM Tris, pH 7.5, 400 mM NaCl, 10 mM imidazole), bound HAT modules were eluted with 250 mM imidazole (20 mM Tris, pH 7.5, 400 mM NaCl, 250 mM imidazole). Flag-GCN5 was purified with anti-Flag M2 affinity gel with overnight incubation at 4°C. After washing the column with IP500 buffer (20 mM Tris, pH 7.9, 0.1% NP-40, 5 mM MgCl2, 10% glycerol, 500 mM KCl), elution was done with 2 mg/ml Flag peptide in IP100 buffer (20 mM Tris, pH 7.9, 0.1% NP-40, 5 mM MgCl2, 10% glycerol, 100 mM KCl). Purified GCN5, ATACHAT or SAGAHAT modules were loaded on either GST- or GST–VP16-bound beads (prepared as described earlier) and incubated for 2 h at 4°C, and then washed with five times NETN150 buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40). Then, 25 μl of resin was diluted with 10 μl of IP100 buffer; out of this slurry, 20 μl was mixed with 6 μl of 5× Laemmli buffer and boiled, one-fourth of the volume was loaded (∼14% of beads) and bound proteins were resolved by SDS–PAGE. Proteins were visualized by western blot analyses with the following antibodies (as indicated in the corresponding figures): anti-GCN5 2GC2C11 mouse mAb (60), anti-TADA3 #2678 rabbit pAb (24) and anti-SGF29 #2461 pAb (24). All binding measurements were repeated in triplicate.
HAT assays and corresponding western blot assays
Either the ATACHAT or SAGAHAT module was incubated with Gal4–VP16 or Gal4-DBD along with 2 nM of nucleosomes in T75 buffer with 300 μM Ac-CoA. Reactions were done at increasing concentrations of the ATACHAT or SAGAHAT, or at different time points. Reaction was quenched with 5 mM CoA before adding 6× SDS loading buffer. Samples were incubated in 95°C for 5 min before loading onto a 16% SDS–PAGE gel. Gels were then transferred onto a membrane and H2A-Cy5-labeled nucleosomes were fluorescently imaged to control for loading. Membranes were incubated with anti-H3K9ac (abcam 4441) and anti-H4K5ac (abcam 51997) antibodies at 1:1000 concentrations at 4°C overnight. The same aliquot of each antibody was used for each triplicate experiment to reduce variations in quantifying histone acetylation with and without either Gal4–VP16 or Gal4-DBD. The membranes were then probed with horseradish peroxidase-conjugated secondary antibodies and developed with enhanced chemiluminescence western blotting substrate (Thermo Scientific). Blots were visualized with the GE Amersham™ Imager 680. The protein bands were quantified using the densitometry analysis in ImageJ software (NIH, Washington, DC, USA). Statistical analysis was performed using GraphPad Prism software. Two-tailed, unpaired t-test with Welch’s correction was conducted using Prism 9 software. Error bars in figures represent standard error of the mean. The time course experiments were fit to the exponential function C(1 − e−t/τ), where C is the western blot signal for long times and τ is the characteristic time for the acetylation reactions. The uncertainty of τ was determined from the covariance matrix of the weighted fit.
Results
Human ATAC and SAGA HAT modules interact with unmodified nucleosomes independent of the Ac-CoA cofactor
While ATAC and SAGA complexes are reported to interact with histone tails of nucleosomes (10), the mechanism of ATACHAT and SAGAHAT interactions with nucleosomes remains elusive. To characterize human ATACHAT and SAGAHAT interactions with nucleosomes, Fluorescence Anisotropy (FA) was performed using a fixed concentration of unmodified nucleosomes and increasing concentrations of either the ATACHAT or SAGAHAT. Recombinant nucleosomes were prepared with a Widom 601 NPS with a Gal4 binding site that starts at the eighth base pair and extends 26 bp into the nucleosome (Figure 1B). The Gal4 binding site is included for TF binding studies described below. On the side proximal to the Gal4 binding site, the nucleosome was labeled with a fluorescein fluorophore that is attached to a 30-bp DNA extension at the eighth base pair outside of the nucleosome, while the side of the nucleosome that is distal to the Gal4 binding site contained a 75-bp DNA extension. As either the ATACHAT or SAGAHAT was titrated, an increase in polarization of the fluorescein fluorophore was observed, indicative of binding (58). We report HAT module binding to nucleosomes as the S1/2, the concentration of the HAT module required for half of the nucleosomes to be bound. FA measurements were fit to a binding isotherm with the ATACHAT binding with an S1/2 of 50 ± 2 nM and reaching saturating conditions around 100 nM (Figure 1D and Supplementary Table S1), while the SAGAHATS1/2 was 25.0 ± 0.3 nM, reaching saturating conditions around 200 nM (Supplementary Figure S6A). To determine the impact of acetylation catalysis on the binding of the HAT modules to the nucleosome, we next added Ac-CoA, which is required for the acetylation activity of ATAC and SAGA (62). Titrations with Ac-CoA revealed that this cofactor and the acetylation activity of the HAT modules do not significantly impact the interaction of ATACHAT and SAGAHAT modules with nucleosomes (Figure 1D and Supplementary Figure S6A). Overall, these results indicate that ATACHAT and SAGAHAT modules strongly bind unmodified nucleosomes independent of the cofactor Ac-CoA and TFs.
Previous studies have shown that the budding yeast SAGA complex interacts with both the nucleosomes and linker DNA that extends out from the nucleosome (63). To investigate whether linker DNA is important for ATACHAT and SAGAHAT binding to nucleosomes, we prepared nucleosomes with the 147-bp 601 NPS that was 5′ end labeled with fluorescein. We carried out ATACHAT and SAGAHAT titrations using FA to determine binding (Supplementary Figure S7A). We find that ATACHAT and SAGAHAT bind 147-bp nucleosomes with an S1/2 of 130 ± 30 and 50 ± 10 nM, respectively (Supplementary Figure S7B). This indicates that both ATACHAT and SAGAHAT do not require linker DNA to bind nucleosomes. We also carried out binding titrations with the 30-bp DNA that is the linker DNA in the sample that contains a nucleosome within 252 bp of DNA (Supplementary Figure S7C). We find evidence for binding above 500 nM of either HAT module, which indicates that the HAT modules weakly interact with DNA alone. Taken together, these measurements imply that both HAT modules largely interact with nucleosome core particles; however, the linker DNA does slightly enhance the binding of the HAT module.
Binding of ATAC and SAGA HAT modules to the nucleosome does not significantly alter the wrapping of DNA into the nucleosome
Histone H3 tails are primary targets for acetylation by ATAC and SAGA and extend away from the nucleosome, where the DNA exits. Therefore, we next investigated whether the binding of ATACHAT or SAGAHAT would change the amount of DNA wrapped into the nucleosome. To this end, we prepared nucleosomes labeled with both Cy3 and Cy5 fluorophores as we have done previously for other H3 tail binding domains (57,64). The DNA was labeled with Cy3, so it was located at the same locations as fluorescein, while the Cy5 was attached to histone H2AK119C (Figure 1B and C). These labeling positions undergo efficient energy transfer when the nucleosome is fully wrapped, while the FRET efficiency reduces significantly as the nucleosome partially unwraps (65). We carried out titrations with ATACHAT (Figure 1E) and SAGAHAT (Supplementary Figure S6B) over the concentration range they bind nucleosomes. We found that the FRET reduction is minimal over the concentration range that they bind nucleosomes revealing that ATACHAT and SAGAHAT binding does not directly impact nucleosome unwrapping.
