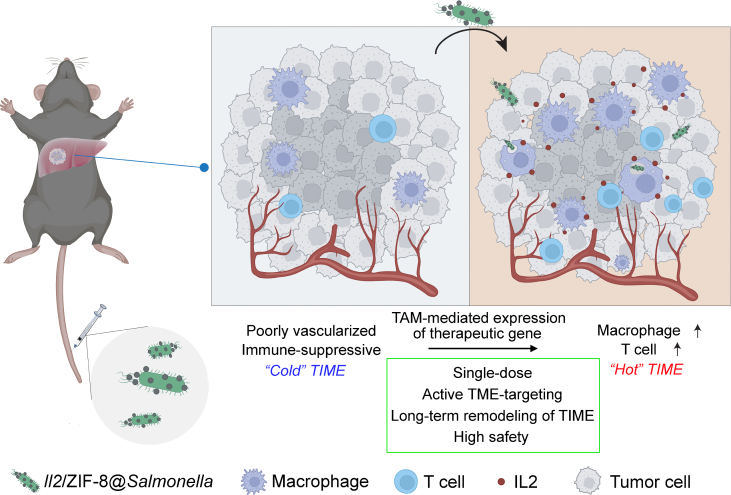
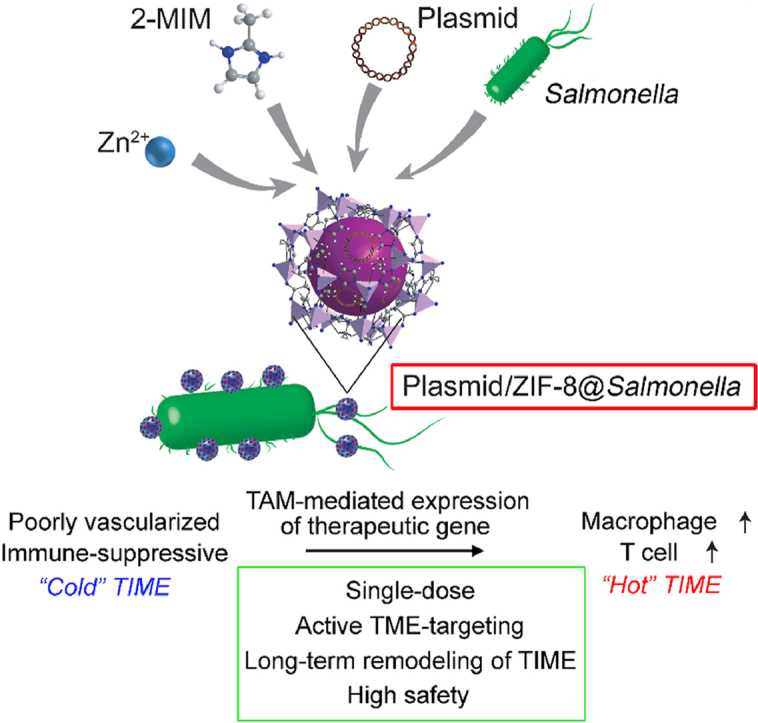
Abstract
Specific tumor-targeted gene delivery remains an unsolved therapeutic issue due to aberrant vascularization in tumor microenvironment (TME). Some bacteria exhibit spontaneous chemotaxis toward the anaerobic and immune-suppressive TME, which makes them ideal natural vehicles for cancer gene therapy. Here, we conjugated ZIF-8 metal-organic frameworks encapsulating eukaryotic murine interleukin 2 (Il2) expression plasmid onto the surface of VNP20009, an attenuated Salmonella typhimurium strain with well-documented anti-cancer activity, and constructed a TME-targeted Il2 delivery system named Il2/ZIF-8@Salmonella. Both in vitro and in vivo experiments demonstrated that Il2/ZIF-8@Salmonella maintained the tumor-targeting feature of bacteria, and could be effectively phagocytosed by intratumoral macrophages, thus leading to the expression and secretion of IL2 in TME. The detailed analysis of tumor immune microenvironment (TIME) showed that one dose of combinatorial Il2/ZIF-8@Salmonella achieved synergistic actions on a potent remodeling of TIME, marked by the activation of cytotoxic T cells and M1-polarization of macrophages in TME, thus leading to significant anti-tumor effects in melanoma, orthotopic hepatocellular carcinoma, and pulmonary metastasis models. More importantly, Il2/ZIF-8@Salmonella exhibited high safety to major organs and hematopoietic systems. Taken together, we report a novel plasmid/ZIF-8@Salmonella system that simultaneously achieves effective TME-targeted delivery of therapeutic gene, as well as synergistic re-activation of TIME.
Key words: Bacteria-mediated cancer therapy, VNP20009, ZIF-8, Gene delivery, Tumor microenvironment, Tumor immune microenvironment, Interleukin 2, Cancer immune therapy
Graphical abstract
We report a novel plasmid/ZIF-8@Salmonella system which simultaneously achieves effective tumor microenvironment-targeted delivery of therapeutic gene, as well as synergistic re-activation of tumor immune microenvironment.
1. Introduction
The attempts to combat cancer by bacteria could be traced to Coley's toxins consisting of heat-inactivated Streptococcus pyogenicus and Serratia marcescens since the late 19th century, originating from the surgeon's occasional observations of spontaneous tumor regression following severe bacterial infection1. An attenuated Salmonella typhimurium strain, VNP20009, is featured by deep tissue penetration, selective intratumoral proliferation, sustained anti-cancer effect, and antibiotic-dependent reversibility2,3. However, its clinical translation encountered stagnation due to unsatisfactory response rate at safe dosage during phase I trail4. There is an urgent need to improve its therapeutic efficacy and safety profile.
The active tumor-targeting behavior of therapeutic bacteria is mostly attributed to the hypoxic and immune-suppressive niche in the tumor microenvironment (TME), which favors the settlement and proliferation of anaerobic or facultative aerobe microbes5. However, immune-escaping signals gradually dominate TME as tumors progress6, and Bacteria-mediated Cancer Therapies (BCTs) suppress tumor growth by tilting the balance of tumor and immune system in favor of anti-tumor immune response. For tumor cells, cytotoxic bacteria lead to cytolysis, followed by the release of tumor-specific antigens and immune-activating molecules7,8. For immune cells, bacteria activate them towards a pro-inflammatory status by pathogen-associated molecule pattern (PAMP), thus simultaneously potentiating their cytotoxicity against tumor cells9. However, the anti-cancer immune responses induced by BCTs are seriously dampened by immune-suppressive TME, which could partially explain the disparity of therapeutic effects in murine models and human patients10. Therefore, targeted remodeling of the tumor immune microenvironment (TIME) is a reasonable strategy to overcome its adverse influences on BCTs.
