Abstract
Background
Lichen sclerosus (LS) is a chronic inflammatory disease affecting skin and mucosal tissues, particularly external genitalia, with a risk of cancer. Its etiology is unknown, possibly involving immune dysregulation and inflammation.
Methods
Study used DNA methylation (DNAme) and single-cell RNA sequencing (scRNA-seq) to compare LS with normal skin. A detailed DNAme profile of LS was created, analyzing differentially methylated probes (DMPs) and cell type-specific DMPs. EpiSCORE deconvolution and immune infiltration analyses identified altered cell types in LS. Immunohistochemistry confirmed cellular changes. Enrichment analysis identified significantly altered pathways, and cell communication analysis described interactions among altered cell types in LS.
Results
DNA methylation patterns generally distinguished LS from normal skin, with a few exceptions. Data analysis showed that T cells significantly increased and fibroblasts decreased in LS. Immunohistochemical staining confirmed the changes in T cells. Enrichment analysis of DMPs indicated significant impacts on fibroblast-related processes and key immune pathways. The COLLAGEN signal was the most prominent in the cell communication. The CD99-CD99 interaction was the strongest between T cells and fibroblasts.
Conclusion
Combining DNAme and scRNA-seq data revealed changes in cellular composition and immune pathways in LS, enhancing understanding of its pathogenesis and highlighting potential therapeutic targets and diagnostic markers.
Keywords: lichen sclerosus, DNA methylation, immune, T cells
Introduction
Lichen sclerosus (LS), first documented by Hallopeau in 1887,1 is a chronic inflammatory condition affecting the skin and mucous membranes, characterized by depigmentation and atrophic changes, with an associated risk of malignant transformation.2 Predominantly involving the external genitalia of both sexes, LS can lead to profound sexual and urinary dysfunction.3,4 Early manifestations typically include white patches or depigmentation on the genital skin, which may or may not be accompanied by erythema, fissures, scarring, or structural contraction and fusion.2 Presently, the etiology and pathogenesis of LS remain elusive, with implications pointing towards genetic predisposition, immunological dysregulation, inflammatory responses, viral infections, dysbiosis of the gut microbiota, and chronic irritation. Genetic susceptibility, immune dysfunction, and abnormalities in collagen metabolism are deemed pivotal in pathophysiology.5–8
Immune dysregulation and aberrant collagen proliferation and synthesis play pivotal roles in the progression of LS, with T cells and fibroblasts identified as the principal cellular components involved. Immunohistological findings confirm the infiltration of lymphocytes in the dermis of LS, with 30% to 50% of LS vulvar biopsies across all disease stages containing T cells harboring monoclonal rearrangements of the T-cell receptor gamma (TCRγ) gene.9–12 Within the immune response, LS exhibits upregulation of pro-inflammatory cytokines (IL-1, IL-7, IL-15, IFN-γ, TNF-α) compared to anti-inflammatory cytokines (TNF-β), indicating LS is mediated by a T-helper type 1 (Th1)-specific process.5,13,14 The pathophysiological processes of LS are jointly influenced by genetic and environmental factors, where the triggering of Th-1 mediated autoimmunity leads to a state of chronic inflammation, subsequently activating signaling pathways involved in fibroblast and collagen metabolism.15 Regulatory T cells (Tregs) act as crucial modulators of skin fibroblast activation, whereby alterations in Treg numbers induce spontaneous activation of dermal fibroblasts, upregulation of fibrogenic gene expression, and dermal fibrosis, further driving LS progression.16 In comparison to non-LS tissues, LS exhibits lower uniformity in collagen and increased structural disarray, suggesting alterations in the physical tissue properties of LS may be attributed to increased abundance and structural changes in collagen.17
Epigenetics encompasses functional modifications within the genome beyond DNA sequences, which may lead to alterations in gene expression or cellular phenotypes, thereby contributing to the onset of various diseases. DNA methylation (DNAme) represents the most prevalent epigenetic modification. In patients with LS of the penis and vulva, genome-wide hypermethylation is a frequent occurrence, with similar rates observed between genders, underscoring the critical role of epigenetic modifications in the pathogenesis of LS.18,19 Studies have revealed that aberrant expression of 5-hydroxymethylcytosine and isocitrate dehydrogenase (IDH) enzymes in vulvar squamous cell carcinoma associated with LS further influences methylation patterns.18 Alterations in DNAme may serve as early indicators of LS progression to malignancy. For instance, LS tissues exhibit significantly elevated methylation levels in genes such as ZNF582 and SST, suggesting their potential as prognostic biomarkers for vulvar LS.20 Beyond initiating disease, DNAme exerts a pivotal influence throughout disease progression by regulating gene expression. In LS patients, hypermethylation-induced inactivation of the IRF6 gene may constitute a key mechanism underlying cancer development.21 Epigenetic regulation by miR-155 also plays an integral role in the pathological progression of LS. Overexpression of miR-155 has been shown to suppress the expression and activity of tumor suppressor genes FOXO3 and CDKN1B, promoting fibroblast proliferation in vulvar LS and exacerbating disease progression through inhibition of the FOXO signaling pathway.22 These findings indicate that DNAme and other epigenetic mechanisms changes orchestrate multi-layered regulatory roles in the initiation, progression, and exacerbation of LS, providing profound insights into the complex interplay of epigenetic alterations in this disease.
At the transcriptomic level, studies utilizing bulk RNA sequencing (RNA-Seq) have further elucidated changes in gene expression within vulvar LS lesions, particularly highlighting enhanced T-cell responses and suppressed neutrophil activity, thereby providing profound insights into the immune dysregulation associated with LS.23 Research by Pilatz et al demonstrated significantly increased expression of genes such as BMP2 and its corresponding receptors, MMP1/9, TIMP1, CCL5, IL-4, TGF-β2, and their respective receptors in male children with LS and phimosis, suggesting the involvement of multiple immune and cellular signaling pathways.24 In LS-related single-cell RNA sequencing (scRNA-seq) studies, emphasis has been placed on the transcriptional changes of immune cells and fibroblasts within LS.25,26 scRNA-seq offers a novel avenue for exploring gene expression and intercellular interactions in LS-related cells; however, there remains a paucity of analyses integrating DNAme with single-cell transcriptomics to uncover the more comprehensive epigenetic mechanisms underlying LS pathogenesis.
In this study, we integrated DNAme and scRNA-seq technologies to systematically analyze the DNAme landscape, cellular composition alterations, and aberrant immune and collagen-associated signaling in LS tissues, thereby identifying novel therapeutic targets and diagnostic biomarkers. These findings not only deepen the understanding of LS pathogenesis but also provide a theoretical foundation for the development of precision therapeutic strategies.
Methods
Patients and Clinical Information
From September 2021 to December 2022, eight male patients diagnosed with LS were enrolled at Beijing Jishuitan Hospital. Table 1 summarizes their comprehensive clinical information, including disease duration and surgical methods. The inclusion criteria were as follows: clinical diagnosis consistent with the typical manifestations of LS, characterized by white skin alterations in the genital area, accompanied by symptoms such as pruritus, atrophy, bleeding, erosion, and sclerosis. Following surgical procedures, including oral mucosa repair, circumcision, or urethral meatus reconstruction, the diagnosis of LS was confirmed through histopathological analysis, which revealed characteristic morphological features such as a lymphocyte-predominant lichenoid inflammatory infiltrate in the dermis, vacuolization of the basal epithelium, and/or homogenization of the papillary dermis.2,27
Table 1.
Clinical Characteristics of LS Patients
| Sample ID | Disease Time (years) | Length (cm) | Surgery | Recurrence |
|---|---|---|---|---|
| P1 | 20 | 10 | Oral mucosal repair | N |
| P2 | 3 | 0 | Circumcision | N |
| P3 | 13 | 0 | Circumcision | N |
| P4 | 10 | 15 | Oral mucosal repair | Y |
| P5 | 7 | 5 | Oral mucosal repair | Y |
| P6 | 1 | 0.5 | Urethral meatus reconstruction | N |
| P7 | 10 | 10 | Oral mucosal repair | N |
| P8 | 5 | 14 | Oral mucosal repair | Y |
The exclusion criteria included patients who did not exhibit typical LS symptoms, such as the absence of the hallmark white skin alterations or pruritus, or lesions located outside the commonly affected genital or anal areas. Additionally, patients diagnosed with other conditions, such as lichen planus, vitiligo, psoriasis, mucous membrane pemphigoid, or genital intraepithelial neoplasia, were excluded.
LS tissue samples were collected from the foreskin of enrolled patients and preserved immediately after excision. To ensure DNA quality and sample integrity, each specimen was either snap-frozen in liquid nitrogen or stored in RNAlater (Thermo Fisher) to minimize degradation. Prior to analyses, all samples were stored at −80°C for long-term preservation.
