Abstract
Hypoxic ischemic encephalopathy (HIE) is a brain injury that occurs in 1 ~ 5/1000 term neonates. Accurate identification and segmentation of HIE-related lesions in neonatal brain magnetic resonance images (MRIs) is the first step toward identifying high-risk patients, understanding neurological symptoms, evaluating treatment effects, and predicting outcomes. We release the first public dataset containing neonatal brain diffusion MRI and expert annotation of lesions from 133 patients diagnosed with HIE. HIE-related lesions in brain MRI are often diffuse (i.e., multi-focal), and small (over half the patients in our data having lesions occupying <1% of the brain volume (including ventricles)). Segmentation for HIE MRI data is remarkably different from, and arguably more challenging than, other segmentation tasks such as brain tumors with focal and relatively large lesions. We hope that this dataset can help fuel the development of MRI lesion segmentation methods for HIE and small diffuse lesions in general.
Subject terms: Diagnostic markers, Databases
Background & Summary
Accurate identification of brain injuries in neonatal brain magnetic resonance images (MRI)1–3 is crucial to improve clinical care of neonates with hypoxic-ischemic encephalopathy (HIE), a brain disease that occurs in around 1 ~ 5/1000 term-born infants around birth4,5. HIE affects around 750,000 term-born neonates every year worldwide4,5, costing about $2 billion/year in the US alone, let alone family burdens. Therapeutic hypothermia, the current clinical treatment of HIE, can reduce mortality and morbidity in high-income countries. Nevertheless, around 1/3 of patients still die or develop neurocognitive deficits by 2 years of age. MRI is used in over 50% of the >100 ongoing HIE-related clinical trials worldwide6, for evaluating treatment effects7–9, and helping discover clinical10–12, biochemical10,13–15, and serum16–18 biomarkers. Accurate identification of brain lesions in neonatal brain MRIs1–3 is needed for disease prognosis, a better understanding of the neural basis of disease progression, and more timely evaluations of novel therapeutic effects.
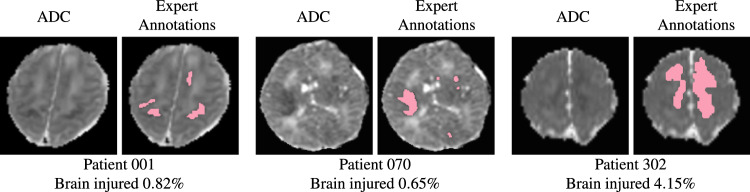
HIE lesions are often diffuse (i.e., multi-focal), and small; hence, algorithms that have shown great promise in segmenting big and focal lesions, such as brain tumors and acute strokes, often encounter challenges when directly applied to MRIs of HIE patients. Indeed, many (over half) patients had lesions occupying <1% of brain volume, as shown in Fig. 1. As a result, the segmentation accuracy measured by the Dice overlap with U-Net19 and other state-of-the-art machine/deep learning algorithms on HIE remains at around 0.520, whereas Dice is over 0.8 when segmenting brain tumors21,22.
Fig. 1.
Lesions associated to hypoxic ischemic encephalopathy (HIE) are typically diffuse (i.e., multi-focal) and small. Here we show two representative images for 3 HIE patients. For each patient, in the left panel: apparent diffusion coefficient (ADC) maps that are clinically used to identify HIE lesions; in the right panel: manually-annotated lesions (shown in pink) overlaid on the ADC map. We listed the percentage of the whole brain volume (including ventricles) being injured at the bottom (i.e., lesion volume divided by the whole brain volume).
A major hurdle in developing algorithms for small diffuse lesions, such as HIE lesions, is the lack of public data. Public data with expert annotations of lesions have fueled the advancement of machine learning algorithms to segment brain tumors23, stroke lesions24, multiple sclerosis lesions25,26, and numerous other diseases in the brain or other organs27,28. However, to date, there is no public MRI data with expert annotations available for HIE lesions.
We present BOston Neonatal Brain Injury Dataset for Hypoxic Ischemic Encephalopathy (BONBID-HIE), an open-source, comprehensive, and representative MRI dataset for HIE. This paper introduces the first part of the BONBID-HIE data. This release contains raw and derived diffusion parameter maps, as well as manually-annotated lesion masks, for 133 HIE patients. Our data was from Massachusetts General Hospital. It includes MRIs from different scanners (Siemens 3T and GE 1.5T), different MRI protocols, and from patients of different races/ethnicities and ages (0-14 days postnatal age). Part I of our data release (this paper) focuses on lesion detection, while Part II (a follow-up paper) will focus on clinical, treatment, and neurologic outcome data for further developing prognostic biomarkers.
Our data contains voxel-wise annotation of HIE lesions beyond the brain region-level rough localization of abnormalities for two reasons. First, extractions of subsequent features, such as lesion volumes, lesion geometry, within-lesion signal heterogeneity, within-lesion histogram analysis, will require voxel-level identification of lesions. Such features are expected to offer additional information for predicting the neurological outcomes by 2 years of age, which is the ultimate goal for the design of prognosis biomarkers. Second, it is possible to derive brain-regional-level injuries from the voxel-wise lesion detection
Methods
Study Approval and Data Security
This work was approved by Institutional Review Boards (IRBs): IRB-P00025916 (PI: Y. Ou) at Boston Children’s Hospital and IRB-2015P001651 (PI: Y. Ou) at Massachusetts General Hospital. Consent was waived because the retrospective nature of this work: many of the patients have already completed follow-up, live at a great distance from the hospitals, or the contact information is not up to date. Data were kept in encrypted folders on password-protected computers within the two hospitals’ firewalls. Also, anonymization included removing identifiable information (medical record number, date of birth, name, physician’s name, visit dates, etc.) in the clinical report and imaging header. Patient names were replaced by MGHNICU_001, MGHNICU_002,… Brain images have undergone defacing to remove the risk of 3D rendering that may recognize the facial features.