Cytokines are immune-regulating molecules that could be adopted for TME remodeling. For example, Interferon α and interleukin-2 (IL2) could enhance T cell infiltration and activation in solid tumors, and their recombinant proteins have been clinically applied as adjuvant cancer therapy for decades11. However, systemic infusion of recombinant cytokines has the drawbacks of short dosing intervals associated with fast renal clearance, flu-like syndromes and gastrointestinal reactions due to systemic immune effects12. Recently, TME-targeted delivery of cytokines has been achieved by nanoparticles of various forms in pre-clinical studies, with significantly improved pharmacokinetic characteristics13,14. Meanwhile, the delivery strategies of cytokine genes, instead of recombinant proteins, are also intensively studied, which further optimizes the dosing frequency, biological activity, and manufacture process for cytokine-mediated cancer therapies15. There have been some attempts to combine BCTs and pro-inflammatory cytokines via prokaryotic expression plasmids, and it is worthwhile to rationally integrate BCTs and cytokine-mediated cancer therapy through eukaryotic expression plasmids16.
Metal-organic frameworks (MOFs) are a category of porous nanoparticles composed of self-assembly organic ligands (linkers) and metal ions (nodes) such as Zinc17. MOFs have the advantages of large loading capacity, mild synthesis condition, high biocompatibility, good stability in neutral condition, and acid-responsive disintegration18. Systemically circulating MOFs could be passively enriched to TME by enhanced permeability and retention (EPR) effects associated with impaired vascular endothelial and blood-lymphatic circulation in tumors19. However, MOF-mediated specific delivery of therapeutic agents into the poorly vesiculated core of tumors remains challenging. Combining MOFs and BCTs by biomimetic mineralization, a process in which living organisms translate mineral elements into their biological matrix20, might overcome this problem by bacteria's intrinsic tropism towards such hypoxic areas in tumor.
In this work, we proposed a TME-targeted gene delivery system, plasmid/ZIF-8@Salmonella, in which ZIF-8 particles containing plasmids are loaded onto live VNP20009. Plasmid/ZIF-8@Salmonella could be enriched in TME, and efficiently phagocytosed by tumor-associated macrophages (TAMs), thus leading to the expression and secretion of therapeutic proteins. One dosage of plasmid/ZIF-8@Salmonella carrying murine interleukin-2 (Il2) gene exhibited significantly improved tumor-suppressive effects in both melanoma and orthotopic hepatocellular carcinoma (HCC) models, compared to monotherapy of either Il2/ZIF-8 particles or Salmonella, while exhibiting low toxicity to major organs as well as hematopoietic system. Mechanistically, Il2/ZIF-8@Salmonella increased IL2 level in TME, leading to the re-activation of anti-tumor immune responses, marked by increased number of cytotoxic T lymphocytes (CTL) and macrophages, as well as a higher activation level of CTL and M1-polarization of macrophages.
2. Materials and methods
2.1. Cell lines and animals
B16/F10, Hepa1-6, RAW264.7 cell lines were obtained from Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). These cell lines were maintained with DMEM (Biological Industries Ltd., #01-052-1A, Kibbutz Beit-Haemek, Israel) or RPMI-1640 (Biological Industries Ltd., #01-100-1A, Kibbutz Beit-Haemek, Israel) supplemented with 10% fetal bovine serum (FBS) (Biological Industries Ltd., #04-001-1ACS, Kibbutz Beit-Haemek, Israel), and 1% penicillin streptomycin (Meilunbio Co., Ltd., #MA0110, Dalian, China) at a humidified incubator (Thermo Fisher Scientific Ltd., FORMA STERI-CYCLE i160, Waltham, MA, USA) with 5% CO2 at 37 °C. Bone marrow-derived macrophages (BMDMs) were collected from the femurs of 8-week-old C57BL/6 mice according to an established protocol and cultured in BMDM complete medium composed of RPMI-1640 medium supplemented with 10% FBS, 1% penicillin streptomycin, 20 ng/mL mGM-CSF (R&D Systems Ltd., #415-ML-020, Minneapolis, MN, USA)21.
Salmonella Typhimurium strain VNP20009 was obtained from American Type Culture Collection (#202165, Manassas, VA, USA) and cultured with Luria‒Bertani (LB) agar plates and LB Broth at 37 °C. The bacteria density in LB Broth was measured according to its optical density (OD) at 600 nm with spectrophotometer (Thermo Fisher Scientific Ltd., Multiskan FC, Waltham, MA, USA). Cells were harvested for further applications at the exponential growth phase.
Six-week-old male C57BL/6 mice were obtained from Ziyuan Experimental Animal Technology (Hangzhou, China) and kept in a specific pathogen free facility in Hangzhou Normal University. The facility followed a 12-h light/dark cycle at controlled room temperature of 72 °F and a relative humidity of 60%.
2.2. Plasmid preparation
Firstly, pcDNA3.1-Il2-3 × Flag-C (NCBI Reference Sequence: NM_008366.3, REPOBIO Co., Ltd., Guangzhou, China) was transformed into competent Escherichia coli cells. Later, a single colony of transformed cells was inoculated into Kanamycin+ (Sangon Biotech Co., Ltd., #A101-03, Shanghai, China) LB broth and cultured at 37 °C with constant shaking overnight. Plasmids were extracted by Miniprep kit (Sangon Biotech Co., Ltd., #DR0202050, Shanghai, China) and quantified by Ultramicro-ultraviolet spectrophotometer (Thermo Fisher Scientific Ltd., Nanodrop One, Waltham, MA, USA) following the instructions of the manufacturer and stored at −20 °C for further usage.
2.3. Synthesis and characterization of ZIF-8, Il2/ZIF-8, ZIF-8@Salmonella and Il2/ZIF-8@Salmonella
For the synthesis of ZIF-8 particles, 0.192 mg/mL ZnSO4‧6H2O (Macklin Biochemical Co., Ltd., #Z820749, Shanghai, China) aqueous solution was dispersed into an equal volume of 1.9 mg/mL 2-MIM (TCI America Ltd., #M0345, Portland, OR, USA) aqueous solution drop by drop with constant agitation, and gently stirred for 1 min. ZIF-8 particles formed after standing for 15 min. Then, the particles were collected by centrifugation at 1000×g for 15 min and washed with ultrapure water three times (Eppendorf Ltd., Centrifuge 5425R, Hamburg, Germany). For the synthesis of Il2/ZIF-8 particles, the procedure remained the same except for adding pcDNA3.1-Il2-3 × Flag-C during the mixture preparation.