This study was approved by the Medical Ethics Committee of Beijing Jishuitan Hospital (202109–04), and all patients provided written informed consent. The study protocol complies with the principles outlined in the Declaration of Helsinki.
DNA Methylation (DNAme) Data Acquisition
Tissue DNA was isolated using the QIAmp DNA Mini Kit (Qiagen, Hilden, Germany) and quantified with a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Total DNA > 0.5 μg was selected, and all samples were utilized for DNA methylation data collection. Following the manufacturer’s instructions, DNA was subjected to bisulfite conversion using the EZ DNA kit (Zymo Research) and subsequently hybridized onto the Infinium MethylationEPIC BeadChip (850 K, Illumina). These microarrays were scanned by Guardian Technology Co. Ltd. on an Illumina HiScan SQ scanner.
Differentially Methylated Probe (DMP) Analysis
The DNA methylation raw data were loaded utilizing the “champ.load” function and subsequently transformed into β-values. The latter denote the proportions of fluorescence intensities between methylated and unmethylated probes, spanning from 0 to 1. The β-value ascends in concordance with the level of methylation. This function was also employed for the initial probe scrutiny and QC (champ.QC). Probes meeting any of the ensuing criteria were excised: (1) p > 0.01; (2) < 3 beads in ≥ 5% of all samples; (3) non-CpG; (4) multihits; (5) underlying SNPs; and (6) situated on the X or Y chromosome. The winnowed probe set was then deployed in the ensuing analyses. The Beta Mixture Quantile Expansion (BMIQ) normalization was implemented with the intent to eradicate technical variances and biases within DNA methylation array data. A Principal Component Analysis (PCA) was executed utilizing the “prcomp” function within the ‘stats’ package of R. PCA served to truncate the dimensionality of the high-dimensional methylation site matrix into a configuration comprised of merely a handful of principal components. This limited array of principal components can adeptly encapsulate the variance across all methylation sites, thus elucidating the disparities between samples.
The DMPs is screened by the method of empirical Bayesian estimation, and the P-value is usually calculated by the empirical Bayesian method. The Δβ-value denotes the arithmetic disparity between the mean β-value of the LS group and that of the normal control for an individual probe. Correction of P-values (P_adj) for multiple comparisons was enacted via the Benjamini-Hochberg procedure. The DMP thresholds were established as P_adj < 0.05 and log|FC| > 0.2. Hyper DMPs (GSE151600: Hyper_DMPsLSvsG1; GSE229848: Hyper_DMPsLSvsG2) exhibited logFC > 0.2, whereas hypo DMPs (GSE151600: Hypo_DMPsLSvsG1; GSE229848: Hypo_DMPsLSvsG2) were characterized by logFC < 0.2. Annotation of DMP genes was undertaken employing the default annotation of the “ChAMP” package in R. The reference genome utilized was hg19.
Single Cell Transcriptome Cell Type Identification and Cell Communication Analysis
We imported the single-cell transcriptomic data from the four normal skin samples in GSE173205 into the R package Seurat (version 4.2.0).28 Within Seurat, we excluded specific cells predicated on their feature count, mitochondrial transcript percentage, and RNA counts. Concretely, we eliminated cells exhibiting fewer than 200 or more than 5000 expressed genes, cells with mitochondrial transcript percentages surpassing 25%, and cells with RNA counts below 1000.
Following data pre-processing, we applied the “SCTransform” methodology to normalize and discern the most variable features across each sample. “SCTransform” circumvents certain limitations inherent in conventional normalization protocols, such as the incorporation of pseudo-counts and logarithmic transformations. To effectively attenuate batch effects across multiple samples, we adopted the anchor-based integration strategy integrated within Seurat 3. Specifically, we utilized the “FindIntegrationAnchors” and “IntegrateData” functions to amalgamate gene expression matrices from five distinct samples. For linear transformation, the “ScaleData” function was enlisted to standardize the data.
The top 3000 genes displaying substantial variability were extracted for PCA, and the 30 most pertinent principal components were employed for cluster analysis. The “FindClusters” function (resolution = 1) was deployed to delineate the clusters, subsequently visualized using the Uniform Manifold Approximation and Projection (UMAP) technique. Cell type characterization was predicated on gene functionality and the expression of established markers cataloged within the CellMarker database.29
Intercellular communication was assessed employing the R package “CellChat”, by scrutinizing the manifestation of ligand-receptor pairs. To delve deeper, the “Secreted Signaling”, “ECM-Receptor”, and “Cell-Cell Contact” segments of the database were specifically chosen, while cell types consisting of less than 100 cells were effectively excluded. Furthermore, interactions amidst disparate cell types were also examined.
Deconvolution Analysis of Bulk DNAme and Cell Type-Specific DMPs Analysis Based on scRNA-seq
Utilizing the EpiSCORE R package based on single-cell RNA-seq tissue atlas, we aim to generate tissue-specific DNAme reference matrices, thereby enabling the quantification of cell type proportions and cell type-specific differential methylation signals within extensive tissue datasets.30 Employing 15,877 cells from the scRNA-seq of four normal skin samples sourced from the GSE173205 dataset as our input, we construct a gene expression reference matrix with maximal marker specificity, encompassing 526 marker genes. Subsequently, EpiSCORE compares the expression-based reference gene matrix with two available datasets containing paired gene expression and DNAme data (SCM2 and RMAP), culminating in the construction of a human skin reference DNAme matrix comprising 173 genes. This input matrix is then utilized to estimate the cellular composition within our bulk DNAme dataset. CellDMC was then used to identify cell type-specific DMPs (P_adj < 0.05).31
Immune Infiltration Analysis
For an intricate deconvolution of the cellular composition inherent in bulk LS samples, we employed the reference-based analysis MethylCIBERSORT. MethylCIBERSORT relies upon DNA methylome-based reference data to deduce distinct cellular constituents, encompassing cancer cells, CD14-positive, CD19-positive, CD56-positive and CD8-positive cells, T regulatory cells, CD4-positive effector cells, eosinophils, fibroblasts, and neutrophils.32,33 Methylation patterns within the sample of interest are juxtaposed with deposited cell-type-specific and defining DNA methylomes. The execution of MethylCIBERSORT analysis adhered to the respective protocols. Succinctly, EPIC array IDAT sets were imported into the R package “ChAMP” to conduct quality assessments, normalization, and acquisition of beta values. Utilizing the “MethylCIBERSORT” R package, a composite file was constructed, comprising beta values for comparative analysis against a reference matrix. This reference repository encompassed signature methylation beta values derived from well-characterized cell types. Following the generation of the composite file, both mixture and reference files were uploaded onto the CIBERSORT portal and subjected to deconvolution (https://cibersortx.stanford.edu/).
Enrichment Analysis
The intersecting sites of Hyper_DMPsLSvsG1 and Hyper_DMPsLSvsG2 are documented in Supplementary Table 1, while the intersecting sites of Hypo_DMPsLSvsG1 and Hypo_DMPsLSvsG2 are delineated in Supplementary Table 2. These probes are respectively annotated to 524 and 1579 genes (Supplementary Table 3). The immune-related gene dataset that was confirmed in earlier studies to be implicated in the natural immune process was downloaded from the IMMPORT database (https://www.immport.org/shared/). Following the annotation, the intersection of annotated genes with immune-related genes yields hypermethylated immune-related genes and hypomethylated immune-related genes (Supplementary Table 4). Subsequently, the hypermethylated and hypomethylated immune-related genes were subjected to functional analysis using ClueGO v. 2.5.9 in Cytoscape v. 3.9.1. “GO-ImmuneSystemProcess” ontologies/pathways were selected for gene ontology (GO) enrichment, forming a network where p < 0.05, with all other parameters set to default.
We performed GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses utilizing the “clusterProfiler” package in R.34 Enrichment analyses for both GO (P_adj < 0.05) and KEGG (P value < 0.05) were conducted on cell-specific DMPs of fibroblasts and T cells.
Statistical Analysis
The screening of DMPs employs empirical Bayesian estimation method, with P-values typically computed using empirical Bayesian approach. The P-values are corrected for multiple comparisons using the Benjamini-Hochberg procedure (P_adj). The comparison of cellular compositions between the LS group and the control group was conducted using the Wilcoxon test. Gene enrichment analysis employed the Hypergeometric test as the hypothesis test, with p-values subjected to multiple testing correction using the Benjamini-Hochberg method (also known as FDR correction). Within the Seurat R package, bilateral Wilcoxon rank-sum tests were implemented to compute marker genes for each cluster, assessing differential gene expression. Bonferroni correction was employed to adjust p-values based on the total number of features in the dataset.