Overview
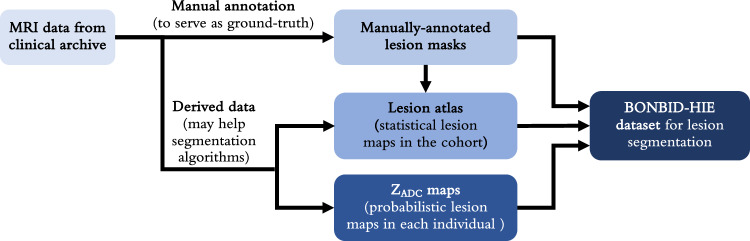
Figure 2 illustrates the overall data archiving process. MRI data for this dataset were acquired from MGH. Manual annotations were performed to create lesion masks for each individual patient, which served as the ground truth. These lesion masks were then aggregated to form a lesion atlas, representing statistical lesion maps across the cohort. Concurrently, the collected MRI data were processed to generate ZADC maps, which were probabilistic lesion maps for each individual. In the following sections, we will discuss each step involved in creating the BONBID-HIE dataset in detail.
Fig. 2.
A schematic diagram showing the steps performed on the BONBID-HIE data for release.
Retrospective Data Collection
Data was retrospectively collected from MGH. Inclusion criteria were: (1) term-born; (2) clinical diagnosis of HIE; (3) initially treated at MGH between 2001 and 2018; (4) no comorbidities such as hydrocephalus or congenital syndromes; and (5) high-quality MRI acquired in Day 0-14 after birth (visually checked by RW, AF, YO). Exclusion criteria were: (1) excessive motion artifacts or missing images; or (2) primary perinatal stroke, focal artery ischemic stroke, or hemorrhage.
Clinical characteristics and demographic information were retrospectively gathered from the electronic health records (EHRs). The clinical variables included maternal information during pregnancy and delivery, as well as infant information. More detail can be found in the “Data Records” Section.
MRI data was downloaded from MGH Radiology Department clinical archives using the mi2b2 search engine29. MRIs were acquired on either a GE 1.5T Signa scanner (N=52, scanned during 2001-2012), or, a Siemens 3T TrioTim or PrismaFit scanner (N=81, scanned during 2012-2018). Diffusion tensor sequences on all scanners had the protocol as follows: Time of Repetition (TR) = 7500–9500 ms, Time of Echo (TE) = 80–115 ms, and b = 1000 s/mm2. The GE 1.5T scanner had resolution 1.5 × 1.5 × (2.0–4.0) mm3 and (6–60) diffusion directions, while the Siemens 3T scanner had a resolution 2 × 2 × 2 mm3 and (25–60) diffusion directions. Apparent diffusion coefficient (ADC) maps were directly generated by the scanners (with the Advantage Windows Workstation for GE scanners30,31 and with Syngo software for Siemens scanners32).
MRI Pre-processing
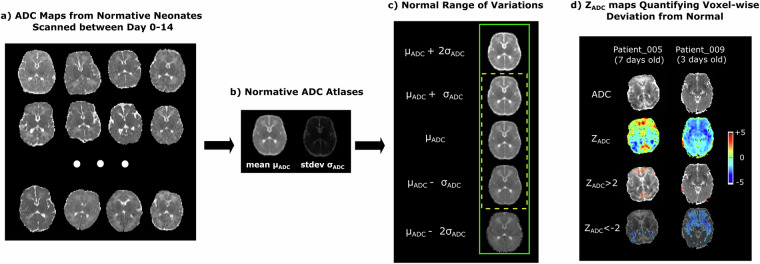
Besides the raw NIfTI image as converted from the DICOM files, we also generated several processed images. The pre-processing steps included: N4 bias correction33, field of view normalization34, multi-atlas skull stripping for the ADC maps35, and deformable registration of each patient’s ADC map to a normative 0-14 day neonatal brain ADC atlas36, by the Deformable Registration via Attribute Matching and Mutual-Saliency weighting (DRAMMS) software37, which has been extended-validated for lifespan ages in various MRI sequences38. This normative ADC atlas was constructed from ADC maps of 13 healthy individuals acquired 0-14 days after birth (Fig. 3a) with our extensively-validated MRI analysis pipeline34,35,37,38. All software packages used in this pre-processing pipeline are publicly available and have been validated in processing both research and clinical MRI scans across ages39–43.
Fig. 3.
The generation and concept of ZADC maps. (a) Examples of ADC maps from normative subjects, which were warped into the same space using unbiased group-wise DRAMMS registration to generate (b) the mean and standard deviation ADC atlases. (c) One (yellow dashed box) or two (green solid box) standard deviations above and below the mean ADC atlas define the normal ranges of voxel-wise ADC variations. (d) Our novel ZADC map quantifies voxel-wise deviations from the mean ADC map in (b). The cool/warm colors in (d) represent voxels with ADC values lower/higher than the mean ADC at the same anatomic location, according to the scale bar on the right.
Multi-Expert Consensus Annotation of Lesions As Ground-Truth
HIE lesions had been qualitatively described in radiology reports that were part of the clinical flow. In this clinical process, ADC maps were used as the primary images, in addition to structural MRIs, to identify HIE abnormalities44–46. To convert the description of lesions in the radiology report into voxel-by-voxel expert annotation, we used a two-step expert consensus approach. First, HIE lesions were manually annotated as a binary mask on the ADC maps in the patient’s raw image space, using the MRICroN software, according to the neuroradiology reports. This was done primarily by a clinical fellow (YS; >3 years of experience in general medicine and >1.5 years of neuroradiology training by practicing neuroradiologists at BCH specifically for HIE-related neuroimaging interpretation). The annotation protocol asked the primary annotator to label voxels as the expert deemed abnormal by his/her expertise, only in those brain regions mentioned in the clinical radiology report, and to leave uncertainties or disagreements for the subsequent expert consensus process. The annotations started from the axial slice and were subsequently modified in the coronal and sagittal planes for the 3D integrity of lesion regions. Second, in those 27 patients where uncertainties or disagreement between the primary annotator (YS) and the clinical radiology report occurred, we created a consensus lesion mask based on discussions among three more experienced pediatric neuroradiologists (CJ, SS, and PEG; >5, >5, and >20 years of experience practicing clinical pediatric neuroradiology), in a simple majority manner. This single set of lesion annotations on the ADC maps, the first for HIE, reflects the collective decision among the neuroradiologists who read the images at the time of clinical care, the primary annotator (YS), and 3 more experienced pediatric neuroradiologists (CJ, SS, PEG). This multi-expert consensus annotation process is less biased than a single-expert annotation. Our ongoing work on a 21-site HIE dataset is using more resource- and expertise-demanding multi-expert independent approach.