For the synthesis of ZIF-8@Salmonella, 107 CFU VNP20009 was washed with ultrapure water three times and resuspended in 300 μL of water containing 108 μg 2-MIM followed by gentle stirring for 1 min. Subsequently, 300 μL of aqueous solution containing 10.8 μg ZnSO4·6H2O was dispersed into the VNP20009/2MIM mixture drop by drop with constant agitation. The mixture was stirred for 1 min. ZIF-8@Salmonella particles formed after standing for 15 min. Then, the particles were collected by centrifugation at 1000×g for 15 min and washed with ultrapure water three times (Eppendorf Ltd., Centrifuge 5425R, Hamburg, Germany). For the synthesis of Il2/ZIF-8@Salmonella, 0.75 μg plasmids (1011 copies) were added into the VNP20009/2-MIM mixture. The remaining steps were the same as ZIF-8@Salmonella.
The morphology, surface charge, and particle size were analyzed by scanning electron microscope (Carl Zeiss Ltd., Supra 55, Jena, Germany) and dynamic light scatterer (Malvern, Zetasizer Nano ZS, Shanghai, China).
2.4. Agarose gel electrophoresis of Il2/ZIF-8 and Il2/ZIF-8@Salmonella
Agarose gel electrophoresis was used to evaluate the amount of plasmid encapsulated by ZIF-8 particles. In brief, Il2/ZIF-8 suspension was centrifuged at 1000×g for 30 min (Eppendorf Ltd., Centrifuge 5425R, Hamburg, Germany), and supernatants were loaded onto 0.8% agarose gels and subject to electrophoresis at 100 V for 50 min Il2/ZIF-8 and Il2/ZIF-8@Salmonella precipitants were washed with ultrapure water and treated with 24 mg/mL EDTA (Sinopharm Chemical Reagent Co., Ltd., #10009617, Shanghai, China) to release the plasmids by lysing ZIF-8 particles, followed by electrophoresis. Later, the gels were visualized by 260 nm UV (Clinx Co., Ltd., ChemiScope 6200, Shanghai, China).
2.5. Organ distribution assay of Salmonella, ZIF-8@Salmonella and Il2/ZIF-8@Salmonella
To study the in vivo distribution of bacteria, B16/F10 cells (105 cells per mouse) were inoculated subcutaneously into the right flanks of C57BL/6 mice. Il2/ZIF-8@Salmonella, ZIF-8@Salmonella, and Salmonella were injected via tail vein at a dose of 107 CFU/mouse when tumor volume reached approximately 400 mm3. Afterward, the mice were sacrificed on Day 3 or 10 after bacteria injection. Livers, spleens, and tumors were harvested. The excised organs and tumor tissues were weighted and homogenized in sterile PBS by Dounce tissue grinders, and the supernatants were collected after removing tissue debris by 200×g centrifugation (Eppendorf Ltd., Centrifuge 5425R, Hamburg, Germany). Then, the volume of supernatants was quantified. Finally, a series of gradient dilution was performed for the collected supernatants, and 100 μL of the diluted supernatants were spread on LB plates and cultured at 37 °C for 12 h, and the number of bacterial colonies was quantified for the plates of suitable dilution ratio.
2.6. In vivo distribution analysis of Dir, DiR/ZIF-8, and DiR/ZIF-8@Salmonella
DiR/ZIF-8 and DiR/ZIF-8@Salmonella were synthesized following method mentioned above, and plasmids were replaced by Dir (US Everbright Inc., #D4006, Suzhou, China) for fluorescent imaging assay. 5 μg DiR was used for 107 CFU of bacteria. Hepa1-6 cells (1 × 105 cells/mouse) were inoculated subcutaneously at the flank of C57BL/6 mice to construct HCC xenograft model. When the xenograft volumes reached approximately 500 mm3, mice were randomly divided into three groups (n = 5). Free Dir, DiR/ZIF-8, and DiR/ZIF-8@Salmonella (107 CFU/mouse) were intravenously administrated at a dose of 5 μg DiR/mouse. Fluorescent images (750 nmEx/780 nmEm) of anesthetized mice were obtained by a fluorescent imaging instrument for live animals (Biospace Lab Ltd., PHOTON IMAGER™ OPTIMA, Nesles-la-Vallée, France) at 0.5, 2, 6, 24, and 48 h after dosing. One mouse was sacrificed at 24 and 48 h in each group, and the hearts, livers, spleens, lungs, kidneys, intestines and xenografts were harvested, rinsed with saline, and further imaged by PHOTON IMAGER™ OPTIMA (Biospace Lab Ltd.).
2.7. In vitro particle uptake efficacy assay
For in vitro particle uptake assay by flow cytometry, Il2 plasmids were fluorescently labeled with propidium iodide (PI) (BD Biosciences Ltd., #550825, Franklin Lakes, NJ, USA) at the ratio of 1:1 (w/w). Il2/ZIF-8 and Il2/ZIF-8@Salmonella particles were synthesized following method mentioned above. Cells were seeded on 6-well plates at the density of 3 × 105 cells/well and cultured overnight. Then cells were incubated with various particles in antibiotic-free media for 2 h. For each well, 1 × 1010 copies of plasmids, 12 μg ZIF-8, and/or 106 CFU of Salmonella were used.
For flow cytometry assay, cells were trypsinized and harvested by 200×g centrifugation (Eppendorf Ltd., Centrifuge 5425R, Hamburg, Germany). Cell pellet was washed with chilled PBS three times, resuspended and filtered through 35 μm nylon mesh. The signals of PI (488 nmEX/617 nmEm) were analyzed by flow cytometer (Beckman Coulter Ltd., Moflo Atstrios Eqs, Brea, CA, USA).
For fluorescent microscope imaging, cells were rinsed with chilled PBS three times and immediately fixed with 4% paraformaldehyde for 1 h at 4 °C. Then, cells were mounted with ProLong™ Glass Antifade Mountant (Invitrogen Ltd., #P36980, Carlsbad, CA, USA). After the sample had been cured, cells were imaged by a confocal laser scanning microscope (OLYMPUS Co., Ltd., FV3000RS, Tokyo, Japan).
For the application of endocytosis inhibitors, RAW264.7 cells were pre-treated with solvent control, 450 μmol/L Amiloride hydrochloride (MedChemExpress Ltd., #HY-B0285A, Monmouth Junction, NJ, USA), 2 μmol/L Chlorpromazine (MedChemExpress Ltd., #HY-B0407A, Monmouth Junction, NJ, USA), 50 μmol/L Nystatin (MedChemExpress Ltd., #HY-17409, Monmouth Junction, NJ, USA), or 3 μmol/L Chloroquine phosphate (MedChemExpress Ltd., #HY-17589, Monmouth Junction, NJ, USA) for 4 h before the addition of plasmid, Il2/ZIF-8, or Il2/ZIF-8@Salmonella particles. Then, the cells were used for subsequent particle uptake assays.
