Abstract
Recent advances in gene editing and precise regulation of gene expression based on CRISPR technologies have provided powerful tools for the understanding and manipulation of gene functions. Fusing RNA aptamers to the sgRNA of CRISPR can recruit cognate RNA-binding protein (RBP) effectors to target genomic sites, and the expression of sgRNA containing different RNA aptamers permit simultaneous multiplexed and multifunctional gene regulations. Here, we report an intracellular directed evolution platform for RNA aptamers against intracellularly expressed RBPs. We optimize a bacterial CRISPR-hybrid system coupled with FACS, and identified high affinity RNA aptamers orthogonal to existing aptamer-RBP pairs. Application of orthogonal aptamer-RBP pairs in multiplexed CRISPR allows effective simultaneous transcriptional activation and repression of endogenous genes in mammalian cells.
Subject terms: Synthetic biology, Functional genomics
Recent advances in gene editing and precise regulation of gene expression based on CRISPR technologies have provided powerful tools for the understanding and manipulation of gene functions. Here the authors develop an intracellular evolution platform to identify novel CRISPR-associated RNA aptamers for intracellular proteins.
Introduction
Genome editors use programmable sequence-specific nucleases1–3 to generate DNA double-strand breaks (DSBs) at predetermined sites and introduce DNA insertions/deletions by non-homologous end joining, homology-directed repair, or microhomology-mediated end joining4–7. The emergence of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas systems has accelerated the development of DSB-dependent and DSB-independent genome editing technologies8–14. CRISPR consists of two components: a CRISPR-associated nuclease (Cas, e.g., Cas9) and an artificial single guide RNA (sgRNA). Repurposing of CRISPR by inactivating the nuclease domain (e.g., catalytically dead Cas9, dCas9)15,16 and covalently tethering functional effectors to dCas9 have enabled precise gene modifications, such as base editors17–21 and prime editor9,22, epigenetic editing23–27 such as methylation28–31 and acetylation32, and gene expression regulation33–36 including activation37–40 and repression41,42. Besides modifying dCas9, the sgRNA scaffold can also be adapted to accommodate incorporations of RNA aptamers43 to directly recruit RNA-binding proteins (RBP) to the target site38,44–47. Multiplexed gene regulation events in the same cell can be coordinated by expressing sgRNA scaffolds carrying orthogonal aptamers and recruiting different numbers and types of effectors to the target region44,48. Despite the great potential, there is only a limited set of mutually orthogonal aptamer-RBP pairs that function intracellularly, posing a significant constraint on the utility of multiplexed CRISPR for simultaneous multifunctional genome editing and transcriptional modulation.
RNA aptamers with specificity and high affinity are traditionally generated by a repetitive in vitro selection process termed SELEX (systematic evolution of ligands by exponential amplification)49,50. Classic SELEX protocols typically consist of four steps: target incubation, wash-off unbound sequences, elution, and amplification of the bound sequences. Although SELEX is well-established, it is considered as a time-consuming and labor-intensive method. Most importantly, in vitro selected aptamers often fail to bind to targets in their natural biological environment. To overcome this limitation, in vivo selection strategies have been proposed as an alternative to SELEX for discovering aptamers for intracellular applications. For example, Liu et al. reported a yeast three-hybrid system (Y3H)-based selection strategy to identify RNA and protein binding partners intracellularly51–54. We took inspiration from Y3H and aptamer-mediated CRISPR-dCas9 systems to develop CRISPR-hybrid to intracellularly evolve functional RNA aptamers as part of the sgRNA. In a CRISPR-hybrid system, the dCas9 binds to a promoter region upstream to a gene encoding a fluorescent reporter. The sgRNA scaffold is extended with a randomized RNA aptamer pool sequence, which is challenged to recruit the target protein that is fused to a transcriptional activator for reporter expression. Our approach implements fluorescent-activated cell-sorting (FACS) to simultaneously isolate cell populations carrying the functional aptamer from unbound species. Using this method, we identified an RNA aptamer-RBP pair orthogonal to all existing pairs when expressed in bacteria and mammalian cells. We demonstrated that simultaneous transcription activation and repression of different genes could be achieved multiplexed using multiplex sgRNAs carrying orthogonal aptamer-RBP pairs. It is noteworthy that parallel to our work, Zhang et al. recently reported another CRISPR/Cas-based screening system for RNA aptamers targeting the extracellular receptor binding domain of the spike glycoprotein of SARS-CoV-255, but this study fell short of identifying aptamers for intracellular targets.
Results
Design and validation of the CRISPR-hybrid system
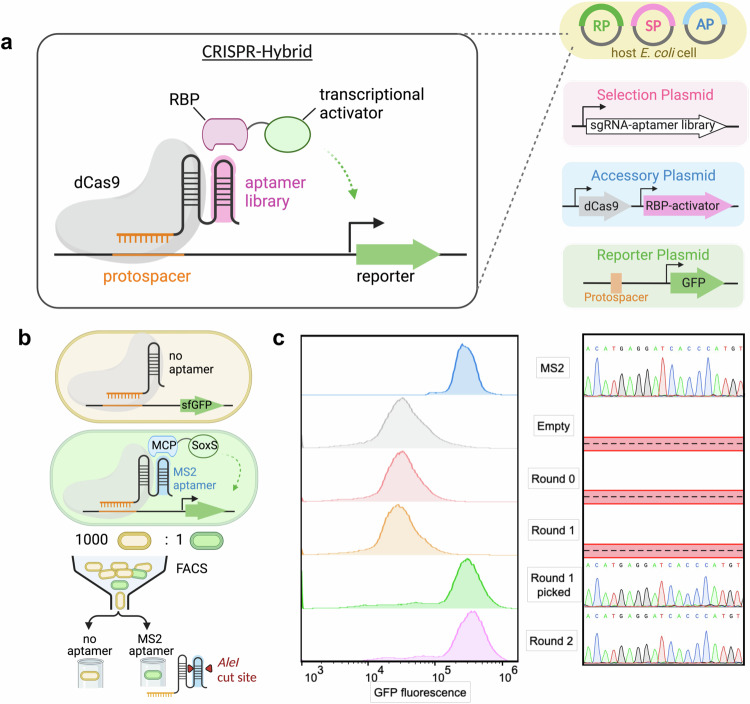
The CRISPR-hybrid system consists of four components: a dCas9 protein, a sgRNA scaffold extended with one copy of aptamer to be evolved (hereafter as the sgRNA-aptamer library chimera), an RBP-transcriptional activator fusion protein, and a gene encoding a selection marker or a fluorescent reporter (Fig. 1a). Our platform relies on the ability of the sgRNA-aptamer library chimera to recruit the target RBP to be proximal to the gene encoding the selection marker/fluorescent reporter such that its transcription is activated. We distributed these components into three plasmids (SP, AP, and RP) carrying orthogonal origins of replication and antibiotic resistance cassettes such that the individual components of the selection system can be independently varied.
Fig. 1. CRISPR-hybrid in vivo aptamer selection method.
a Overview of the selection strategy. b Schematic of the FACS-based mock selection. c FACS histogram overlays of mock populations (left), showing significant enrichment of cell population with MS2 after two rounds, and Sanger Sequencing of selection plasmids (right) in each round.
To maximize the sensitivity of the transcriptional activation, we used a known MS2- bacteriophage coat protein (MCP) aptamer-RBP pair and the sfGFP gene that encodes the super-fold GFP reporter to optimize parameters including (1) sgRNA binding site, (2) sgRNA binding site-promoter distance, (3) target RBP-transcription activator linker length, (4) protein expression level, (5) reporter gene ribosome-binding site (Supplementary Fig. 1a–e). After initial optimizations, a 5.4-fold transcription activation was achieved for the optimized construct that contains the Escherichia coli RNA polymerase omega subunit (RpoZ) as the transcriptional activator, a G6’ sgRNA binding at 16 base pairs upstream of -35 signal of the J23107 promoter, and promoter from Streptococcus pyogenes to drive dCas9 transcription.
Next, we sought to couple transcriptional activation with a reporter suitable for intracellular evolution of aptamers, and antibiotic-based survival selection was first attempted. Using the MS2-MCP pair, we introduced various antibiotic resistance genes upstream to the sfGFP gene and screened the resistance of the transformed cells to antibiotics. We identified a kanamycin-resistant gene, in which cells that contain the CRISPR-hybrid construct except the aptamer survived up to 1.0 mg/ml kanamycin, while introducing the MS2 aptamer to these cells led to survival up to 1.5 mg/ml kanamycin (Supplementary Fig. 2a). To test the efficacy of this system, we designed and executed a mock selection to enrich a MS2-incorporated sgRNA (“MS2-sgRNA”) from a sgRNA scaffold lacking any aptamers (“empty sgRNA”) at 1:1000 ratio against kanamycin. In one round, we observed ≥403-fold enrichment of the MS2 gene (Supplementary Fig. 2b). However, subsequent rounds of selection with identical or higher concentrations of kanamycin did not result in further enrichment of the MS2 gene. We attributed the lack of productive enrichment to the low degree of transcription activation and the high background caused by RpoZ-independent transcription. These results suggest that the dynamic range of the RpoZ-based construct is inadequate for an effective survival-based aptamer selection system.
