Abstract
The 12-lead electrocardiogram (ECG) is inexpensive and widely available. Whether conditions across the human disease landscape can be detected using the ECG is unclear. We developed a deep learning denoising autoencoder and systematically evaluated associations between ECG encodings and ~1,600 Phecode-based diseases in three datasets separate from model development, and meta-analyzed the results. The latent space ECG model identified associations with 645 prevalent and 606 incident Phecodes. Associations were most enriched in the circulatory (n = 140, 82% of category-specific Phecodes), respiratory (n = 53, 62%) and endocrine/metabolic (n = 73, 45%) categories, with additional associations across the phenome. The strongest ECG association was with hypertension (p < 2.2×10-308). The ECG latent space model demonstrated more associations than models using standard ECG intervals, and offered favorable discrimination of prevalent disease compared to models comprising age, sex, and race. We further demonstrate how latent space models can be used to generate disease-specific ECG waveforms and facilitate individual disease profiling.
Subject terms: Risk factors, Predictive markers
Introduction
The modern resting electrocardiogram (ECG) utilizes waveform data generated from surface electrodes to represent cardiac activation and impulse conduction1. Introduced in the early 1900s, the original ECG was primarily used for arrhythmia detection, but its diagnostic utility expanded rapidly to include the identification of coronary artery disease and other cardiac structural abnormalities2,3. It has now become clear that non-cardiac diseases, from electrolyte derangements to central nervous system pathology, also cause characteristic changes in the ECG waveform4–7.
Recent advances in machine learning have revealed that the ECG contains diagnostic and prognostic information that extends beyond traditional clinical interpretation8–11. Low-dimensional representations of ECGs constructed from deep learning models can detect cardiac diseases such as left ventricular dysfunction and paroxysmal atrial fibrillation for patients in sinus rhythm12,13. Other models have demonstrated predictive power beyond the cardiovascular system, estimating factors such as age, sex, serum potassium, and one-year mortality with high accuracy7,14–16. However, the full extent of human diseases that may become manifest on the surface ECG remains unknown.
Modern electronic health record (EHR)-based technology has made available large, detailed datasets with rich phenotypic information, enabling large-scale disease-based association testing17–19. Phenome-wide association studies (PheWAS) facilitate high-throughput association testing between predictor variables and multiple disease states using electronically ascertained diagnostic codes. Diseases are commonly represented by Phecodes, or standardized, aggregated groupings of International Classification of Disease (ICD) codes20–22.
In the present study, we sought to harness the inferential capabilities of deep learning models and the analytic power of PheWAS to comprehensively assess the array of disease states that manifest in the ECG waveform. Given the growing number of consumer-based wearable devices capable of recording single-lead ECGs, we performed parallel analyses using both traditional 12-lead ECGs and data derived only from lead I, a common vector used for consumer-based ECG recording23. Specifically, we trained deep learning models known as denoising autoencoders to encode 12- and single-lead ECGs within a latent space using a large primary care sample. We selected the autoencoder model because it is unsupervised and optimized to learn ECG waveform features alone, without additional information regarding patient demographics or clinical outcomes. We then used the position of ECG representations in the latent space to perform high-throughput association testing with roughly 1600 prevalent and incident diseases across three independent datasets spanning over 150,000 individuals. We further demonstrate how latent space modeling can be used to display characteristic ECG features for detectable conditions and to clinically profile individual patients.
Results
Study sample and autoencoder development
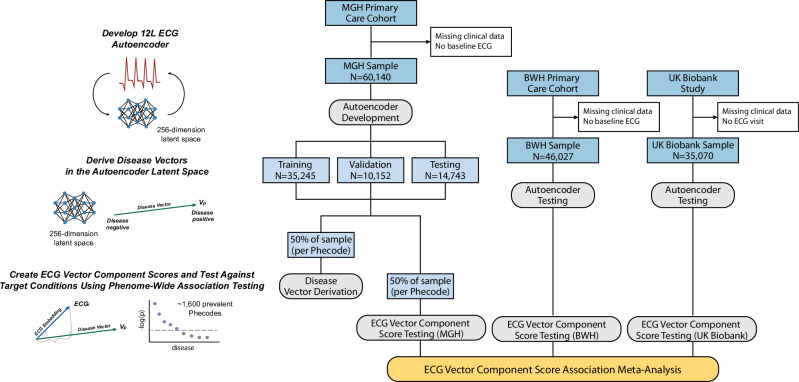
Our study utilized three nonoverlapping, independent, datasets spanning over 150,000 individuals, each of which contained individual-level demographic and clinical information, including 12-lead ECGs (Fig. 1). Two datasets were taken from the Community Care Cohort Project (C3PO), a previously established cohort comprising adults aged ≥ 18 years who received longitudinal primary care within the Mass General Brigham network between 2000–2018, which is linked by a common EHR data warehouse24. The Massachusetts General Hospital (MGH) C3PO dataset included 60,140 primary care patients and the Brigham and Women’s Hospital (BWH) C3PO dataset included 46,027 primary care patients. The third dataset comprised 35,070 participants from the UK Biobank, a prospective national biorepository that enrolled individuals aged 40-60 years between 2006–2010, with deep phenotyping data, baseline questionnaires, and linkage to electronic health record data25. Characteristics of participants by dataset are provided in Table 1.
Fig. 1. Study overview.
Flow diagram of autoencoder and phenotype vector derivation for latent space phenome-wide association studies (PheWAS), conducted in parallel for both 12-lead and single-lead electrocardiogram (ECG) models. We trained an autoencoder to encode and reconstruct 12- and single-lead ECGs using the Massachusetts General Hospital (MGH) subset of the Community Care Cohort Project (C3PO) dataset (MGH-C3PO). We tested the autoencoder in three test sets without modification: a) an MGH-C3PO holdout set, b) the independent Brigham and Women’s Hospital (BWH) subset of C3PO (BWH-C3PO), and c) the UK Biobank prospective cohort study. To assess for associations with disease, we derived phenotype vectors using labeled ECGs from 50% of the MGH-C3PO dataset, and projected those vectors onto each test set without modification. For every individual in each test set, we calculated the projected component, or the position along each phenotype vector (hereafter termed “vector component score”), and tested associations between vector component scores and corresponding Phecodes. We performed sample-level PheWAS in each of the three datasets and then meta-analyzed the results.
Table 1.
Patient characteristics by dataset
| Covariate | Mean (± SD) or n (%) | ||
|---|---|---|---|
| MGH (n = 60,140) | BWH (n = 46,027) | UKBB (n = 35,070) | |
| Age at ECG | 51.6 (17.2) | 54.8 (15.9) | 63.6 (7.6) |
| Female sex | 31,366 (52.2%) | 27,345 (59.4%) | 18,144 (51.7%) |
| Racea | |||
| White | 46,127 (76.7%) | 30,973 (67.3%) | 33,924 (96.7%) |
| Black | 3876 (6.4%) | 6066 (13.2%) | 234 (0.7%) |
| Asian | 2759 (4.6%) | 1177 (2.6%) | 472 (1.4%) |
| Hispanic | 3201 (5.3%) | 3724 (8.1%) | – |
| Other | 2542 (4.2%) | 1785 (3.9%) | 339 (1.0%) |
| Not reported | 1635 (2.7%) | 2302 (5.0%) | 101 (0.3%) |
| Year ECG performed (median, IQR) | 2005 (2001, 2012) | 2008 (2003, 2013) | 2018 (2016, 2019) |
SD standard deviation
aRace/ethnicity information is reported differently across datasets. The groupings above have been generated from available ascertainments.
For both 12- and single-lead models, autoencoders were developed using ECGs from the MGH-C3PO dataset, including 35,245 for training, 10,152 for validation, and 6674 for testing. The best-performing architecture contained over 11 million neurons and used mish activations, 2 dense convolutional blocks (each with 5 layers of convolutions per block), a 71 timestep convolutional kernel, and layer normalization, with 256 neurons in the fully connected layer (Supplementary Figs 1-2). The models were trained to reconstruct a median waveform corresponding to a single PQRST cycle (Supplementary Fig. 3). To assess the accuracy of reconstruction in each of the three datasets, we pooled voltages across test set ECGs and compared them to the voltages generated from reconstructions. For each dataset, we assessed the average per-voltage Pearson correlation coefficient and the 95% confidence interval based on 1000 bootstrap resamplings. We selected this method, as opposed to comparing per-person pooled voltages across test set and reconstructed ECGs, to assess the model’s global performance in ECG reconstruction rather than its performance reproducing ECGs from specific individuals within the datasets. We observed high-fidelity reconstruction of novel ECGs with Pearson correlation coefficients of 0.9956 (95% CI 0.9931–0.9972) in the MGH-C3PO testing set; 0.9916 (95% CI 0.9853–0.9950) in BWH-C3PO; and 0.9526 (95% CI 0.9427–0.9617) in the UK Biobank.