2.8. In vitro IL2 expression efficacy assay
Cells were seeded on 6-well plates at the density of 3 × 105 cells/well and cultured overnight. Then the cells were treated with plasmids, ZIF-8, Il2/ZIF-8, ZIF-8@Salmonella and Il2/ZIF-8@Salmonella, and PolyJet™ DNA In Vitro Transfection Reagent (SignaGen Laboratories Ltd., #SL100688, Frederick, MD, USA) was used as a positive control for plasmid transfection efficacy. For each well, 1 × 1010 copies of plasmids, 12 μg ZIF-8, and/or 106 CFU of Salmonella were used. Cells were treated with various regimes in antibiotic-free media for 2 h. Then, the media were discarded. Cells were rinsed twice with sterile PBS and cultured with fresh complete medium with 1% penicillin streptomycin for 48 h. Then, the cells were subject to qRT-PCR, flow cytometry or fluorescent microscope imaging assays to analyze Il2 expression level.
For qRT-PCR assay, primers used were as follows: Actb forward primer (TTGATGGCAACAATCTCCAC); Actb reverse primer (CGTCCCGTAGACAAAATGGT); Il2 forward primer (CACCCACTTCAAGCTCCAC); Il2 reverse primer (TTGAGTCAAATCCAGAACAT). Detailed protocol was described in supporting information.
For flow cytometry assay, APC-conjugated anti-mouse IL2 antibody (Biolegend Ltd., #503809, San Diego, CA, USA) was used. Detailed protocol was described in supporting information.
For fluorescent microscope imaging assay, cells were imaged by FV3000RS confocal laser scanning microscope (OLYMPUS Co., Ltd.), and images of serial sections were compressed into one image to show the overall expression level of IL2. Detailed protocol was described in Supporting Information Materials and Methods.
2.9. The in vivo evaluation of anti-tumor efficacy in melanoma model, orthotopic HCC model, and melanoma pulmonary metastasis model
All animal experiments complied with the ethics of laboratory animals and approved by the Animal Ethics Committee of Hangzhou Normal University (HSD20221101, HSD20221003).
For the therapeutic efficacy assay in the xenograft model, B16/F10 (105 cells per mouse) were injected subcutaneously into the right flanks of C57BL/6 mice. Mice were divided into the required number of groups (n = 6) when tumor volume reached approximately 200 mm3. One single dose of Il2/ZIF-8@Salmonella (107 CFU/mouse), ZIF-8@Salmonella, Il2/ZIF-8, ZIF-8, Il2 plasmid (1011 copies/mouse), Salmonella or PBS were injected through the caudal vein for each group. The minor radius (a, mm) and the major radius (b, mm) of tumors were measured by calipers every day, and the volumes (V, mm3) were calculated as shown in Eq. (1):
| V = a2 × b × 0.52 | (1) |
At the end of experiments, mice were sacrificed. Tumors were harvested and weighted. A part of the tumor tissues was fixed in 10% formalin and subsequently analyzed by immunohistochemistry, and the rest was cryopreserved for RNA analysis. The RNA extraction and Il2 qPCR were conducted following procedures mentioned above.
For the therapeutic efficacy assay in the orthotopic HCC model, Hepa1-6 cells (Hepa1-6-luc) stably expressing luciferase (5 × 104 cells per mouse) were inoculated with 50% Matrigel (Corning Inc., #354248, Corning, NY, USA) into the left liver lobe under the capsule. On Day 5 after tumor inoculation, the mice were randomly divided into 4 groups (n = 6), and a single dose of Il2/ZIF-8@Salmonella (107 CFU/mouse), Il2/ZIF-8, Salmonella (103 CFU/mouse) or PBS was injected through the caudal vein for each group. On Day 16 after tumor inoculation, D-Luciferin potassium salt (Yeasen Biotechnology Co., Ltd., #40901ES03, Shanghai, China) was injected intraperitoneally, and the anesthetized mice were imaged by PHOTON IMAGER™ OPTIMA (Biospace Lab Ltd.). Then, the mice were sacrificed, and the tumors were resected and fixed in 10% formalin and subsequently analyzed by immunohistochemistry.
Melanoma pulmonary metastasis model was established by intravenous injection of B16/F10 (105 cells per mouse) via tail vein, and the dosing scheme was same with orthotopic HCC model.
2.10. Immunohistochemical (IHC) staining
The resected tumors were fixed with 10% formalin, embedded in paraffin and sectioned. IHC staining was performed according to standard histological procedures, with Ki67 antibody (Abclonal Technology Co., Ltd., #A20018, Wuhan, China), cleaved Caspase-3 antibody (Cell Signaling Technology Inc., #9664S, Danvers, MA, USA), CD8α antibody (Santa Cruz Biotechnology Ltd., #sc-7970, Dallas, TX, USA), CD4 antibody (Abcam Ltd., #ab288724, Cambridge, UK), F4/80 antibody (Abcam Ltd., #Αb111101, Cambridge, UK), and IL2 antibody (Proteintech Inc., #60306-1-lg, Rosemont, IL, USA). The slides were viewed and imaged by SLIDEVIEW™ imaging system (OLYMPUS Co., Ltd., VS200, Tokyo, Japan).
2.11. Liver, kidney, spleen and hematological toxicity analysis
To thoroughly measure the toxicity to the liver, kidney and spleen for various formulations, melanoma-bearing mice were randomly divided into 8 groups (n = 3). Mice received one single dose of Il2/ZIF-8@Salmonella (107 CFU/mouse), ZIF-8@Salmonella (107 CFU/mouse), Il2/ZIF-8, ZIF-8, Il2 plasmid (1011 copies/mouse), Salmonella (103 CFU/mouse) Salmonellahigh (107 CFU/mouse) or PBS by tail-intravenous injection. Peripheral blood was collected by retro-orbital puncture on Day 8 after treatment, which were kept at room temperature for 4 h and centrifuged at 1000 g (Eppendorf Ltd., Centrifuge 5425R, Hamburg, Germany) to extract serum for the subsequent analysis of Alanine Aminotransferase (ALT), Aspartate Aminotransferase (AST), Crea and Urea concentrations by Laboratory Animal Resources Center, Hangzhou Normal University. Then, the mice were sacrificed, and the livers, kidneys and spleens were harvested and fixed in 10% formalin and subsequently analyzed by Hematoxylin and eosin (H&E) staining. The slides were viewed and imaged by SLIDEVIEW™ imaging system (OLYMPUS Co., Ltd.).
To measure the hematological toxicity, melanoma-bearing mice were randomly divided into 7 groups (n = 4). Mice received one single dose of Il2/ZIF-8@Salmonella (107 CFU/mouse), ZIF-8@Salmonella (107 CFU/mouse), Il2/ZIF-8, ZIF-8, Il2 plasmid (1011 copies/mouse), Salmonella (103 CFU/mouse) or PBS by tail-intravenous injection. Peripheral blood was collected into anticoagulant tubes by retro-orbital puncture on Day 3 after treatment, and immediately analyzed by blood routine examination in Laboratory Animal Resources Center, Hangzhou Normal University.