To circumvent this issue, we changed the RBP-fused transcriptional activator from RpoZ to SoxSR93A, which has been shown to enhance transcription activation in CRISPR constructs45 (Supplementary Fig. 3a). Indeed, this modification resulted in a 20-fold increase in sfGFP expression after the sgRNA binding site and RNA polymerase-promoter affinity were further optimized (Supplementary Fig. 3b, c). We also confirmed that the enhanced reporter activation arose from the specific recruitment of MCP-SoxS by the MS2 aptamer (Supplementary Fig. 3d). Consistent with the improved reporter activation, cells consisting of the SoxS-based construct showed resistance to up to 5 mg/mL of kanamycin (Supplementary Fig. 4a). We repeated mock selection by challenging a 1000:1 mixture of cells containing empty sgRNA and MS2-sgRNA to activate an upstream anti-kanamycin selection marker and a downstream sfGFP reporter. Despite higher enrichment (≥583-fold) of the MS2 gene was observed (Supplementary Fig. 4b), the library still predominantly consisted of inactive variants. These results indicated that the antibiotics survival selection approach may become limited for intracellular evolution of aptamers due to the high background activation.
During the antibiotic selection, we noticed that the survived cells can be distinguished as two separate populations on sfGFP fluorescence histograms (Supplementary Fig. 4c). Cells containing MS2 exhibited higher levels of fluorescence, while cells lacking the aptamer exhibited only background levels of fluorescence. Inspired by this finding, we explored an alternative method of fluorescence-based selection using fluorescence-activated cell sorting (FACS) (Supplementary Fig. 4d–f). We also removed the anti-kanamycin selection marker upstream to the sfGFP gene to achieve more reliable transcription activation of the sfGFP gene cassette (Fig. 1b). Furthermore, cells were grown at 37 °C for 17 hours for maximal sfGFP reporter gene expression (Supplementary Fig. 5a). To maximize cell viability, the post-sorting cells were not directly amplified; instead, they were lysed after sorting and the DNA fragments encoding the enriched sgRNA-aptamer chimera were PCR amplified, followed by assembly with selection plasmid backbone and reintroduced into selection strain carrying appropriate accessory and reporter plasmids for subsequent rounds (Supplementary Fig. 5b). This strategy allows each round of CRISPR-Hybrid selection to be completed within two days, as illustrated in Supplementary Fig. 5c.
For the mock selection via FACS, we applied stringent selection pressure by sorting the top 5 out of every 100,000 cells expressing the most fluorescence, and collecting them in at least 1 mL of rich media. Gating strategy is detailed in Supplementary Fig. 6. A total of 108 cells were sorted in each round. Selection plasmids before and after the mock selection were digested with SacI and AleI, which specifically cleaves plasmid backbone and the MS2 gene, respectively, suggesting that the MS2 gene was successfully enriched by ≥700-fold after only two rounds of FACS-based mock selection (Supplementary Fig. 7a). FACS histogram overlays showed the progress of the selection with an evident appearance of a fluorescence-increased cell population after two rounds (Fig. 1c left). Twenty colonies were sequenced in each round and the MS2 gene was observed in the fluorescence-increased population (Fig. 1c right). These results validated that our designed intracellular selection platform using FACS can strongly enrich active binders from mixtures that predominantly contain inactive or less active variants.
Initial intracellular selection
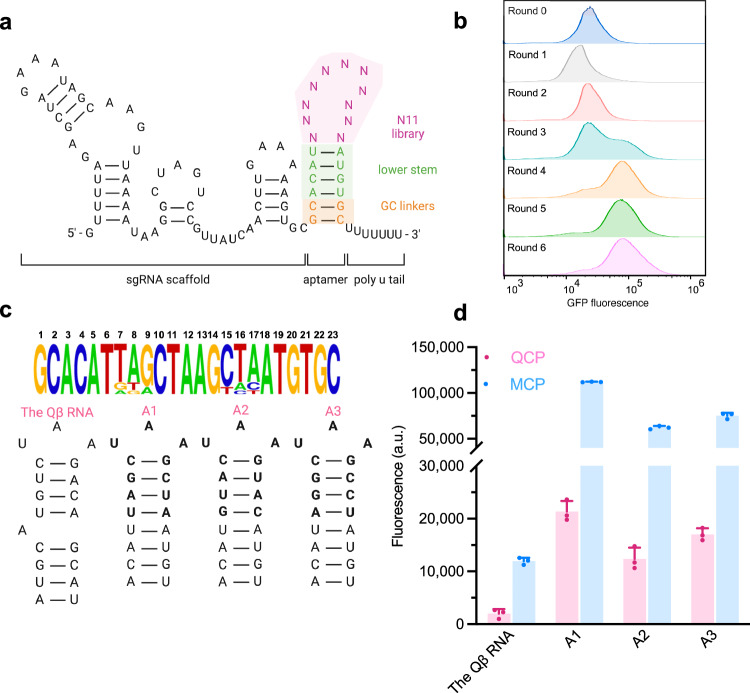
We then sought to discover aptamers for intracellularly expressed proteins using the intracellular selection strategy. To this end, we generated an aptamer library derived from the MS2 aptamer by randomizing 11 nucleotides in the upper stem and loop regions, while keeping the lower stem of MS2 intact (Fig. 2a, Supplementary Fig. 8). We reason that this design maintained the stability of the hairpin and could cause minimal disruption to sgRNA scaffold structures46. The theoretical diversity of the library is 411 = 4.2 × 106 unique sequences, and we obtained ~5 × 108 transformants, providing a diversity coverage greater than 100-fold. As a proof-of-principle, we first applied this library to the selection with the well-characterized target MCP. We screened ~109 cells on a single round of FACS. Roughly 25% of population exhibited higher fluorescence post sorting, and a second population emerged after only one round by setting the sorting gate to include top 7 cells out of every 100,000 cells (Supplementary Fig. 7b left). It is noteworthy that a less stringent sorting gate (top 45 cells out of every 100,000 cells) failed to further enrich the high fluorescent population, suggesting that a tighter sorting gate is critical to provide sufficient selection pressure necessary for a successful round. After four rounds of selection, the Sanger Sequencing revealed the most abundant sequence to be the MS2 gene, the consensus aptamer for MCP (Supplementary Fig. 7b right).
Fig. 2. Initial intracellular selection of N11 aptamer library.
a Design of N11 aptamer library. b FACS histogram overlays of six rounds selection of the aptamer library in cells expressing QCP-SoxS. c Analysis of the 1% most-abundant sequences from round 6 (n = 15) is shown above, with the relative abundance of each base at each randomized position. The secondary structures of the Qβ RNA and the top three hits for QCP are shown below. d GFP fluorescence measurements of aptamers binding to QCP and MCP. Values are mean ± s.d. (n = 3 independent replicates).
To evolve new RNA-RBP pairs, we next applied our selection system and library N11 to discover RNA aptamers for bacteriophage Qβ coat protein (QCP). QCP represents a unique RBP with an RNA binding interface distinct from that of MCP56, but shares a similar RNA binding mode57. The loop and upper region of the original consensus RNA sequence responsible for QCP binding in the Qβ phage (hereafter denoted as the Qβ RNA), adopts a conformation that is remarkably similar to that of MS2 upon protein recognition (Supplementary Fig. 9a). In addition, the +8 A of the Qβ RNA interacts with an adenosine-binding pocket in QCP56 that resembles a similar structure in MCP. It is not surprising that the Qβ RNA displayed strong binding to MCP intracellularly in CRISPR activation constructs (Supplementary Fig. 9b), although it did not display strong interactions with MCP in vitro58–60. Notably, a SELEX-selected aptamer for QCP60, Qβ-SELEX, exhibited minimal to no binding activity to QCP in E. coli cells, despite strong binding in the in vitro assays reported previously60. These results suggest that these in vitro aptamer-RBP interactions failed to translate into reliable and specific intracellular activities, and that the development of intracellularly active and aptamers for QCP that is orthogonal to existing aptamer-RBP pairs is highly desirable.
We first screened ~109 cells on FACS and collected the top 7 cells out of every 100,000 cells each round. Significant enrichment was evident after four rounds, with a distinct population displaying increased fluorescence (Fig. 2b). Further enrichments were modest in the next two rounds. We then subjected the recovered aptamer pools from each round to high-throughput DNA sequencing (HTS) analysis. Sequences are QC-filtered, counted for abundance, normalized to total number of reads per million (RPM) and ranked according to decreasing abundance. We compared the genotypic frequencies in each round of selection by plotting RPM values of one round against the immediate next round (Supplementary Fig. 10). We observed a group of sequences that is shifted below the diagonal and a large group that is shifted notably upward, indicating successful aptamer evolution. These results matched with the fluorescence histograms displaying round-to-round enrichment (Fig. 2b). The scatter plots of round 5 and 6 showed a vast majority of the sequences clustered closely to the diagonal, suggesting these two populations are highly similar, as expected from observing their corresponding fluorescence histograms (Supplementary Fig. 10, Fig. 2b). Analysis of the top 1% most-abundant sequences displayed an overrepresented sequence motif with a UAA loop and CG base pair in the first position of the upper stem, which resembled the Qβ RNA61 (Fig. 2c). We analyzed three most-abundant hits (A1 to A3), and all were unique sequences containing UAA loop, 1CG base pair, and three different base pairs in the remaining upper stem region. Validation of A1 to A3 using a GFP reporter, including the Qβ RNA as reference, showed that selected aptamers induced stronger transcriptional activation than the Qβ RNA (Fig. 2d). These results confirmed that efficient binding to QCP requires a three-nucleotide loop and four base pairs upper stem61.