Phenotype vector derivation and ECG projection
For phenotype vector derivation, we mapped 50% of ECGs from the MGH-C3PO dataset to a library of 1,866 Phecodes, grouped across 17 disease categories (e.g., circulatory system, endocrine/metabolic, genitourinary)21. Labeled ECG encodings were used to derive phenotype vectors within the latent space. We then encoded the remaining unlabeled ECGs across the three datasets (n = 30,070 for MGH-C3PO; n = 46,027 for BWH-C3PO; and n = 35,070 for UK Biobank) and determined the position of each ECG along each phenotype vector to generate disease-specific vector component scores. Scores were scalar values ranging [-10.1, 8.5] in MGH, [-10.3, 8.3] in BWH, and [-7.8, 5.6] in the UK Biobank (Supplementary Table 1). Representations of age, sex, and body mass index in the latent space are shown in Supplementary Fig. 4, and a schematic summarizing the phenotype vector concept is displayed in Supplementary Fig. 5.
Latent space PheWAS
A PheWAS was then performed in the remaining MGH-C3PO samples. To assess the validity of results prior to downstream analysis, we performed empiric perturbation testing by randomly reclassifying samples based on Phecode presence or absence. We generated quantile-quantile plots for MGH-C3PO 12-lead results as well as random Phecode reclassification levels of 10%, 20%, and 100%. Supplementary Fig. 6 demonstrates that, as expected, the number of significant associations decreases as the degree of random reclassification increases.
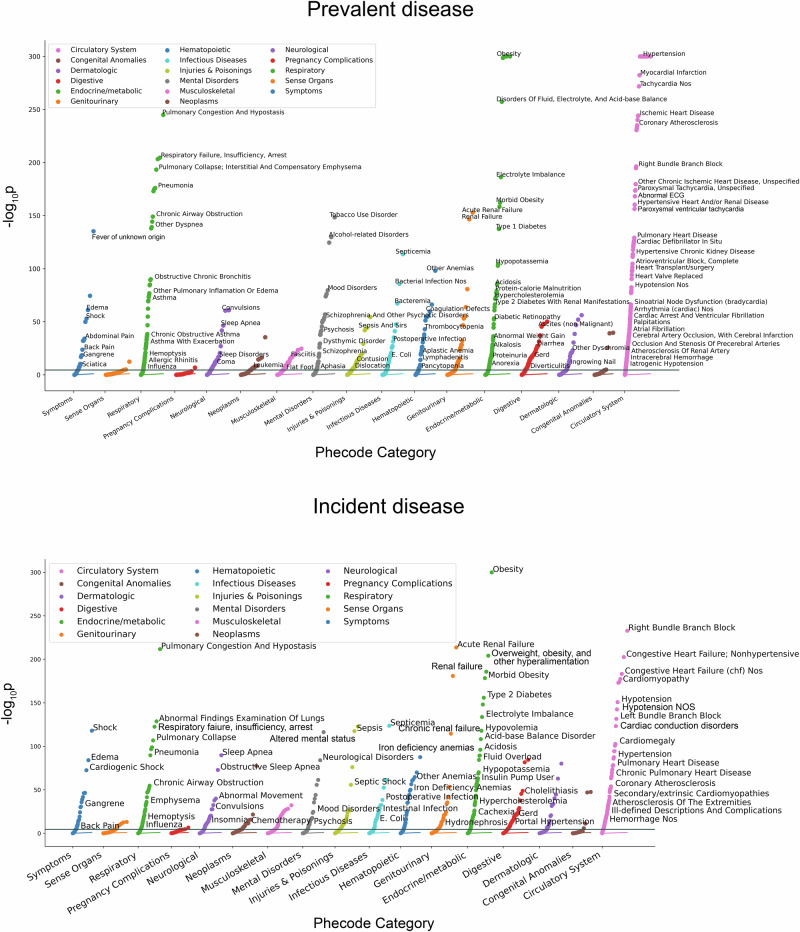
The PheWAS was then performed in each of the independent validation sets. After filtering out Phecodes with less than 100 combined cases, those present in only one dataset, and/or those for which model convergence failed, we meta-analyzed the study-specific results for 1595 Phecodes for the 12-lead model and 1,600 Phecodes for the single-lead model (Fig. 2, Supplementary Tables 2–3). Using a Bonferroni-corrected two-sided p-value of 3.1 × 10-5 (0.05/1595 and 0.05/1600), we observed significant associations between latent space position and disease status for 643 Phecodes in the 12-lead model (40% of overall Phecodes) and 565 Phecodes in the single-lead model (35%), respectively. The circulatory system category comprised the greatest enrichment of significant associations (n = 140, or 82% of Phecodes in this category for the 12-lead model, and n = 139, or 81% for the single-lead model). Enrichment was also observed within the respiratory (n = 53, 62% of category-specific Phecodes for the 12-lead model; n = 46, 54% for the single-lead model) and endocrine/metabolic categories (n = 73, 45% of category-specific Phecodes for the 12-lead model; n = 72, 44% for the single lead model; Fig. 2).
Fig. 2. Latent space phenome-wide association study results for the 12-lead electrocardiogram autoencoder model.
Panels depict phenome-wide association study results for the 12-lead electrocardiogram autoencoder. Top panel depicts existing disease associations, and bottom panel incident disease associations. Each Phecode tested for association is represented as a single point on the plot. The x-axis represents the phenotype category and the y-axis represents the -log10(p value) for the association test.
For the 12-lead model, the latent space positions for the Phecodes for hypertension, including “hypertension” (odds ratio [OR] per 1-point increase in vector component score 1.24, 95% CI 1.23–1.26, p < 2.2×10-308) and “essential hypertension” (OR 1.24, 95% CI 1.23–1.26, p < 2.2×10-308) showed the strongest associations (i.e., smallest p-values), followed by “cardiomyopathy” (OR 1.75, 95% CI 1.71–1.79, p < 2.2×10-308). The strongest associations with non-cardiac Phecodes included “obesity” (OR 1.30, 95% CI 1.29–1.32, p < 2.2×10-308), “diabetes mellitus” (OR 1.26, 95% CI 1.24–1.27, p = 5.9×10-304), “disorders of fluid, electrolyte, and acid-base balance” (OR 1.28, 95% CI 1.26–1.30, p = 5.5×10-258), and “pulmonary congestion and hypostasis” (OR 1.59, 95% CI 1.55–1.63, p = 1.2×10-245). For the single-lead model, the latent space positions for the Phecodes for cardiomyopathy, including “cardiomyopathy” (OR 1.22, 95% CI 1.21–1.23, p < 2.2×10-308) and “primary/intrinsic cardiomyopathies” (OR 1.24, 95% CI 1.23–1.26, p < 2.2×10-308) showed the strongest associations, followed by “congestive heart failure NOS” (OR 1.22, 95% CI 1.21-1.23, p < 2.2×10-308).
When ranked by effect estimate, we observed associations with substantial effect sizes across all disease categories, but in particular for cardiac Phecodes (e.g., “Cardiac defibrillator in situ” OR 2.18, 95% CI 2.04–2.32, p = 6.83×10-124) (Table 2). Similar patterns were observed using the single-lead model (Supplementary Table 4). We additionally identified unexpected and highly robust relationships, including tobacco use disorder (12-lead: OR 1.19, 95% CI 1.18–1.21, p = 1.0×10-149; single-lead: OR 1.08, 95% CI 1.08-1.09, p = 1.9×10-131), fever of unknown origin (OR 1.18, 95% CI 1.16–1.19, p = 5.5×10-136; OR 1.07, 95% CI 1.07–1.08, p = 1.2×10-104), and non-alcoholic liver disease (OR 1.17, 95% CI 1.15–1.20, p = 8.0×10-51; OR 1.05, 95% CI 1.05–1.06, p = 1.9×10-39). Importantly, effects were generally consistent across datasets, including in the UK Biobank, which had an overall lower prevalence of disease (individual association results shown in Supplementary Tables 2-3).
Table 2.