KAT2A, ATAC and SAGA HAT modules efficiently bind to the VP16 AD
Activators are known to interact with coactivator complexes. In the yeast SAGA complex, VP16 AD has been shown to specifically interact with the Tra1 subunit (66). Less is known about possible activator interactions with the mammalian ATACHAT or SAGAHAT modules; nevertheless, it has been shown that the human SAGA complex can bind to VP16 AD or to Myc (40,59). To investigate whether the ATACHAT or SAGAHAT modules directly interact with the VP16 AD, we tested their binding abilities to the activation domain of VP16 when fused to GST (Figure 2A). The GST–VP16 fusion protein was immobilized on glutathione-agarose resin in parallel with GST, as a negative control. ATACHAT and SAGAHAT within SF9 cell extract were able to bind to the GST–VP16 fusion protein, but not, or only very weakly, to GST alone (Figure 2B and C). This indicates both HAT modules can specifically bind to the VP16 AD in the presence of numerous other factors in the insect cell extract. To further validate these interactions, we carried out additional binding measurements with purified HAT modules (Supplementary Figure S5). Again, both ATACHAT and SAGAHAT bind to the GST–VP16 fusion protein, while they bound very weakly or not at all to GST alone. These binding studies together reveal that both HAT modules directly interact with the VP16 AD.
To provide information about the HAT module subunit that interacts with the AD, we examined whether KAT2A alone could bind the VP16 AD. Similar to the results obtained with purified HAT modules, recombinantly purified KAT2A is able to bind directly to the VP16 AD (Supplementary Figures S4 and S5). These results indicate that the HAT modules of ATAC and SAGA can specifically interact with the VP16 AD and this interaction is possibly through a direct interaction between VP16 AD and KAT2A, which is the shared enzymatic subunit of ATAC and SAGA HAT modules.
Gal4–VP16 binds its target site within nucleosomes via the nucleosome site exposure model
Having determined that ATACHAT and SAGAHAT can efficiently bind the VP16 AD, we proceeded to investigate the binding of the well-established model TF Gal4–VP16 within nucleosomes alone (53). The Gal4–VP16 fusion TF contains the first 147 amino acids of the Gal4 TF (Gal4-DBD), which contains the DNA binding and dimerization domains, and is fused to the highly potent transcription VP16 AD (Figure 3A) (53). These measurements provide critical information for later measurements that include both Gal4–VP16 and either ATACHAT or SAGAHAT.
Figure 3.
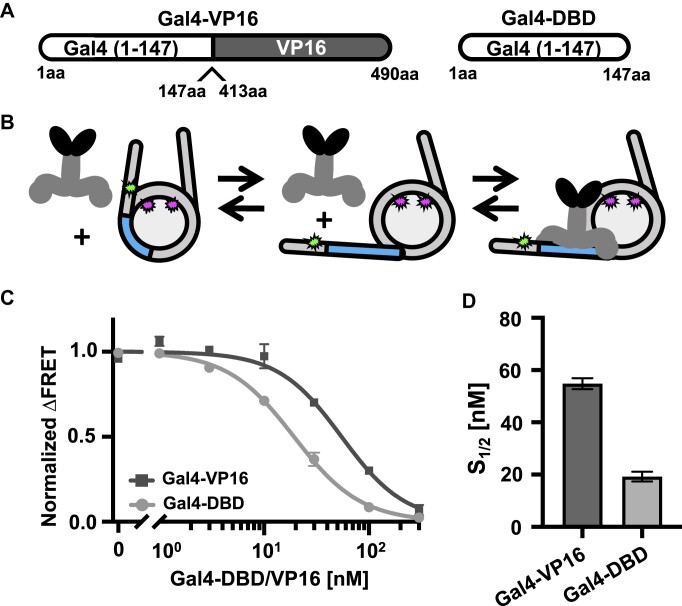
TF interactions with nucleosomes. (A) Schematic of TFs: Gal4–VP16 and Gal4-DBD. Gal4–VP16 contains the DNA binding domain and dimerization domain of Gal4 fused to the VP16 AD from the herpes simplex virus. Gal4-DBD contains the DNA binding domain and dimerization domain of Gal4. (B) Schematic of Gal4–VP16 binding equilibrium to a partially unwrapped nucleosome. (C) Normalized ΔFRET (the asymptote of the beginning of the Hill plot fit is set to 1 and the asymptote of the ending of the fit is set to 0) of Gal4–VP16 (dark gray squares) and Gal4-DBD (light gray circles) binding nucleosomes at increasing amounts. Gal4–VP16 binds with an S1/2 of 55 ± 2 nM and Gal4-DBD binds with an S1/2 of 19 ± 1 nM. (D) Bar plot of S1/2 is determined from the weighted fits to binding isotherms in panel (C). Each FRET value was determined in triplicate and the uncertainty is determined from the standard error of the three measurements. The uncertain of each S1/2 value was determined from the covariance matrix of the weighted fit.
To detect Gal4–VP16 binding to nucleosomes, we used the Cy3–Cy5-labeled nucleosomes that were used to detect whether the HAT modules directly induce nucleosome unwrapping (Figure 1A and B), which has been previously used to detect TF binding within nucleosomes (44,49,67). As mentioned earlier, the nucleosomes contained a 19-bp Gal4 binding site that starts at the eighth base pair and extends 26 bp into the Widom 601 NPS (Figure 1B). This positions the Gal4 binding site within the nucleosome, near where the DNA exits the nucleosome, and is reminiscent to positions of TF binding sites located in vivo (41–43). As Gal4–VP16 is titrated with these Cy3–Cy5-labeled nucleosomes, Gal4–VP16 binds to its site within the nucleosome via the nucleosome site exposure mechanism (Figure 3B). This mechanism involves the nucleosome partially unwrapping, which transiently exposes the Gal4–VP16 target site. Gal4–VP16 then can bind to the site trapping the nucleosome in a partially unwrapped state. This results in a significant reduction in FRET efficiency.
We carried out FRET efficiency measurements of Gal4–VP16 titrations. As expected, the FRET efficiency is reduced as Gal4–VP16 traps the nucleosome in a partially unwrapped state (Figure 3C). We fit the FRET efficiency measurements to a binding isotherm to determine the binding half saturation concentration, S1/2, which is interpreted as an apparent KD. We find that Gal4–VP16 binds to the target sequence within the nucleosome with an S1/2 of 55 ± 2 nM (Figure 3D). To control for the impact of the VP16 AD, we also carried out titrations with Gal4-DBD alone and determined an S1/2 of 19 ± 1 nM. This shows that Gal4–VP16 is able to bind to its target site within nucleosomes. To determine whether this binding is site specific, we carried out Gal4–VP16 titrations with nucleosomes that did not contain the Gal4 binding site (Supplementary Figure S8). As previously reported for Gal4-DBD (68), Gal4–VP16 does not reduce the FRET efficiency, which indicates that Gal4–VP16 binds site specifically within the nucleosome. This binding appears to follow the site exposure model (4) even with the additional steric bulk of the VP16 activator, although with a 3-fold higher apparent KD.