Unexpectedly, A1 to A3 also exhibited strong binding preference for MCP (Fig. 2d). We hypothesize that affinity for MCP was most likely because these motifs contain the MS2 lower stem that was not randomized in the library. To dissect key sequence-activity relationships among evolved aptamers, we selected the most abundant sequence A1, and introduced nucleotide mutations in the -5 and -7 position of loop region to profile its specificity for QCP using a GFP reporter (Supplementary Fig. 11a, b). Reduced binding to both QCP and MCP were observed with all variants. The interactions between -5A variant aptamers and the bacteriophage coat proteins were weakened likely due to loss of protein contacts in the adenosine-binding pocket, which is consistent with previous findings that -5A is a crucial base for QCP binding56,58,61.
To examine the contribution of MS2 lower stem sequence to the specificity of A1, we systematically mutated each base pair to the remaining three possibilities and tested the transcriptional activation of the sfGFP reporter. We hypothesize that slight modifications in the lower stem could possibly tune A1 selectivity while preserving activity to QCP. Indeed, all the variants showed significant lower sfGFP expression with MCP-SoxS (Supplementary Fig. 11c), while the expression of sfGFP with QCP-SoxS remained constant except for variants 2GC, 2CG, 2UA, and 3 AU, which showed lower sfGFP expression with QCP-SoxS. Further rational mutation included addition of a bulge A between -11U and -12U to mimic the bulge nucleotide in the Qβ RNA. This variant, named as A1 + A, exhibited significant improvement in specificity, and it is the only variant displaying a stronger transcriptional activation with QCP-SoxS over MCP-SoxS. Furthermore, co-crystal structure of QCP and the Qβ RNA showed that a flexible loop region from Q54 to N61 in QCP interacts with the lower stem of the Qβ RNA. This loop is longer and differs in sequence from that of MCP56,62. These findings, together with the results of rational mutation, suggest that an additional selection focusing on the lower stem of the aptamer may provide an opportunity to differentiate the binding of the aptamer to QCP from MCP and improve specificity.
Second intracellular selection
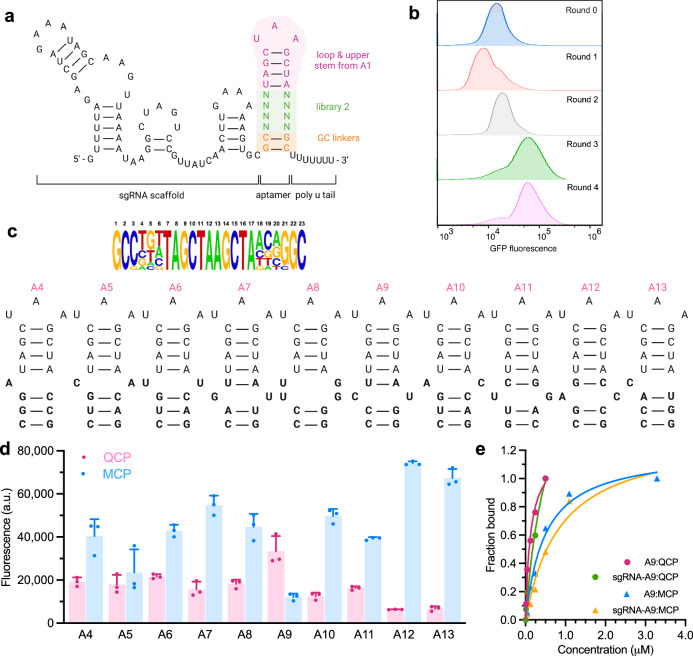
As a starting point of the second intracellular selection toward QCP binding, we randomized eight bases in the lower stem of the first-generation aptamer A1. Transformation of cells with this library produced 5 × 108 transformants, ensuring a ~ 104-fold coverage of the library with a theoretical diversity of 6.5 × 104 (Fig. 3a). Four rounds of selections were completed using FACS (Fig. 3b). Sequence pools recovered from each round of sorting were analyzed with HTS. The sequences in Round 3 and 4 were distributed along the diagonal of the scattered plot (Supplementary Fig. 12), suggesting that these two populations are highly similar. In contrast to the first library, an overrepresented sequence was not observed in the second selection (Fig. 3c). A large number of sequences were present in comparable abundances (Supplementary Fig. 12); therefore, we analyzed sequences in the top 10 most-abundant hits of rounds 3 and 4, A4-A13, and observed enrichment of bulge bases in the first and second positions of the lower stem. Of the ten unique sequences, three contained single bulged nucleotide, and three contained a mismatched base pair on the first position of the lower stem. Three sequences contained a mismatched base pair on the second position (Fig. 3c). No sequence contained more than one mismatched base pair. We profiled the activity of A4 to A13 with QCP-SoxS and MCP-SoxS to see if they favor their respective target QCP over MCP. The specificity profile of all the motifs exhibited lower transcriptional activation with the off-target MCP-SoxS than A1 (Fig. 3d). Notably, aptamer A9 exhibited significantly higher specificity for QCP versus MCP. A9 contains unpaired CU bases occupying the second position of the lower stem, followed by two CG base pairs stabilizing the formation and rigidity of the aptamer. Binding affinities characterized by surface plasmon resonance (SPR) confirmed that A9 binds strongly to QCP with a dissociation constant (KD) of 10.2 nM, compared to a weaker affinity to MCP KD of 95.1 nM (Fig. 3e, Supplementary Fig. 14). A9 extended with sgRNA scaffold exhibited even higher binding affinity and specificity to QCP (KD = 6.21 nM) compared to MCP (KD = 132 nM) (Fig. 3e), further validating the selectivity of A9 for QCP over MCP observed intracellularly. These results imply that the A9-QCP pair and the MS2-MCP pair may be orthogonal in E. coli cells. To test their orthogonality, we designed the sgRNA-A9 chimera to activate an sfGFP reporter, while sgRNA-MS2 chimera activated an RFP reporter simultaneously, when either QCP-SoxS or MCP-SoxS was expressed in the cells. Indeed, we only observed negligible cross-binding activities as specific reporters were activated with corresponding aptamer-RBP pair (Supplementary Fig. 13).
Fig. 3. Second intracellular selection of N8 aptamer library.
a Design of N8 aptamer library. b Overlaid histograms show aptamer library was fully enriched after four rounds of FACS for QCP. c Analysis of 1% most-abundant sequences from round 4 (n = 15) is shown, with the relative abundance of each base at each randomized position. The secondary structures of top 10 hits for QCP are also shown. d Fluorescence measurements of top ten aptamers binding to QCP and MCP. e SPR characterizations of A9 and sgRNA-A9 binding to QCP and MCP. All values are mean ± s.d. (n = 3 independent replicates).