Top associations by effect size across disease category
| Disease groupinga | N events | Odds ratio (95% CI)b | p |
|---|---|---|---|
| Circulatory system | |||
| Cardiac defibrillator in situ | 514 | 2.18 (2.04–2.32) | 6.83 x 10-124 |
| Bundle branch block | 1679 | 2.16 (2.07–2.25) | 1.11 x 10-303 |
| Other hypertrophic cardiomyopathy | 127 | 2.13 (1.78–2.54) | 4.21x10-17 |
| Left bundle branch block | 927 | 2.12 (2.03–2.22) | 1.13x10-244 |
| Right bundle branch block | 868 | 2.08 (1.98–2.18) | 5.80x10-197 |
| Congenital anomalies | |||
| Cardiac shunt/ heart septal defect | 1303 | 1.32 (1.25–1.39) | 3.16x10-26 |
| Cardiac congenital anomalies | 2876 | 1.22 (1.18–1.26) | 7.76x10-40 |
| Cardiac and circulatory congenital anomalies | 3632 | 1.21 (1.18–1.24) | 2.38x10-40 |
| Spondylolisthesis, congenital | 478 | 1.17 (1.09–1.25) | 1.28x10-05 |
| Dematologic | |||
| Decubitus ulcer | 479 | 1.44 (1.33–1.55) | 2.05x10-20 |
| Chronic ulcer of leg or foot | 1410 | 1.36 (1.30–1.42) | 1.29x10-47 |
| Chronic ulcer of skin | 2155 | 1.30 (1.25–1.34) | 8.35x10-53 |
| Cellulitis and abscess of foot, toe | 1204 | 1.29 (1.22–1.36) | 7.20x10-22 |
| Cellulitis and abscess of trunk | 1513 | 1.28 (1.22–1.33) | 3.81x10-31 |
| Digestive | |||
| Portal hypertension | 426 | 1.51 (1.40–1.63) | 4.01x10-28 |
| Acute and subacute necrosis of liver | 251 | 1.40 (1.25–1.56) | 2.04x10-09 |
| Complications of gastrostomy, colostomy and enterostomy | 181 | 1.36 (1.19–1.54) | 3.60x10-06 |
| Liver replaced by transplant | 162 | 1.35 (1.18–1.53) | 9.43x10-06 |
| Cirrhosis of liver without mention of alcohol | 1078 | 1.33 (1.28–1.39) | 7.05x10-38 |
| Endocrine/Metabolic | |||
| Cachexia | 176 | 1.85 (1.64–2.09) | 4.30x10-23 |
| Alkalosis | 224 | 1.76 (1.58–1.95) | 1.27x10-26 |
| Diabetes type 1 with peripheral circulatory disorders | 158 | 1.75 (1.54–1.98) | 1.91x10-18 |
| Type 1 diabetes with ophthalmic manifestations | 297 | 1.60 (1.47–1.75) | 1.54x10-25 |
| Acidosis | 1239 | 1.52 (1.45–1.58) | 1.08x10-85 |
| Genitourinary | |||
| Nephritis and nephropathy in diseases classified elsewhere | 481 | 1.46 (1.37–1.56) | 5.80x10-29 |
| Renal dialysis | 1001 | 1.38 (1.32–1.44) | 1.92x10-47 |
| Acute renal failure | 3700 | 1.37 (1.34–1.40) | 1.43x10-153 |
| End stage renal disease | 709 | 1.36 (1.29–1.44) | 4.39x10-29 |
| Kidney replaced by transpant | 734 | 1.35 (1.28–1.43) | 3.39x10-26 |
| Hematopoietic | |||
| Anemia in chronic kidney disease | 447 | 1.51 (1.40–1.63) | 5.40x10-27 |
| Secondary thrombocytopenia | 610 | 1.51 (1.41–1.61) | 2.32x10-35 |
| Deficiency anemias | 367 | 1.33 (1.22–1.46) | 4.84x10-10 |
| Aplastic anemia | 712 | 1.31 (1.24–1.39) | 4.52x10-21 |
| Acquired hemolytic anemias | 249 | 1.30 (1.18–1.44) | 6.93x10-08 |
| Infectious diseases | |||
| Gram positive septicemia | 388 | 1.57 (1.45–1.71) | 8.88x10-28 |
| Infection with drug-resistant microorganisms | 468 | 1.42 (1.32–1.52) | 1.28x10-21 |
| Bacteremia | 1373 | 1.41 (1.35–1.46) | 6.06x10-68 |
| Septicemia | 2585 | 1.40 (1.36–1.44) | 1.84x10-114 |
| Methicillin-sensitive Staphylococcus aureus | 892 | 1.39 (1.32–1.46) | 6.48x10-36 |
| Injuries and poisonings | |||
| Septic shock | 241 | 1.48 (1.35–1.63) | 1.78x10-16 |
| Systemic inflammatory response syndrome (SIRS) | 151 | 1.45 (1.25–1.69) | 1.33x10-06 |
| Subarachnoid hemorrhage (injury) | 162 | 1.44 (1.23–1.68) | 3.21x10-06 |
| Sepsis | 926 | 1.43 (1.36–1.50) | 8.79x10-43 |
| Sepsis and SIRS | 1028 | 1.42 (1.35-1.49) | 7.02x10-45 |
| Mental disorders | |||
| Alcoholic liver damage | 683 | 1.48 (1.40–1.56) | 2.80x10-46 |
| Delirium due to conditions classified elsewhere | 1375 | 1.39 (1.34–1.45) | 1.13x10-57 |
| Altered mental status | 1783 | 1.32 (1.28–1.37) | 8.45x10-52 |
| Alcoholism | 4669 | 1.31 (1.28–1.34) | 2.71x10-125 |
| Alcohol-related disorders | 5982 | 1.28 (1.26–1.31) | 1.71x10-130 |
| Musculoskeletal | |||
| Panniculitis | 109 | 1.47 (1.25–1.74) | 5.12x10-06 |
| Osteitis deformans and osteopathies associated with other disorders classified elsewhere | 273 | 1.33 (1.22–1.45) | 9.86x10-11 |
| Infective connective tissue disorders | 159 | 1.32 (1.18–1.48) | 1.96x10-06 |
| Acute osteomyelitis | 383 | 1.32 (1.22–1.42) | 1.32x10-12 |
| Unspecified osteomyelitis | 812 | 1.26 (1.20–1.33) | 4.84x10-20 |
| Neoplasms | |||
| Hodgkin’s disease | 450 | 1.38 (1.28–1.49) | 6.23x10-16 |
| Bone marrow or stem cell transplant | 327 | 1.30 (1.19–1.42) | 4.18x10-09 |
| Secondary malignancy of bone | 546 | 1.16 (1.09–1.23) | 1.87x10-06 |
| Secondary malignancy of respiratory organs | 686 | 1.15 (1.09–1.22) | 2.49x10-06 |
| Cancer of other lymphoid, histiocytic tissue | 1882 | 1.14 (1.11–1.18) | 4.63x10-17 |
| Neurological | |||
| Coma | 280 | 1.42 (1.27–1.58) | 1.14x10-10 |
| Peripheral autonomic neuropathy | 285 | 1.32 (1.21–1.43) | 7.19x10-11 |
| Cerebral degeneration, unspecified | 373 | 1.30 (1.19–1.42) | 6.35x10-09 |
| Encephalitis, non-infectious | 256 | 1.29 (1.17–1.43) | 7.74x10-07 |
| Other paralytic syndromes | 556 | 1.27 (1.18–1.37) | 1.07x10-09 |
| Pregnancy complications | |||
| Hypertension complicating pregnancy, childbirth, and the puerperium | 1282 | 1.15 (1.09–1.22) | 1.55x10-07 |
| Respiratory | |||
| Obstructive chronic bronchitis | 1182 | 1.71 (1.62–1.80) | 8.56x10-91 |
| Respiratory failure | 1881 | 1.65 (1.60–1.71) | 9.38x10-174 |
| Emphysema | 2471 | 1.64 (1.58–1.71) | 1.05x10-138 |
| Pneumonitis due to inhalation of food or vomitus | 919 | 1.60 (1.52–1.68) | 9.01x10–76 |
| Pulmonary congestion and hypostasis | 3653 | 1.59 (1.55–1.63) | 1.24x10–245 |
| Sense organs | |||
| Other nondiabetic retinopathy | 237 | 1.24 (1.13–1.36) | 8.18x10-06 |
| Blindness and low vision | 976 | 1.15 (1.08–1.23) | 2.16x10-05 |
| Dizziness and giddiness (Light-headedness and vertigo) | 14603 | 1.05 (1.04–1.07) | 3.94x10-13 |
| Symptoms | |||
| Cardiogenic shock | 287 | 1.71 (1.57–1.87) | 5.34x10-33 |
| Shock | 656 | 1.60 (1.51–1.70) | 9.88x10–54 |
| Gangrene | 302 | 1.49 (1.36–1.63) | 1.47x10–17 |
| Rhabdomyolysis | 185 | 1.28 (1.15–1.42) | 7.23x10–06 |
| Fever of unknown origin | 13114 | 1.18 (1.16–1.19) | 5.46x10–136 |
aDisplayed are significant associations with the top 5 largest effect sizes within each disease category. In cases where there are fewer than 5 significant associations, all significant associations are shown.