2.12. Flow cytometry analysis of immune cells in tumor tissues and spleens
B16/F10 (105 cells per mouse) were injected subcutaneously into the right flanks of C57BL/6 mice. Mice were divided into the required number of groups (n = 3) when tumor volume reached approximately 400 mm3. One single dose of Il2/ZIF-8@Salmonella (107 CFU/mouse), ZIF-8@Salmonella (107 CFU/mouse), Il2/ZIF-8, ZIF-8, Il2 plasmid (1011 copies/mouse), Salmonella (103 CFU/mouse) or PBS was administered by tail-intravenous injection. Then tumor tissues and spleens were harvested on Day 3 after treatment. Cell preparation protocol was described in supporting information. The immune cells were stained by the following antibodies: Brilliant Violet 605™ anti-mouse CD45 (BioLegend Ltd., #103140, San Diego, CA, USA), FITC anti-mouse F4/80 (BioLegend Ltd., #123107, San Diego, CA, USA), APC anti-mouse/human CD11b (BioLegend Ltd., #101212, San Diego, CA, USA), PerCP/Cyanine5.5 anti-mouse CD3ε (BioLegend Ltd., #100328, San Diego, CA, USA), KIRAVIA Blue 520™ anti-mouse CD4 (BioLegend Ltd., #100478, San Diego, CA, USA), PE/Cyanine7 anti-mouse CD8a (BioLegend Ltd., #100722, San Diego, CA, USA), APC anti-mouse CD69 (BioLegend Ltd., #104513, San Diego, CA, USA), PE/Cyanine7 anti-mouse CD206 (BioLegend Ltd., #141719, San Diego, CA, USA), PE anti-Nos2 (iNOS) (BioLegend Ltd., #696805, San Diego, CA, USA), Zombie NIR™ Fixable Viability Kit (BioLegend Ltd., #423106, San Diego, CA, USA), and TruStain FcX™ anti-mouse CD16/32 (BioLegend Ltd., #101319, San Diego, CA, USA) following the manufacturer's instructions. Then, cells were analyzed by flow cytometry (Beckman Coulter Ltd., Moflo Atstrios Eqs).
2.13. Single-cell RNA sequencing by 10x Genomics chromium GEX and data analysis
Microdroplet generation, single cell encapsulation and nucleic acid capture were performed by a Single cell preparator (M20 Genomics Co., Ltd., VITAcruizer V1.0, Hangzhou, China). RNA library containing P5 and P7 adapter was prepared and purified by VITApiloteHigh-Throughput Eukaryotic Single-Cell Transcriptome Kits (Frozen) (M20 Genomics Co., Ltd., #R20122124, Hangzhou, China). Then the library was sequenced by high-throughput sequencer (Illunima Inc., Novaseq 6000, San Diego, CA, USA). VITAseer Software and Data Platform from M20 Genomics were used for data analysis.
10x Genomics scRNA-seq gene expression raw sequencing data were processed using the CellRanger software v.7.1.0 (10x Genomics Ltd., Pleasanton, CA, USA) and 10X human transcriptome GRCh38-2020-A as reference. Sequential bioinformatic analysis procedure was described in Supporting Information.
2.14. Statistical analysis
Data were analyzed with GraphPad Prism software version 8 (GraphPad Software Ltd., Boston, MA, USA). Grouped data of two independent groups were compared using Student’ t test. Data were expressed as mean ± standard deviation (SD). Statistical significance was confirmed when P < 0.05. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant.
3. Results
3.1. The design, preparation, characterization and in vivo distribution of Il2/ZIF-8@Salmonella
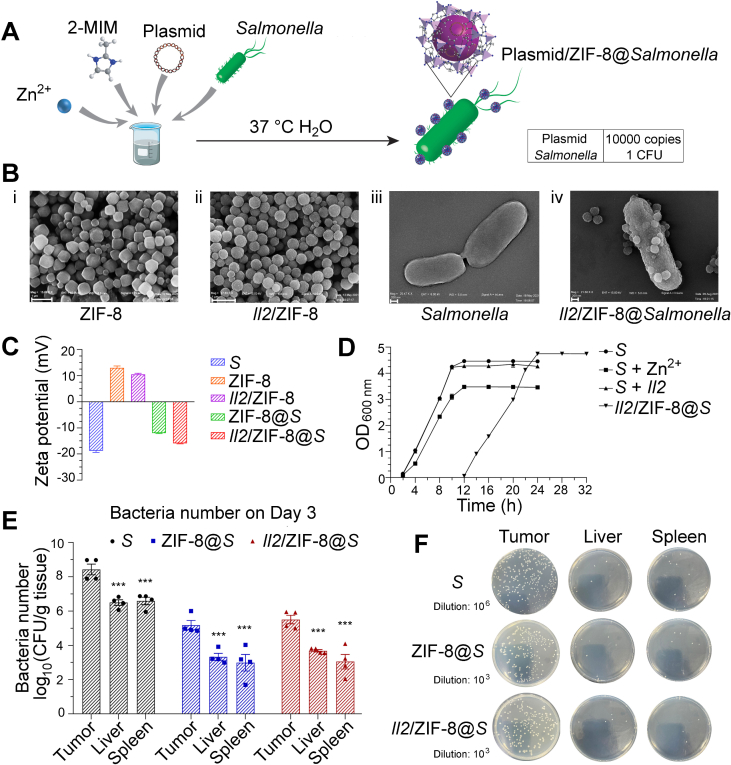
pcDNA-Il2 plasmid was encapsulated into ZIF-8 coating layer on the surface of Salmonella (VNP20009) via the one-pot biomimetic mineralization method at a ratio of 10,000 plasmids to 1 bacterium in water at 37 °C (Fig. 1A). The ratio of plasmids to ZIF-8 was 1–150 (w/w), at which the maximal amount of plasmid was encapsulated into ZIF-8 frames while maintaining the dispersion of ZIF-8 as nanoparticles as shown by gel electrophoresis analysis (Supporting Information Fig. S1A and B). Plasmid encapsulation efficiency was higher than 95% in this combination (Fig. S1C). The morphology of ZIF-8 particles and Salmonella was characterized with transmission electron microscopy. As shown in Fig. 1B, pure ZIF-8 particles were mainly regular dodecahedrons, and ZIF-8 particles containing plasmids turned into rough spheres. Salmonella displayed a smooth surface and low contrast, while Il2/ZIF-8@Salmonella exhibited a rough surface and high contrast. A thin layer of plasmid/ZIF-8 particles was attached to the surface of bacteria, with a few protrusions of round plasmid/ZIF-8 spheres (Fig. 1B). Consistently, the negative surface potential of Salmonella reduced slightly after the loading of ZIF-8 particles carrying positive charge (Fig. 1C).