Generation of ZADC Maps for Each Patient– Derived to Aid Automated Lesion Segmentation
Neuroradiologists identify acute brain injury from HIE as regions with low ADC values. Low ADC values represent a reduced water diffusion, which occurs in the first week after birth due to ischemic necrosis resulting from the hypoxic ischemic insult6. However, a dilemma is, what ADC value is considered abnormally low versus just low within the normal variation? The normal variations of ADC values differ across brain regions36,47, making this question difficult even for experienced neuroradiologists. For example, a voxel with an ADC value of 800 ( × 10−6mm2/s) may be considered normal at one brain region, whereas another voxel with an ADC value of 900 ( × 10−6mm2/s) may be considered lesioned at another brain region, if the normal ranges of ADC variations in the two brain regions are 700–900 and 950–1100 (same unit), respectively.
To address this dilemma, we have developed ZADC maps to normalize and make ADC values comparable across brain voxel locations39. First, a normative ADC atlas was generated from scans of 13 normative neonates (Fig. 3a). This atlas quantifies the mean ADC values and standard deviation at every voxel36 (Fig. 3b), and hence the normal range of variations at each voxel (Fig. 3c). Then, we converted each patient’s ADC map (first row, Fig. 3d) into a ZADC map (second row, Fig. 3d). The ZADC maps compared the patient’s ADC value at each voxel to the normal variations at the corresponding voxel in the atlas. In a nutshell, ZADC maps quantify how many standard deviations away a patient’s ADC value at a voxel is from the normal mean at the same anatomical location.
Specifically, a deformation (D) was computed, which mapped every voxel x in the patient’s ADC map to its anatomically-corresponding location D(x) in the atlas space. The normal range of ADC variation per voxel was defined by the mean μ(D(x)) and standard deviation σ(D(x)) denoted for that voxel across all healthy neonates. Finally, the patient’s ADC value Ix at voxel x was converted to a Z value: Zx = (Ix − μ(D(x)))/σ(D(x)). We calculated the ZADC map, which resides in the patient’s raw ADC image space, for each patient. This offers an option for developing anatomy-aware lesion segmentation algorithms48.
It is important to note that ZADC maps were not part of the scanner-generated images, nor stored in scanner or PACS. ZADC maps were not used in expert consensus annotation of lesions, which serve as ground truth for computer-assisted lesion detection. ZADC maps were generated in post-processing steps, to aid the computer-assisted lesion detection. Results from ZADC maps were validated against expert consensus of lesion annotation for the accuracy of computer-assisted lesion detection algorithms. Algorithm developers have options to use or not use ZADC maps in their lesion segmentation algorithms.
Construction of Statistical Lesion Atlases for the Cohort
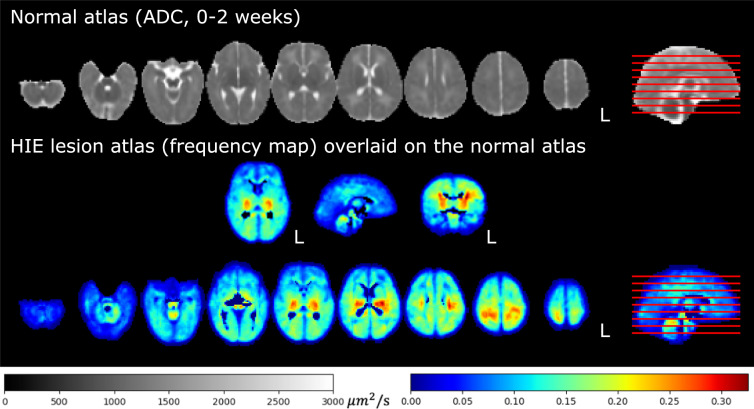
The same deformation field that was computed by the non-rigid registration from the patient’s skull-stripped ADC map to the normal ADC atlas was used to transform the binary brain lesion maps of each patient into the normal neonatal ADC atlas space37. The transformed binary lesion masks were then summed and divided by the total number of patients at each voxel. This led to a statistical lesion atlas that quantifies voxel-wise frequency, or probability, of HIE lesions in our cohort, as illustrated in Fig. 4.
Fig. 4.
Statistical lesion atlas quantifying the voxel-wise lesion frequency in our cohort of N = 133 patients in the normal 0-14 days ADC atlas space.
The normal neonatal ADC atlas is part of the release, to offer an option to algorithm developers. An algorithm for HIE lesion detection can opt to use the normal ADC atlas to contrast HIE-related abnormality, or choose not to use this atlas. We would clarify that the HIE cohort, on which the lesion detection algorithms are to be developed, only contains data from patients with clinically confirmed HIE diagnosis.
Data Records
The dataset is available on Zenodo (https://zenodo.org/records/10602767)49. All data has been made publicly available under the CC BY license (https://creativecommons.org/licenses/by/4.0/legalcode).
Dataset Characteristics
Table 1A lists the demographics and clinical characteristics of mothers and neonates. Maternal information includes demographics (age at delivery, race), birth mode (C-section or vaginal), and complications during pregnancy and delivery. Neonatal information includes demographics (age at MRI scan, gestational age at birth, birth weight, head circumference, sex), birth conditions (1/5/10-minute Apgar scores, lowest pH value in umbilical cord), treatment (hypothermia or not), and complications in the neonatal intensive care unit (NICU), including seizure (yes/no), the length of stay (in days), the use of endotracheal tube (ETT, yes/no), and the administration of total parenteral nutrition (TPN, yes/no). In each row, we also listed the number of patients who had such information available.
Table 1.