bOdds ratios per 1-standard deviation increase in vector component score (see text)
ECG intervals PheWAS
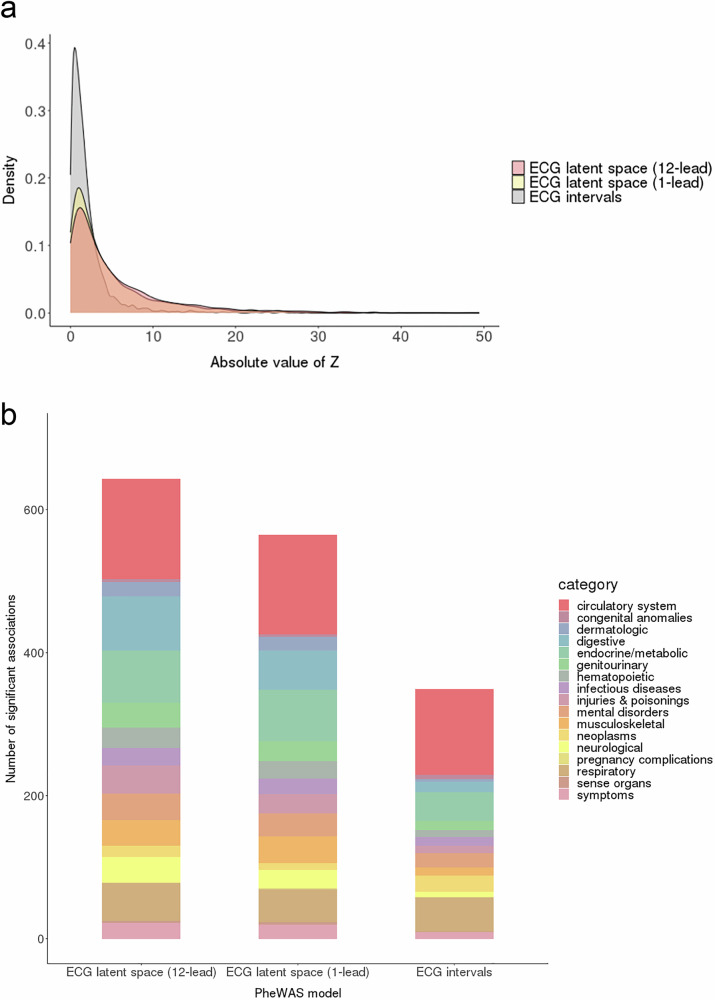
Meta-analyses of the ECG intervals PheWAS included 1607 Phecodes for the PR interval; 1607 Phecodes for the QRS duration; and 1605 Phecodes for the QT interval. In comparing the ECG intervals and latent space models, we restricted to Phecodes that were present in all meta-analyses (n = 1584). Using the smallest meta-analyzed p-value across any ECG interval for each Phecode, we observed fewer significant associations for the intervals models relative to both the 12-lead and single-lead latent space models, both overall and within disease categories (Fig. 3 and Supplementary Tables 5-7). Forest plots summarizing the associations for the top Phecodes are displayed in Supplementary Fig. 7.
Fig. 3. Significant associations in the latent space and electrocardiogram intervals phenome-wide association studies.
Panel a displays the test statistic distribution (absolute z-score) for the ECG term in the meta-analyzed phenome-wide association study (PheWAS), stratified by modeling approach. Results are displayed for the 12-lead and 1-lead electrocardiogram (ECG) latent space models, as well as the ECG intervals model. Panel b demonstrates the number of significantly associated Phecodes, defined as those exceeding a Bonferroni-corrected two-sided p value of 3.1 × 10-5 (0.05 divided by 1584, the number of unique Phecodes included across all meta-analyses). For the intervals model, a result was considered significant if the meta-analyzed p value for any of the tested ECG intervals (PR, QRS, QT) exceeded the significance threshold. When compared to the ECG intervals model, the latent space models yield a greater number of significant associations, both overall and across disease categories.
Discrimination of Phecode diseases
We then compared discrimination using the area under the receiver operator characteristic curve (AUC) across logistic regression models in which Phecodes having significant associations with the ECG in the primary meta-analysis were regressed on age, sex, and race, with or without an additional term for ECG vector component scores. We generally observed substantial increases in discrimination for models that included the ECG vector component score term as compared to models that did not, particularly for circulatory system (median difference in AUC, interquartile range for MGH-C3PO 0.031, 0.016–0.065; BWH-C3PO 0.024, 0.010–0.06; UKB 0.0053, 0.00086–0.011) and respiratory (MGH-C3PO 0.044, 0.023–0.072; BWH-C3PO 0.028, 0.011–0.058; UKB 0.0015, 0.0019–0.0056) Phecode categories, though additional improvements in discrimination were observed for other Phecode categories as well (Supplementary Fig. 8). The top and bottom five conditions per category based on AUC improvement with incorporation of the ECG latent space are shown in Supplementary Tables 8-9. Substantial AUC improvement persisted when the ECG latent space was compared to models including standard ECG intervals (top 50 conditions with the largest AUC improvement in BWH test set shown in Supplementary Table 10).
Latent space incident disease PheWAS
In an exploratory analysis using the 12-lead ECG model, we observed significant associations between the vector component scores and incident disease for 457 out of 1370 tested Phecodes (33.4%). Similar to the analyses focused on existing conditions, we observed greatest enrichment for associations among circulatory (n = 107, 74% of Phecodes in this category), endocrine/metabolic (n = 60, 52% of category-specific Phecodes) and digestive (n = 53, 38% of category-specific Phecodes) conditions. Significant associations included considerable effect sizes for a variety of conditions including incident paroxysmal ventricular tachycardia (hazard ratio [HR] per 1-point increase in vector component score 1.61, 95% CI 1.53–1.70, p = 2.58×10-65), end stage renal disease (HR 1.31, 95% CI 1.23–1.40, p = 8.49×10-18), and respiratory failure (HR 1.35, 95% CI 1.30–1.40, p = 2.16×10-52). Association results are summarized in Fig. 2 and listed in detail in Supplementary Tables 11–12.
Model-based, disease-specific median waveforms
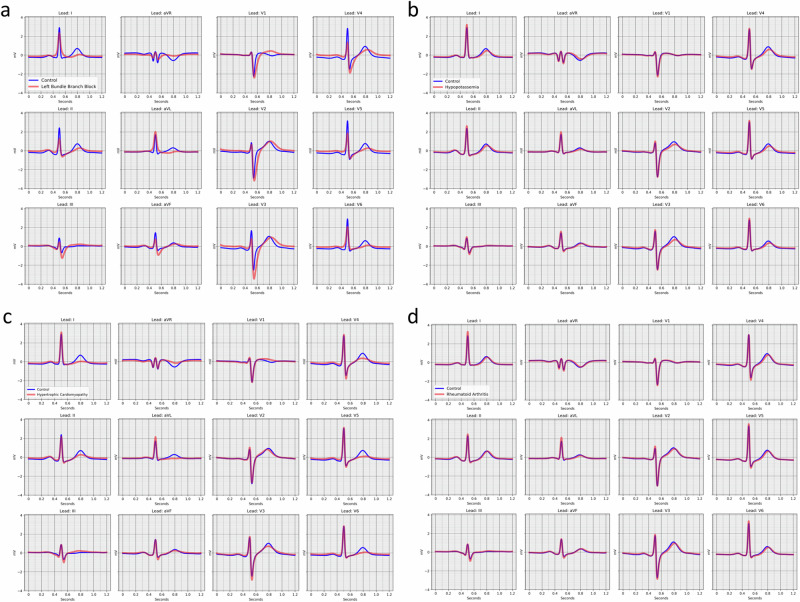
For certain diseases with well-characterized ECG manifestations, model-derived features were consistent with expectations. For example, median waveforms generated from the left bundle branch block disease-positive centroid demonstrated QRS widening, smaller initial r waves in the right-sided precordial leads (V1–V3), and R wave slurring in the left-sided leads (I, aVL, V5, V6)26 relative to median waveforms generated from the disease-negative centroid (Fig. 4). For hypokalemia, model-derived features included decreased T wave amplitude and relative QT prolongation27 (Fig. 4). In other instances, however, reconstructed disease case and control ECGs appeared morphologically similar visually despite highly significant differences in projected components from the latent space models. For example, model-derived ECG reconstructions of hypertrophic cardiomyopathy were notable for broad, flattened T-waves, particularly in the left-sided leads, which are more subtle than the classical well-defined ECG manifestations of hypertrophic cardiomyopathy (e.g., prominent precordial voltages, repolarization abnormalities/T-wave inversions, pathologic Q waves28) (Fig. 4). Likewise, rheumatoid arthritis, an inflammatory condition associated with higher risk of cardiovascular disease which had a strong ECG vector component score association but has no clinically characteristic ECG signature, was notable for subtle differences in T wave morphology and QT interval (Fig. 4). Overall findings suggest that the latent space is sensitive to subtle manifestations of disease, while the model-derived ECG reconstructions are conservative and may not visually replicate all hallmarks of disease.
Fig. 4. Model-based, disease-specific ECG reconstructions.
Median waveform reconstructions for centroids reflecting individuals without (blue) and with (red) left bundle branch block in panel a, hypokalemia (hypoptassemia) in panel b, hypertrophic cardiomyopathy in panel c, and rheumatoid arthritis in panel d.