Figure 1.
The design, preparation, characterization and in vivo distribution of Il2/ZIF-8@Salmonella. (A) The one-pot preparation method of Il2/ZIF-8@Salmonella via biomimetic mineralization. (B) Representative TEM images of ZIF-8 particles, scale bar = 1 μm (i), Il2/ZIF-8 particles, scale bar = 1 μm (ii), Salmonella, scale bar = 200 nm (iii), and Il2/ZIF-8@Salmonella, scale bar = 200 nm (iv). (C) Zeta potential of Salmonella (abbreviated as “S” in this figure and successive figures), ZIF-8 particles, Il2/ZIF-8 particles, ZIF-8@Salmonella, and Il2/ZIF-8@Salmonella. n = 3. Data were expressed as mean ± SD. (D) Cell proliferation curves of Salmonella, the mixture of Zi2+ and Salmonella, the mixture of pcDNA-Il2 plasmid and Salmonella, and Il2/ZIF-8@Salmonella. n = 3. Data were expressed as mean ± SD. (E) Live bacteria titers in the melanoma tumor tissues, livers and spleens on Day 3 after the intravenous injection of Salmonella, ZIF-8@Salmonella, and Il2/ZIF-8@Salmonella at the dosage of 107 CFU/mouse. n = 4. Data were expressed as mean ± SD. Students' t test was carried out using intratumoral titer as control for each treatment regime. ∗∗∗P < 0.001. (F) Representative images of bacterial colonies produced by homogenized melanoma tumor tissues, livers, and spleens on Day 3 after the intravenous injection of Salmonella, ZIF-8@Salmonella and Il2/ZIF-8@Salmonella at the dosage of 107 CFU/mouse.
Cell growth analysis showed that the loading of ZIF-8 particles onto bacteria led to a 10-h retardation for the logarithmic growth phase, without significant reduction of proliferation rate as shown by similar slopes for Salmonella, ZIF-8@Salmonella, and Il2/ZIF-8@Salmonella (Fig. 1D). The in vivo bacterial distribution was analyzed by measuring bacterial titer in livers, spleens and tumors on Day 3 and 10 after intravenous administration of 107 CFU Salmonella/mouse, ZIF-8@Salmonella, and Il2/ZIF-8@Salmonella. The results showed that both ZIF-8@Salmonella and Il2/ZIF-8@Salmonella had a 100-fold enrichment in tumors compared to livers and spleens, which was similar to free Salmonella (Fig. 1E and F, Supporting Information Fig. S2A and B). The bacteria numbers and distribution patterns were similar between Day 3 and 10, and such self-limiting behavior indicated the safety of Il2/ZIF-8@Salmonella-mediated cancer therapy.
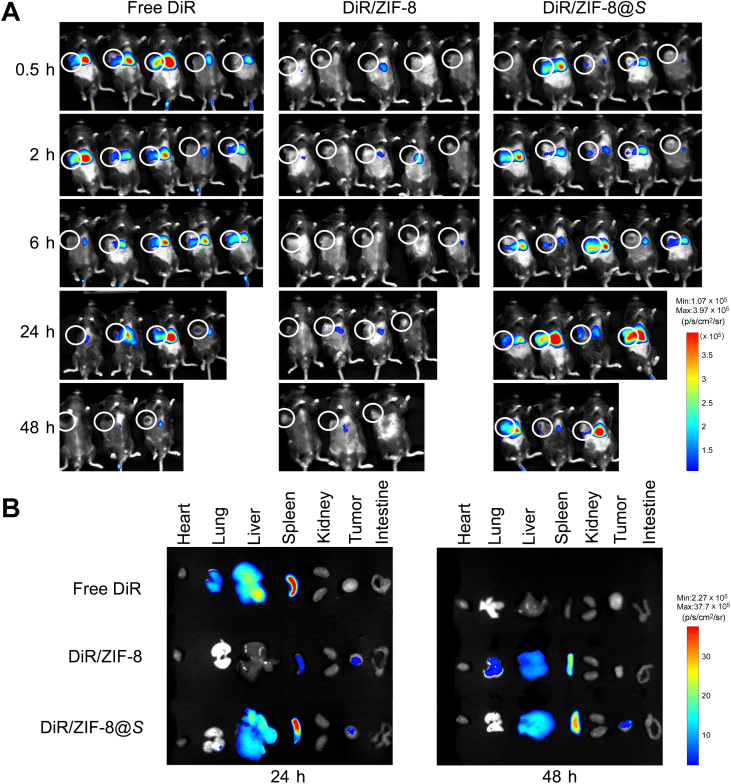
Fluorescent in vivo imaging assay showed that DiR/ZIF-8@Salmonella exhibited the most durable localization in tumors, compared to free DiR and DiR/ZIF-8 particles (Fig. 2A), and the fluorescent signals of dissected tumors and major organs showed a similar trend at 24 and 48 h after dosing (Fig. 2B). Dir/ZIF-8@Salmonella started to accumulate at tumors after 2 h post-treatment, and such enrichment remained till 48 h. In vitro stability assays indicated that the structural integrity of Il2/ZIF-8@Salmonella particles could maintain stable during the required circulation time to reach tumors (Fig. S2C and S2D).
Figure 2.
In vivo distribution assay of DIR/ZIF-8@Salmonella. (A) Fluorescent imaging of mice bearing HCC xenografts at 0.5, 2, 8, 24, and 48 h after the intravenous administration of free DIR, DIR/ZIF-8, or DIR/ZIF-8@Salmonella particles. n = 5. One of the mice from each group were sacrificed for fluorescent imaging of major organs and tumors at 24 and 48 h. (B) Fluorescent imaging of resected major organs and tumors at 24 and 48 h after the intravenous administration of particles.
Taken together, Il2/ZIF-8 particles could be loaded onto the surface of Salmonella, and such surface decoration did not significantly alter the growth and tumor-targeting behavior of Salmonella.
3.2. In vitro analysis of gene delivery efficiency of Il2/ZIF-8@Salmonella in macrophages and tumor cells
According to previous literature, the intratumoral enrichment of Salmonella is mostly attributed to its intrinsic taxis towards the hypoxic and immune-suppressive microenvironment, as well as the dysplastic tumor vascular with leaky endothelium facilitating bacterial invasion22. Once Salmonella settles in TME, macrophages are the major type of cells involved in its clearance by phagocytosis23. Therefore, Il2/ZIF-8@Salmonella was aimed to deliver target genes to macrophages in tumors. Here, we performed in vitro analysis to validate whether Il2/ZIF-8@Salmonella could be phagocytosed by macrophages and achieve gene expression.