The transcription activator Gal4–VP16 significantly enhances the acetylation of nucleosomes by the HAT modules of ATAC and SAGA
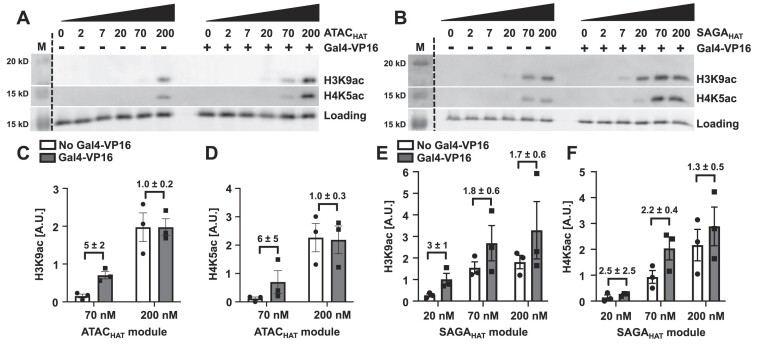
Having determined that (i) the ATACHAT and SAGAHAT interact directly with nucleosomes, (ii) the VP16 AD directly interacts with ATACHAT and SAGAHAT, and (iii) Gal4–VP16 binds efficiently within nucleosomes via the site exposure model, we decided to investigate the influence of Gal4–VP16 on the ability of ATACHAT and SAGAHAT to acetylate nucleosomes within histone H3 and H4 tails. Nucleosomes were incubated for 30 min with an increasing concentration of either ATACHAT or SAGAHAT, Ac-CoA and either with or without Gal4–VP16. The HAT module concentrations were increased over the concentration at which they bind nucleosomes (Figure 1). A Gal4–VP16 concentration of 160 nM was used to ensure saturated binding since it is ∼2.5-fold higher than the S1/2 measured by the above FRET measurements (Figure 3D). Western blot assays were used to quantify the acetylation of the H3 tail at lysine 9 (H3K9) and the H4 tail at lysine 5 (H4K5) with and without Gal4–VP16 (Figure 4). To control for nucleosome loading, we relied on Cy5 fluorescence, which is attached to H2A at K119C.
Figure 4.
Gal4–VP16 influences the acetylation of both the ATAC and SAGA HAT modules. Acetylation-coupled western blot assay of the ATACHAT (A) and SAGAHAT (B) modules at increasing concentrations with and without Gal4–VP16 at the indicated PTM sites. The fluorescence of Cy5-labeled histone H2A within the nucleosome was used to quantify nucleosome loading. Acetylation efficiency was tested by western blot using antibodies recognizing either histone H3K9ac or H4K5ac. Images are representative of three independent experiments with similar results (n = 3). Two-tailed, unpaired t-test with Welch’s correction was conducted using Prism 9 software. Error bars represent standard error of the mean. (C, D) Bar plots of panel (A). (E, F) Bar plots of panel (B).
We find that Gal4–VP16 enhances acetylation of both H3K9 and H4K5 by ATACHAT (Figure 4A, C-D). At sub-saturating concentrations of 70 nM ATACHAT, Gal4–VP16 induced 5-fold and 6-fold increase in the acetylation of H3K9 and H4K5, respectively (Figure 4C and D, and Supplementary Table S2). In contrast, the impact of Gal4–VP16 on acetylation by SAGAHAT was less dramatic (Figure 4B, E-F). At the sub-saturating concentration of 20 nM SAGAHAT, Gal4–VP16 increased acetylation activity at H3K9 by 3-fold (Figure 4E). In contrast, at 20 nM of SAGAHAT, Gal4–VP16 did not induce a statistically significant increase in acetylation of H4K5, while a statistically significant change in acetylation of H4K5 (2.2-fold) with 70 nM SAGAHAT and Gal4–VP16 was observed. This indicates that Gal4–VP16 has a modest impact on H4K5 acetylation by SAGAHAT.
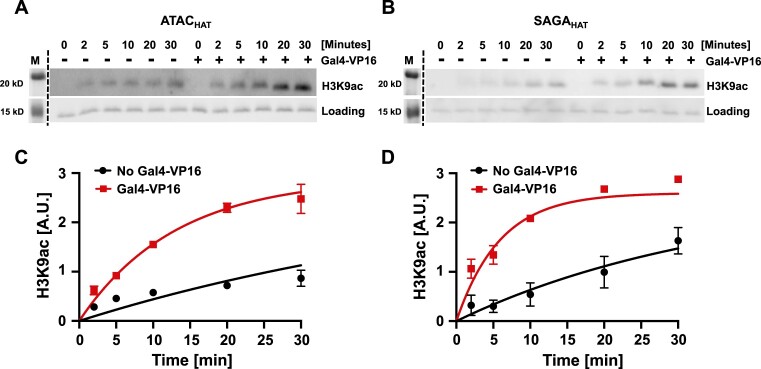
To further quantify the impact of Gal4–VP16 on the acetylation activity of ATACHAT and SAGAHAT, we determined the effective rate of acetylation at limiting concentrations of each HAT module. Nucleosomes were incubated for 2, 5, 10, 20 or 30 min with limiting concentration of either ATACHAT (50 nM) or SAGAHAT (20 nM) at their S1/2, Ac-CoA and either with or without saturating concentration of Gal4–VP16 (160 nM). The different time points were quenched with CoA and then SDS–PAGE loading buffer. The level of H3K9ac was then quantified by western blot analysis (Figure 5). We find that Gal4–VP16 increased the rate of H3K9 acetylation by 4.6 ± 0.2 and 5.1 ± 0.1 for ATACHAT and SAGAHAT (Supplementary Table S3). This confirms that Gal4–VP16 increases the acetylation activity of both ATACHAT and SAGAHAT.
Figure 5.
The acetylation kinetics of nucleosomes at H3K9 by ATACHAT and SAGAHAT. Acetylation assay by the ATACHAT (A) or SAGAHAT (B) module at increasing time with and without Gal4–VP16 at H3K9ac. The fluorescence of Cy5-labeled histone H2A within the nucleosome was used to observe nucleosome loading. Acetylation efficiency was tested by western blot using antibodies recognizing histone H3K9ac. Images are representative of three independent experiments with similar results (n = 3). Error bars represent standard error of the mean. (C) Graph of panel (A). (D) Graph of panel (B). The time courses were fit to a single exponential as described in the ‘Materials and methods’ section to determine characteristic time, τ, for the acetylation reaction.
These observed differences in acetylation by ATACHAT and SAGAHAT at sub-saturating concentrations are consistent with in vivo studies, where SAGA targets H3 at K9 and K14 (16,20,21), while ATAC appears to acetylate both histones H3 and H4 (15,22–26). Interestingly, at saturating concentrations of either ATACHAT or SAGAHAT, Gal4–VP16 no longer impacts acetylation activity at either H3K9 or H4K5. These results are consistent with the conclusion that Gal4–VP16 facilitates recruitment of the HAT modules to nucleosomes, but once the HAT module fully occupies the nucleosome, this transcription activator no longer enhances acetylation. In addition, the difference in the impact of Gal4–VP16 on acetylation by ATACHAT could in part be due to the lower starting activity of ATACHAT as compared to SAGAHAT, allowing Gal4–VP16 to have a larger impact on ATACHAT acetylation activity. Overall, these results indicate that Gal4–VP16, which directly interacts with both HAT modules and nucleosomes that contain the Gal4 binding site, facilitates acetylation of both the H3 and H4 tails by both HAT modules, although Gal4–VP16 has a larger impact on ATACHAT targeted acetylation.