Immunohistochemistry Staining
All samples were embedded in paraffin blocks. The tissue sections were subjected to routine embedding, processing and staining with hematoxylin and eosin for histological diagnosis. Pathology slides were reviewed to identify areas of interest which appeared pathognomonic for LS (Figure 1). Before further processing immunostaining, the diagnosis of specimens was confirmed again by a senior pathologist. Immunohistochemistry staining was performed for markers of inflammation (CD4 and CD8). Immunohistochemical staining was performed with an automated immunostainer (Autostainer 720, Labvision, San Diego, CA) according to standard heat-induced epitope retrieval and the avidin-biotin-peroxidase complex method. Simultaneously, appropriate positive and negative control sections were used. Positive immunostaining was characterized by brown cytoplasmic staining under a microscope.
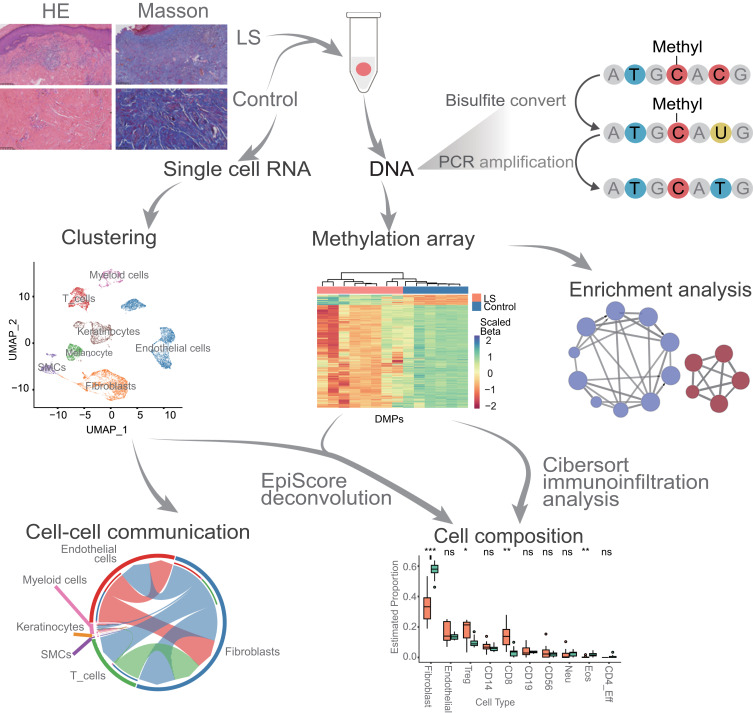
Figure 1.
Flow chart of all analysis. We conducted integrated analysis by combining DNA methylation data from eight LS patients with publicly available datasets of normal skin DNA methylation (G1_Control16 and G2_Control6) and single-cell transcriptome data. Following differential methylation analysis between LS and G1_Control16 and G2_Control6 respectively, we performed EpiScore-based cell type deconvolution analysis, immune infiltration analysis, and enrichment analysis to characterize changes in cell composition and alterations in disease-associated immune pathways (ns, P value ≥ 0.05; *, P value < 0.05; **, P value < 0.01; ***, P value < 0.001). Additionally, at the single-cell transcriptome level, we conducted cell communication analysis on cell types showing significant differences in LS to further understand the pathophysiological mechanisms of LS.
Data Accessibility
Eight DNAme datasets pertaining to LS have been deposited in the Genome Sequence Archive of the Beijing Institute of Genomics, Chinese Academy of Sciences, under the accession number PRJCA025881.35 These datasets are readily accessible to the public via https://ngdc.cncb.ac.cn/omix/release/OMIX006366. Moreover, publicly accessible datasets, namely GSE151600 (G1_Control16) and GSE229848 (G2_Control6), which have produced DNAme data for eight and six healthy skin tissues respectively, can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151600 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE229848. We also utilized the public dataset GSE173205 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173205), which comprises single-cell data from 10X Genomics for four healthy control skin tissues.
Result
DNA Methylation Profiling for Classical LS in Comparison with Normal Skin
We performed DNAme analysis from eight cases of classical LS skin samples (Methods) ”. The mean age was 49.4 ± 22.7 years, the disease duration averaged 8.6 ± 6.1 years, and the stenotic segment length measured 6.8 ± 6.3 cm (Table 1). Compared to healthy skin, histological examination via Hematoxylin and Eosin (HE) staining in cases of LS unveils escalated fibrosis and fibroplasia, concomitant with pronounced infiltration of inflammatory cellular constituents (Figure 1). Masson’s trichrome staining, when applied to LS specimens, demonstrating collagenous fiber hyperplasia, augmented density, or disarrayed disposition (Figure 1). Meanwhile, two public DNA methylation datasets from normal skin were retrieved from the NCBI GSA database (Methods) taken as controls (G1_Control16 and G2_Control6) for comparison with our LS dataset. Considering that batch effect correction when combining datasets from three studies may be too extensive to reduce the biological signals, we performed the comparisons with the two controls respectively.
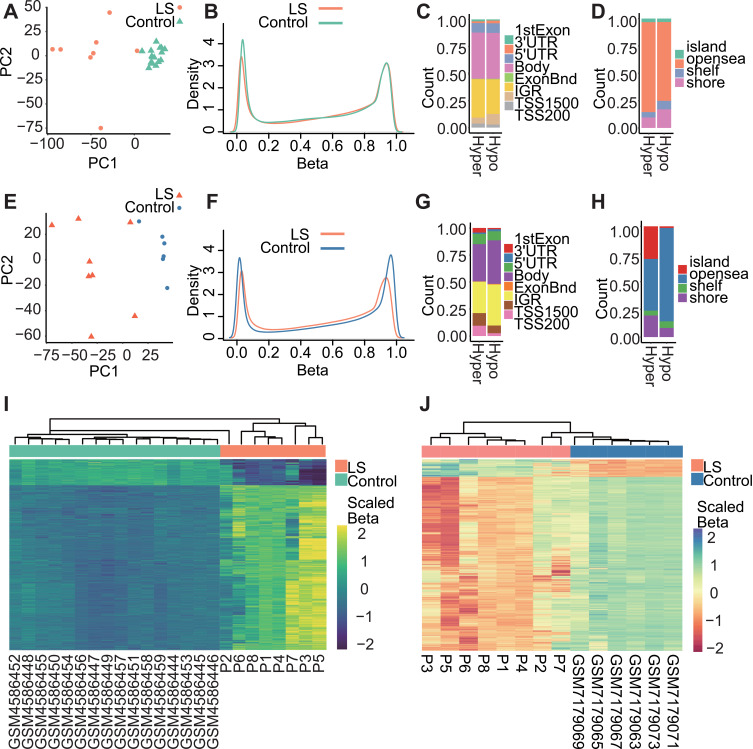
Following quality control procedures, PCA analysis for LS and G1_Control16 were based on 743,785 CpG probes to show overall differences between these two groups (PCA, Figure 2A). PC1 and PC2 explained 65% variations in samples which were clustered according to the disease status. Most of all, LS exhibited decreased loci with lower methylation levels (Hypo_DMPsLSvsG1, Figure 2B). Therefore, most of DMPsLSvsG1 were hyper-methylated 29,605 out of 34,291, (Hyper_DMPsLSvsG1, 86.33%), and 4686 hypo-methylation (Hypo_DMPsLSvsG1, 13.67%). In terms of genomic annotation (Figure 2C and D), hyper- and hypo-methylation loci exhibited comparable distributions across gene regions. Specifically, the highest frequency of DMPs was observed within the gene body region (Figure 2C). Regarding CpG islands, the greatest abundance of differential methylation sites was found within the “opensea” category (Figure 2D). Overall, the heatmap demonstrates that 34,291 DMPsLSvsG1 exhibit significant differences in their methylation levels (P_adj < 0.05, Empirical Bayesian, Methods), allowing for a relatively effective differentiation between LS and controls.
Figure 2.
Differential methylation profiles between LS and control group. (A) PCA of DNAme in 24 samples revealed overall differences between LS and G1_Control16 samples. (B) β-value density distribution between LS and G1_Control16 samples. (C) Hyper_DMPsLSvsG1 and Hypo_DMPsLSvsG1 distributions in genomic features. (D) Hyper_DMPsLSvsG1 and Hypo_DMPsLSvsG1 distributions in CpG islands. (E) PCA of DNAme in 14 samples revealed overall differences between LS and G2_Control6 samples. (F) β-value density distribution between LS and G2_Control6 samples. (G) Hyper_DMPsLSvsG2 and Hypo_DMPsLSvsG2 distributions in genomic features. (H) Hyper_DMPsLSvsG2 and Hypo_DMPsLSvsG2 distributions in CpG islands. (I) Hyper_DMPsLSvsG1 and Hypo_DMPsLSvsG1 methylation levels in LS and G1_Control16 samples. Scaled colour bar = β-value. (J) Hyper_DMPsLSvsG2 and Hypo_DMPsLSvsG2 methylation levels in LS and G2_Control6 control samples. Scaled colour bar = β-value.