Cohort characteristics (N=133).
| A. Demographics and Clinical Characteristics | ||
|---|---|---|
| Maternal Information | ||
| Maternal age at delivery (years) | 29.5 ± 6.7 | N=133 |
| Race | White (43), Black or African American (7), Hispanic or Latino (15), Multi Race (5), Unknown (57), Other (6) | N=133 |
| Delivery | C-section (78), Vaginal (55) | N=133 |
| Antepartum hemorrhage | Yes (29), No (104) | N=133 |
| Thyroid dysfunction | Yes (5), No (128) | N=133 |
| Pre-eclampsia | Yes (9), No (124) | N=133 |
| Fetal decels | Yes (72), No (61) | N=133 |
| Shoulder dystocia | Yes (8), No (125) | N=133 |
| Chorioamnionitis | Yes (20), No (108) | N=133 |
| Emergency c-section | Yes (69), No (58) | N=133 |
| Neonatal Information | ||
| Age at scan (days) | 3.9 ± 2.7 | N=133 |
| Gestational age at birth (weeks) | 39.1 ± 1.9 | N=133 |
| Birth weight (g) | 3321.9 ± 615.9 | N=133 |
| Infant head circumference (cm) | 34.2 ± 1.4 | N=85 |
| Sex | Male (74), Female (59) | N=133 |
| 1-minute APGAR scores | 1.9 ± 1.7 | N=133 |
| 5-minute APGAR scores | 4.2 ± 2.3 | N=132 |
| 10-minute APGAR scores | 5.3 ± 2.1 | N=118 |
| Lowest pH value in umbilical cord | 7.00 ± 0.20 | N=129 |
| Therapeutic hypothermia before MRI? | Yes (86), No (47) | N=133 |
| Endotracheal tube (ETT) in NICU | Yes (78), No (47) | N=125 |
| Total parenteral nutrition (TPN) in NICU | Yes (111), No (21) | N=133 |
| Seizures NICU | Yes (64), No (69) | N=133 |
| Length of stay in NICU (days) | 12.10 ± 9.92 | N=133 |
| B. Lesion Characteristics (N=133) | |
|---|---|
| Whole-brain lesion volume – minimum | 0 mm3 (0% of the brain injured) |
| Whole-brain lesion volume - 25th percentile | 441.96 mm3 (0.10% of the brain injured) |
| Whole-brain lesion volume - median | 2765.63 mm3 (0.63% of the brain injured) |
| Whole-brain lesion volume - 75th percentile | 24264.27 mm3 (4.86% of the brain injured) |
| Whole-brain lesion volume - maximum | 412120.00 mm3 (82.59% of the brain injured) |
| C. Number of Patients by Percentage of Lesions in the Brain (N=133) | |
|---|---|
| [0%, 1%) of the brain being injured | 55.64% (N=74) |
| [1, 5%) of the brain was injured | 19.55% (N=26) |
| [5, 10%) of the brain was injured | 5.26% (N=7) |
| [10, 20%) of the brain was injured | 4.51% (N=6) |
| [20, 50%) of the brain was injured | 8.27% (N=11) |
| [50, 100]% of the brain was injured | 6.77% (N=9) |
| D. Scanner (N=133) | |
|---|---|
| GE 1.5T | 39% (N=52) |
| Siemens 3T | 61% (N=81) |
Table 1B quantifies the distribution of the absolute lesion volumes (in mm3) and relative lesion volume (percentage of the brain being injured). Here, the relative lesion volume was calculated by the volume of the expert-annotated lesion regions divided by the algorithm-extracted whole brain volumes (including ventricles) 34,35. The median lesion volume accounted for 0.63% of the whole brain volume. This confirms that over half of the patients had less than 1% of the brain being injured. Table 1C further calculates the distribution of the relative lesion volumes (by the percentage of the brain volume being lesioned). The absolute and relative volumes were both computed in the patient’s raw ADC image space. The minimum lesion was 0 mm3, which is a common issue in HIE – mild HIE cases may not show explicit lesions in neonatal MRIs50–52.
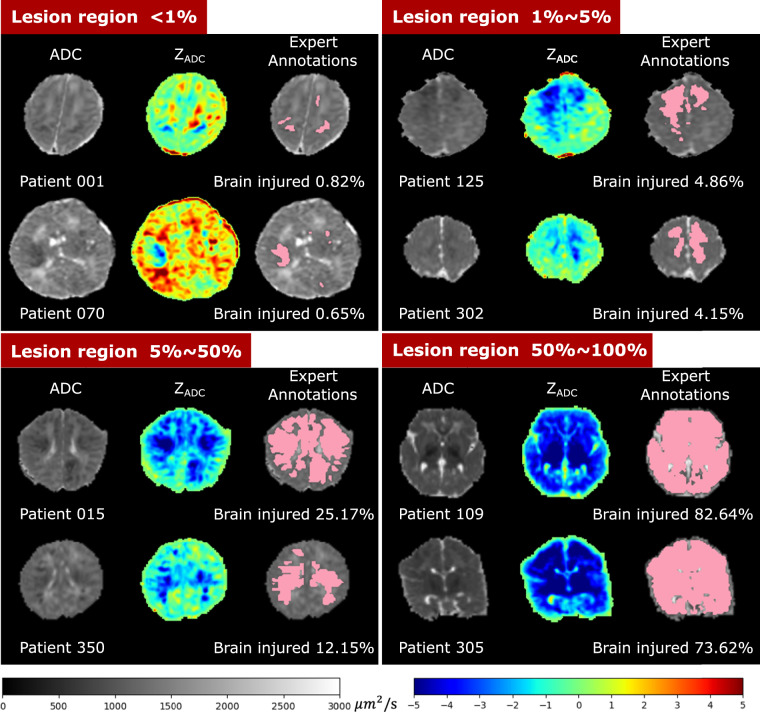
Figure 5 shows the ADC map, ZADC map, and expert annotations of example patients with different HIE lesion percentages. Two patients are shown in each of the four groups: those with lesions occupying < 1% (upper left panel), 1–5% (upper right panel), 5–50% (lower left panel), and 50–100% (lower right panel) of the whole brain volume. Overall, around 1 in 2 (55.64%) patients had HIE lesions occupying less than 1% of their brain volume, and 3 in 4 patients (75.19%) patients had lesions occupying less than 5% of their brain. This confirms that HIE lesions detectable in the diffusion MRI in our cohort are often small.