Patient report card prototype
To demonstrate the potential for the ECG to serve as a digital biomarker for disease status, we generated a prototype of an ECG-based patient report for select Phecodes (Table 3). In this illustrative example, an ECG from a 65-year-old female is projected onto the phenotype vectors for select circulatory system diseases, and the positions relative to the whole cohort along vectors from the disease-negative to disease-positive centroids are reported. These diseases were selected based on clinical relevance and the potential to cause substantial morbidity if undetected, including myocardial infarction, ventricular tachycardia, and heart failure29–33.
Table 3.
Patient report prototype for a 65-year-old female
| Phecode | Probability of disease (95% CI)* | Background prevalence (95% CI) |
|---|---|---|
| Hypertension | 62.1% (60.7–63.4) | 46.8% (45.9–47.6) |
| Diabetes | 22.0% (20.9–23.1) | 15.4% (14.8–16.0) |
| Hyperlipidemia | 51.1% (50.0–52.2) | 42.9% (42.0–43.7) |
| Cardiomyopathy | 16.3% (14.4–18.3) | 5.6% (5.2–6.0) |
| Heart failure with preserved ejection fraction | 2.6% (1.9–3.6) | 0.9% (0.8–1.1) |
| Myocardial infarction | 23.5% (20.8–26.3) | 8.0% (7.6–8.5) |
| Paroxysmal ventricular tachycardia | 6.9% (5.3–9.0) | 2.0% (1.7–2.3) |
| Mitral and aortic valve stenosis | 2.1% (1.5–2.8) | 0.8% (0.6–0.9) |
*The probability of disease is estimated as the disease prevalence among all individuals in the MGH-C3PO test set with a vector component score value greater than or equal to the individual’s value. The corresponding disease prevalence in all of the MGH-C3PO test set is depicted as a reference. The report reflects diseases that were selected for illustrative purposes based on statistical significance as well as clinical relevance.
CI = confidence interval.
Discussion
Here, we highlight the use of autoencoder deep learning models to encode and reconstruct 12- and single-lead ECGs in order to generate a multidimensional latent space encoding ECG waveform features with demonstrable relevance to risk of disease across the spectrum of human conditions. Indeed, our results demonstrate robust associations between ECG waveform patterns and conditions spanning the full spectrum of human disease. When compared to standard ECG intervals, the autoencoder-based latent space models reveal a substantially greater number of associations, suggesting that the rich representations afforded by the architectural complexity of the deep learning model provide more information about disease status than routinely ascertained ECG intervals.
Autoencoder models have been used previously in fields such as linguistics and image processing to better understand input information. Prior studies on image representation have demonstrated that attribute labeling (e.g., headshot photograph categorized as “wearing glasses” versus “no glasses”) can aid interpretation of the machine-learned latent space, as images with similar attributes form interpretable semantic clusters. Cluster centroids are spatially defined using attribute vectors, and any unlabeled image projection can be relationally defined by its position along the vector, based solely on the image characteristics interpreted by the model. These studies further demonstrate that the reconstruction of an input image (e.g., person without glasses) can be modified by movement along an attribute vector, generating a novel image with features that are more aligned with the corresponding centroid pair (e.g., the same photograph reconstructed with glasses)34,35. Although autoencoders have recently been applied to the ECG primarily in efforts to create more interpretable deep learning pipelines36–39, our work extends prior models by leveraging attribute, or in this case phenotype, labeling to explore disease-state information across the full spectrum of human conditions, to define a range of both cardiac and non-cardiac conditions detectable by the ECG.
By applying autoencoder-based technology to clinical ECG data, our results yield several implications. First, we demonstrate how latent space modeling can be used for the discovery of novel information contained within the ECG. Specifically, we apply phenotype labeling to explore disease-state information contained in 12- and single-lead ECGs. By reconstructing median waveforms from latent space centroids, we visually represent disease-based patterns identified by deep learning models, which in some cases confirm expectations and in other cases reflect subtle waveform manifestations potentially below the level of human detection. Furthermore, although our primary aim was to investigate the detection of existing disease, our secondary analyses suggest that autoencoder representations may additionally possess utility for the prediction of incident disease. In the future, samples acquired prior to and after a given diagnosis may facilitate the derivation of “disease progression vectors,” allowing visualization of waveform evolution over time. We submit that the methodology of mapping clinical status onto the latent space may have implications far beyond the ECG, extending to other modalities (e.g., laboratory testing, imaging results) individually or in combination, thereby greatly expanding the clinical utility of existing, easily acquired diagnostics.
Second, while ECG-based deep learning models have been developed previously, studies have predominantly focused on disease-specific risk prediction within the cardiovascular system8,9,12,13. In taking a more global analytic approach, we have identified a potential role for the ECG-based classification of non-cardiac disease. Specific conditions for which further study may be particularly high yield include diseases not classically associated with ECG findings but each independently supported by prior studies (e.g., type 2 diabetes40,41, sleep apnea42–46, chronic liver disease/cirrhosis15,47, and renal failure48), as well as diseases with previously undescribed associations (e.g., fever of unknown origin, tobacco use disorder). Improvements in disease discrimination were particularly enriched for conditions commonly encountered during critical illness with clear ECG manifestations (e.g., acid-base disorders, sepsis, shock, arrhythmias), consistent with recent work demonstrating the particular value of ECG-based deep learning in critical care populations49.
Third, we demonstrate the potential for personalized and scalable disease detection with ECG-based latent space modeling. Using data derived from large samples, we construct a complex architectural environment informed by disease status, in which each ECG encoding represents a single individual and occupies a unique position in the latent space. Projection of new ECGs from independent individuals can therefore be used to generate likelihoods of disease at scale. As latent space modeling approaches are refined with data from larger and more diverse samples, we anticipate that the utility for disease-based classification will grow. Our patient report card is illustrative in nature, and future studies are warranted to prospectively evaluate the specific test characteristics of latent space proximity in discriminating disease status, and quantify the degree to which discrimination may be affected by treatment effects. We submit that the approach we outline will have particular value for detecting diseases in which screening may be cumbersome, inaccurate, or expensive, and for which early disease manifestations may be highly morbid (e.g., aortic aneurysm and valvular heart disease). Importantly, although our approach possesses several potential advantages when compared to training a large number of individual disease-specific classifiers (e.g., the requirement to train and implement only a single model) or a single large multi-task disease classifier (e.g., simpler architecture, lower requirements on model capacity, no dependence on a varying frequency of disease labels), we do not claim our approach is necessarily superior to alternative modeling strategies. Rather, we submit our unsupervised modeling strategy is better suited to broad risk profiling at scale. Indeed, the robust performance of our single-lead model for prevalent disease detection highlights the potential utility for screening large populations, particularly given the widespread emergence of consumer-based wearable and handheld devices with ECG recording capabilities23.
Our study presents certain limitations. First, since our primary aim was to develop and apply an autoencoder-based approach to systematically identify conditions whose presence may be detectable on the ECG, rather than develop a model to predict future disease, we derived phenotype vectors among individuals with known disease, and of varying durations. Although training among individuals with known disease may enrich for more severe cases, such an approach is customary in the development of disease classification models12,50. We acknowledge prospective validation would be required to confirm performance among individuals in whom disease status is unknown at the time of ECG acquisition. Second, we used single linear probes to define phenotype vectors. Although the use of linear probes to interpret latent spaces is common and has theroretical support51, future work is warranted to assess whether methods capable of leveraging potentially non-linear relations (e.g., use of multiple vectors or non-linear probes) may result in improved performance. Third, improvements in discrimination with the incorporation of autoencoder information were substantial in MGH and BWH but more modest in the UK Biobank, which is likely due to differences in sample composition (i.e., healthier) and lower event rates, although lower autoencoder reconstruction accuracy or varying informativeness of ECG features across datasets are additional potential contributing factors. Fourth, to standardize the phase of the cardiac cycle and minimize the effects of artifacts, we encoded ECGs as median waveforms. Such an approach may result in the loss of some information related to R-R regularity and subtle beat-to-beat changes in morphology, including ectopic beats. However, we do demonstrate that our median samples encode heart rate information. Fifth, we did not compare our approach to the analysis of the raw median ECG beat. However, we submit our autoencoder approach provides retains specific advantages over use of the raw ECG beat (e.g., flexibility, computational efficiency), and possesses the potential to extend to modalities where use of the raw signal may be computationally infeasible (e.g., multi-modality imaging). Sixth, we offer the patient report card prototype as a demonstration of the concept of how an ECG autoencoder model could be applied to broadly classify risk of present but potentially undiagnosed disease. We acknowledge that certain factors, such as unclear actionability of intermediate probabilities of disease, require further investigation before clinical implementation. Seventh, although we adjusted our models for basic factors likely to confound all potential disease associations (e.g., age, sex, race), given the number of associations tested we cannot exclude residual confounding or quantify the degree to which associations may be driven by clinical factors encoded by the ECG, and therefore our findings should not be used to infer causal relations. Eighth, our autoencoder approach does not provide a straightforward translation from ECG signal to disease risk, and is not suitable for quantitative model interpretation methods designed for trained supervised models (e.g., CNN Explanations Framework for ECG Signals)52. However, we do plot median samples which demonstrate changes consistent with clinical expectations. Ninth, our findings are subject to selection bias due to the requirement for a 12-lead ECG obtained for clinical purposes in MGH and BWH. However, we note the continued value of the autoencoder latent space in the UK Biobank, where ECGs were obtained prospectively as part of a research protocol.