Trapping the nucleosome in a partially unwrapped state enhances the ATAC HAT module acetylation activity but does not enhance the SAGA HAT module acetylation activity
The increase in HAT module acetylation activity by Gal4–VP16 could occur by two non-mutually exclusive mechanisms. The first mechanism is that the direct interaction of the VP16 AD helps stabilize the HAT module binding to the nucleosome, which in turn enhances the acetylation activity. The second mechanism is that Gal4–VP16 traps the nucleosome in a partially unwrapped state, increasing the exposure of the H3 and H4 tails for HAT module binding and acetylation. To determine the impact of nucleosome unwrapping alone, we carried out the above-described acetylation activity measurements with Gal4-DBD alone, which does not contain the VP16 AD. Gal4-DBD reduces the FRET efficiency as it binds its target site within the nucleosome, demonstrating that it traps the nucleosome in a partially unwrapped state similarly to Gal4–VP16 (Figure 3). This allowed the determination of the impact of the DBD of a TF trapping the nucleosome in an unwrapped state on HAT module acetylation activity without the confounding impact of interactions with an AD.
As with Gal4–VP16, nucleosomes were incubated for 30 min with an increasing concentration of either ATACHAT or SAGAHAT, Ac-CoA and either with or without 70 nM Gal4-DBD, which ensures saturating binding of nucleosomes by Gal4-DBD (Figure 3). Western blot assays were again used to quantify the acetylation at H3K9 and H4K5. We observe minimal acetylation activity at H3K9 below 70 nM of ATACHAT and 20 nM SAGAHAT (Supplementary Figure S9). Interestingly at 70 nM of ATACHAT, which is near the apparent KD, the acetylation activity of H3K9 is increased 4-fold by Gal4-DBD. At the saturating concentration of 200 nM ATACHAT, Gal4-DBD no longer influences the acetylation activity at H3K9 (Supplementary Figure S9 and Supplementary Table S4). In contrast, we find that Gal4-DBD does not induce a statistically significant change in the acetylation activity of ATACHAT at H4K5. For SAGAHAT, we found that Gal4-DBD did not enhance the acetylation activity of SAGAHAT at either H3K9 or H4K5 (Supplementary Figure S9 and Supplementary Table S4).
To further verify the impact of Gal4-DBD on the acetylation activity of ATACHAT and SAGAHAT, we carried out kinetic measurements of H3K9 acetylation. As with Gal4–VP16, nucleosomes were incubated for 2, 5, 10, 20 and 30 min with sub-saturating concentrations of either ATACHAT (50 nM) or SAGAHAT (20 nM), Ac-CoA and either with or without 60 nM Gal4-DBD (Supplementary Figure S10), which ensures saturating binding of nucleosomes by Gal4-DBD (Figure 3). These kinetic measurements show that Gal4-DBD increased by 2.7 ± 0.4 the rate of H3K9 acetylation by ATACHAT, while Gal4-DBD did not impact the rate of H3K9 acetylation by SAGAHAT (Supplementary Table S3).
These results can be explained by considering the structure of the nucleosome. The H3 tail extends out from the nucleosome where the DNA exits the nucleosome, which positions the H3 tail such that a partial nucleosome unwrapping could impact its accessibility. In contrast, the H4 tail extends out from the face of the histone octamer, which is at a position where partial nucleosome unwrapping is not likely to enhance H4 tail accessibility. Interestingly, these findings also suggest that trapping the nucleosome in a partially unwrapped state with Gal4-DBD provides ATACHAT additional access to the H3 tail to acetylate H3K9 but does not impact H3 tail accessibility to SAGAHAT. This points to a possible mechanism where a TF can differentially target histone H3 tail acetylation by the tested two HAT modules.
ATAC and SAGA HAT modules enhance TF accessibility independent of their acetylation activity
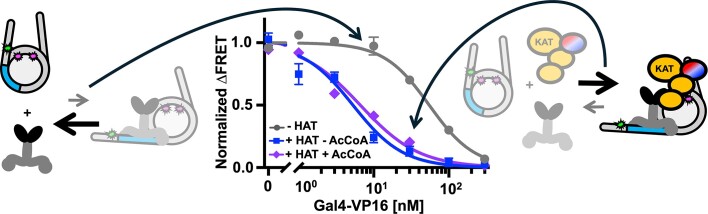
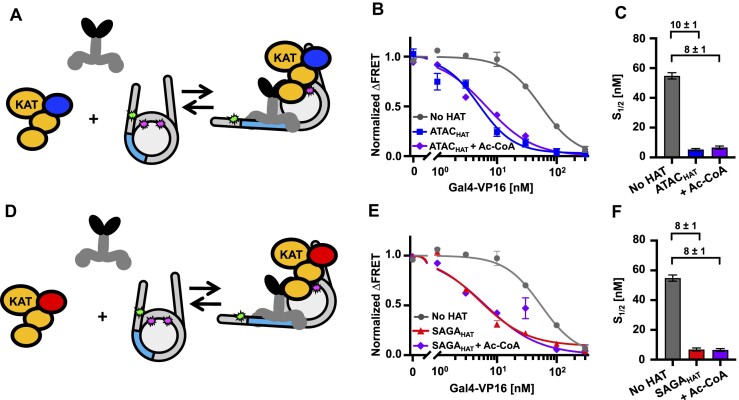
Coactivator complexes are reported to be recruited to promoter regions by TFs to aid Pol II transcription initiation (69). However, the mechanisms by which coactivator complexes and TFs function together is still not fully understood. In the acetylation assays, we showed that the addition of Gal4–VP16 can significantly enhance the acetylation function of the studied HAT modules. This finding raised the following question: Do the HAT modules in turn influence the accessibility of Gal4–VP16 to the nucleosome? To investigate how ATACHAT and SAGAHAT function with a TF to target nucleosomes, we used ensemble FRET efficiency measurements to quantify Gal4–VP16 binding within partially unwrapped nucleosomes in the presence or absence of either the ATACHAT or SAGAHAT (Figure 6A and D). By comparing the Gal4–VP16 S1/2 for binding to nucleosomes with and without the HAT modules, we determined the impact of the HAT modules on the site accessibility for Gal4–VP16 occupancy at its site within a partially unwrapped nucleosome.
Figure 6.
ATACHAT and SAGAHAT influence nucleosome accessibility to Gal4–VP16 binding independent of Ac-CoA. (A) Model of Gal4–VP16 and the ATACHAT module binding to a nucleosome. (B) Normalized ΔFRET of Gal4–VP16 binding nucleosomes with increasing amounts. Gal4–VP16 binding to nucleosomes at an S1/2 of 55 ± 2 nM (dark gray circles), Gal4–VP16 binding to nucleosomes with the addition of 200 nM ATACHAT at an S1/2 of 5.3 ± 0.7 nM (dark blue squares) and Gal4–VP16 binding to nucleosomes with the addition of 300 μM Ac-CoA at an S1/2 of 7 ± 1 nM (purple diamonds). (C) Bar plot of the S1/2 from panel (B). (D) Model of Gal4–VP16 and the SAGAHAT module binding to a nucleosome. (E) Normalized ΔFRET of Gal4–VP16 binding to nucleosomes at increasing amounts. Gal4–VP16 binding to nucleosomes at an S1/2 of 55 ± 2 nM (dark gray circles), Gal4–VP16 binding to nucleosomes with the addition of 150 nM SAGAHAT at an S1/2 of 7 ± 1 nM (red triangles) and Gal4–VP16 binding to nucleosomes with the addition of 300 μM Ac-CoA at an S1/2 of 6.5 ± 0.9 nM (purple diamonds). (F) Bar plot of S1/2 values that are determined from the weighted fits to binding isotherms in panel (E). Each FRET value was determined in triplicate and the uncertainty is determined from the standard error of the three measurements. The uncertainty for each S1/2 value is determined from the covariance matrix of the weighted fits in panel (E).