Fig. 5.
Visualization of patients with different lesion percentages. In every patient, the left image is the ADC map (skull stripped) with range of ADC values designated by the gray scale bar, the middle is the computed ZADC map with range of Z-scores designated by the rainbow scale bar, and the right image is the expert-annotated lesion regions (pink) overlaid on the ADC map. Percentages of injury were calculated by the volume of the expert-annotated lesion regions divided by algorithm-extracted whole brain volumes (including ventricles).
Data structure and file formats
All medical imaging files were exported from the Picture Archiving and Communication System (PACS) and converted into the NIfTI format. Segmentation masks created by expert annotations were also saved in NIfTI format. Corresponding scanner metadata from the Digital Imaging and Communications in Medicine (DICOM) header in the .json file format is provided with the released dataset. All data in the BONBID-HIE dataset was separated into a training dataset (N=89) and a test dataset (N=44). Both the training and testing contain data from both scanners (GE 1.5T Signa and Siemens 3T Trio). The split between the training and testing dataset has been performed (RB, YO) so that both sets include a similar variance of HIE lesion patterns as shown in Table 1C.
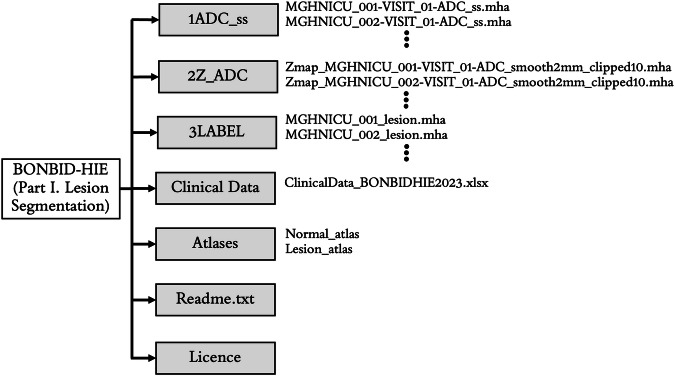
The data is organized in the format shown in Fig. 6. BONBID-HIE provides, per patient: (i) 1ADC_ss: skull stripped ADC map; (ii) 2Z_ADC: ZADC map; (iii) 3LABEL: expert lesion annotations; and (iv) clinical data: clinical variables as written in Table 1A. There is also (v) Atlases: a folder for the normal and lesion atlases; (vi) a readme.txt file: a text file to provide information on this data organization; and (vii) the license file of the BONBID-HIE dataset.
Fig. 6.
Folder structure of the BONBID-HIE dataset (Part I. Lesion Segmentation).
Technical Validation
Representativeness of patient cohort
Our data is representative of HIE cohorts in the developed countries. At least three characteristics of our data agree with documented clinical knowledge about HIE.
Lesion distribution in space agrees with clinical knowledge
Our statistical lesion atlas in Fig. 4 shows that HIE lesions can occur anywhere in the brain. The regions most frequently injured included the basal ganglia, internal capsules, thalamus, perirolandic cortex/subcortical white matter, temporal lobes, cerebral white matter, brainstem, and vermis (red, orange, and yellow regions in Fig. 4). This lesion atlas map coincides with clinical knowledge of brain regions often vulnerable to HIE injuries2,6,13. Indeed, HIE-related injuries in these regions have been key criteria in expert MRI scoring systems, which are used to assess the severity of HIE. Examples include the NICHD Neonatal Research Network (NRN)2, the Barkovich53, the Weeke-deVeries3, and the Trivedi54 scoring systems. In addition, lesions appeared in less than 35% of the patients at any given voxel, according to the color bar in this figure. This confirms the clinical knowledge that HIE lesions are diffuse, spatially distributed, and almost half to two-thirds of the HIE patients have no or minimal injuries on diffusion imaging, at least in patients in the USA51,52.
Lesion distribution in time agrees with clinical knowledge
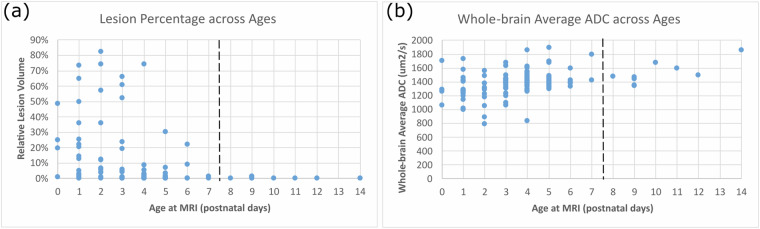
Figure 7(a) shows the percentage of the whole brain volume being lesioned at different postnatal ages. The lesion percentage in the ADC maps came down to almost 0 in the 9 patients who underwent MRI scans after postnatal day 7. This agrees with the clinical knowledge that HIE-related lesions are more detectable in ADC maps during 0-7 postnatal days or in T1/T2-weighted images than in relatively later scans (after postnatal day 7)2,46,52.
Fig. 7.
Representativeness of our cohort for (a) lesion distribution across ages; and (b) whole-brain ADC values across ages. In both panels, each dot denotes a patient in our cohort.
ADC evolvement with age agrees with clinical knowledge
Figure 7(b) shows the whole-brain average ADC values of all patients (each dot is one patient). In normal cohorts, ADC values drop rapidly in the early postnatal life (see Figures 4 and 5 in36, and Figures 3 and 4 in47). However, the presence of HIE-related abnormalities disrupted this trend – HIE patients undergoing earlier MRIs (0-7 postnatal days) had decreased ADC values in a larger precentage of the brain (Fig. 7(a)), so the ADC values in 0-7 postnatal days were at similar or even lower levels than ADC values in 7-14 postnatal days among HIE patients (Fig. 7(b)). This has also been documented in HIE literature55,56.