Characterizations of QCP aptamer A9
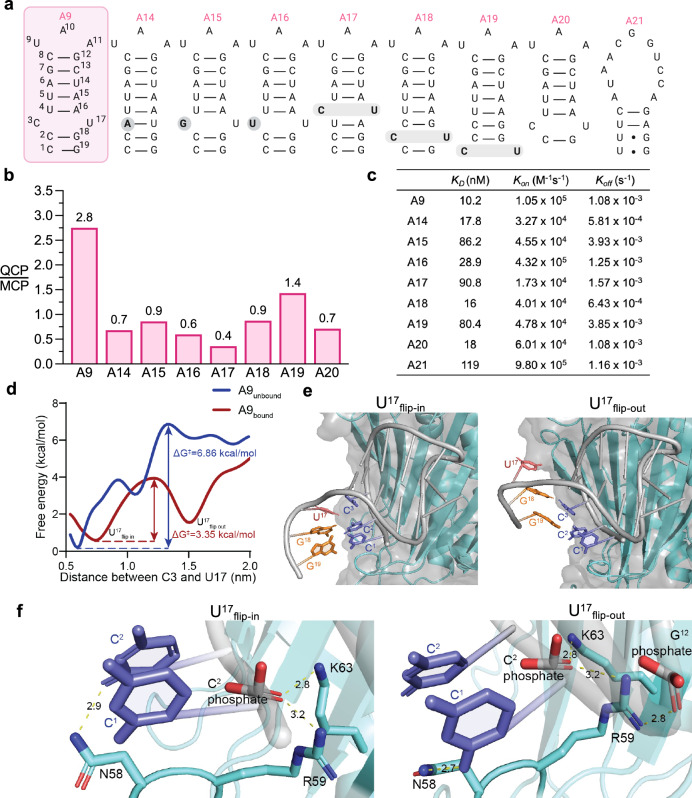
We hypothesize that the unpaired C3-U17 bulge in the lower stem of A9 is critical for protein recognition and specificity. To analyze the importance of sequence identity and position of the unpaired CU in relation to the binding affinity and the intracellular activity of A9, we generated variants A14 to A20 by systematically mutating the unpaired CU bulge of A9 (Fig. 4a). Profiling the transcriptional activation of the sfGFP reporter in E. coli showed that A14 to A20 all had markedly reduced specificity for QCP over MCP compared to A9 (Fig. 4b). Consistently, A14 (KD = 17.8 nM), A16 (KD = 28.9 nM), and A18 (KD = 16.0 nM) were found to have lower in vitro affinities to QCP than A9 (Fig. 4c, Supplementary Fig. 14). In addition, A15 (KD = 86.2 nM), A17 (KD = 90.8 nM), and A19 (KD = 80.4 nM) exhibited significantly lower in vitro affinities to QCP than A9 (Fig. 4c, Supplementary Fig. 14). Interestingly, A20, a variant of A9 with 1 bp truncated at the bottom of the lower stem, also had a reduced affinity (KD = 18 nM) to QCP, suggesting that A9 is the optimal binding sequence (Fig. 4c). Finally, A21, an A9-scrambled sequence, exhibited a weak in vitro affinity to QCP (KD = 119 nM) by SPR (Fig. 4c), but showed no activity in the transcriptional activation assay (Supplementary Fig. 15).
Fig. 4. Structure-activity characterizations of the QCP aptamer A9.
a Secondary structures of the A9 and its variants. b Selectivity of A9 variants with rationally designed mutations including removal of bulge nucleotide, different bulge sequence identities, and varied unpaired CU positions. The binding activities of these variants are measured using a fluorescent reporter and plotted as ratios of QCP over MCP. c Binding properties of A9 variants and QCP by SPR. d Free energy profile of A9 U17 nucleobase flipping out of the secondary structure, with ΔG‡ = 6.86 kcal/mol in free solution and ΔG‡ = 3.35 kcal/mol when A9 is bound to the protein. e Selected structures of A9 bound to QCP from metadynamics simulations. Left, structure of A9-QCP complex when U17 flipped in. Right, the structure of A9-QCP complex when U17 flipped out. f Residues interactions and distance in selected structures with U17 flipped in and out. Left, when U17 flips in, N58 forms a hydrogen bond with C2, R59 and K63 interact with C2 phosphate. Right, when U17 flips out, N58 forms a hydrogen bond with the exposed C1, R59 is stabilized by interactions with both C2 and G12 phosphate groups. K63 interacts with C2 phosphate.
It is noteworthy that the alterations of the unpaired C-U bulge were found to mainly impact kon: variants containing changes to the unpaired C-U bulge all showed lower kon in the SPR assay (Fig. 4c). The high kon suggests that the unpaired C-U bulge of A9 promotes rapid association with QCP.
To better understand why A9 was capable of strongly and selectively binding to QCP, we conducted a computational study of the A9-QCP interaction. Our initial hypothesis proposed that the C-U mismatch created a bulge in the RNA secondary structure, allowing nucleobase C3 to establish an aromatic ring-stacking interaction with Y89 of QCP like the bulged A nucleobase in the Qβ RNA from PDB ID 4L8H. However, this interaction could not fully explain the increased activity and selectivity over the Qβ RNA since it already exists in the Qβ RNA, which has low QCP selectivity. Thus, unique structural features of A9 was likely causing additional nucleobase-QCP interactions that contribute to sequence-based selectivity observed in the experiments.
While initial modeling predictions of the secondary structure of A9 did indeed predict a C-U bulge due to base mismatch, the energetic benefit of maintaining hydrophobic π-π stacking in the RNA structure outweighed the penalty of lacking proper hydrogen bonding between C and U bases (Supplementary Fig. 16a-b). We then conducted molecular dynamics (MD) simulations of A9 alone, as well as A9 docked into the QCP. Metadynamics, which was used to better sample configurations along the distance between the mismatched C and U bases, revealed a key difference63,64.
The lowest energy structure of the A9-QCP complex was captured when C3 and U17 of A9 remained stacked within the RNA backbone at a distance of 0.74 nm (Fig. 4d). In this conformation, N58, R59 and K63 of QCP interact with A9. Specifically, N58 forms a hydrogen bond with C2 of A9, while R59 and K63 interact with the C2 phosphate in the backbone (Fig. 4e left Fig. 4f left). However, as the distance between C3 and U17 of A9 increased, QCP-bound A9 reached another stable conformation with U17 flipped out and a C3-U17 distance of 1.5 nm, after overcoming a modest energy barrier of ΔG‡ = 3.35 kcal/mol (Fig. 4d). In this alternative conformation, U17 flips out of the RNA stem, allowing C3 to base pair with the lower G18, and C2 to interact with G19. This shift leaves C1 exposed, forming a hydrogen bond with N58 in the aforementioned flexible loop in QCP. R59 is now stabilized by interactions with both C2 phosphate and G12 phosphate groups. K63 retains its interaction with C2 phosphate group and remains unchanged between the flip-in and flip-out conformations (Fig. 4e right Fig. 4f right). To validate the simulation results, we generated three single-point mutants (N58A, R59A, and K63A) and one double mutant (N58A/R59A) of QCP. Functional assay assessing their ability to activate the sfGFP reporter in E. coli revealed reduced activity for all mutants, confirming the essential role of these residues in A9 binding (Supplementary Fig. 16c).
In contrast, when A9 is not bound to QCP, the activation energy required for U17 to flip from an “in” to an “out” conformation is significantly higher (ΔG‡ = 6.86 kcal/mol) (Fig. 4d). The decrease in the energy barrier upon A9 binding to QCP highlights the structural flexibility and adaptability of A9 in its interaction with QCP, helping to explain its high kon upon binding to QCP.
Orthogonal RNA-protein recruitment in mammalian cells
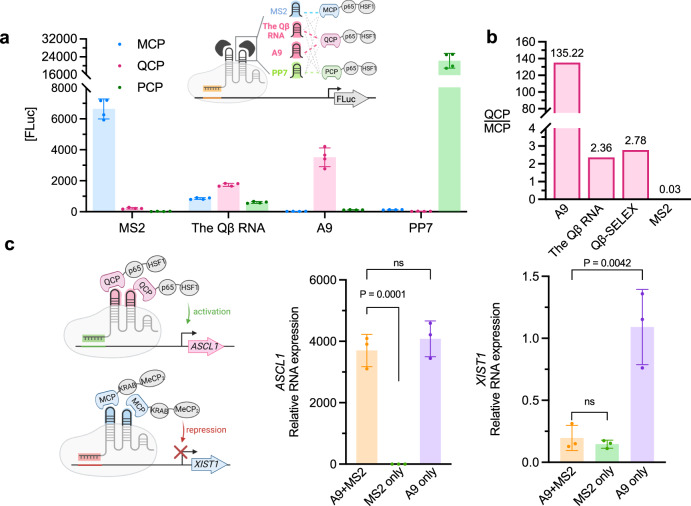
Orthogonal aptamer-protein interactions in multiplexed CRISPR genome editing are critical for reducing crossbinding between components and minimizing off-target effects. To determine whether there is crossbinding between aptamers and non-cognate proteins in multiplexed CRISPR genome editing, we expressed three previously reported consensus RBP-recognizing RNA sequences (MS2, the Qβ RNA, and PP7) and A9 in HEK293T mammalian cells containing either MCP, QCP or bacteriophage PP7 coat protein65 (PCP) fused with mammalian transcriptional activators P65 and HSF1 for luciferase reporter ([FLuc]) activation (Fig. 5a). No crossbinding was detected between PP7 and non-cognate proteins. While no crossbinding was found between MS2 and PCP, modest crossbinding was observed between MS2 and QCP. In contrast, significant crossbinding was found between the Qβ RNA and MCP and PCP, suggesting that this previously reported QCP aptamer is not orthogonal to other aptamer-RBP pairs when applied in vivo. Intriguingly, A9 mediated robust [FLuc] activation with QCP over MCP by 135-fold (Fig. 5a, b), indicating that this RNA aptamer enriched through our intracellular selection method exhibits high specificity for the cognate target in mammalian cells. In contrast, the Qβ RNA, as well as Qβ-SELEX, exhibited significantly lower activation and selectivity of less than 3-fold (Fig. 5b). The strong activation of the reporter gene by orthogonal modules (MS2-MCP, A9-QCP, and PP7-PCP) demonstrates the potential for simultaneous, locus-specific, independent transcriptional regulation of multiple genes.