In the other comparison (G2_Control6), methylation levels of 720,903 filtered CpG probes discerned LS samples from normal controls (PCA, Figure 2E). The methylation density curves in both comparisons highlighted the common tendency in decreasing of hypomethylation probes in LS samples, while the latter comparison also revealed a more significant reduction of hypermethylation probes (Figure 2F). Hence, most DMPsLSvsG2 were hypomethylated, accounting for over 90.82% DMPsLSvsG2 (16,879 out of 18,585 DMPs). In addition, 9.18% were Hyper_DMPsLSvsG2 (1706). Their genomic distribution was consistent with the previous comparison, which also demonstrated a higher frequency of DMPs within the “gene body” region and the predominant “opensea” category in CpG islands (Figure 2G and H). Notably, sample P2 collected from a patient were shown to be similar to normal samples in both comparison (Figure 2I and J).
In both comparative analyses, despite demonstrating consistency between results, the first comparison (G1_Control16) reveals a notable resemblance among normal samples in the PCA plot, clustering closely together, while the diseased samples exhibit a comparatively scattered distribution possibly due to their heterogeneity (Figure 2A and E). However, in the second comparative analysis (G2_Control6), while normal samples also exhibit substantial divergence, the overall methylation density curve illustrates a significantly greater contrast compared to normal samples (Figure 2F). In subsequent analyses, we prioritize the first comparative analysis as the primary basis, utilizing the second analysis as supportive evidence for our conclusions.
Cell Type Deconvolution Revealed the Increase of T Cells and Reduction of Fibroblasts in LS
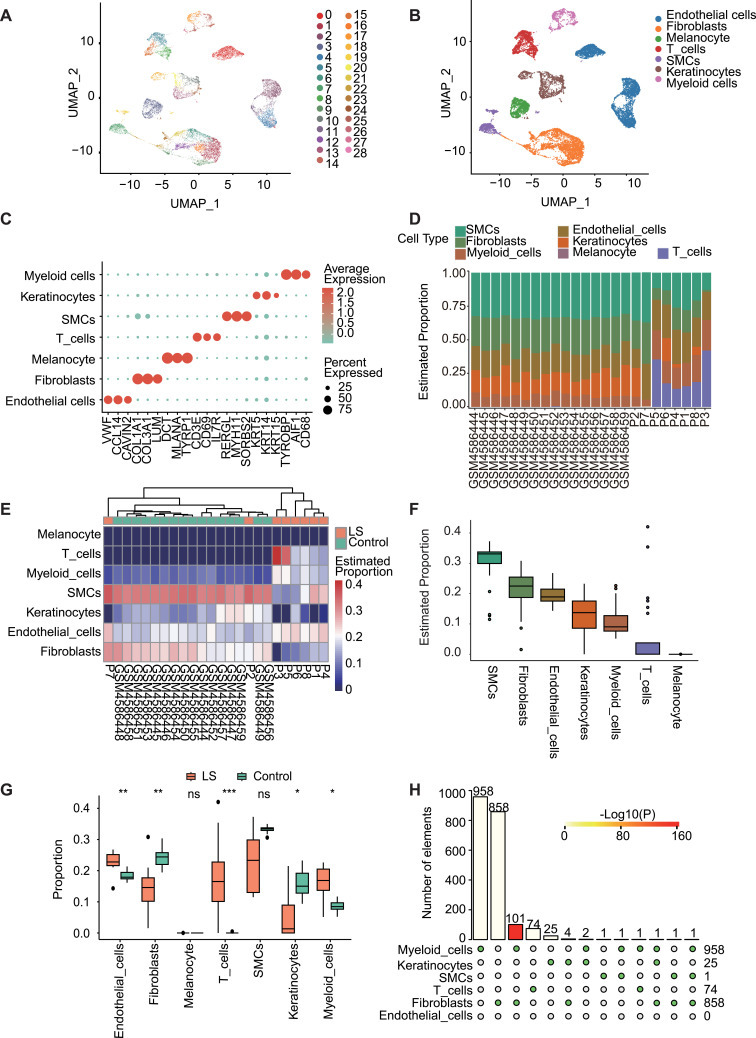
In order to investigate changes in cellular composition within the lesions, we conducted deconvolution analysis on DNA methylation data using single-cell transcriptome data. A total of 15,877 qualified cells were identified from four normal skin scRNA-seq datasets, which formed 29 distinct cell clusters (Figure 3A). They were further categorized into seven cell types based on known marker genes (Figure 3B and C): endothelial cells (n=4634, VWF, CCL14, CAVIN2); fibroblasts (n=4364, COL1A1, COL3A1, LUM); melanocytes (n=1292, DCT, MLANA, TYRP1); T cells (n=1718, CD3E, CD69, IL7R); smooth muscle cells (SMCs, n=979, RERGL, MYH11, SORBS2); keratinocytes (n=1877, KRT5, KRT14, KRT15); myeloid cells (n=1013, TYROBP, AIF1, CD68).
Figure 3.
Analysis of skin scRNA-seq data to obtain cell proportions and cell-specific DMPs between LS and G1. (A) Uniform manifold approximation and projection (UMAP) plot of 15,877 cells, color-coded by associated cluster. Each point depicts a single cell. (B) All cells were identified as seven different cell types, and (C) corresponding marker genes were determined for each cell type: endothelial cells (VWF, CCL14, CAVIN2); fibroblasts (COL1A1, COL3A1, LUM); melanocytes (DCT, MLANA, TYRP1); T cells (CD3E, CD69, IL7R); smooth muscle cells (RERGL, MYH11, SORBS2); keratinocytes (KRT5, KRT14, KRT15); myeloid cells (TYROBP, AIF1, CD68). Scaled color bar = average expression, Size of the point = percent expressed. Proportions of the seven cell types in the sample are shown on the right. (D) Bar chart of the proportion distribution of the seven cells types in each sample of LS and G1. (E) Heat map of the proportion distribution of seven cell types in each sample of LS and G1. Colour bar = estimated proportion. (F) In all samples of LS and G1, the proportion distribution of seven kinds of cells. (G) The box plot displays the proportions of different cell types between the LS and G1 sample groups. Significant changes between the two groups were assessed using the Wilcoxon test (ns, P value ≥ 0.05; *, P value < 0.05; **, P value < 0.01; ***, P value < 0.001). (H) Distribution of specific DMPs in LS and G1 cells.
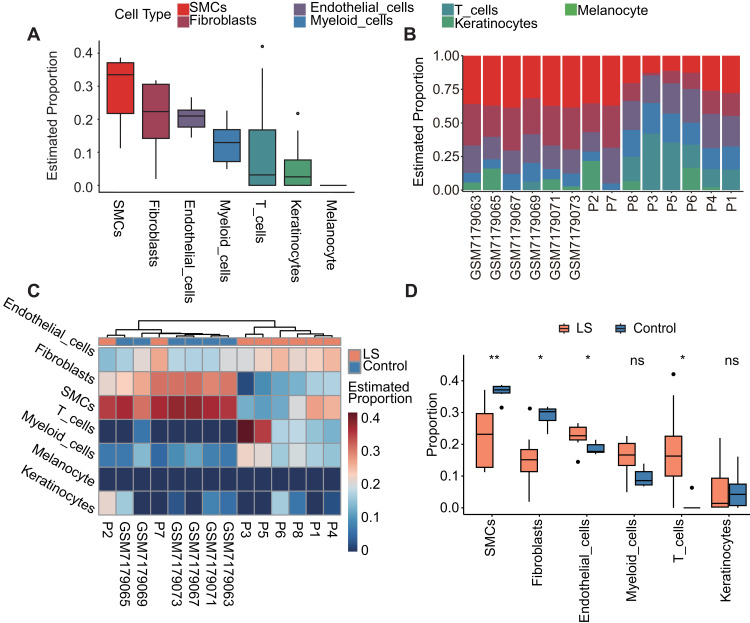
The order of top three cell components was the same in LS vs G1 (Figure 3D and F) and LS vs G2 (Figure 4A and B) analysis: SMCs, fibroblasts, endothelial cells. The rank 4–6 components were the also the same with a slightly different order: LS vs G1, keratinocytes, myeloid cells, and T cells; LS vs G2, myeloid cells, T cells, and keratinocytes. Consistent with the deconvolution results only based on cell-specific DNA makers, the analysis combined with scRNA-seq date also identified the significant decrease of fibroblasts in LS (LS vs G1, p < 0.01, Wilcoxon test, Figure 3G; LS vs G2, p < 0.05, Figure 4D) and the significant reduction of T cells (LS vs G1, p < 0.001, Figure 3G; LS vs G2, p < 0.05, Figure 4D). In addition, in the clustering results between LS samples and normal samples in G1 and G2 datasets, samples P2 and P7 showed similarity to the cellular composition of normal tissues (LS vs G1, Figure 3E; LS vs G2, Figure 4C).