Utility of ADC maps and ZADC maps in Automated Lesion Segmentation
The only ground truth for lesion segmentation in our released data is the expert consensus of lesion annotations. Our data release includes ZADC maps as an option to algorithm developers. To demonstrate the utility of the computed ZADC maps, we compared the accuracies of using ADC or ZADC maps for lesion segmentation. We attempted simple thresholding of ADC and ZADC maps, at several threshold values, for segmenting HIE lesions. Although simple, thresholding-based segmentation accuracy is a strong indicator for segmentation accuracies in more sophisticated machine/deep learning algorithms57. For ADC maps, we used different thresholds ranging from 800-1100 μm2/s, as suggested in the literature46,58–61. For ZADC maps, we used thresholds -1.5, -2, and -2.5. Voxels in patient MRIs with ADC values 1.5 to 2.5 standard deviations below the average ADC values from healthy controls were considered abnormally low and hence, lesioned. The choice of thresholds around -2 in ZADC maps was also based on the hypothesis of a normal distribution of ADC values at each voxel across subjects36,39.
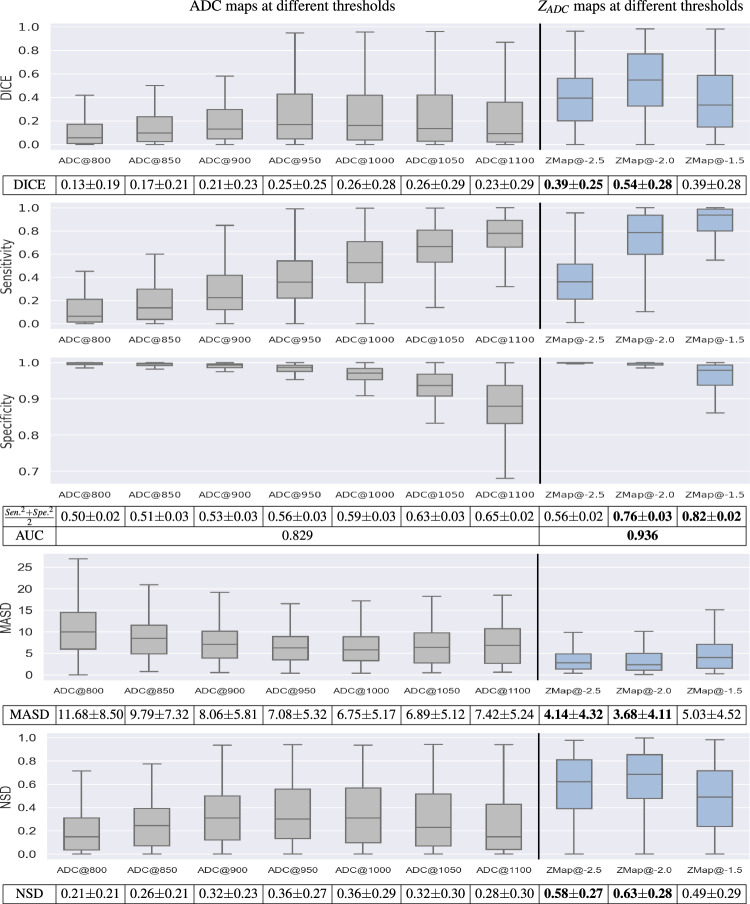
We evaluated the accuracy of these maps compared to expert-annotated ground-truth masks using the Dice coefficient, sensitivity, specificity, Mean Average Surface Distance (MASD)62,63, and Normalized Surface Distance (NSD)63. These evaluation metrics were selected based on64. Results for Dice, sensitivity, specificity and area under the receiver-operating characteristic curve (AUC) are shown in Fig. 8. Here, the gray boxplots are the accuracy measurements when ADC maps were thresholded between 800 and 1100 μm2/s (50 μm2/s intervals). The blue boxplots are the accuracy measurements when the ZADC maps were thresholded at -1.5, -2, and -2.5. Figure 8 demonstrates: (i) both ADC and ZADC have values in helping segment the HIE-related lesions, since the specificity from simple naive thresholding-based segmentations was comparable to those from machine learning-based algorithms20, although the Dice and sensitivity were lower; (ii) ZADC maps thresholded at -2, the most intuitive and straightforward threshold value, yielded the highest Dice (0.54 ± 0.28), followed by ZADC maps thresholded at -2.5 (Dice 0.39 ± 0.25); (iii) ZADC maps thresholded at -1.5 yielded the highest (0.82 ± 0.02), quantifying the balance between sensitivity and specificity compared with any ADC thresholds, followed by ZADC maps thresholded at -2 (0.76 ± 0.03); and (iv) overall, across all thresholds, ZADC maps showed a higher area under the curve (AUC: 0.936). Meanwhile, accuracies measured by MASD (lower MASD for a higher accuracy) and NSD (higher NSD for a higher accuracy), confirmed that ZADC map’s superior accuracy than ADC maps. This shows that ZADC maps – anatomy-normalized ADC images – carry the potential to improve lesion detection accuracy compared to ADC maps.
Fig. 8.
Accuracy of thresholding-based lesion segmentation on ADC and ZADC maps with different threshold values, measured by Dice, sensitivity, sepecificity, , AUC, MASD and NSD. Bold texts in the tables beneath the figure panels highlight the two scenarios with the lowest MASD scores (lower is more accurate lesion detection), or the two scenarios with the highest Dice, AUC and NSD metrics (higher is more accurate lesion detection).
Acknowledgements
This work was funded, in part, by the Harvard Medical School and Boston Children’s Hospital through Early Career Development Fellowship, Thrasher Research Fund Early Career Awards, NIH R21NS121735, R61NS126792, and R03HD104891.
Author contributions
Study Design: R.B., S.V.B., A.N.F., R.L.H., P.E.G., Y.O.; Data Collection: S.V.B., R.J.W., A.N.F., R.L.H., P.E.G., Y.O.; Data Analysis: R.B., Y.S., S.B.V., R.J.W., A.N.F., C.J.C., S.S., Y.Z., R.L.H., P.E.G., Y.O.; Manuscript Writing: R.B., S.V.B., A.N.F., R.L.H., P.E.G., Y.O.