In conclusion, we demonstrate how latent space modeling can be used to organize and better understand disease-related information contained within currently available diagnostics. The corresponding analysis demonstrates that the ECG waveform contains a wealth of disease-state information beyond the circulatory system, with the potential to detect hundreds of prevalent conditions and even stratify the risk of incident disease. Future studies are warranted to prospectively validate the ability of the ECG-based autoencoder latent space to facilitate scalable disease profiling.
Methods
Study subjects
Two of the three datasets included were derived from the Community Care Cohort Project (C3PO), a previously established cohort comprising over 500,000 adults aged ≥ 18 years who receive longitudinal primary care at one of eleven hospitals within the Mass General Brigham network, which is linked by a common EHR data warehouse24. C3PO datasets included a cohort from the Massachusetts General Hospital (MGH-C3PO dataset) and a cohort from the Brigham and Women’s Hospital (BWH-C3PO dataset). The third, external dataset was derived from the UK Biobank, a prospective community-based cohort study comprising adults aged 40–60 years at enrollment between the years 2006–2010 from the United Kingdom25. The present analysis includes the subsets of individuals in each dataset with at least one 12-lead ECG performed within three years prior to the start of follow-up (C3PO) or who had a 12-lead ECG performed during at least one study visit (UK Biobank). Use of MGB and UK Biobank (application 7089) data were approved by the MGB Institutional Review Board. The UK Biobank was approved by the UK Biobank Research Ethics Committee (reference number 11/NW/0382). All UK Biobank participants provided written informed consent.
ECG autoencoder model and latent space derivation
We trained densely connected convolutional autoencoders to encode and reconstruct 12- and single-lead ECGs. In general, autoencoders consists of an encoder, which maps a high-dimensional input into a lower-dimensional latent space, and a decoder, which reconstructs the original data from the latent space representation (Supplementary Fig. 1). Autoencoders are trained to encode variance present within the original data into the latent space, which encourages the model to minimize differences between the original data and its reconstruction. Both the 12-lead and single-lead autoencoders were trained and validated using subsets of ECGs from the MGH-C3PO cohort. The models were then tested in an MGH-C3PO holdout set as well as two true holdout datasets, including BWH-C3PO and the UK Biobank.
To standardize the phase of the cardiac cycle across all individuals while minimizing the effects of signal artifact (e.g., baseline drift, transient noise), we encoded ECGs as median waveforms by segmenting 10-second ECG recordings into 1200 millisecond windows, sampling 600 voltage timepoints per window, and performed piecewise linear interpolation to generate R-R adjusted medians53,54. Median waveforms therefore represent the aggregate morphology of at least one cardiac cycle from each lead (Supplementary Fig. 2). When assessed in 1,000 randomly sampled individuals from the UK Biobank, heart rate was easily recovered from the median waveform (r = 0.94, 95% CI 0.93–0.95), demonstrating little loss in heart rate information. The 12-lead model utilized median waveforms generated from all available leads (i.e., 12 waveforms per ECG), while the single-lead model utilized only the median waveform generated from lead I. In the following analysis, the term ECG generally refers to the median waveform.
Models were trained using one-dimensional convolutions over voltage-time series, corresponding to 7,200 voltage timepoints for 12-lead ECGs and 600 voltage timepoints for single leads. For ECGs with incomplete voltage data (i.e., less than 10 seconds recorded from each lead), we used zero padding, converting non-available data into zeros (Supplementary Table 13). The mean squared error per voltage timepoint across the full ECG was minimized, as demonstrated in Eq. (1):
| 1 |
In C3PO, ECGs were excluded if the acquisition date was greater than three years prior to the start of clinical follow up, defined for each individual as the time of the second primary care visit of the earliest qualifying pair24. Only one ECG per individual was represented; for patients with multiple ECGs, the most recent was used.
The neural net architecture was a variant of Densenet, featuring several densely connected convolutional blocks operating at different time resolutions55. Architecture hyperparameters, including width, depth, activation, normalization, and regularization were chosen via Bayesian hyperparameter optimization56.
To assess the accuracy of reconstruction in each of the three datasets, we pooled voltages across test set ECGs and compared this to the voltages generated from reconstructions. For each dataset, we assessed the average per-voltage Pearson correlation coefficient and the 95% confidence interval based on 1000 bootstrap resamplings. We selected this method, as opposed to comparing per-person pooled voltages across test set and reconstructed ECGs, to assess the model’s global performance in ECG reconstruction rather than its performance reproducing ECGs from specific individuals within the datasets (Supplementary Fig. 3).
Phenotype definitions
ICD codes in each of the three datasets were mapped to a publicly available Phecode library (https://phewascatalog.org/Phecodes_icd10cm)21. As previously described, Phecodes distinguish cases from controls using hierarchical groupings of ICD 9 and 10 codes to better define clinically meaningful disease phenotypes. Only prevalent Phecodes were used, i.e., all corresponding ICD codes had been entered into the patient’s chart prior to the ECG acquisition date. For certain Phecodes, participants without that Phecode but with very similar ICD codes were excluded from serving as controls to avoid biasing results, as described previously (e.g., in association testing for the myocardial infarction case group, patients were removed from serving as controls if they had ICD codes corresponding to a list of disease exclusions, including angina or other evidence of ischemic heart disease)20,21.
Phenotype vector derivation
If a given disease, represented by a Phecode, has a significant impact on the ECG, we expect ECG encodings from individuals with the disease to distribute to a different location in the autoencoder-derived latent space relative to ECG encodings from individuals without the disease. In contrast, if the disease has little impact on the ECG, or if the ECG encoding does not adequately capture disease-relevant features, then we expect there to be no significant relationship between the position of the ECG encoding in latent space and the presence or absence of disease.
To quantify this expectation, we define the highest density of ECG encodings labeled as having the disease (“disease-positive centroid” for cases) and the highest density of ECG encodings labeled as not having the disease (“disease-negative centroid” for controls). Each Phecode is therefore spatially represented by its centroid pair, and the line that connects them is referred to as the phenotype vector (Supplementary Fig. 5). Uniform manifold approximation and projections (UMAPs) were also generated to visually assess ECG encodings for age, sex, and body mass index.
During autoencoder training/latent space derivation, we labeled a subset of ECGs from the MGH-C3PO dataset as disease cases or controls based on the presence or absence of the corresponding Phecode in the patient’s EHR. ECG encodings from the derivation set were then used to define centroid pairs and phenotype vectors for each Phecode within the latent space. The phenotype vector correlation matrix is displayed in Supplementary Fig. 9.
ECG projections
Any ECG encoded within the latent space, including encodings from unlabeled samples, can be projected onto any phenotype vector, and the relative position along the phenotype vector can be used to assess how closely related the unlabeled ECG encoding is to encodings that were used to define the disease positive centroid for that Phecode.
We then sought to externally validate our approach to classify the presence of disease in samples independent of autoencoder and phenotype vector derviation. After deriving phenotype vectors in the MGH-C3PO training set, we projected unlabeled ECG encodings from three test sets independent of autoencoder and phenotype vector derviation: a) the 50% holdout component of the MGH-C3PO cohort, b) BWH-C3PO, and c) the UK Biobank, onto the phenotype vectors. There was no modification of the autocoder or the phenotype vectors in the application of the model to the test sets.
We scaled the entire space of ECG encodings to have norm and standard deviation of one. Each phenotype vector was normalized to have length one. The high-dimensional spatial relatedness of each ECG and Phecode was quantified by each sample’s component in the direction of a given phenotype vector. As illustrated in Supplementary Fig. 10, each ECG encoding (“ECG embedding”, ECGi) projects onto each phenotype vector, Vp. The projected component, (“componentip”) is calculated from the angle between the ECG encoding and the phenotype vector, scaled by the length of the ECG, as displayed in Eq. (2). Thus, the projected component signifies the latent space position of a single individual along a single phenotype vector and therefore represents a disease-specific “vector component score”.
| 2 |
We used analogous methods for both 12- and single-lead models. In the single-lead model, the autoencoder-derived latent space, phenotype vector derivation, and ECG projections were based only on median waveform data derived from lead I.