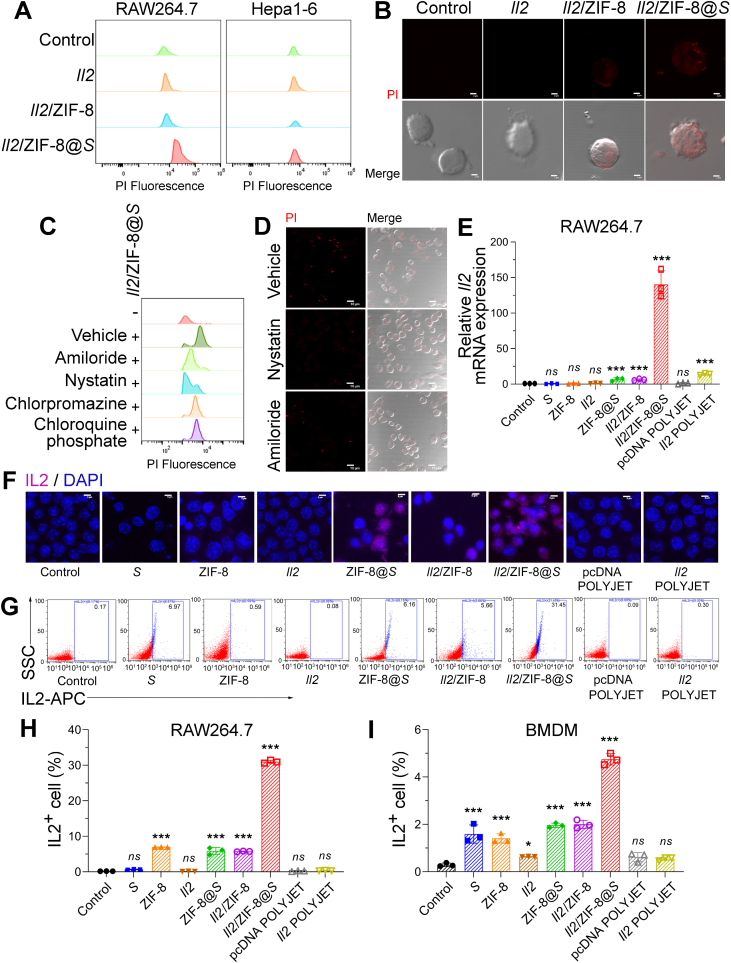
Firstly, Il2 plasmids were fluorescently labeled by propidium iodide (PI), and the uptake efficacy of free plasmids, Il2/ZIF-8 particles, and Il2/ZIF-8@Salmonella after 2-h co-incubation with cells. The flow cytometry assay showed that Il2/ZIF-8@Salmonella was intensively phagocytosed by RAW264.7 cells (murine macrophage cell line), while Hepa1-6 (murine HCC cell line) did not show obvious fluorescent signals in any treatment group (Fig. 3A). Similarly, Il2/ZIF-8@Salmonella formed obvious fluorescent dots in RAW264.7 cells in confocal microscopy imaging experiment (Fig. 3B). Then, RAW264.7 cells were pre-treated with Amiloride, Nystatin, Chlorpromazine, and Chloroquine phosphate, which were inhibitors against macropinocytosis, caveolae-mediated endocytosis, Clathrin-mediated endocytosis and endomorphic maturation respectively, and Amiloride and Nystatin profoundly impaired the uptake rates of Il2/ZIF-8@Salmonella as documented by both flow cytometry assay (Fig. 3C) and fluorescent imaging (Fig. 3D), suggesting that Il2/ZIF-8@Salmonella mainly entered macrophages via macropinocytosis and caveolae-mediated endocytosis.
Figure 3.
In vitro analysis of gene delivery efficiency of Il2/ZIF-8@Salmonella in macrophages and tumor cells. (A) Flow cytometry analysis of RAW264.7 and Hepa1-6 cells after the incubation with PI-labeled Il2 plasmid, Il2/ZIF-8 particles, and Il2/ZIF-8@Salmonella particles. (B) Fluorescent (upper) and merged DIC (lower) images of RAW264.7 cells after the incubation with PI-labeled Il2 plasmid, Il2/ZIF-8 particles, and Il2/ZIF-8@Salmonella particles. Red dots indicated the signals of PI. Scale bars = 2 μm. (C) Flow cytometry analysis of RAW264.7 incubated with Il2/ZIF-8@Salmonella particles and indicated inhibitors. (D) Fluorescent (upper) and merged DIC (lower) images of RAW264.7 incubated with Il2/ZIF-8@Salmonella particles and indicated inhibitors. Red dots indicated the signals of PI. Scale bars = 10 μm. (E) qPCR analysis of RAW264.7 cells incubated with indicated particle compositions. n = 3. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated cells as control for each treatment regime. ∗∗∗P < 0.001; ns, not significant. (F) Merged fluorescent images showing IL2 expression in RAW264.7 cells incubated with indicated particle compositions. Pink represented the signals of APC-labeled anti-mouse IL2 antibody. Blue represented DAPI signals. Scale bars = 5 μm. (G–I) Representative images (G) and histograms (H, I) for the flow cytometry analysis of IL2 expression in RAW264.7 (G, H) and BMDM (I) cells incubated with indicated particle compositions. n = 3. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated cells as control for each treatment regime. ∗∗∗P < 0.001; ns, not significant.
Later, in vitro gene delivery efficiency was analyzed after a 2-h co-incubation of cells and various preparations followed by a 48-h culture allowing gene expression, with a commercially available liposome-based plasmid transfection reagent as positive control (Supporting Information Fig. S3A). Apoptosis assay showed that the viability of RAW264.7 remained stable after the treatment of Salmonella or Il2/ZIF-8@Salmonella (Fig. S3B). As shown by qPCR assay, RAW264.7 exhibited significantly enhanced Il2 expression after the treatment of Il2/ZIF-8@Salmonella, compared to other preparations and liposome-based transfection reagent (Fig. 3E). On the contrary, the treatment of Il2/ZIF-8@Salmonella led to a minimal increase of Il2 expression in Hepa1-6 and B16/F10, two murine cancer cell lines, while liposome-based transfection reagent achieved potent Il2 expression in these cell lines (Fig. S3C and S3D), suggesting that plasmid/ZIF-8@Salmonella might be a selective vehicle for macrophage-targeted gene therapy. Moreover, the expression of Il2 was further validated by immunofluorescent staining or flow cytometry assay in RAW264.7 and bone marrow-derived macrophage (BMDM). In consistence with previous experiments, the treatment of Il2/ZIF-8@Salmonella resulted in a significantly elevated ratio of IL2+ cells in both RAW264.7 (Fig. 3F‒H, Fig. S3E) and BMDM (Fig. 3I, Fig. S3F) cells.