Figure 4.
Cell proportion of LS vs G2: deconvolution analysis of DNA methylation data using scRNA-seq. (A) In all samples of LS and G2, the proportion distribution of six kinds of cells. (B) Bar chart of the proportion distribution of the six cells types in each sample of LS and G2. (C) Heat map of the proportion distribution of six cell types in each sample of LS and G2. Colour bar = estimated proportion. (D) The box plot displays the proportions of different cell types between the LS and G2 sample groups. Significant changes between the two groups were assessed using the Wilcoxon test (ns, P value ≥ 0.05; *, P value < 0.05; **, P value < 0.01).
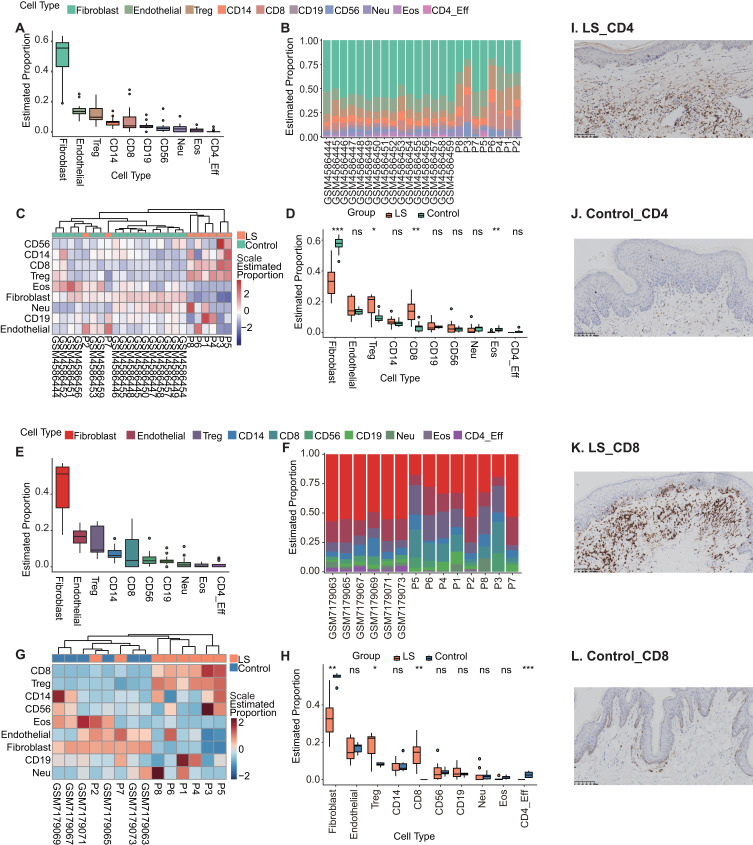
Further in-depth analysis of the alterations in additional immune cell populations, we performed the immune infiltration analysis based on methylation profiles revealed the proportions of 10 immune cell types across the samples (Methods). Consistent to previous analysis, fibroblasts dominated the cellular composition in both LS and normal skins (LS vs G1, Figure 5A and B; LS vs G2, Figure 5E and F). Cluster analysis of the heatmap depicting the proportions of different cell types in each sample demonstrates that the majority of LS samples cluster together, whereas the proportions of various cell types in P2, P7, and normal skin samples exhibit greater similarity (LS vs G1, Figure 5C; LS vs G2, Figure 5G). In comparison with both normal controls, LS samples showed significant reduction in fibroblasts (p < 0.001, Wilcoxon test, Methods; LS vs G1, Figure 5D and LS vs G2, Figure 5H), and significant increase in Treg cells (p < 0.05), CD8 T cells (p < 0.01). Besides, LS vs G1 comparison also showed increase of eosinophils in disease tissues (p < 0.01, Figure 5D), while LS vs G2 comparison demonstrated the reduction of CD4_eff (p < 0.001, Figure 5H).
Figure 5.
Immune infiltration analysis revealed the proportions of ten immune cell types across the samples. (A) In all samples of LS and G1, the proportion distribution of ten immune cell types. (B) Bar chart of the proportion distribution of the ten immune cell types in each sample of LS and G1. (C) Heat map of the proportion distribution of ten immune cell types in each sample of LS and G1. Colour bar = Scale estimated proportion. (D) The box plot displays the proportions of different immune cell types between the LS and G1 sample groups. Significant changes between the two groups were assessed using the Wilcoxon test (ns, P value ≥ 0.05; *, P value < 0.05; **, P value < 0.01; ***, P value < 0.001). (E) In all samples of LS and G2, the proportion distribution of ten immune cell types. (F) Bar chart of the proportion distribution of the ten immune cell types in each sample of LS and G2. (G) Heat map of the proportion distribution of ten immune cell types in each sample of LS and G2. Colour bar = Scale estimated proportion. (H) The box plot displays the proportions of different immune cell types between the LS and G2 sample groups. Significant changes between the two groups were assessed using the Wilcoxon test (ns, P value ≥ 0.05; *, P value < 0.05; **, P value < 0.01; ***, P value < 0.001). (I) Immunohistochemical staining of T cell CD4-positive in LS samples. (J) Immunohistochemical staining of T cell CD4-positive in controls. (K) Immunohistochemical staining of T cell CD8-positive in LS samples. (L) Immunohistochemical staining of T cell CD8-positive in controls.
We then evaluated the level of expression of inflammation markers including CD4 and CD8 (Figure 5I and L). The proportion of T cells staining positive for CD4 and CD8 were significantly higher in LS samples (Figures 5I and 6K) than those without LS (Figures 5J and 6L).
Figure 6.
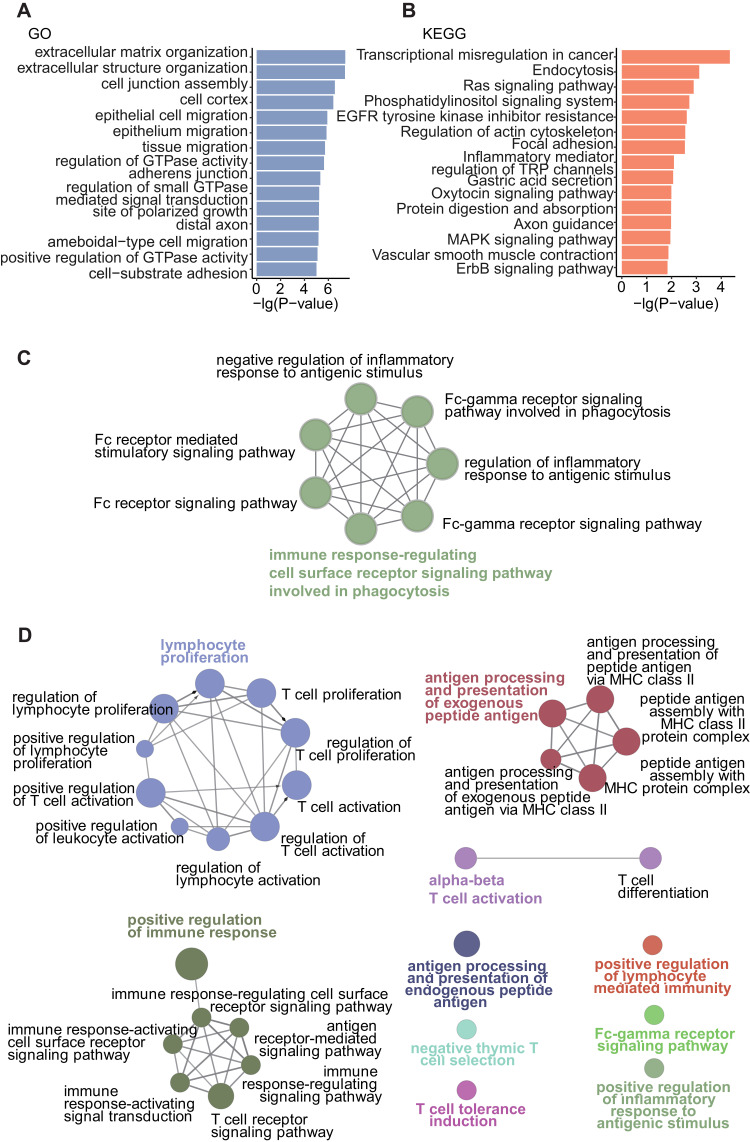
The affected biological processes in fibroblasts and key immune-related pathways in LS. (A) Fifteen most significant GO enriched pathways of fibroblast DMP genes. (B) Fifteen most significant KEGG enriched pathways of fibroblast DMP genes. (C) The network of GO enriched pathways of hypermethylated immune-related genes. (D) The network of GO enriched pathways of hypomethylated immune-related genes.