Fig. 5. Orthogonal RNA-protein recruitments in HEK293T cells.
a No significant crossbinding is observed for MS2, PP7 and CRISPR-hybrid aptamer A9 with non-cognate RBPs, while evident crossbinding is observed for the Qβ RNA (n = 4 technical replicates). b A9 is highly specific for cognate target QCP over MCP, compared to the Qβ RNA and the MS2 aptamer, and an in vitro-selected aptamer Qβ-SELEX. Binding activities are measured using luciferase reporter, (n = 4 technical replicates), and measurements are plotted in ratios of QCP over MCP, as well as MCP over QCP. c sgRNA-aptamer chimeras mediated simultaneously activation and repression at endogenous genes in HEK293T cells, measured by RT-qPCR. Fold of changes are calculated over cells missing both A9 and MS2 sgRNA chimeras. (n = 3 independent replicates). Values in a and c are mean ± s.d. ns, not significant. P values were determined by one-tailed paired Student’s t-test.
Multifunctional transcriptional modulation in human cells
Next, we sought to explore the application of A9 in aptamer-mediated, multifunctional CRISPR transcriptional control in human cells. We designed multiplex sgRNA-A9 chimera targeting the ASCL1 gene for activation with QCP-p65-HSF1, while another sgRNA-MS2 chimera simultaneously targeting the XIST1 gene for repression with orthogonal MCP-KRAB-MeCP2 in HEK293T cells. We observed simultaneous significant activation of ASCL1 (3701-fold) and 88% repression of XIST1 measured by RT-qPCR. These folds of change are similar to those observed when single genes are targeted by the respective activation or repression construct (Fig. 5c). Therefore, thanks to its orthogonality to existing aptamer-RBP pairs, the A9-QCP pair could be utilized in multiplexed CRISPR for simultaneous multifunctional modulation of endogenous genes.
Discussion
Over the last ten years, a wide range of CRISPR technologies have emerged for gene manipulation, epigenetic functionalization, and transcriptional regulation. Among them, fusing effector proteins directly to the Cas protein allows the resulting CRISPR machinery to direct these effector proteins to multiple sites of the same gene or multiple genes at once35,37,40. Although they can be used to target multiple genetic loci simultaneously, these methods are often limited to applying one regulatory function (e.g., activation or repression) at a time. On the other hand, recruiting effector proteins via aptamer-RBP recognition enabled multiplexed and multifunctional gene manipulations44,48. However, there are only a limited set of aptamer-RBP pairs that can function orthogonally and intracellularly, e.g., MS2-MCP and PP7-PCP. The scarcity of orthogonal intracellular aptamer-RBP pairs imposes severe constraints on the CRISPR-mediated multifunctional manipulations of the genome and the epigenome. This work has expanded the scope of aptamer-RBP toolkit for CRISPR transcription regulators by establishing an intracellular directed evolution platform for sgRNA-fused aptamers. Applying this selection strategy, we successfully identified A9, a highly specific aptamer for QCP with an in vitro KD of 10 nM.
A major challenge with implementing aptamers identified by SELEX for in vivo applications is that the aptamers enriched from selections in simple buffers in vitro often fail to function properly in vivo, as a result of the non-specific interactions in the highly crowded and complex intracellular environment. For example, we found the original binding RNA sequence of QCP, the Qβ RNA, or the existing QCP aptamer discovered by in vitro SELEX, Qβ-SELEX, exhibited low intracellular activity and low crossbinding with other proteins (Fig. 5a). To overcome this challenge, herein we directly evolve aptamers intracellularly using CRISPR-hybrid. Recognition of aptamer library by RBP of interest in a CRISPR-hybrid construct recruited the transcriptional activator and enhanced the expression of a GFP reporter, allowing cells carrying the functional aptamer hits to be enriched using FACS. We performed a mock selection with 1:1000 mixtures of cells carrying functional and non-functional aptamers, and observed >960-fold enrichment after two rounds of sorting. When applied to intracellular selection of aptamers from a diverse (>106) library, the CRISPR-hybrid construct successfully enriched new aptamer sequences targeting bacteriophage coat proteins.
Orthogonality of the aptamer-RBP pairs is key to enable multiplexed and multifunctional manipulations of the genome and the epigenome. To improve the specificity of the evolved aptamer, we performed two intracellular selections of RNA libraries that originated from the MS2 aptamer and randomized the upper stem/loop region and the lower stem region, respectively. After the second selection, we observed top ten hits displayed significant lower crossbinding to MCP; and notably, we successfully obtained an aptamer, A9, that displays a stronger binding activity and preference to QCP than MCP. In vitro SPR affinity characterization confirmed the specificity of A9 to QCP. Importantly, although A9 exhibited modest affinity to MCP in vitro, it showed no crossbinding to MCP and PCP in a CRISPR-hybrid construct in mammalian cells, suggesting that the A9-QCP pair is orthogonal to other known aptamer-RBP pairs in genome editing applications in vivo. To demonstrate the utility of orthogonal aptamer-RBP pairs in multiplexed CRISPR, we designed sgRNAs carrying A9 and MS2 to simultaneously activate and repress endogenous genes through the recruitment of QCP-p65-HSF1 activator and MCP-KRAB-MeCP2, respectively. Simultaneous transcriptional activation and repression at respective targeted genes was successfully achieved.
Beyond bacteriophage coat proteins, the intracellular selection system developed in this work can be used to discover aptamers capable of recruiting endogenous effector proteins, such as transcription factors, epigenetic editors and readers, kinases and phosphatases, DNA/RNA repair enzymes, translocation regulators, etc. Doing so would eliminate the need for the delivery of exogenous RBP-effector fusion proteins and allow CRISPR genomic and epigenomic editing system to recapitulate the endogenous processes and pathways more accurately. Furthermore, compared to genes encoding the exogenous fusion, DNA elements encoding aptamers are much smaller and easier to deliver by size-sensitive vectors such as adeno-associated virus (AAV).
Despite these advantages, an intracellular aptamer selection system also has its own intrinsic limitations. As an example, unless high-frequency mutation to the evolving aptamer library is generated within the cell itself, the diversity of the library is limited by the transformation efficiency (108–109 in E. coli). While such a library diversity could be sufficient for some protein targets (e.g., QCP), targeting large intracellular proteins may require a greater number of randomized bases and higher library diversity. In such cases, in vitro pre-enrichment of the initial library using purified proteins via SELEX may become necessary to predispose the library for the desired target and productively reduce library diversity prior to the intracellular selection procedure.
In summary, we demonstrated that the intracellular aptamer evolution platform based on the CRISPR-hybrid construct is a promising strategy for discovering highly specific aptamers functional in a wide range of CRISPR-related technologies and other intracellular applications. The intracellular evolved aptamers are particularly suited for multiplexed and multifunctional editing of the genome and the epigenome. Going forward, the application of this approach to identify aptamers capable of recognizing and recruiting endogenous effector proteins will greatly expand our ability to interrogate complex networks of genes and their regulating proteins.
Methods
Molecular cloning
DNA oligos longer than 60 bases were purchased from Integrated DNA Technologies (IDT), while shorter oligos were purchased from Azenta Life Sciences. For CRISPR-hybrid selection, the MS2 aptamer in pCD061 (Addgene #113315) was removed to make pQS14.3 for aptamer library construction. Selection plasmids were modified from pCD185 (Addgene #113317). For mammalian cell assays, dCas9 was cloned from plasmid #156501 (Addgene), p65-HSF1 was cloned from plasmid #61423 (Addgene), and KRAB-MeCP2 was cloned from plasmid #167902 (Addgene). sgRNA-chimeras were modified from plasmid #61424 (Addgene). PCR was performed using Q5 High-Fidelity DNA polymerase from New England Biolabs (NEB M0491L). Genes were either synthesized as bacterial codon-optimized gBlocks fragments (IDT) or amplified by PCR from listed sources. All PCR products were purified using MinElute PCR Purification Kit (Qiagen, Inc 28004) to 30 μL final volume and quantified using a NanoDrop™ One/OneC Microvolume UV-Vis Spectrophotometer (Thermo Scientific). Gibson Assembly was performed using NEBuilder HiFi DNA Assembly Master Mix (NEB E2621L) according to the manufacturer’s instructions. The hybridized constructs were transformed into electrocompetent E. coli Turbo cells (NEB C2984H) or TOP10 cells (Invitrogen C4040-20) according to the manufacturer’s instructions. Transformants were recovered in 1 mL Super Optimal broth with Catabolite repression (S.O.C.) medium for 1 h at 37 °C, 250 r.p.m. Transformants were selected on Luria Bertani (LB) agar plates supplemented with appropriate antibiotic(s). Individual clones were inoculated in LB for overnight growth and plasmids were purified using GeneJET Plasmid Miniprep Kit (Thermo Scientific K0503) and verified by Sanger sequencing (Azenta Life Sciences).