We used HAT module concentrations of 200 and 150 nM for ATACHAT or SAGAHAT, respectively, to ensure that the HAT modules were at concentrations that efficiently bind nucleosomes (Figure 1D, and Supplementary Figure S6A). We carried out these measurements without Ac-CoA to eliminate the impact of histone acetylation on Gal4–VP16 binding. In the presence of ATACHAT, the binding affinity of Gal4–VP16 increased by 10-fold, where the S1/2 lowered from 55 ± 2 to 5.3 ± 0.7 nM (Figure 6B and C, and Supplementary Table S5). Likewise, the addition of SAGAHAT increased the ability of Gal4–VP16 to bind by 8-fold, where the S1/2 of Gal4–VP16 lowered to 7 ± 1 nM (Figure 6E and F, and Supplementary Table S5). We then repeated these measurements with 300 μM Ac-CoA to determine how the HAT modules’ histone acetylation activity influences the impact of the HAT modules on Gal4–VP16 binding. We found that the addition of Ac-CoA did not statistically impact the binding affinity of Gal4–VP16 (Figure 6C and F), which implies that the order of magnitude increases in Gal4–VP16 binding within the nucleosome by ATACHAT and SAGAHAT are independent of the cofactor, Ac-CoA and the HAT module’s ability to acetylate histones.
Since VP16 AD directly interacts with both HAT modules, we investigated whether the VP16 AD is required for the acetylation-independent increase in Gal4–VP16 binding by determining whether the HAT modules facilitate Gal4-DBD binding and by how much. We carried out Gal4-DBD titrations with and without ATACHAT or SAGAHAT to determine the change in the S1/2 of Gal4-DBD binding (Supplementary Figure S11 and Supplementary Table S5). We found that ATACHAT increased Gal4-DBD binding by ∼2.5-fold, where the S1/2 decreased from 19 ± 1 to 7.6 ± 0.8 nM, while SAGAHAT increases Gal4-DBD binding by ∼4-fold, where the S1/2 decreased to 4.9 ± 0.3 nM. We also investigated the impact of Ac-CoA and found that this increase in Gal4-DBD binding was not significantly altered by the acetylation activity of either HAT module. This implies that the enhanced binding of Gal4-DBD by both HAT modules does not depend on their cofactor or their acetylation activity.
These differences in S1/2 induced by each HAT module allow for the determination of the change in the TF binding free energy since ΔΔGTF = −kBT ln(S1/2,HAT/S1/2,no HAT) (70). So, based on the S1/2 measurements, ATACHAT and SAGAHAT induce a ΔΔGGal4–VP16 of −2.3kBT and −2.0kBT for Gal4–VP16 binding, respectively, while they induce a ΔΔGGal4-DBD of −0.9kBT and −1.4kBT for Gal4-DBD binding. A comparison of the ΔΔGGal4–VP16 to ΔΔGGal4-DBD reveals that the VP16 activator provides an additional reduction in binding free energy of −1.4kBT with ATACHAT, while for SAGAHAT the VP16 activator provides an additional −0.6kBT. This suggests that the VP16 activator has an ∼2-fold stronger interaction with the ATACHAT than the SAGAHAT module.
In combination, these results indicate that the VP16 AD is important for the order of magnitude increase of Gal4–VP16 binding induced by either ATACHAT or SAGAHAT. This is likely due to direct interactions between VP16 and the HAT modules. However, the more modest increase in binding of Gal4 DBD alone indicates that while the HAT modules do not directly induce nucleosome unwrapping (Figure 1E and Supplementary Figure S6B), they can help stabilize Gal4 binding even without directly interacting with an AD, perhaps by preventing rewrapping. Overall, these findings indicate that not only do transcription-activating TFs, such as Gal4–VP16, enhance ATACHAT and SAGAHAT acetylation, but these HAT modules also facilitate TF binding independent of their acetylation activity.
Discussion
This study provides quantitative and mechanistic insight into how the HAT modules of two important coactivators, ATAC and SAGA, function with a transcription-activating TF to target and open chromatin. Our results reveal that both ATACHAT and SAGAHAT have two distinct yet synergistic functions: (i) coordinate with TFs to acetylate histones to open chromatin and (ii) target the same TFs to its binding site within the nucleosome. These observations also provide insight into how both the ATAC and SAGA full complexes function with a TF. KAT2A and other SAGA subunits have been reported to interact with transcription-activating TFs (34), while ATAC subunit(s) that directly interacts with TFs has yet to be characterized. Interestingly, our in vitro binding experiments suggest that KAT2A in both ATACHAT and SAGAHAT is likely to contribute to TF binding, and consequently to the recruitment of these HAT modules, and/or their holo complexes, to activate genes. In addition, the SAGA holo complex contains the TRRAP subunit that also interacts with TFs (36). In a recent screen that tested for the interactions of 109 TFs with other proteins by proximity-dependent biotinylation and affinity purification mass spectrometry measurements, the authors described that the endogenous ATAC complex interacts with E2F1; ELF1, HNF4a, KLF3, KLF12, MYC; PPARγ, TYY1 TFs, while endogenous SAGA complex was shown to interact with KLF6, MYC and MYOD TFs (71). Our results indicate that the ATAC and SAGA HAT modules may contribute to these TF interactions, which could help target the ATAC and/or SAGA holo complexes to promoters and enhancers.
A recent cryo-EM study reported that upon nucleosome binding to the budding yeast SAGA complex, the HAT module’s structure cannot be resolved (15). It was proposed that nucleosome binding to SAGA displaces the HAT module. One possibility is that TFs are needed to help stabilize the HAT module even within the context of the full SAGA complex. It is also possible that the HAT module does not bind the nucleosome in a distinct orientation, so cryo-EM is not able to visualize the HAT module bound to the nucleosome. In addition, these HAT modules could also function outside of the endogenous ATAC and SAGA holo complexes (34,35). Thus, it is conceivable that TFs can directly target the HAT modules, free or incorporated in their respective holo complexes, to TF target sites allowing the ATACHAT and SAGAHAT modules or their holo complexes to function as transcription coactivators.
Our studies of the impact of Gal4–VP16 versus Gal4-DBD on the acetylation activities of ATACHAT and SAGAHAT allowed us to decouple the impact of the VP16 activator direct interaction with the HAT modules and the impact of the TF trapping the nucleosome in a partially unwrapped state. The combined results of Gal4–VP16 and Gal4-DBD indicate that a TF with an AD that directly interacts with the HAT module enhances ATACHAT acetylation of the H3 tail by two mechanisms: (i) it facilitates recruitment of ATACHAT module through direct interactions with the AD and (ii) it increases ATACHAT accessibility to the histone tail through partial nucleosome unwrapping. In contrast, only the first mechanism appears to be important for ATACHAT acetylation of the H4 tail, which as mentioned earlier is consistent with the location of the H4 tail within the nucleosome. For SAGAHAT, trapping the nucleosome in a partially unwrapped state does not impact its acetylation activity, implying that only the first mechanism has a significant impact on SAGAHAT acetylation activity of both the H3 and H4 tails. These results highlight differences between the two HAT modules, which could be important in how ATACHAT and SAGAHAT target distinct chromatin locations.