Code availability
No custom code was generated for this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Randy L. Hirschtick, P. Ellen Grant, Yangming Ou.
Contributor Information
Rina Bao, Email: rina.bao@childrens.harvard.edu.
Yangming Ou, Email: yangming.ou@childrens.harvard.edu.
References
- 1.Rutherford, M. et al. Magnetic resonance imaging in hypoxic-ischaemic encephalopathy. Early Human Development86, 351–360 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Shankaran, S. et al. Brain injury following trial of hypothermia for neonatal hypoxic–ischaemic encephalopathy. Archives of Disease in Childhood-Fetal and Neonatal Edition97, F398–F404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weeke, L. C. et al. A novel magnetic resonance imaging score predicts neurodevelopmental outcome after perinatal asphyxia and therapeutic hypothermia. The Journal of Pediatrics192, 33–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham, E. M., Ruis, K. A., Hartman, A. L., Northington, F. J. & Fox, H. E. A systematic review of the role of intrapartum hypoxia-ischemia in the causation of neonatal encephalopathy. American Journal of Obstetrics and Gynecology199, 587–595 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Lee, A. C. et al. Intrapartum-related neonatal encephalopathy incidence and impairment at regional and global levels for 2010 with trends from 1990. Pediatric Research74, 50–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss, R. J. et al. Mining multi-site clinical data to develop machine learning mri biomarkers: application to neonatal hypoxic ischemic encephalopathy. Journal of Translational Medicine17, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrera, T. I. et al. Outcomes of preterm infants treated with hypothermia for hypoxic-ischemic encephalopathy. Early Human Development125, 1–7 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Inder, T. E. et al. Randomized trial of systemic hypothermia selectively protects the cortex on mri in term hypoxic-ischemic encephalopathy. The Journal of Pediatrics145, 835–837 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Rogers, E. E. et al. Erythropoietin and hypothermia for hypoxic-ischemic encephalopathy. Pediatric Neurology51, 657–662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natarajan, G., Pappas, A. & Shankaran, S. Outcomes in childhood following therapeutic hypothermia for neonatal hypoxic-ischemic encephalopathy (HIE). In Seminars in Perinatology, vol. 40, 549–555 (Elsevier, 2016). [DOI] [PMC free article] [PubMed]
- 11.Ramaswamy, V. et al. Systematic review of biomarkers of brain injury in term neonatal encephalopathy. Pediatric Neurology40, 215–226 (2009). [DOI] [PubMed] [Google Scholar]
- 12.van Laerhoven, H., de Haan, T. R., Offringa, M., Post, B. & van der Lee, J. H. Prognostic tests in term neonates with hypoxic-ischemic encephalopathy: a systematic review. Pediatrics131, 88–98 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Laptook, A. R. et al. Effect of therapeutic hypothermia initiated after 6 hours of age on death or disability among newborns with hypoxic-ischemic encephalopathy: a randomized clinical trial. JAMA318, 1550–1560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shankaran, S. et al. Effect of depth and duration of cooling on death or disability at age 18 months among neonates with hypoxic-ischemic encephalopathy: a randomized clinical trial. JAMA318, 57–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Mara, K. & Weiss, M. D. Dexmedetomidine for sedation of neonates with HIE undergoing therapeutic hypothermia: a single-center experience. American Journal of Perinatology Reports8, e168–e173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massaro, A. N. et al. Serum biomarkers of mri brain injury in neonatal hypoxic ischemic encephalopathy treated with whole-body hypothermia: a pilot study. Pediatric Critical Care Medicine14, 310–317 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massaro, A. N. et al. Plasma biomarkers of brain injury in neonatal hypoxic-ischemic encephalopathy. The Journal of Pediatrics194, 67–75 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Douglas-Escobar, M. & Weiss, M. D. Biomarkers of hypoxic-ischemic encephalopathy in newborns. Frontiers in Neurology3, 144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 234–241 (Springer, 2015).
- 20.Murphy, K. et al. Automatic quantification of ischemic injury on diffusion-weighted mri of neonatal hypoxic ischemic encephalopathy. NeuroImage: Clinical14, 222–232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menze, B. H. et al. The multimodal brain tumor image segmentation benchmark (brats). IEEE Transactions on Medical Imaging34, 1993–2024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakas, S. et al. Advancing the cancer genome atlas glioma mri collections with expert segmentation labels and radiomic features. Scientific Data4, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakas, S. et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv preprint arXiv:1811.02629 (2018).