Luciferase assay in bacterial cell
Reporter plasmid was co-transformed with accessory and selection plasmids of interest into electrocompetent S1030 cells (Addgene #105063) and recovered using Davis rich media66 (DRM). Transformants were plated onto 1.8% agar-2x YT plates supplemented with antibiotics. After overnight growth at 37°C, single colonies were incubated in 2 mL DRM supplemented with antibiotics for 16 h at 37°C, 250 r.p.m. Cultures were diluted 1000-fold in a 96-well deep well plate supplemented with antibiotics and inducers isopropyl-β-D-thiogalactoside (IPTG) (Invitrogen 15529019), or anhydrotetracycline (ATc) (Thermo Scientific J66688-MB) to induce protein expression when necessary. After growth for 5 h at 37°C, 200 µL of culture was transferred to a 96-well black wall, clear bottom plate (Costar COS3603), and the A600nm and luminescence for each well was measured on a plate reader (BioTek, Inc.). The A600nm of a well containing only media was subtracted from each sample to obtain a correct A600nm value. The raw luminescence value of each well was then divided by that well’s corrected A600nm value to obtain the luminescence value normalized to cell density. Each experiment was completed in at least biological triplicate, and the error bars shown reflect the standard deviations of the measurements. gRNA sequences are listed in Supplementary Table 1.
Flow cytometry
E. coli MG1655 K12 cells (Addgene #37854) transformed with reporter, accessory and or selection plasmids were inoculated in 3 mL LB medium supplemented with appropriate antibiotics and grown for 17 h at 37°C, 250 r.p.m. Cells were pelleted by centrifugation for 5 min at 17,900 x g, followed by one wash with phosphate-buffered saline, pH 7.4 (PBS) (Fisher Scientific BP243820). Cells were then diluted 50-fold in PBS and analyzed on Becton Dickinson Accuri C6 Plus (BD Biosciences). To enrich for single cells, a side scatter threshold trigger (SSC-H) was applied. To gate for single bacterial cells, we first selected events that appeared on the center of the FSC-A vs. SSC-A plot, then selected events along the diagonal of the FSC-H vs FSC-A plot. Events that appeared on the edges of the fluorescence histogram were excluded.
Cell culture
HEK293T cells (ATCC CRL-3216) were maintained at 37 °C and 5% CO2 in DMEM-high glucose, GlutaMAX Supplement, pyruvate (Gibco 10569010) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Corning 35010CV). All references to DMEM below refer to the complete medium described here.
Transient gene expression
HEK293T cells were plated seeded approximately 500,000 cells per well in 6-well plates and transfected the next day. HEK293T cells were co-transfected with plasmids encoding dCas9-P2A-Puro (500 ng), a mix containing equal masses of sgRNA-targeting endogenous genes, QCP-p65-HSF1 (500 ng), and MCP-KRAB-MeCP2 (500 ng). Sequences of the sgRNA are provided in Supplementary Table 2. Transfection complexes were prepared using Lipofectamine 3000 (Invitrogen L3000015), following the manufacturer’s instructions. Cells were treated with 3.5 μg/ml puromycin at 24 h post-transfection. Media with puromycin was refreshed after 24 h, and cells were collected 72 h post-transfection.
Quantitative RT-PCR
Cells were washed once with PBS, and total RNA was isolated with Trizol (Invitrogen 15596026), following the manufacturer’s protocol. cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen 18080085), according to the manufacturer’s protocol, priming from anchored oligo-dT20, random hexamers (Invitrogen 12577011). qRT-PCR was performed using Universal SYBR Green Master Mix (Abclonal RK21203) and gene-specific primers (Supplementary Table 3). cDNA template-less was used as a negative control. Bulk gene expression measurements were normalized to a GAPDH internal control (Supplementary Table 3), and fold-changes were calculated against no sgRNA control group. qPCR primers are listed in Supplementary Table 3.
Luciferase assays in mammalian cell
The sgRNA target sequence is listed in Supplementary Table 2. HEK293T cells were plated at approximately 2.0 × 104 cells per well in a 96-well plate and cultured for 24 h at 37 °C in 5% CO2. The cells were then transfected with Lipofectamine 3000 in Opti-MEM (Gibco 31985070) according to the manufacturer’s protocol. Plasmids encoding for dCas9, RBP-p65-HSF1, sgRNA and luciferase reporter were transfected at a 1:1:1:1 ratio. The total amount of DNA was 0.1 μg per well. Bioluminescence measurements were obtained 48 h post-transfection using Dual-Glo Luciferase Kit (Promega E2920) with plate reader.
Directed evolution of RNA aptamers
Library construction
The single-stranded DNA (ssDNA) libraries were synthesized by IDT, using a customized recipe (A: 25%, C: 25%, G: 25%, T: 25%) for random regions. Oligonucleotides are listed in Supplementary Table 4. ssDNA libraries were purified by 6% TBEU-Urea gel electrophoresis (Invitrogen EC6865BOX) with ZR small-RNA PAGE Recovery Kit (Zymo Research R1070), and eluted with nuclease-free water. Plasmid pQS14.3, which only contains sgRNA scaffold without aptamer insertion, was used as template to generate aptamer libraries. Piece A was amplified with primers QS94 and QSL2 or QSL5 for library N11 and N10, respectively. Piece B was amplified with primers QS91 and QS92. Primerless overlap extension PCR (OEPCR) was performed in 16 ×50 µL reactions (10% DMSO, 20 cycle number) containing 100 ng piece A and equimolar piece B. Piece C was amplified with primers QS95 and QS96. All PCR products were treated with DpnI (NEB R0176S) at 37 °C for 2 h to digest any residual template plasmid, followed by purification on 1.5% TAE-agarose gels using QIAquick gel extraction kit (Qiagen, Inc. 28704). Aptamer libraries were constructed with piece C and OEPCR product (1:4 molar ratio) by Gibson Assembly and ethanol-precipitated with yeast-tRNA (Invitrogen AM7119) to transform into electrocompetent Top10 cells. Freshly electroporated cells were recovered in 10 mL S.O.C. medium for 1 h at 37 °C, 250 r.p.m., followed by 17 h growth in 150 mL LB supplemented with carbenicillin (100 µg mL−1). More than 108 transformants were obtained to ensure library coverage. Plasmid library was isolated with the ZymoPURE II Plasmid Midiprep Kit (Zymo Research D4200) for following transformation into the appropriate MG1655 K12 (+ accessory and reporter plasmids) electrocompetent cells.
Electrocompetent strain preparation
Electrocompetent E. coli MG1655 K12 cells were transformed with reporter plasmid and accessory plasmid of interest, depending on the target RNA-binding protein. Single clones were inoculated in 10 mL LB media supplemented with chloramphenicol (25 µg mL−1) and kanamycin (50 µg mL−1) for 16 h at 37 °C, 250 r.p.m. Next day, culture was diluted 100-fold in 1 L Super Optimal Broth (SOB) media and grown under identical condition until it reached mid-log-phase (OD600 = 0.5–0.7). Cells were pelleted in three JA10 tubes centrifuged at 5000 x g for 15 min at 4 °C. The media was immediately decanted and the interior of the tubes was wiped with a few Kimwipes to remove residual media and salts. Each cell pellet was quickly resuspended in 15 mL of pre-chilled, sterile filtered 15% glycerol in MilliQ purified water using a serological pipette and combined into one tube with ~300 mL of 15% glycerol. The cells were centrifuged and washed an additional two times. After the last wash, the interior of the tube was wiped with Kimwipes to remove residual glycerol solution. The pellet with resuspended in 1.5 mL 15% glycerol and split into 50 µL aliquots, which were flash-frozen using liquid N2 bath and quickly transferred to -80 °C for storage. More than 108 transformants is usually obtained to ensure aptamer library coverage. In addition, empty electrocompetent MG1655 K12 cells prepared by this method typically yielded 109–1010 cfu per µg of plasmid DNA and enable simultaneous transformation of all three plasmids.
Antibiotic selection
Selection plasmids with and without MS2 aptamer were separately transformed into MG1655 K12 (+ accessory MCP and reporter plasmids) electrocompetent cells, and selected on LB-agar plates supplemented with appropriate antibiotics. A single colony from these two cell types was inoculated in 3 mL LB for 17 h at 37 °C, 250 r.p.m. After measuring culture’s optical density (OD600), cells were mixed in ratios of 1:1000 MS2 aptamer: no aptamer. A total of 107 cells were challenged by plating them onto a 150 mm petri dish (Fisher Scientific) containing LB-agar, plasmid maintenance antibiotics (100 µg mL−1 carbenicillin and 25 µg mL−1 chloramphenicol), and a concentration of kanamycin pre-determined to be above the MIC of the MG1655 strain lacking aptamer component of the CRISPR-hybrid system. Plates were incubated at 37 °C for 17-24 h and surviving colonies were harvested with 10 mL LB medium, centrifuged and plasmids were purified via miniprep. The enriched selection plasmids were extracted using 1% TAE-agarose gel and re-transformed into MG1655 K12 (+ accessory MCP and reporter plasmids) electrocompetent cells for next round.