Both the absolute S1/2 and the changes in S1/2 are important values for considering how ATACHAT and SAGAHAT impact TF binding within nucleosomes. Both HAT modules induced a reduction in the S1/2 for Gal4–VP16 that is larger than the reduction for Gal4-DBD. However, Gal4–VP16 has a higher initial S1/2 than Gal4-DBD. These combined differences result in similar final S1/2 values for both Gal4–VP16 and Gal4-DBD. This reveals how TFs that initially bind at different concentration can be regulated by either HAT module to bind at similar concentrations. In addition to considering the absolute S1/2 values, the change in S1/2 provides important insight as well. The reduction in the S1/2 by the HAT module is the maximum concentration range the HAT module can regulate TF occupancy. Thus, for a TF with an AD that does not interact with the HAT module, the HAT module can impact TF binding over a range of 2.5–4-fold, while for a TF with an AD that interacts with the HAT module, the HAT module can impact TF binding by 10-fold. Overall, this comparison suggests that details of the interaction of the AD with the HAT module are important for how the HAT module influences the binding of the wide range of different TFs.
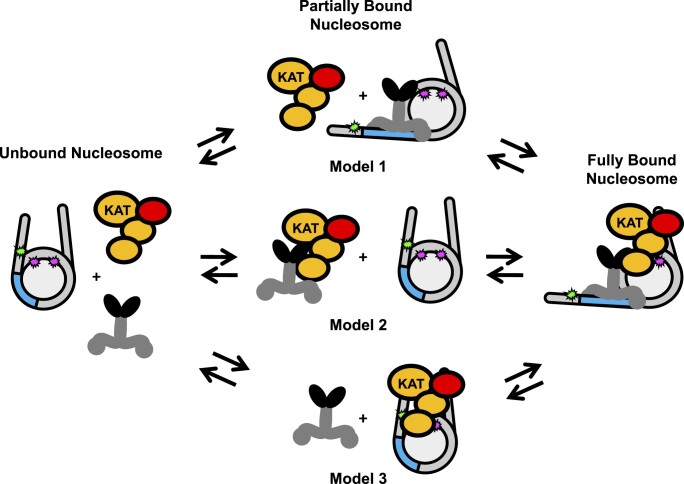
The new function of ATACHAT and SAGAHAT modules identified in this study by in vitro experiments, where they help target a TF to its recognition site within a nucleosome, is highly synergistic with their well-established function to acetylate histones. By helping the TF target their binding sites within nucleosomes, ATACHAT and SAGAHAT enhance the targeting for their own HAT activity. This targeting could happen through three non-exclusive models (Figure 7), where the TF and nucleosome bind first (Model 1), the TF and the HAT module bind first (Model 2) or the HAT module and the nucleosome bind first (Model 3). Since we found that Gal4–VP16, HAT modules and nucleosomes all interact strongly with each other, it is possible that all three pairwise interactions are possible pathways to the nucleosome being bound by both Gal4–VP16 and a HAT module. Our current studies do not differentiate between these pathways. However, a previous study reported that the suppression of Gal4-DBD at its site within the nucleosome is influenced by both a reduced binding rate and an accelerated dissociation rate (68). This suggests that the HAT module could influence both the rate of Gal4–VP16 binding and/or dissociation. Future studies are needed to investigate how the binding and dissociation rates are changed, and which pathway(s) (see Figure 7) are involved in these HAT modules increasing Gal4–VP16 occupancy by an order of magnitude.
Figure 7.
Models of HAT module and TF binding to a partially unwrapped nucleosome. Model 1: Gal4–VP16 first binds to its recognition sequence and then recruits the HAT module. Model 2: Gal4–VP16 and the HAT module interact with each other before binding the nucleosome together. Model 3: the HAT module interacts with the nucleosome first, allowing Gal4–VP16 to bind at a higher affinity. Since the impact of the HAT module on TF binding does not depend on the HAT acetylation activities, acetylation is not indicated in the model. However, histone acetylation will occur when the HAT module is bound to the nucleosome, which includes the fully bound nucleosome and the intermediate state of Model 3.
Interestingly, recent molecular and genetic studies revealed that many histone-modifying enzymes have functions independent of their HAT or methyltransferase activities. Many of the histone modifications deposited by these enzymes, previously considered to be crucial for transcriptional activation, have been demonstrated to function for gene expression independent of their catalytic activities (33,72,73). Our discovery of the new HAT-independent function of the ATAC and SAGA HAT modules in targeting TFs to their recognition sites within chromatin in a HAT-independent manner suggests that the newly discovered function of these chromatin-modifying complexes may be even more important than their given enzymatic activities. Future experiments will be required to verify whether the HAT-independent targeting role of these complexes also operates in living organisms.
Supplementary Material
Acknowledgements
We thank the members of the Poirier, Tora and Berger labs for helpful discussions and comments. We are grateful the Parthun lab and the Musier-Forsyth labs for help with the western blots.
Author contributions: K. Chesnutt, M.G.P. and L.T. conceived and designed the research. K. Chesnutt carried out the FA, western blot and FRET experiments. K. Chesnutt and K. Cox prepared the nucleosome samples. G.Y. and M.D. carried out the pull-down experiments. C.T. and G.G. prepared the ATAC and SAGA HAT modules. M.G.P. and L.T. supervised the study. K. Chesnutt, M.G.P. and L.T. wrote the first draft. All authors edited and finalized the manuscript.
Contributor Information
Kristin V Chesnutt, Ohio State Biochemistry Program, Ohio State University, 191 W. Woodruff Ave. Columbus, OH, 43210, USA.
Gizem Yayli, Institut de Génétique et de Biologie Moléculaire et Cellulaire, 1 Rue Laurent Fries 67400 Illkirch, France; Centre National de la Recherche Scientifique, UMR 7104, 1 Rue Laurent Fries 67400, Illkirch, France; Institut National de la Santé et de la Recherche Médicale, U1258, 1 Rue Laurent Fries 67400, Illkirch, France; Université de Strasbourg, 1 Rue Laurent Fries 67400 Illkirch, France.
Christine Toelzer, School of Biochemistry, University of Bristol, 1 Tankard's Close, Bristol BS8 1TD, UK.
Mylène Damilot, Institut de Génétique et de Biologie Moléculaire et Cellulaire, 1 Rue Laurent Fries 67400 Illkirch, France; Centre National de la Recherche Scientifique, UMR 7104, 1 Rue Laurent Fries 67400, Illkirch, France; Institut National de la Santé et de la Recherche Médicale, U1258, 1 Rue Laurent Fries 67400, Illkirch, France; Université de Strasbourg, 1 Rue Laurent Fries 67400 Illkirch, France.
Khan Cox, Department of Physics, Ohio State University, 191 W. Woodruff Ave. Columbus, OH 43210, USA.
Gunjan Gautam, School of Biochemistry, University of Bristol, 1 Tankard's Close, Bristol BS8 1TD, UK.
Imre Berger, School of Biochemistry, University of Bristol, 1 Tankard's Close, Bristol BS8 1TD, UK.
László Tora, Institut de Génétique et de Biologie Moléculaire et Cellulaire, 1 Rue Laurent Fries 67400 Illkirch, France; Centre National de la Recherche Scientifique, UMR 7104, 1 Rue Laurent Fries 67400, Illkirch, France; Institut National de la Santé et de la Recherche Médicale, U1258, 1 Rue Laurent Fries 67400, Illkirch, France; Université de Strasbourg, 1 Rue Laurent Fries 67400 Illkirch, France.
Michael G Poirier, Ohio State Biochemistry Program, Ohio State University, 191 W. Woodruff Ave. Columbus, OH, 43210, USA; Department of Physics, Ohio State University, 191 W. Woodruff Ave. Columbus, OH 43210, USA; Department of Chemistry & Biochemistry, Ohio State University, Columbus, OH43210, USA.