Affected Biological Processes in LS Fibroblast and Crucial Immune-Related Pathways
We then identified DMPs in each cell types in LS and G1 samples being combined with scRNA-seq data. In all, 958 DMPs were identified in myeloid cells, 858 in fibroblasts, 74 in T cells, 25 in keratinocytes, 1 in smooth muscle cells, and none in endothelial cells (P_adj < 0.05, Hypergeometric test, Figure 3H, Methods). Fibroblast DMPs affected 479 genes, which were significantly enriched in “extracellular matrix organization” (P_adj = 8.22E-05, Hypergeometric test, Methods), “extracellular structure organization” (P_adj = 8.22E-05), “cell junction assembly” (P_adj = 0.000386), “epithelial cell migration” (P_adj = 0.001148), “epithelium migration” (P_adj = 0.001148), and “Inflammatory mediator regulation of TRP channels” (P value = 0.008023) (Figure 6A and B). Whereas, other cell type specific DMPs were not adequate for enrichment analysis.
Furthermore, common DMPs in LS vs G1 and LS vs G2 comparisons resolved 903 hypermethylated probes (Supplementary Table 1), and 3473 hypomethylated probes (Supplementary Table 2). They were annotated to 524 and 1579 genes, respectively (Supplementary Table 3). Particularly, certain methylated sites associated with cutaneous Th1 immune-related genes, such as IFNG, CD28, and TLA.36–38 Among immune-related genes (Supplementary Table 4), there were 41 hypermethylated ones, which exhibit significance in pathways such as “immune response-regulating cell surface receptor signaling pathway involved in phagocytosis” (P_adj < 0.001) and “Fc receptor signaling pathway” (P_adj < 0.001) following immune-related pathway enrichment analysis (Figure 6C and Supplementary Table 5). Whereas, 132 hypomethylated immune-related genes were significantly enriched in “lymphocyte proliferation” (P_adj < 0.001), “positive regulation of immune response” (P_adj<0.001), “antigen processing and presentation of exogenous peptide antigen” (P_adj < 0.001), and “alpha-beta T cell activation” (P_adj = 0.01) following immune-related pathway enrichment analysis (Figure 6D and Supplementary Table 6).
Intercellular Communication and Interactions Between Fibroblasts and T Cells
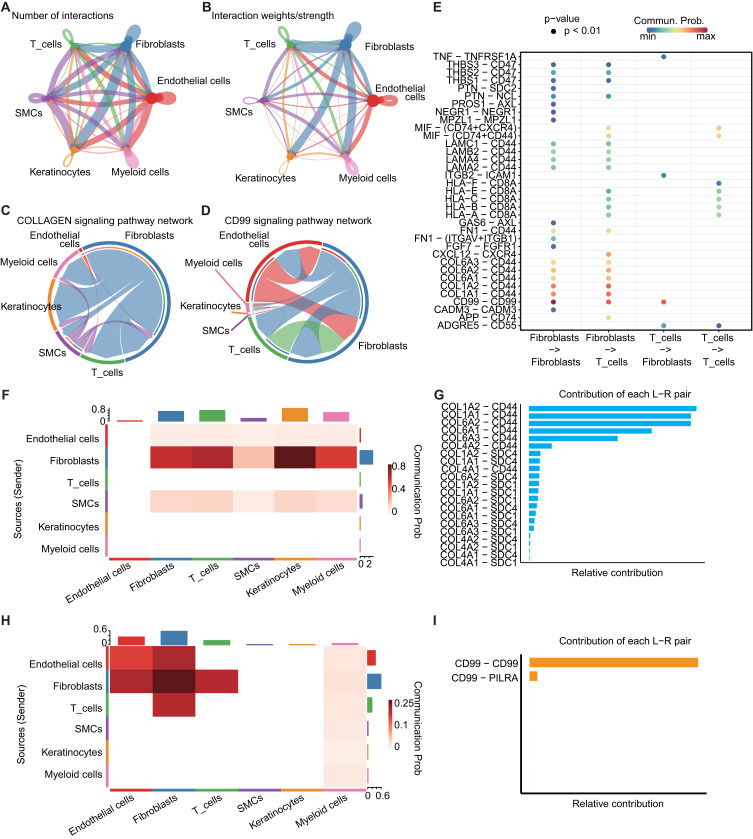
Cellular interaction analysis revealed a total of 53 cell communication signals. We delineated both the quantity and intensity of intercellular communication in normal skin (Figure 7A and B). Notably, the significance of the COLLAGEN signal was the highest (p < 0.001, Figure 7C). Collagen proteins are abundantly distributed in the skin, playing a pivotal role. Ligand signals typically originate from collagen-coding genes, with receptor genes including CD44, SDC1, and SDC4. Fibroblasts predominantly serve as the main senders, while other cells receive the signals (Figure 7F and G). In the interaction between fibroblasts and T cells, the CD99-CD99 signal exhibited high intensity (Figure 7E), with CD99 serving as the ligand gene and CD99 and PILRA as the receptor genes (Figure 7H and I).
Figure 7.
Intercellular communication and interactions between fibroblasts and T Cells. (A) Number of interactions network among cells. The thickness of the lines represents the number of interactions. (B) Interaction weight network among cells. The thickness of the lines represents the interaction weight. (C) Chord plot showing the inferred intercellular communication network of COLLAGEN signaling. (D) Chord plot showing the inferred intercellular communication network of CD99 signaling. (E) Probability of communication between fibroblast and T cell ligand-receptor pairs in cell communication results. p-value < 0.01, Scaled color bar = Communication Probability. (F) Heatmap of communication probability in COLLAGEN signaling, scaled color bar = Communication Probability. (G) Ligand-receptor pairs included in the COLLAGEN signaling and their relative contribution. (H) Heatmap of communication probability in CD99 signaling, scaled color bar = Communication Probability. (I) Ligand-receptor pairs included in the CD99 signaling and their relative contribution.
Discussion
Here, we conducted an analysis of the DNA methylation chip data from LS patients, comparing it with samples from two publicly available datasets of normal skin specimens. We integrated this analysis with skin scRNA-seq. We observed that certain individuals within the LS samples, notably P2 and P7, exhibited methylation profiles more akin to those of normal skin (Figure 2I and J). Furthermore, our immune infiltration analysis and scRNA-based deconvolution results revealed a cellular composition more closely resembling that of normal skin (Figure 3C and G). Clinical data indicated that neither of the two patients experienced narrowing recurrence postoperatively, despite variations in other clinical information, suggesting a milder condition (Table 1). As the molecular classification of diseases based on DNA methylation matures, our study hints at the potential value of subtyping LS at the molecular level of DNA methylation.39 This, in turn, unveils the prospect of certain gene methylation abnormalities serving as markers for early or late events in LS or cancer.19,40
In LS, there is a discernible alteration in the composition of T cells and fibroblasts,5,11,17,22,41 a phenomenon our research corroborates. Immunoinfiltration analysis and scRNA-based deconvolution analysis reveal fluctuations in the cellular composition of LS, characterized primarily by a significant increase in T-cell proportions, alongside a marked decrease in fibroblasts (Figure 3D and H). Recent scRNA-seq studies on LS corroborate our findings, indicating an expansion of T-cells and a reduction in fibroblasts in LS.25,26 In the immunohistochemical staining results of CD4 and CD8, a significant increase of T cells in the LS was observed, further supporting our data analysis findings. Within our investigation, we unearthed noteworthy hypomethylated sites corresponding to genes associated with skin Th1 immune factors, such as IFNG, CD28, and ITLA.36–38 Enrichment analysis further unveiled immune pathways intertwined with T cell function (Figure 6C and D).
The reduced proportion of fibroblasts in LS does not appear to conflict with the increased or aberrant collagen deposition. Despite the decrease in fibroblast numbers, it is possible that the remaining fibroblasts are in a highly active state, with collagen accumulation resulting from impaired collagen degradation.42,43 Simultaneously, alterations in the immune microenvironment may modify the organization of collagen fibers, leading to collagen disarray.44 Among the fibroblast-specific differentially methylated sites, we identified significant enrichment in collagen-related pathways (Figure 6A), suggesting that these DNAme changes may regulate fibroblast activity, maintaining their active state. At the scRNA-seq level, the predominant signals observed in the skin are those associated with collagen. Ligand genes typically encode collagen, while receptor genes include CD44, SDC1, and SDC4 (Figure 7C). This underscores the pivotal role of collagen-related pathways in preserving the integrity of skin homeostasis.