Association Testing by Latent Space PheWAS
For both 12- and single-lead models, we performed a PheWAS in each dataset using a logistic regression model to assess the strength of the relationship between a given disease-specific vector component score and the presence of the target disease state (using Phecode presence or absence as the outcome variable). In this way, the odds ratio represents the adjusted odds for the presence of disease for every 1-point increase in the vector component score. For the UK Biobank dataset, the model was adjusted for age, sex, and race. For the MGH-C3PO holdout set and the BWH-C3PO dataset, the model was additionally adjusted for ECG acquisition date and the amount of zero padding (as the degree of missing voltage data may represent other confounders such as ECG quality, ECG machine used, hospital location, etc.).
For both latent space models, we performed a fixed-effects inverse variance weighted meta-analysis, filtering for Phecodes that were present in at least two datasets with at least 100 combined cases. Coefficients corresponding to each phenotype vector were pooled across datasets. For comparison, we generated a separate model based on ECG intervals, including the PR interval, QRS duration, and QT interval. We chose an intervals-based model as our comparator because it utilizes routinely ascertained, standardized, and automated measurements known to have disease-based prognostic implications57–61. As above, we performed an intervals PheWAS in each dataset and meta-analyzed results. We then compared the number of significant associations between the latent space models and the ECG intervals model, using a Bonferroni-corrected two-sided p-value of 0.05 divided by the number of common Phecodes across all meta-analyses. For the intervals model, we considered a result significant if the p-value for any interval met the significance threshold. Metanalyses were performed using the R package meta. Python was used to create a visual summary of meta-analyzed data, grouped according to disease category. To estimate the potential added value for disease detection, we assessed the difference in discrimination of individual Phecodes by calculating the difference in the area under the curve (AUC) for the logistic regression models described above with versus without ECG vector component scores, among Phecodes with Bonferroni-corrected significant associations with the component model in the primary meta-analysis. In a secondary analysis, we repeated AUC estimation using models comparing the ECG component vectors to standard ECG intervals (PR interval, RR interval, QRS duration, and QT interval).
Although our primary goal was to leverage latent space models to assess the degree to which existing diseases may be detectable using ECG waveform, we explored the potential for the ECG latent space to predict the risk of incident disease using an analogous approach to that outlined above, except using Cox proportional hazards models rather than logistic regression models. Individuals with a Phecode diagnosis present at baseline (ie., start of follow-up) were excluded from incident analyses of that Phecode, and a Phecode event was defined as the first instance of any component of the given Phecode definition. In the C3PO datasets, person-time began at the start of follow-up and ended at the earliest of an outcome event, death, last encounter in the EHR, or August 31, 2019. In the UK Biobank, person-time began at the ECG study visit and ended at the earliest of outcome event, death, or last follow-up. The date of the last follow-up in the UK Biobank was March 31, 2021 for individuals enrolled in England and Scotland, and February 28, 2018 for individuals enrolled in Wales.
Phenotype-based ECG reconstructions
To better understand which ECG features may have contributed to Phecode segregation in latent space, we generated disease-specific median waveforms. Specifically, we decoded ECGs from disease-positive centroids and overlayed the resultant median waveforms on ECGs decoded from the corresponding disease-negative centroids.
Patient report card
One advantage of latent space modeling is the ability to incorporate an enormous amount of ECG- and EHR-based data in a multidimensional environment. The multidimensional nature of the model allows for simultaneous assessment of an ECG encoding’s proximity to all Phecode centroids. We sought to demonstrate the associated potential for personalized and scalable disease reporting by converting the position of an ECG encoding for a single individual into an estimated probability of disease. The probability of disease is estimated as the disease prevalence among all individuals in the MGH-C3PO test set with a vector component value greater than or equal to the individual’s value. The exact method was used to estimate 95% confidence intervals.
Supplementary information
Acknowledgements
Dr. Lubitz previously received support from NIH grants R01HL139731 and R01HL157635, and American Heart Association 18SFRN34250007. Dr. Anderson is supported by NIH grants R01NS103924 and U01NS069763 and American Heart Association grants 18SFRN34250007 and 21SFRN812095. Dr. Weng is supported by National Institutes of Health (NIH) grant 1R01HL139731. Dr. Choi is supported by the NHLBI BioData Catalyst Fellows program. Dr. Ellinor is supported by the NIH (1R01HL092577, K24HL105780), AHA (18SFRN34110082) and by MAESTRIA (965286). Dr. Lau is supported by the NIH (K23HL159243), the American Heart Association (853922), and the Massachusetts Life Sciences Center. Dr. Pirruccello is supported by the NIH (K08HL159346). Dr. Ho is supported by NIH grants R01HL134893, R01HL140224, R01HL160003, and K24HL153669. Dr. Khurshid is supported by NIH grant K23HL169839 and American Heart Association (23CDA1050571).
Author contributions
S.F.F., S.K., R.A.V., and X.W. contributed equally and are co-first authors. S.F.F., R.A.V., and S.A.L. conceived of the study. S.F.F., S.K., X.W., N.D., P.D.A., L.C.W., S.H.C., C.R., and P.S. contributed to study design, modeling, and statistical analysis. S.K., S.F.F., R.A.V., and S.A.L. drafted the manuscript. J.P.P., E.S.L., A.P., C.D.A., M.M., P.B., P.T.E., J.E.H., and S.A.L. performed critical reviews. All authors discussed the results, contributed to the final work, and have provided final approval of the completed version.
Data availability
The Mass General Brigham source data are not publicly available because they are electronic health records. Making the data publicly available without additional consent or ethical could compromise privacy. Source data from the UK Biobank are available to qualified investigators via application at https://www.ukbiobank.ac.uk.
Code availability
Data processing scripts underlying the current analyses, including the ECG autoencoder, are available at https://github.com/broadinstitute/ml4h/tree/master/model_zoo/ECG_PheWAS. The JEDI data processing pipeline underlying C3PO is available at https://github.com/broadinstitute/jedi-public.
Competing interests
Dr. Lubitz is a full-time employee of Novartis Institutes for Biomedical Research as of July 18, 2022. Dr. Lubitz has received sponsored research support from Bristol Myers Squibb, Pfizer, Boehringer Ingelheim, Fitbit, Medtronic, Premier, and IBM, and has consulted for Bristol Myers Squibb, Pfizer, Blackstone Life Sciences, and Invitae. Dr. Anderson receives sponsored research support from Bayer AG and Massachusetts General Hospital and has consulted for ApoPharma. Dr. Weng receives sponsored research support from IBM to the Broad Institute. Dr. Ellinor has received sponsored research support from Bayer AG and IBM Health, and he has consulted for Bayer AG, Novartis and MyoKardia. Dr. Batra, Dr. Reeder and Dr. Friedman have received sponsored research support from Bayer AG and IBM Health. Dr. Ho and Dr. Khurshid have received sponsored research support from Bayer AG. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sam F. Friedman, Shaan Khurshid, Rachael A. Venn, Xin Wang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41746-024-01418-9.