Data availability
The experimental datasets are included either in this manuscript or the supplemental information, or are available from the authors upon request.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health [R35 GM139564 and R01 GM131626 to M.G.P. and L.T., T32 GM118291 and T32 GM144293 to K. Chesnutt]; Agence Nationale de la Recherche [ANR-19-CE11-0003-02, ANR-20-CE12-0017-03, ANR-PRCI-19-CE12-0029-01 and ANR-22-CE11-0013-01_ACT to L.T.]; Initiative d'exce llence, Université de Strasbourg [ANR-10-IDEX-0002]; Structuration de la formation par la recherche dans les initiatives d'excellence, STRAT’US project [ANR-20-SFRI-0012]; L'École Universitaire de Recherche, Integrative Molecular and Cellular Biology [ANR-17-EURE-0023]; Wellcome Trust [106115/Z/14/Z to I.B.]; Nancy Byrd Johnson and Howard M. Johnson Endowed Fund (to K. Chesnutt). Funding for open access charge: National Institutes of Health [R35 GM139564].
Conflict of interest statement. None declared.
References
- 1. Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 2. Richmond T.J., Davey C.A.. The structure of DNA in the nucleosome core. Nature. 2003; 423:145–150. [DOI] [PubMed] [Google Scholar]
- 3. Tan S., Davey C.A.. Nucleosome structural studies. Curr. Opin. Struct. Biol. 2011; 21:128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Polach K.J., Widom J.. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995; 254:130–149. [DOI] [PubMed] [Google Scholar]
- 5. Robinson P.J.J., Rhodes D.. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr. Opin. Struct. Biol. 2006; 16:336–343. [DOI] [PubMed] [Google Scholar]
- 6. Lowary P.T., Widom J.. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998; 276:19–42. [DOI] [PubMed] [Google Scholar]
- 7. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yun M., Wu J., Workman J.L., Li B.. Readers of histone modifications. Cell Res. 2011; 21:564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Helmlinger D., Tora L.. Sharing the SAGA. Trends Biochem. Sci. 2017; 42:850–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Helmlinger D., Papai G., Devys D., Tora L.. What do the structures of GCN5-containing complexes teach us about their function?. Biochim. Biophys. Acta Gene Regul. Mech. 2021; 1864:194614. [DOI] [PubMed] [Google Scholar]
- 11. Spedale G., Timmers H.T.M., Pijnappel W.W.M.P.. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012; 26:527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Herbst D.A., Esbin M.N., Louder R.K., Dugast-Darzacq C., Dailey G.M., Fang Q., Darzacq X., Tjian R., Nogales E.. Structure of the human SAGA coactivator complex. Nat. Struct. Mol. Biol. 2021; 28:989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee K.K., Sardiu M.E., Swanson S.K., Gilmore J.M., Torok M., Grant P.A., Florens L., Workman J.L., Washburn M.P.. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol. Syst. Biol. 2011; 7:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Papai G., Frechard A., Kolesnikova O., Crucifix C., Schultz P., Ben-Shem A.. Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature. 2020; 577:711–716. [DOI] [PubMed] [Google Scholar]
- 15. Wang H., Dienemann C., Stützer A., Urlaub H., Cheung A.C.M., Cramer P.. Structure of the transcription coactivator SAGA. Nature. 2020; 577:717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Riss A., Scheer E., Joint M., Trowitzsch S., Berger I., Tora L.. Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) coactivator complexes enhance the acetyltransferase activity of GCN5. J. Biol. Chem. 2015; 290:28997–29009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yayli G., Bernardini A., Mendoza Sanchez P.K., Scheer E., Damilot M., Essabri K., Morlet B., Negroni L., Vincent S.D., Timmers H.T.M.et al.. ATAC and SAGA co-activator complexes utilize co-translational assembly, but their cellular localization properties and functions are distinct. Cell Rep. 2023; 42:113099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kusch T., Guelman S., Abmayr S.M., Workman J.L.. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 2003; 23:3305–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muratoglu S., Georgieva S., Pápai G., Scheer E., Enünlü I., Komonyi O., Cserpán I., Lebedeva L., Nabirochkina E., Udvardy A.et al.. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 2003; 23:306–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonnet J., Wang C.-Y., Baptista T., Vincent S.D., Hsiao W.-C., Stierle M., Kao C.-F., Tora L., Devys D.. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 2014; 28:1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feller C., Forné I., Imhof A., Becker P.B.. Global and specific responses of the histone acetylome to systematic perturbation. Mol. Cell. 2015; 57:559–571. [DOI] [PubMed] [Google Scholar]
- 22. Ciurciu A., Komonyi O., Pankotai T., Boros I.M.. The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol. Cell. Biol. 2006; 26:9413–9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guelman S., Kozuka K., Mao Y., Pham V., Solloway M.J., Wang J., Wu J., Lill J.R., Zha J.. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol. Cell. Biol. 2009; 29:1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagy Z., Riss A., Fujiyama S., Krebs A., Orpinell M., Jansen P., Cohen A., Stunnenberg H.G., Kato S., Tora L.. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell. Mol. Life Sci. 2010; 67:611–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pankotai T., Komonyi O., Bodai L., Ujfaludi Z., Muratoglu S., Ciurciu A., Tora L., Szabad J., Boros I.. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol. Cell. Biol. 2005; 25:8215–8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suganuma T., Gutiérrez J.L., Li B., Florens L., Swanson S.K., Washburn M.P., Abmayr S.M., Workman J.L.. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. Mol. Biol. 2008; 15:364–372. [DOI] [PubMed] [Google Scholar]
- 27. Fournier M., Orpinell M., Grauffel C., Scheer E., Garnier J.-M., Ye T., Chavant V., Joint M., Esashi F., Dejaegere A.et al.. KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nat. Commun. 2016; 7:13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirsch C.L., Coban Akdemir Z., Wang L., Jayakumaran G., Trcka D., Weiss A., Hernandez J.J., Pan Q., Han H., Xu X.et al.. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev. 2015; 29:803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krebs A.R., Karmodiya K., Lindahl-Allen M., Struhl K., Tora L.. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol. Cell. 2011; 44:410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nagy Z., Riss A., Romier C., le Guezennec X., Dongre A.R., Orpinell M., Han J., Stunnenberg H., Tora L.. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol. Cell. Biol. 2009; 29:1649–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Orpinell M., Fournier M., Riss A., Nagy Z., Krebs A.R., Frontini M., Tora L.. The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 2010; 29:2381–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stegeman R., Spreacker P.J., Swanson S.K., Stephenson R., Florens L., Washburn M.P., Weake V.M.. The spliceosomal protein SF3B5 is a novel component of Drosophila SAGA that functions in gene expression independent of splicing. J. Mol. Biol. 2016; 428:3632–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fischer V., Plassard D., Ye T., Reina-San-Martin B., Stierle M., Tora L., Devys D.. The related coactivator complexes SAGA and ATAC control embryonic stem cell self-renewal through acetyltransferase-independent mechanisms. Cell Rep. 2021; 36:109598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soffers J.H.M., Li X., Saraf A., Seidel C.W., Florens L., Washburn M.P., Abmayr S.M., Workman J.L.. Characterization of a metazoan ADA acetyltransferase complex. Nucleic Acids Res. 2019; 47:3383–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Torres-Zelada E.F., Stephenson R.E., Alpsoy A., Anderson B.D., Swanson S.K., Florens L., Dykhuizen E.C., Washburn M.P., Weake V.M.. The Drosophila Dbf4 ortholog Chiffon forms a complex with Gcn5 that is necessary for histone acetylation and viability. J. Cell Sci. 2019; 132:jcs214072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheon Y., Kim H., Park K., Kim M., Lee D.. Dynamic modules of the coactivator SAGA in eukaryotic transcription. Exp. Mol. Med. 2020; 52:991–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McMahon S.B., Van Buskirk H.A., Dugan K.A., Copeland T.D., Cole M.D.. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998; 94:363–374. [DOI] [PubMed] [Google Scholar]