Notably, among the fibroblast-specific differentially methylated sites, we also observed significant enrichment in pathways related to “inflammatory mediators’ regulation of TRP channels” (Figure 6B). This suggests the potential for interaction between fibroblasts and immune cells, particularly the markedly increased T-cells, a cellular communication relationship that has not been prominently emphasized in previous scRNA-seq studies of LS.25,26 In our study, the scRNA-seq-based cellular communication analysis revealed that the CD99 signaling between T-cells and fibroblasts (CD99-CD99/PILRA) was the most pronounced (Figure 7E). CD99 is a glycoprotein primarily expressed in immune cells, playing a crucial role in immune cell adhesion, migration, and development under physiological conditions. Previous studies have also highlighted the significant function of CD99 in the skin. CD99 contributes to maintaining homeostasis and initiating early differentiation of the skin.45 The overexpression of CD99 has been identified as a marker for virus-positive Merkel cell carcinoma.46 In the skin of systemic sclerosis, CD99 is known to be overexpressed in dermal fibroblasts.47 CD99 can mediate monocyte transendothelial migration and lymphocyte recruitment into inflamed skin in mice.48 Additionally, studies have indicated that anti-CD99 antibodies can inhibit the recruitment of T cells to inflamed skin and the formation of edema.49 These findings underscore the pivotal role of CD99 signaling in the inflammatory processes of the skin, offering compelling evidence for its potential as a therapeutic target to intervene in the inflammation-related mechanisms of LS.
Limitations
While this study offers novel insights into disease mechanisms, it has some limitations. The small sample size may limit the statistical power and generalizability of the findings. Expanding the cohort to include individuals at different disease stages would enhance reliability, though comparisons with healthy controls partially address this issue. Incomplete clinical data, such as treatment history and lifestyle factors, may also impact the interpretation of DNA methylation profiles and cellular composition. A more comprehensive dataset could better link molecular changes with clinical outcomes. Despite correction efforts, technical variability between DNA methylation and single-cell RNA-seq platforms remains a potential source of bias, highlighting the need for validation using larger, multi-center datasets. Future studies incorporating advanced technologies like spatial multi-omics and real-time in vivo imaging could reveal intercellular interactions and tissue dynamics with greater precision. These approaches would provide deeper insights into disease mechanisms. Despite these challenges, the study improves the understanding of the disease, and addressing these limitations through larger cohorts and multi-omics integration will further advance this field.
Conclusion
This study integrates DNAme data with scRNA-seq to conduct an in-depth, multi-dimensional analysis of the pathological characteristics of LS. We observed that certain patients exhibited DNAme profiles akin to those of normal skin, suggesting that DNAme may serve as a potential biomarker for early disease diagnosis. In terms of DNAme-based deconvolution of cellular composition, LS patients demonstrated a marked increase in T-cell proportions, coupled with a notable reduction in fibroblast presence. The DNAme landscape of LS revealed significant associations with immune-related and collagen-related pathways. Moreover, cellular communication analysis revealed a pronounced enhancement of CD99 signaling between T-cells and fibroblasts. As a critical marker in immune cells, CD99 may play a pivotal role in both immune regulation and the pathological changes observed in LS. Consequently, targeting CD99 signaling could offer novel therapeutic strategies for the treatment of LS.
Funding Statement
This study was supported by grants from the Beijing JST Research Funding (ZR-202120), Beijing Municipal Natural Science Foundation (No. 7222073) and the Higher Education Discipline Innovation Project (111 Project No. B13003). These funders were not involved in any aspect of the present study.
Abbreviations
DNAme, DNA methylation; DMPs, differentially methylated probes; GO, gene ontology;; KEGG, Kyoto Encyclopedia of Genes and Genomes; LS, Lichen sclerosus; PCA, Principal Component Analysis; Tregs, Regulatory T cells; scRNA-seq, single-cell RNA sequencing; SMCs, smooth muscle cells.
Data Sharing Statement
The datasets supporting the conclusions of this article are available in the Genome Sequence Archive. Each sample’s files are hyperlinked to the dataset on https://ngdc.cncb.ac.cn/omix/release/OMIX006366.
Ethics Approval and Consent to Participate
This study was conducted in accordance with the Declaration of Helsinki. All enrolled patients provided written informed consent, and the research protocol was approved by the Medical Ethics Committee of Beijing Jishuitan Hospital (202109-04).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Hallopeau H. Du lichen plan et particulierement desa forme atrophique. Ann derm Syph. 1887;8:790. [Google Scholar]
- 2.Kirtschig G, Kinberger M, Kreuter A, et al. EuroGuiderm guideline on lichen sclerosus-introduction into lichen sclerosus. J Eur Acad Dermatol Venereol. 2024;38(10):1850–1873. doi: 10.1111/jdv.20082 [DOI] [PubMed] [Google Scholar]
- 3.Hu N, Zou Y, Deng X, Zhang L, Zhai Z, Yin R. Photodynamic therapy for male genital lichen sclerosus with urethral stricture-Case report. Photodiag Photodyn Ther. 2024;45:103947. doi: 10.1016/j.pdpdt.2023.103947 [DOI] [PubMed] [Google Scholar]
- 4.Ngo M, Chang H, Carr C, Barnard A, Reisch J, Mauskar MM. Patient questionnaire responses predict severe clinical signs in patients with vulvar lichen sclerosus: a retrospective cohort study. J Am Acad Dermatol. 2024;91(3):516–518. doi: 10.1016/j.jaad.2024.04.046 [DOI] [PubMed] [Google Scholar]
- 5.De Luca DA, Papara C, Vorobyev A, et al. Lichen sclerosus: the 2023 update. Front Med Lausanne. 2023;10:1106318. doi: 10.3389/fmed.2023.1106318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran DA, Tan X, Macri CJ, Goldstein AT, Fu SW. Lichen Sclerosus: an autoimmunopathogenic and genomic enigma with emerging genetic and immune targets. Int J Biol Sci. 2019;15(7):1429–1439. doi: 10.7150/ijbs.34613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay S, Arnold JD, Malayil L, et al. Potential role of the skin and gut microbiota in premenarchal vulvar lichen sclerosus: a pilot case-control study. PLoS One. 2021;16(1):e0245243. doi: 10.1371/journal.pone.0245243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regauer S. Immune dysregulation in lichen sclerosus. Eur J Cell Biol. 2005;84(2–3):273–277. doi: 10.1016/j.ejcb.2004.12.003 [DOI] [PubMed] [Google Scholar]
- 9.Lukowsky A, Muche JM, Sterry W, Audring H. Detection of expanded T cell clones in skin biopsy samples of patients with lichen sclerosus et atrophicus by T cell receptor-gamma polymerase chain reaction assays. J Invest Dermatol. 2000;115(2):254–259. doi: 10.1046/j.1523-1747.2000.00040.x [DOI] [PubMed] [Google Scholar]
- 10.Regauer S, Reich O, Beham-Schmid C. Monoclonal gamma-T-cell receptor rearrangement in vulvar lichen sclerosus and squamous cell carcinomas. Am J Pathol. 2002;160(3):1035–1045. doi: 10.1016/s0002-9440(10)64924-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arena S, Ieni A, Currò M, et al. Immunohistological analysis of lichen sclerosus of the foreskin in pediatric age: could it be considered a premalignant lesion? Biomedicines. 2023;11(7). doi: 10.3390/biomedicines11071986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regauer S, Liegl B, Reich O, Beham-Schmid C. Vasculitis in lichen sclerosus: an under recognized feature? Histopathology. 2004;45(3):237–244. doi: 10.1111/j.1365-2559.2004.01929.x [DOI] [PubMed] [Google Scholar]
- 13.Terlou A, Santegoets LA, van der Meijden WI, et al. An autoimmune phenotype in vulvar lichen sclerosus and lichen planus: a Th1 response and high levels of microRNA-155. J Invest Dermatol. 2012;132(3 Pt 1):658–666. doi: 10.1038/jid.2011.369 [DOI] [PubMed] [Google Scholar]
- 14.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942 [DOI] [PubMed] [Google Scholar]