Fluorescence-activated cell sorting (FACS)
For mock selection, cells with and without MS2 aptamer were mixed as described above. For library selections, plasmid library was transformed into the appropriate MG1655 K12 (+ accessory and reporter plasmids) electrocompetent cells and grown in 15 mL LB with maintenance antibiotics for 17 h at 37 °C, 250 r.p.m. Cells were harvested by centrifugation for 5 min at 4 °C, 17,900 x g. Cells were washed once and diluted 50-fold in cold PBS to analyze and sort on Becton Dickinson FACSAria cell sorter (BD Biosciences). The sorting gate was set to include library members with fluorescence higher than control, which are cells lacking the aptamer component of CRISPR-hybrid. The top 0.007% (total of 1500 cells) were collected in 1 mL S.O.C. medium. Collected cells were harvested at 17,900 x g for 5 min, and resuspended in 12 µL of PCR-Rescue Buffer (1 mM Triton X-100, 20 mM Tris-HCl, pH 8.0, 2 mM EDTA) for cell lysis. The reaction was incubated at 95 °C for 3 min and cooled to 4 °C. This reaction mixture was used as DNA template for 3 × 50 µL PCRs with primers QS91 and QS96 to amplify the region flanking the aptamer library. PCR products were subsequently purified on 1% TAE-agarose gels using QIAquick Gel Extraction Kit and eluted with 25 µL nuclease-free water. The enriched aptamers were subcloned back into plasmid backbone with the Gibson assembly protocol described above. Top10 transformed cells were grown in 15 mL LB under the same growth conditions as above. This enriched library was transformed again into the appropriate MG1655 K12 (+ accessory and reporter plasmids) electrocompetent cells and re-challenged with the selection condition.
Restriction enzyme digestion analysis
Selection plasmids or PCR products flanking the region of the aptamer library isolated from each round of mock selection were digested under the following conditions: 500 ng DNA, 1 µL of rCutSmart buffer (NEB B6004S), 1 µL of appropriate restriction enzyme (NEB), up to 10 µL of nuclease-free water. The reactions were incubated in a thermal cycler at 37 °C for 30 min, and halted by subsequent heat denaturation at 65 °C for 20 min. DNA was analyzed on a 1% TAE-agarose gel stained with ethidium bromide (Invitrogen 15585011). The relative recovery of MS2 aptamers was quantified in ImageJ (imageJ.nih.gov).
High-throughput sequencing and data processing
QS_seq18 and Qs_seq19 primers (see Supplementary Table 5 for sequences) containing adapters for Amplicon Sequencing were used for PCR amplification of enriched aptamer library plasmids. PCR products were purified by 2% TAE-agarose gel using the QIAquick gel extraction kit, and submitted to Azenta Life Sciences for Amplicon Sequencing. Processing of sequencing data was performed using Python script, available on GitHub. A Q-score filter was applied to analyze the quality scores of each read in the FASTQ files, and any reads below the specific threshold (Q-scores lower than 10) was discarded. A mismatch filter then used to compare each read with the expected sequence in the fixed regions of the sgRNA scaffold, skipping the aptamer pool region containing randomized sequences. The sequences in the randomized region were then extracted and collected in a comma-separated file, along with each read’s count. The most abundant sequences (listed in Supplementary Table 6) were individually validated by reporter assays. Aptamer structures were predicted using Mfold webserver.
Surface plasmon resonance (SPR)
RNA aptamer preparation
Aptamer containing 3’ 24-mer poly (A) sequence was purchased from IDT. RNA was purified with 8% TBE-Urea PAGE gel extraction, recovered with crush and soak method, precipitated with ethanol, washed with 70% ethanol and dissolved in nuclease-free water. Purified RNA was verified once more by alkaline hydrolysis on a TBE-Urea PAGE gel stained with SYBR Gold dye (Invitrogen S11494).
Sensor chip surface generation
Experiments were performed on a OpenSPR (Nicoya) instrument at 23 °C with HBS-EP+ (Cytiva BR100826) as the running buffer. Streptavidin (0.5 μM) was immobilized to ~800 RU in both reference (FC1) and sample (FC2) flow cells on a biotin sensor chip using the Biotin-streptavidin Sensor Kit (Nicoya).
Aptamer binding assay
Aptamers (~1.5 μM, 10 μL) were diluted in running buffer for thermal treatment of heating 95 °C for 3 min and slow cooling to 4°C at 0.1°C s−1 in the MiniAmp thermal cycler (Thermo Scientific). A dilution series of protein analytes were prepared in running buffer and filtered through a 0.2 μm membrane, using a minimum of six concentrations ranging from 0.1 x KD to 100 x KD for a more accurate estimate of the kinetic parameters. Each assay cycle includes a capture, association, dissociation, and regeneration step. The aptamer was further diluted to 0.1 μM in running buffer and captured onto FC2 for 1200 s at 5 μL/min. The target protein was injected over both FCs for 300 s at 30 μL/min, and running buffer was then injected over both FCs for 300 s to monitor target dissociation. The surface was regenerated with 25 mM of NaOH for 30 s at 30 μL/min over both FCs, which removed the captured RNA and protein. The TraceDrawer software (Nicoya) used to process the datasets and analyze interaction kinetics. The FC1 data was first subtracted from FC2 to correct for injection noise, baseline drift, nonspecific surface binding and bulk refractive index changes. This corrected data was fit to a 1:1 binding model.
Bacterial expression and purification of recombinant proteins
BL21 (DE3) competent E. coli cells (NEB C2527I) transformed with plasmid pET22b-MCP-MBP-his6 or pET22b-Qβ-MBP-his6 was inoculated in 10 mL LB medium supplemented with carbenicillin (100 µg mL−1) for overnight culture at 37 °C, 250 r.p.m. On Day 2, the culture was dilute 100-fold in 1 L LB medium supplemented with carbenicillin, and grown to OD600 of ~0.5. Protein expression was induced with 1 mM final IPTG for 16 h at 16 °C, 250 r.p.m. All purification steps were performed at 4 °C with Stock Buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 6 mM BME). Cells were harvested by centrifugation for 15 min at 6000 r.p.m, resuspended in 30 mL Lysis Buffer (Stock Buffer + 10 mM imidazole), and lysed by sonication (5 cycles of 20 s pulse-on, 1 min pulse-off at amplitude 80%). Lysate was homogenized by centrifugation at 14,000 rcf for 30 min. The clarified lysate was incubated with 1.5 mL Ni-NTA resin (Takara 635677) for 1 h with gentle rotation and subsequently applied to an Econo-PacTM chromatography column (Bio-Rad 7321010). The protein-bound resin was washed with 25 mL Wash Buffer (Stock Buffer + 50 mM imidazole), and His-tagged protein was eluted with 16 mL of Elution Buffer 1 (Stock Buffer + 100 mM imidazole) and 8 mL of Elution Buffer 2 (Stock Buffer + 200 mM imidazole). Fractions of 2 mL were collected and analyzed on 4-12% Bis-Tris PAGE gel (Invitrogen NP0323BOX) with NuPAGETM MES SDS running buffer (Invitrogen NP000202) and Coomassie staining. Fractions without impurifies were pooled, buffer exchanged into Dialysis Buffer (Stock Buffer + 5% glycerol), and concentrated with a Amicon Ultra-15 Centrifugal Filter (MilliporeSigma UFC901024), molecular weight cutoff of 10 kDa, at 14,000 rcf for 15 min. Protein concentration was measured by Pierce™ Coomassie (Bradford) Protein Assay Kit (Thermo Scientific A55866). Aliquots were flash-frozen at -80 °C for storage or used directly for in vitro binding assay.
Computational methods
RNA-protein docking
Qβ coat protein was extracted from PDB ID 4L8H, with missing residues added using the GalaxyFill algorithm67. A9 RNA was modeled using FARFAR from Rosetta, with the initial guess for the secondary structure as a C-U bulge and UAA hairpin and all other residues engaged in traditional hydrogen bonding68. Docking was conducted using LightDock, with both protein and RNA treated as rigid69.
Molecular dynamics (MD)
Explicit solvent simulations involved the A9 RNA alone as well as the A9 RNA in complex with the Qβ coat protein. For each, a box of TIP3P water70 was created to simulate the solvent. Zn2+ ions were added to neutralize the charge of the system for Particle-Mesh Ewald (PME) electrostatics71. For equilibration, each structure was minimized and then allowed to couple to a Langevin-dynamics controlled thermostat at 300 K for 1 ns, followed by another 1 ns coupling to a MonteCarlo barostat to maintain a pressure of 1 bar72. Position restraints of 100 kJ/mol/nm for the temperature and pressure coupling, and incrementally lowered. Each lowering of the restraints involved a 1 ns simulation. We then allowed each system to equilibrate for 500 ns. To ensure simulation convergence, the RMSF of RNA/protein atoms were computed and ensured they remained the same between halves of the simulations.
Metadynamics
Well-tempered metadynamics employs a biasing force on a selected collective variable (CV) to sample configurations along the CV that are energetically inaccessible in the timeframe of a normal MD run63. The biasing force can be accounted for and thus the free energy change of the process of interest can be calculated. Our selected CV was the distance between the C3 and U17, constructed by a centroid bond force between the average coordinate of the side chains of the two RNA residues. This biasing force was set to vary this distance from 0.5 nm to 2.0 nm, effectively allowing the bases to remain in the secondary structure or pop out into the solvent. A hill height of 0.3 kJ/mol was used, with a grid width of 0.1 nm. The bias potential was updated every 200 ps, with a bias factor of 1064.