- 38. McMahon S.B., Wood M.A., Cole M.D.. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 2000; 20:556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang N., Ichikawa W., Faiola F., Lo S.-Y., Liu X., Martinez E.. MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim. Biophys. Acta. 2014; 1839:395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu X., Tesfai J., Evrard Y.A., Dent S.Y.R., Martinez E.. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J. Biol. Chem. 2003; 278:20405–20412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. North J.A., Shimko J.C., Javaid S., Mooney A.M., Shoffner M.A., Rose S.D., Bundschuh R., Fishel R., Ottesen J.J., Poirier M.G.. Regulation of the nucleosome unwrapping rate controls DNA accessibility. Nucleic Acids Res. 2012; 40:10215–10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koerber R.T., Rhee H.S., Jiang C., Pugh B.F.. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol. Cell. 2009; 35:889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Michael A.K., Stoos L., Crosby P., Eggers N., Nie X.Y., Makasheva K., Minnich M., Healy K.L., Weiss J., Kempf G.et al.. Cooperation between bHLH transcription factors and histones for DNA access. Nature. 2023; 619:385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luo Y., North J.A., Poirier M.G.. Single molecule fluorescence methodologies for investigating transcription factor binding kinetics to nucleosomes and DNA. Methods. 2014; 70:108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anderson J.D., Widom J.. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000; 296:979–987. [DOI] [PubMed] [Google Scholar]
- 46. Liang S.D., Marmorstein R., Harrison S.C., Ptashne M.. DNA sequence preferences of GAL4 and PPR1: how a subset of Zn2Cys6 binuclear cluster proteins recognizes DNA. Mol. Cell. Biol. 1996; 16:3773–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bernier M., Luo Y., Nwokelo K.C., Goodwin M., Dreher S.J., Zhang P., Parthun M.R., Fondufe-Mittendorf Y., Ottesen J.J., Poirier M.G.. Linker histone H1 and H3K56 acetylation are antagonistic regulators of nucleosome dynamics. Nat. Commun. 2015; 6:10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Makde R.D., England J.R., Yennawar H.P., Tan S.. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010; 467:562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burge N.L., Thuma J.L., Hong Z.Z., Jamison K.B., Ottesen J.J., Poirier M.G.. H1.0 C terminal domain is integral for altering transcription factor binding within nucleosomes. Biochemistry. 2022; 61:625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shimko J.C., North J.A., Bruns A.N., Poirier M.G., Ottesen J.J.. Preparation of fully synthetic histone H3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes. J. Mol. Biol. 2011; 408:187–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Luger K., Rechsteiner T.J., Richmond T.J.. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999; 119:1–16. [DOI] [PubMed] [Google Scholar]
- 52. Hong M., Fitzgerald M.X., Harper S., Luo C., Speicher D.W., Marmorstein R.. Structural basis for dimerization in DNA recognition by Gal4. Structure. 2008; 16:1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sadowski I., Ma J., Triezenbergt S., Ptashne M.. GAL4–VP16 is an unusually potent transcriptional activator. Nature. 1988; 335:563–564. [DOI] [PubMed] [Google Scholar]
- 54. Berger I., Fitzgerald D.J., Richmond T.J.. Baculovirus expression system for heterologous multiprotein complexes. Nat. Biotechnol. 2004; 22:1583–1587. [DOI] [PubMed] [Google Scholar]
- 55. Trowitzsch S., Bieniossek C., Nie Y., Garzoni F., Berger I.. New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol. 2010; 172:45–54. [DOI] [PubMed] [Google Scholar]
- 56. Clegg R.M. Fluorescence resonance energy transfer. Curr. Opin. Biotechnol. 1995; 6:103–110. [DOI] [PubMed] [Google Scholar]
- 57. Tencer A.H., Cox K.L., Di L., Bridgers J.B., Lyu J., Wang X., Sims J.K., Weaver T.M., Allen H.F., Zhang Y.et al.. Covalent modifications of histone H3K9 promote binding of CHD3. Cell Rep. 2017; 21:455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Canzio D., Liao M., Naber N., Pate E., Larson A., Wu S., Marina D.B., Garcia J.F., Madhani H.D., Cooke R.et al.. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature. 2013; 496:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hardy S., Brand M., Mittler G., Yanagisawa J., Kato S., Meisterernst M., Tora L.. TATA-binding protein-free TAF-containing complex (TFTC) and p300 are both required for efficient transcriptional activation. J. Biol. Chem. 2002; 277:32875–32882. [DOI] [PubMed] [Google Scholar]
- 60. Brand M., Moggs J.G., Oulad-Abdelghani M., Lejeune F., Dilworth F.J., Stevenin J., Almouzni G., Tora L.. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001; 20:3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. White J., Brou C., Wu J., Lutz Y., Moncollin V., Chambon P.. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 1992; 11:2229–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tanner K.G., Trievel R.C., Kuo M.-H., Howard R.M., Berger S.L., Allis C.D., Marmorstein R., Denu J.M.. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 1999; 274:18157–18160. [DOI] [PubMed] [Google Scholar]
- 63. Mittal C., Culbertson S.J., Shogren-Knaak M.A.. Distinct requirements of linker DNA and transcriptional activators in promoting SAGA-mediated nucleosome acetylation. J. Biol. Chem. 2018; 293:13736–13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Musselman C.A., Gibson M.D., Hartwick E.W., North J.A., Gatchalian J., Poirier M.G., Kutateladze T.G.. Binding of PHF1 Tudor to H3K36me3 enhances nucleosome accessibility. Nat. Commun. 2013; 4:2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li G., Widom J.. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004; 11:763–769. [DOI] [PubMed] [Google Scholar]
- 66. Bhaumik S.R., Raha T., Aiello D.P., Green M.R.. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004; 18:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li G., Levitus M., Bustamante C., Widom J.. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005; 12:46–53. [DOI] [PubMed] [Google Scholar]
- 68. Luo Y., North J.A., Rose S.D., Poirier M.G.. Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Res. 2014; 42:3017–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fischer V., Schumacher K., Tora L., Devys D.. Global role for coactivator complexes in RNA polymerase II transcription. Transcription. 2019; 10:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Donovan B.T., Luo Y., Meng Z., Poirier M.G.. The nucleosome unwrapping free energy landscape defines distinct regions of transcription factor accessibility and kinetics. Nucleic Acids Res. 2023; 51:1139–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Göös H., Kinnunen M., Salokas K., Tan Z., Liu X., Yadav L., Zhang Q., Wei G.-H., Varjosalo M.. Human transcription factor protein interaction networks. Nat. Commun. 2022; 13:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Morgan M.A.J., Shilatifard A.. Epigenetic moonlighting: catalytic-independent functions of histone modifiers in regulating transcription. Sci. Adv. 2023; 9:eadg6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ciabrelli F., Rabbani L., Cardamone F., Zenk F., Löser E., Schächtle M.A., Mazina M., Loubiere V., Iovino N.. CBP and Gcn5 drive zygotic genome activation independently of their catalytic activity. Sci. Adv. 2023; 9:eadf2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The experimental datasets are included either in this manuscript or the supplemental information, or are available from the authors upon request.