- 15.Corazza M, Schettini N, Zedde P, Borghi A. Vulvar lichen sclerosus from pathophysiology to therapeutic approaches: evidence and prospects. Biomedicines. 2021;9(8). doi: 10.3390/biomedicines9080950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalekar LA, Cohen JN, Prevel N, et al. Regulatory T cells in skin are uniquely poised to suppress profibrotic immune responses. Sci Immunol. 2019;4(39). doi: 10.1126/sciimmunol.aaw2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mora EMM, Champer MI, Huang W, Campagnola PJ, Grimes MD. Collagen is more abundant and structurally altered in lichen sclerosus. Urology. 2023;173:192–197. doi: 10.1016/j.urology.2022.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambichler T, Terras S, Kreuter A, Skrygan M. Altered global methylation and hydroxymethylation status in vulvar lichen sclerosus: further support for epigenetic mechanisms. Br J Dermatol. 2014;170(3):687–693. doi: 10.1111/bjd.12702 [DOI] [PubMed] [Google Scholar]
- 19.Guerrero-Setas D, Pérez-Janices N, Ojer A, Blanco-Fernandez L, Guarch-Troyas C, Guarch R. Differential gene hypermethylation in genital lichen sclerosus and cancer: a comparative study. Histopathology. 2013;63(5):659–669. doi: 10.1111/his.12204 [DOI] [PubMed] [Google Scholar]
- 20.Voss FO, Berkhof J, Duin S, et al. DNA methylation and p53 immunohistochemistry as prognostic biomarkers for vulvar lichen sclerosus. Mod Pathol. 2024;37(9):100553. doi: 10.1016/j.modpat.2024.100553 [DOI] [PubMed] [Google Scholar]
- 21.Rotondo JC, Borghi A, Selvatici R, et al. Hypermethylation-induced inactivation of the IRF6 gene as a possible early event in progression of vulvar squamous cell carcinoma associated with lichen sclerosus. JAMA Dermatol. 2016;152(8):928–933. doi: 10.1001/jamadermatol.2016.1336 [DOI] [PubMed] [Google Scholar]
- 22.Ren L, Zhao Y, Huo X, Wu X. MiR-155-5p promotes fibroblast cell proliferation and inhibits FOXO signaling pathway in vulvar lichen sclerosis by targeting FOXO3 and CDKN1B. Gene. 2018;653:43–50. doi: 10.1016/j.gene.2018.01.049 [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Lv Q, Guo J, Wang J, Pan J. Transcriptome profiling and network analysis provide insights into the pathogenesis of vulvar lichen sclerosus. Front Genet. 2022;13:905450. doi: 10.3389/fgene.2022.905450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilatz A, Altinkilic B, Schormann E, et al. Congenital phimosis in patients with and without lichen sclerosus: distinct expression patterns of tissue remodeling associated genes. J Urol. 2013;189(1):268–274. doi: 10.1016/j.juro.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Zhang J, Jiao D, et al. Single-cell RNA sequencing reveals a unique fibroblastic subset and immune disorder in lichen sclerosus urethral stricture. J Inflamm Res. 2024;17:5327–5346. doi: 10.2147/jir.S466317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun P, Kraus CN, Zhao W, et al. Single-cell and spatial transcriptomics of vulvar lichen sclerosus reveal multi-compartmental alterations in gene expression and signaling cross-talk. bioRxiv. 2024. doi: 10.1101/2024.08.14.607986 [DOI] [Google Scholar]
- 27.Lichen Sclerosus-Presentation KG. Diagnosis and management. Dtsch Arztebl Int. 2016;113(19):337–343. doi: 10.3238/arztebl.2016.0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Lan Y, Xu J, et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–d728. doi: 10.1093/nar/gky900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teschendorff AE, Zhu T, Breeze CE, Beck S. EPISCORE: cell type deconvolution of bulk tissue DNA methylomes from single-cell RNA-Seq data. Genome Biol. 2020;21(1):221. doi: 10.1186/s13059-020-02126-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng SC, Breeze CE, Beck S, Teschendorff AE. Identification of differentially methylated cell types in epigenome-wide association studies. Nat Methods. 2018;15(12):1059–1066. doi: 10.1038/s41592-018-0213-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakravarthy A, Furness A, Joshi K, et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat Commun. 2018;9(1):3220. doi: 10.1038/s41467-018-05570-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2(3):100141. doi: 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen T, Chen X, Zhang S, et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinf. 2021;19(4):578–583. doi: 10.1016/j.gpb.2021.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belpaire A, van Geel N, Speeckaert R. From IL-17 to IFN-γ in inflammatory skin disorders: is transdifferentiation a potential treatment target? Front Immunol. 2022;13:932265. doi: 10.3389/fimmu.2022.932265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy T, Banang-Mbeumi S, Boateng ST, et al. Dual targeting of mTOR/IL-17A and autophagy by fisetin alleviates psoriasis-like skin inflammation. Front Immunol. 2022;13:1075804. doi: 10.3389/fimmu.2022.1075804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaba LC, Cardinale I, Gilleaudeau P, et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J Exp Med. 2007;204(13):3183–3194. doi: 10.1084/jem.20071094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choudhury A, Magill ST, Eaton CD, et al. Meningioma DNA methylation groups identify biological drivers and therapeutic vulnerabilities. Nat Genet. 2022;54(5):649–659. doi: 10.1038/s41588-022-01061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F, Zhang X, Zhang H, et al. Pan-precancer and cancer DNA methylation profiles revealed significant tissue specificity of interrupted biological processes in tumorigenesis. Epigenetics. 2023;18(1):2231222. doi: 10.1080/15592294.2023.2231222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Canady J, Karrer S, Fleck M, Bosserhoff AK. Fibrosing connective tissue disorders of the skin: molecular similarities and distinctions. J Dermatol Sci. 2013;70(3):151–158. doi: 10.1016/j.jdermsci.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 42.Dierckx S, Patrizi M, Merino M, González S, Mullor JL, Nergiz-Unal R. Collagen peptides affect collagen synthesis and the expression of collagen, elastin, and versican genes in cultured human dermal fibroblasts. Front Med Lausanne. 2024;11:1397517. doi: 10.3389/fmed.2024.1397517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abraham D, Lescoat A, Stratton R. Emerging diagnostic and therapeutic challenges for skin fibrosis in systemic sclerosis. Mol Aspects Med. 2024;96:101252. doi: 10.1016/j.mam.2024.101252 [DOI] [PubMed] [Google Scholar]
- 44.Shu LZ, Zhang XL, Ding YD, Lin H. From inflammation to bone formation: the intricate role of neutrophils in skeletal muscle injury and traumatic heterotopic ossification. Exp Mol Med. 2024;56(7):1523–1530. doi: 10.1038/s12276-024-01270-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong YL, Okubo T, Uno E, Suda K, Ishii T. Role of CD99 in regulating homeostasis and differentiation in normal human epidermal keratinocytes. Biochem Biophys Res Commun. 2022;606:108–113. doi: 10.1016/j.bbrc.2022.03.087 [DOI] [PubMed] [Google Scholar]
- 46.Kervarrec T, Tallet A, Miquelestorena-Standley E, et al. Morphologic and immunophenotypical features distinguishing Merkel cell polyomavirus-positive and negative Merkel cell carcinoma. Mod Pathol. 2019;32(11):1605–1616. doi: 10.1038/s41379-019-0288-7 [DOI] [PubMed] [Google Scholar]
- 47.Rabquer BJ, Hou Y, Del Galdo F, et al. The proadhesive phenotype of systemic sclerosis skin promotes myeloid cell adhesion via ICAM-1 and VCAM-1. Rheumatology. 2009;48(7):734–740. doi: 10.1093/rheumatology/kep091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bixel MG, Petri B, Khandoga AG, et al. A CD99-related antigen on endothelial cells mediates neutrophil but not lymphocyte extravasation in vivo. Blood. 2007;109(12):5327–5336. doi: 10.1182/blood-2006-08-043109 [DOI] [PubMed] [Google Scholar]
- 49.Bixel G, Kloep S, Butz S, Petri B, Engelhardt B, Vestweber D. Mouse CD99 participates in T-cell recruitment into inflamed skin. Blood. 2004;104(10):3205–3213. doi: 10.1182/blood-2004-03-1184 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Eight DNAme datasets pertaining to LS have been deposited in the Genome Sequence Archive of the Beijing Institute of Genomics, Chinese Academy of Sciences, under the accession number PRJCA025881.35 These datasets are readily accessible to the public via https://ngdc.cncb.ac.cn/omix/release/OMIX006366. Moreover, publicly accessible datasets, namely GSE151600 (G1_Control16) and GSE229848 (G2_Control6), which have produced DNAme data for eight and six healthy skin tissues respectively, can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151600 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE229848. We also utilized the public dataset GSE173205 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173205), which comprises single-cell data from 10X Genomics for four healthy control skin tissues.
The datasets supporting the conclusions of this article are available in the Genome Sequence Archive. Each sample’s files are hyperlinked to the dataset on https://ngdc.cncb.ac.cn/omix/release/OMIX006366.