Statistical methods
For evaluating reporter expression in bacteria and HEK293T cells, the mean of at least three independent replicated was reported, and error bars represent the standard deviation.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
The authors thank Mitchel Tepe and Zeiyi Huang for feedback on Python scripts for HTS analysis, Dr. Mi Zhou for assistance with mammalian cell culture, Weiqi Qiu for assistance with in vitro transcription, Dr. Yang Wang for discussions on metadynamics, and Professor Abhishek Chatterjee for the helpful discussion. U.M. and M.G. also acknowledge the support of Andromeda computing cluster at Boston College. This work was supported by NIH Director’s New Innovator Award 1DP2HG011027-01 (J.N.) and National Science Foundation Graduate Research Fellowship Program (Q.S.-T.). U.M. is a John Simon Guggenheim Memorial Foundation Fellow. Illustrations were created in BioRender. Su, Q. (2024) https://BioRender.com/x75w983.
Author contributions
Q.S.-T. and J.N. designed the project and prepared the manuscript. J.N. supervised the research. Q.S.-T. conducted the experiments. J.F. completed in vitro studies of A9 variants and in vivo measurements of A9 with QCP mutants. K.F. and P.W. assisted with flow cytometry measurements and cell culture. K.F., P.W. and Y.H. assisted on molecular cloning and protein purification. H.R. edited the Python scripts. P.A. conducted FACS. M.G. and U.M. designed the MD simulations and analyzed the well-tempered metadynamics-enhanced sampling data. M.G. carried out the MD simulations.
Peer review
Peer review information
Nature Communications thanks James Carothers, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided with this paper. The NGS datasets for intracellular selections with N8 and N11 libraries have been deposited in NCBI Sequence Read Archive under accession code PRJNA1206477. Source data are provided with this paper.
Code availability
All scripts used in this manuscript are available on GitHub at https://github.com/JNiulab/CRISPR-hybrid.git.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-55957-0.
References
- 1.Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science326, 1509–1512 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Christian, M. et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics186, 757–761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim, Y. G., Cha, J. & Chandrasegaran, S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. Usa.93, 1156–1160 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibikova, M., Golic, M., Golic, K. G. & Carroll, D. Targeted chromosomal cleavage and mutagenesis in drosophila using Zinc-finger nucleases. Genetics161, 1169–1175 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rouet, P., Smih, F. & Jasin, M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA91, 6064–6068 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, H., La Russa, M. & Qi, L. S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem.85, 227–264 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Cheng, C., Zhou, M., Su, Q., Steigmeyer, A. & Niu, J. Genome editor-directed in vivo library diversification. Cell. Chem. Biol.28, 1109–1118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strecker, J. et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science365, 48–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, S. W., Kim, S., Kim, J. M. & Kim, J.-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol.31, 230–232 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA.109, E2579–E2586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinek, M. et al. A Programmable Dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mali, P. et al. RNA-guided human genome engineering via Cas9. Science339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek, M. et al. RNA-programmed genome editing in human cells. Elife2, e00471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess, G. T. et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods13, 1036–1042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science353, aaf8729 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, Y. et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods13, 1029–1035 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Choi, J. et al. Precise genomic deletions using paired prime editing. Nat. Biotechnol.40, 218–226 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, X.-M., Zhou, J., Mao, Y., Ji, Q. & Qian, S.-B. Programmable RNA N6-methyladenosine editing by CRISPR-Cas9 conjugates. Nat. Chem. Biol.15, 865–871 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson, C., Chen, P. J., Miao, Z. & Liu, D. R. Programmable m6A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat. Biotechnol.38, 1431–1440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakore, P. I., Black, J. B., Hilton, I. B. & Gersbach, C. A. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat. Methods13, 127–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morita, S. et al. Targeted DNA demethylation in vivo using dCas9–peptide repeat and scFv-TET1 catalytic domain fusions. Nat. Biotechnol.34, 1060–1065 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Pulecio, J., Verma, N., Mejía-Ramírez, E., Huangfu, D. & Raya, A. CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell21, 431–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amabile, A. et al. Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell167, 219–232.e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald, J. I. et al. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol. Open5, 866–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vojta, A. et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res.44, 5615–5628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, X. et al. A CRISPR-based approach for targeted DNA demethylation. Cell. Discov.2, 16009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilton, I. B. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol.33, 510–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng, A. W. et al. Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res.26, 254–257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilbert, L. A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bikard, D. et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res.41, 7429–7437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat. Methods10, 973–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng, A. W. et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res.23, 1163–1171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods12, 326–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelles, D. A. et al. Programmable RNA tracking in live vells with CRISPR/Cas9. Cell165, 488–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature500, 472–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shechner, D. M., Hacisuleyman, E., Younger, S. T. & Rinn, J. L. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods12, 664–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zalatan, J. G. et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell160, 339–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong, C., Fontana, J., Patel, A., Carothers, J. M. & Zalatan, J. G. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun.9, 2489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Briner, A. E. et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol. Cell56, 333–339 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Li, C. et al. SWISS: multiplexed orthogonal genome editing in plants with a Cas9 nickase and engineered CRISPR RNA scaffolds. Genome Biol.21, 141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Truong, V. A. et al. CRISPRai for simultaneous gene activation and inhibition to promote stem cell chondrogenesis and calvarial bone regeneration. Nucleic Acids Res.47, e74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature346, 818–822 (1990). [DOI] [PubMed] [Google Scholar]
- 50.Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science249, 505–510 (1990). [DOI] [PubMed] [Google Scholar]
- 51.Wurster, S. E. & Maher, L. J. Selection and characterization of anti-NF-κB p65 RNA aptamers. RNA14, 1037–1047 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wurster, S. E. & Maher, L. J. Selections that optimize RNA display in the yeast three-hybrid system. RNA16, 253–258 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hook, B., Bernstein, D., Zhang, B. & Wickens, M. RNA-protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA11, 227–233 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.SenGupta, D. J. et al. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl Acad. Sci. Usa.93, 8496–8501 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, J. et al. Repurposing CRISPR/Cas to Discover SARS-CoV-2 Detecting and Neutralizing Aptamers. Adv. Sci.10, 2300656 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rumnieks, J. & Tars, K. Crystal structure of the bacteriophage Qβ coat protein in complex with the RNA operator of the replicase gene. J. Mol. Biol.426, 1039–1049 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Spingola, M., Lim, F. & Peabody, D. S. Recognition of diverse RNAs by a single protein structural framework. Arch. Biochem. Biophys.405, 122–129 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Horn, W. T. et al. Structural basis of RNA binding discrimination between bacteriophages Qβ and MS2. Structure14, 487–495 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katz, N. et al. An in vivo binding assay for RNA-binding proteins based on repression of a reporter gene. ACS Synth. Biol.7, 2765–2774 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Hirao, I., Spingola, M., Peabody, D. & Ellington, A. D. The limits of specificity: An experimental analysis with RNA aptamers to MS2 coat protein variants. Mol. Divers.4, 75–89 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Witherell, G. W. & Uhlenbeck, O. C. Specific RNA binding by Q.beta. coat protein. Biochemistry28, 71–76 (1989). [DOI] [PubMed] [Google Scholar]
- 62.Golmohammadi, R., Fridborg, K., Bundule, M., Valegård, K. & Liljas, L. The crystal structure of bacteriophage Qβ at 3.5 å resolution. Structure4, 543–554 (1996). [DOI] [PubMed] [Google Scholar]
- 63.Barducci, A., Bussi, G. & Parrinello, M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys. Rev. Lett.100, 20603 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Laio, A. & Parrinello, M. Escaping free-energy minima. Proc. Natl Acad. Sci. USA.99, 12562–12566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim, F. & Peabody, D. S. RNA recognition site of PP7 coat protein. Nucleic Acids Res.30, 4138 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carlson, J. C., Badran, A. H., Guggiana-Nilo, D. A. & Liu, D. R. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol.10, 216–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coutsias, E. A., Seok, C., Jacobson, M. P. & Dill, K. A. A kinematic view of loop closure. J. Comput. Chem.25, 510–528 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Das, R., Karanicolas, J. & Baker, D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat. Methods7, 291–294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiménez-García, B., Roel-Touris, J. & Barradas-Bautista, D. The LightDock Server: Artificial Intelligence-powered modeling of macromolecular interactions. Nucleic Acids Res.51, W298–W304 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys.79, 926–935 (1983). [Google Scholar]
- 71.Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys.98, 10089–10092 (1993). [Google Scholar]
- 72.Eastman, P. et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol.13, e1005659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this paper. The NGS datasets for intracellular selections with N8 and N11 libraries have been deposited in NCBI Sequence Read Archive under accession code PRJNA1206477. Source data are provided with this paper.
All scripts used in this manuscript are available on GitHub at https://github.com/JNiulab/CRISPR-hybrid.git.