- 24.Maier, O. et al. Isles 2015-a public evaluation benchmark for ischemic stroke lesion segmentation from multispectral mri. Medical Image Analysis35, 250–269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carass, A. et al. Longitudinal multiple sclerosis lesion segmentation: resource and challenge. NeuroImage148, 77–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Commowick, O. et al. Objective evaluation of multiple sclerosis lesion segmentation using a data management and processing infrastructure. Scientific Reports8, 13650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antonelli, M. et al. The medical segmentation decathlon. Nature Communications13, 4128 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J. & Maier-Hein, K. H. nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods18, 203–211 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Murphy, S. N. et al. High throughput tools to access images from clinical archives for research. Journal of Digital Imaging28, 194–204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Provenzale, J. M., Liang, L., DeLong, D. & White, L. E. Diffusion tensor imaging assessment of brain white matter maturation during the first postnatal year. American Journal of Roentgenology189, 476–486 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Dudink, J. et al. Fractional anisotropy in white matter tracts of very-low-birth-weight infants. Pediatric Radiology37, 1216–1223 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forman, C., Wetzl, J., Hayes, C. & Schmidt, M. Compressed sensing: a paradigm shift in mri. MAGNETOM Flash66, 9–13 (2016). [Google Scholar]
- 33.Tustison, N. J. et al. N4ITK: improved n3 bias correction. IEEE Transactions on Medical Imaging29, 1310–1320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou, Y. et al. Field of view normalization in multi-site brain mri. Neuroinformatics16, 431–444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ou, Y. et al. Brain extraction in pediatric adc maps, toward characterizing neuro-development in multi-platform and multi-institution clinical images. NeuroImage122, 246–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ou, Y. et al. Using clinically acquired mri to construct age-specific adc atlases: Quantifying spatiotemporal adc changes from birth to 6-year old. Human Brain Mapping38, 3052–3068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ou, Y., Sotiras, A., Paragios, N. & Davatzikos, C. Dramms: Deformable registration via attribute matching and mutual-saliency weighting. Medical Image Analysis15, 622–639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou, Y., Akbari, H., Bilello, M., Da, X. & Davatzikos, C. Comparative evaluation of registration algorithms in different brain databases with varying difficulty: results and insights. IEEE Transactions on Medical Imaging33, 2039–2065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinto, A. L., Ou, Y., Sahin, M. & Grant, P. E. Quantitative apparent diffusion coefficient mapping may predict seizure onset in children with sturge-weber syndrome. Pediatric Neurology84, 32–38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He, S. et al. Multi-channel attention-fusion neural network for brain age estimation: accuracy, generality, and interpretation with 16,705 healthy mris across lifespan. Medical Image Analysis72, 102091 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He, S., Grant, P. E. & Ou, Y. Global-local transformer for brain age estimation. IEEE Transactions on Medical Imaging41, 213–224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He, S., Feng, Y., Grant, P. E. & Ou, Y. Deep relation learning for regression and its application to brain age estimation. IEEE Transactions on Medical Imaging41, 2304–2317 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khoury, J. E. et al. Maternal childhood maltreatment is associated with lower infant gray matter volume and amygdala volume during the first two years of life. Biological psychiatry global open science2, 440–449 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Douglas-Escobar, M. & Weiss, M. D. Hypoxic-ischemic encephalopathy: a review for the clinician. JAMA Pediatrics169, 397–403 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Wei, R. et al. Prediction of poor outcome after hypoxic-ischemic brain injury by diffusion-weighted imaging: A systematic review and meta-analysis. Plos One14, e0226295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liauw, L., van Wezel-Meijler, G., Veen, S., Van Buchem, M. & van der Grond, J. Do apparent diffusion coefficient measurements predict outcome in children with neonatal hypoxic-ischemic encephalopathy? American Journal of Neuroradiology30, 264–270 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sotardi, S. et al. Voxelwise and regional brain apparent diffusion coefficient changes on mri from birth to 6 years of age. Radiology298, 415 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, L., Wolterink, J. M., Brune, C. & Veldhuis, R. N. Anatomy-aided deep learning for medical image segmentation: a review. Physics in Medicine & Biology66, 11TR01 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Bao, R., Ou, Y. & Grant, E. BOston Neonatal Brain Injury Data for Hypoxic Ischemic Encephalopathy (BONBID-HIE): I. MRI and Manual Lesion Annotation Version V3. Zenodo, https://zenodo.org/records/10602767 (2024).
- 50.Imanishi, T. et al. Brain injury following mild hypoxic-ischemic encephalopathy in neonates–ten-year experience in a tertiary perinatal center. Journal of Perinatology 1–7 (2022). [DOI] [PubMed]
- 51.Li, Y. et al. Mild hypoxic-ischemic encephalopathy (HIE): Timing and pattern of mri brain injury. Pediatric Research 1–6 (2022). [DOI] [PMC free article] [PubMed]
- 52.Chalak, L., Latremouille, S., Mir, I., Sánchez, P. J. & Sant’Anna, G. A review of the conundrum of mild hypoxic-ischemic encephalopathy: Current challenges and moving forward. Early Human Development120, 88–94 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Barkovich, A. J. et al. Prediction of neuromotor outcome in perinatal asphyxia: evaluation of mr scoring systems. American Journal of Neuroradiology19, 143–149 (1998). [PMC free article] [PubMed] [Google Scholar]
- 54.Trivedi, S. B. et al. A validated clinical mri injury scoring system in neonatal hypoxic-ischemic encephalopathy. Pediatric Radiology47, 1491–1499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunt, R. W., Neil, J. J., Coleman, L. T., Kean, M. J. & Inder, T. E. Apparent diffusion coefficient in the posterior limb of the internal capsule predicts outcome after perinatal asphyxia. Pediatrics114, 999–1003 (2004). [DOI] [PubMed] [Google Scholar]
- 56.van der Aa, N. E., Benders, M. J., Vincken, K. L., Groenendaal, F. & de Vries, L. S. The course of apparent diffusion coefficient values following perinatal arterial ischemic stroke. PLoS One8, e56784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He, S., Feng, Y., Grant, P. E. & Ou, Y. Segmentation ability map: Interpret deep features for medical image segmentation. Medical Image Analysis84, 102726 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kushwah, S., Kumar, A., Verma, A., Basu, S. & Kumar, A. Comparison of fractional anisotropy and apparent diffusion coefficient among hypoxic ischemic encephalopathy stages 1, 2, and 3 and with nonasphyxiated newborns in 18 areas of brain. Indian Journal of Radiology and Imaging27, 447–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shibasaki, J. et al. Comparison of predictive values of magnetic resonance biomarkers based on scan timing in neonatal encephalopathy following therapeutic hypothermia. The Journal of Pediatrics239, 101–109 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Al Amrani, F. et al. Early imaging and adverse neurodevelopmental outcome in asphyxiated newborns treated with hypothermia. Pediatric Neurology73, 20–27 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Heursen, E.-M. et al. Prognostic value of the apparent diffusion coefficient in newborns with hypoxic-ischaemic encephalopathy treated with therapeutic hypothermia. Neonatology112, 67–72 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Beneš, M. & Zitová, B. Performance evaluation of image segmentation algorithms on microscopic image data. Journal of Microscopy257, 65–85 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Reinke, A. et al. Common limitations of image processing metrics: A picture story. arXiv preprint arXiv:2104.05642 (2021).
- 64.Maier-Hein, L. et al. Metrics reloaded: recommendations for image analysis validation. Nature Methods 1–18 (2024). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No custom code was generated for this work.