References
- 1.Trobec, R. & Tomašić, I. Synthesis of the 12-lead electrocardiogram from differential leads. IEEE Trans. Inf. Technol. Biomed.15, 615–621 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Barold, S. S. Willem Einthoven and the birth of clinical electrocardiography a hundred years ago. Card. Electrophysiol. Rev.7, 99–104 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Rivera-Ruiz, M., Cajavilca, C. & Varon, J. Einthoven’s string galvanometer: the first electrocardiograph. Tex. Heart Inst. J.35, 174–178 (2008). [PMC free article] [PubMed] [Google Scholar]
- 4.Salvati, M. et al. Electrocardiographic changes in subarachnoid hemorrhage secondary to cerebral aneurysm. Report of 70 cases. Ital. J. Neurol. Sci.13, 409–413 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Surawicz, B. Relationship between electrocardiogram and electrolytes. Am. Heart J.73, 814–834 (1967). [DOI] [PubMed] [Google Scholar]
- 6.Van Mieghem, C., Sabbe, M. & Knockaert, D. The clinical value of the ECG in noncardiac conditions. Chest125, 1561–1576 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Yasin, O. Z. et al. Noninvasive blood potassium measurement using signal-processed, single-lead ecg acquired from a handheld smartphone. J. Electrocardiol.50, 620–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Zaiti, S. et al. Machine learning-based prediction of acute coronary syndrome using only the pre-hospital 12-lead electrocardiogram. Nat. Commun.11, 3966 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cikes, M. et al. Machine learning-based phenogrouping in heart failure to identify responders to cardiac resynchronization therapy. Eur. J. Heart Fail21, 74–85 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Feeny, A. K. et al. Artificial Intelligence and Machine Learning in Arrhythmias and Cardiac Electrophysiology. Circ. Arrhythm. Electrophysiol.13, e007952 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannun, A. Y. et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med25, 65–69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attia, Z. I. et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat. Med25, 70–74 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Attia, Z. I. et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet394, 861–867 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Attia, Z. I. et al. Age and Sex Estimation Using Artificial Intelligence From Standard 12-Lead ECGs. Circ. Arrhythm. Electrophysiol.12, e007284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galloway, C. D. et al. Development and Validation of a Deep-Learning Model to Screen for Hyperkalemia From the Electrocardiogram. JAMA Cardiol.4, 428–436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghunath, S. et al. Prediction of mortality from 12-lead electrocardiogram voltage data using a deep neural network. Nat. Med. (2020) 10.1038/s41591-020-0870-z. [DOI] [PubMed]
- 17.Barak-Corren, Y. et al. Predicting Suicidal Behavior From Longitudinal Electronic Health Records. Am. J. Psychiatry174, 154–162 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Liu, C., Wang, F., Hu, J. & Xiong, H. Temporal Phenotyping from Longitudinal Electronic Health Records: A Graph Based Framework. in Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 705–714 (ACM, Sydney NSW Australia, 2015) 10.1145/2783258.2783352
- 19.Zhao, J. et al. Learning from Longitudinal Data in Electronic Health Record and Genetic Data to Improve Cardiovascular Event Prediction. Sci. Rep.9, 717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denny, J. C. et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol.31, 1102–1110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny, J. C. et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics26, 1205–1210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, M., Quan, Y., Zhou, X.-H. & Zhang, H.-Y. PheWAS-Based Systems Genetics Methods for Anti-Breast Cancer Drug Discovery. Genes (Basel)10, 154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Alusi, M. A., Ding, E., McManus, D. D. & Lubitz, S. A. Wearing Your Heart on Your Sleeve: the Future of Cardiac Rhythm Monitoring. Curr. Cardiol. Rep.21, 158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khurshid, S. et al. Cohort design and natural language processing to reduce bias in electronic health records research. npj Digit. Med.5, 47 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Littlejohns, T. J., Sudlow, C., Allen, N. E. & Collins, R. UK Biobank: opportunities for cardiovascular research. Eur. Heart J.40, 1158–1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan, N. Y., Witt, C. M., Oh, J. K. & Cha, Y.-M. Left Bundle Branch Block: Current and Future Perspectives. Circ. Arrhythm. Electrophysiol.13, e008239 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Levis, J. T. ECG diagnosis: hypokalemia. Perm. J.16, 57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finocchiaro, G. et al. The electrocardiogram in the diagnosis and management of patients with hypertrophic cardiomyopathy. Heart Rhythm17, 142–151 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Wolf, P. A., Abbott, R. D. & Kannel, W. B. Atrial fibrillation: a major contributor to stroke in the elderly. The Framingham Study. Arch. Intern. Med.147, 1561–1564 (1987). [PubMed] [Google Scholar]
- 30.Velagaleti, R. S. et al. Long-term trends in the incidence of heart failure after myocardial infarction. Circulation118, 2057–2062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maron, B. J. et al. Efficacy of implantable cardioverter-defibrillators for the prevention of sudden death in patients with hypertrophic cardiomyopathy. N. Engl. J. Med342, 365–373 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Wang, T. J. et al. Temporal relations of atrial fibrillation and congestive heart failure and their joint influence on mortality: the Framingham Heart Study. Circulation107, 2920–2925 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Tsao, C. W. et al. Heart Disease and Stroke Statistics-2022 Update: A Report From the American Heart Association. Circulation145, e153–e639 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Liu, Y., Jun, E., Li, Q. & Heer, J. Latent Space Cartography: Visual Analysis of Vector Space Embeddings. Comput. Graph. Forum38, 67–78 (2019). [Google Scholar]
- 35.Xiao, L. & Wang, J. LatentVis: Investigating and Comparing Variational Auto-Encoders via Their Latent Space. in, (2020).
- 36.Chen, S., Meng, Z. & Zhao, Q. Electrocardiogram Recognization Based on Variational AutoEncoder. in Machine Learning and Biometrics (eds. Yang, J., Park, D. S., Yoon, S., Chen, Y. & Zhang, C.) (InTech, 2018). 10.5772/intechopen.76434.
- 37.Liu, H., Zhao, Z., Chen, X., Yu, R. & She, Q. Using the VQ-VAE to improve the recognition of abnormalities in short-duration 12-lead electrocardiogram records. Comput. Methods Prog. Biomed.196, 105639 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Jang, J.-H., Kim, T. Y., Lim, H.-S. & Yoon, D. Unsupervised feature learning for electrocardiogram data using the convolutional variational autoencoder. PLoS ONE16, e0260612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van De Leur, R. R. et al. Improving explainability of deep neural network-based electrocardiogram interpretation using variational auto-encoders. Eur. Heart J. Digital Health3, 390–404 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordeiro, R., Karimian, N. & Park, Y. Hyperglycemia Identification Using ECG in Deep Learning Era. Sens. (Basel)21, 6263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, L., Mu, Y., Zhao, J., Wang, X. & Che, H. IGRNet: A Deep Learning Model for Non-Invasive, Real-Time Diagnosis of Prediabetes through Electrocardiograms. Sens. (Basel)20, 2556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, A., Chen, S., Quan, S. F., Powers, L. S. & Roveda, J. M. A deep learning-based algorithm for detection of cortical arousal during sleep. Sleep43, zsaa120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee, D., Dhar, K., Schwenker, F. & Sarkar, R. Ensemble of Deep Learning Models for Sleep Apnea Detection: An Experimental Study. Sens. (Basel)21, 5425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urtnasan, E., Park, J. U., Joo, E. Y. & Lee, K. J. Identification of Sleep Apnea Severity Based on Deep Learning from a Short-term Normal ECG. J. Korean Med Sci.35, e399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, H. et al. Sleep staging from electrocardiography and respiration with deep learning. Sleep43, zsz306 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, H., Guo, P. & Sano, A. Zero-Shot ECG Diagnosis with Large Language Models and Retrieval-Augmented Generation. in Proceedings of the 3rd Machine Learning for Health Symposium vol. 225 650–653 (2023).
- 47.Toma, L. et al. Electrocardiographic Changes in Liver Cirrhosis-Clues for Cirrhotic Cardiomyopathy. Med. (Kaunas.)56, 68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deo, R. et al. Electrocardiographic Measures and Prediction of Cardiovascular and Noncardiovascular Death in CKD. J. Am. Soc. Nephrol.27, 559–569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin, C.-S. et al. AI-enabled electrocardiography alert intervention and all-cause mortality: a pragmatic randomized clinical trial. Nat. Med30, 1461–1470 (2024). [DOI] [PubMed] [Google Scholar]
- 50.Khurshid, S. et al. Deep Learning to Predict Cardiac Magnetic Resonance-Derived Left Ventricular Mass and Hypertrophy From 12-Lead ECGs. Circ. Cardiovasc Imaging14, e012281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alain, G. & Bengio, Y. Understanding intermediate layers using linear classifier probes. Preprint at http://arxiv.org/abs/1610.01644 (2018).
- 52.Maweu, B. M., Dakshit, S., Shamsuddin, R. & Prabhakaran, B. CEFEs: A CNN Explainable Framework for ECG Signals. Artif. Intell. Med115, 102059 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Carreiras, C. et al. BioSPPy - Biosignal Processing in Python. (2015).
- 54.Verweij, N. et al. The Genetic Makeup of the Electrocardiogram. Cell Syst.11, 229–238.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iandola, F. et al. DenseNet: Implementing Efficient ConvNet Descriptor Pyramids. Preprint at http://arxiv.org/abs/1404.1869 (2014).
- 56.Snoek, J., Larochelle, H. & Adams, R. P. Practical Bayesian Optimization of Machine Learning Algorithms. in (2012).
- 57.Brenyo, A. & Zaręba, W. Prognostic significance of QRS duration and morphology. Cardiol. J.18, 8–17 (2011). [PubMed] [Google Scholar]
- 58.Castiglione, A. & Odening, K. [QT Interval and Its Prolongation - What Does It Mean?]. Dtsch Med Wochenschr.145, 536–542 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Chen, X. et al. Incremental changes in QRS duration as predictor for cardiovascular disease: a 21-year follow-up of a randomly selected general population. Sci. Rep.11, 13652 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Estrada, A. H. et al. Diagnostic accuracy of computer aided electrocardiogram analysis in dogs. J. Small Anim. Pr.62, 145–149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rasmussen, P. V. et al. Electrocardiographic PR Interval Duration and Cardiovascular Risk: Results From the Copenhagen ECG Study. Can. J. Cardiol.33, 674–681 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Mass General Brigham source data are not publicly available because they are electronic health records. Making the data publicly available without additional consent or ethical could compromise privacy. Source data from the UK Biobank are available to qualified investigators via application at https://www.ukbiobank.ac.uk.
Data processing scripts underlying the current analyses, including the ECG autoencoder, are available at https://github.com/broadinstitute/ml4h/tree/master/model_zoo/ECG_PheWAS. The JEDI data processing pipeline underlying C3PO is available at https://github.com/broadinstitute/jedi-public.