Taken together, Il2/ZIF-8@Salmonella could be effectively phagocytosed by macrophages and drive the expression of Il2.
3.3. In vivo analysis of therapeutic efficacy of Il2/ZIF-8@Salmonella in murine melanoma model
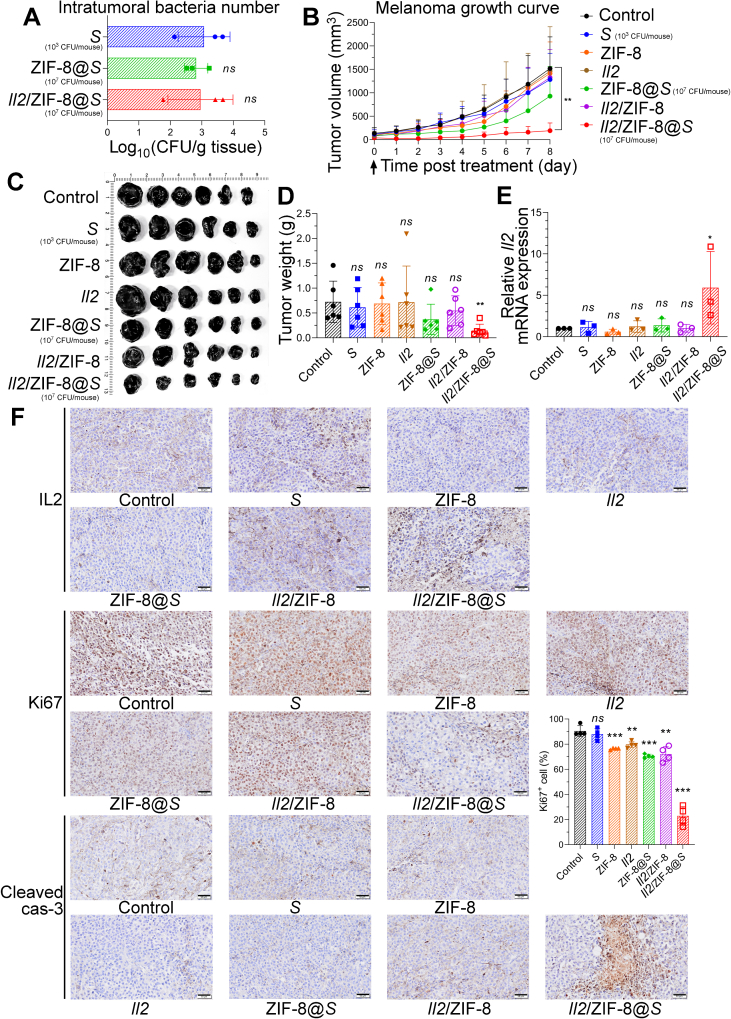
In experiment mentioned above, we noticed an approximately 1000-fold reduction in the intratumoral titer of live bacteria on Day 3 after dosing in mice treated with Il2/ZIF-8@Salmonella compared to free Salmonella without the assembly of ZIF-8 particles (Fig. 1E), partially due to faster macrophage-mediated phagocytosis and a transitory growth retardation (Figure 1, Figure 3A). Therefore, on the premise that an equal number (107 CFU/mice) of bacteria were administrated intravenously, even though Il2/ZIF-8@Salmonella was well tolerated and exhibited obvious anti-tumor activity in murine melanoma model (Supporting Information Fig. S4A‒C), free Salmonella led to sever splenic swelling and lethal toxicity at the same dosage (Fig. S4D and E). Thus, the dosage of free Salmonella was reduced to 103 CFU/mouse, thereby achieving an equal number of intratumoral bacteria in mice treated with Salmonella (103 CFU/mouse), ZIF-8@Salmonella (107 CFU/mouse) and Il2/ZIF-8@Salmonella (107 CFU/mouse) on Day 3 post-dosing (Fig. 4A).
Figure 4.
In vivo analysis of therapeutic efficacy of Il2/ZIF-8@Salmonella in murine melanoma model. (A) Live bacteria titers in the melanoma tumor tissues on Day 3 after the intravenous administration of Salmonella (103 CFU/mouse), ZIF-8@Salmonella (107 CFU/mouse), and Il2/ZIF-8@Salmonella (107 CFU/mouse). n = 3. Data were expressed as mean ± SD. Students' t test was carried out using Salmonella-treated group as control for each treatment regime. ns, not significant. This dosage was used in successive figures unless otherwise noted. (B) The tumor growth curves for mice treated with one dose of various regimes. n = 6. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗∗P < 0.01. (C) The image of resected melanomas. (D) The histogram showing tumor weights for groups treated with indicated regimes. n = 6. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗∗P < 0.01; ns, not significant. (E) The histograms showing the RNA level of Il2 in melanomas treated with indicated regimes. n = 3. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗P < 0.05; ns, not significant. (F) The representative IHC results showing the protein level of IL2, Ki67 and cleaved caspase-3 in melanomas treated with indicated regimes. Scale bars = 50 μm. The percentage of Ki67+ cells was shown as histogram. n = 4. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant.
Afterward, the anti-cancer efficacy was evaluated on the premise of an equal number of intratumoral bacteria in each group. A single dose of Salmonella (103 CFU/mouse), plasmids (1011 copies/mouse), ZIF-8 particles, Il2/ZIF-8 particles, ZIF-8@Salmonella (107 CFU/mouse) or Il2/ZIF-8@Salmonella (107 CFU/mouse) were administered intravenously when melanoma volume reached 200 mm3. Tumor growth curves showed that Il2/ZIF-8@Salmonella exhibited a significantly stronger suppression on melanoma growth compared to free Salmonella or Il2/ZIF-8 particles on the premise of equal intratumoral bacteria titer (Fig. 4B), in consistence with the images and weights of resected melanoma tissues (Fig. 4C and D). Meanwhile, the body weights and spleen volumes remained stable in all treatment groups (Fig. S4F‒S4H). The qPCR assay demonstrated significantly enhanced Il2 expression in tumors treated with Il2/ZIF-8@Salmonella (Fig. 4E). The IHC assays of melanoma tissues showed strongest signals for IL2 and cleaved Caspase-3 (marker for apoptotic cells) in mice treated with Il2/ZIF-8@Salmonella (Fig. 4F). In consistence with remarkable apoptosis, Il2/ZIF-8@Salmonella treatment also led to a significant reduction in Ki67+ (marker for proliferating cell) cells in melanoma (Fig. 4F).
Collectively, Il2/ZIF-8@Salmonella exhibited a potent anti-cancer activity in murine melanoma model.
3.4. The activation of anti-tumor immune responses induced by Il2/ZIF-8@Salmonella in murine melanoma model
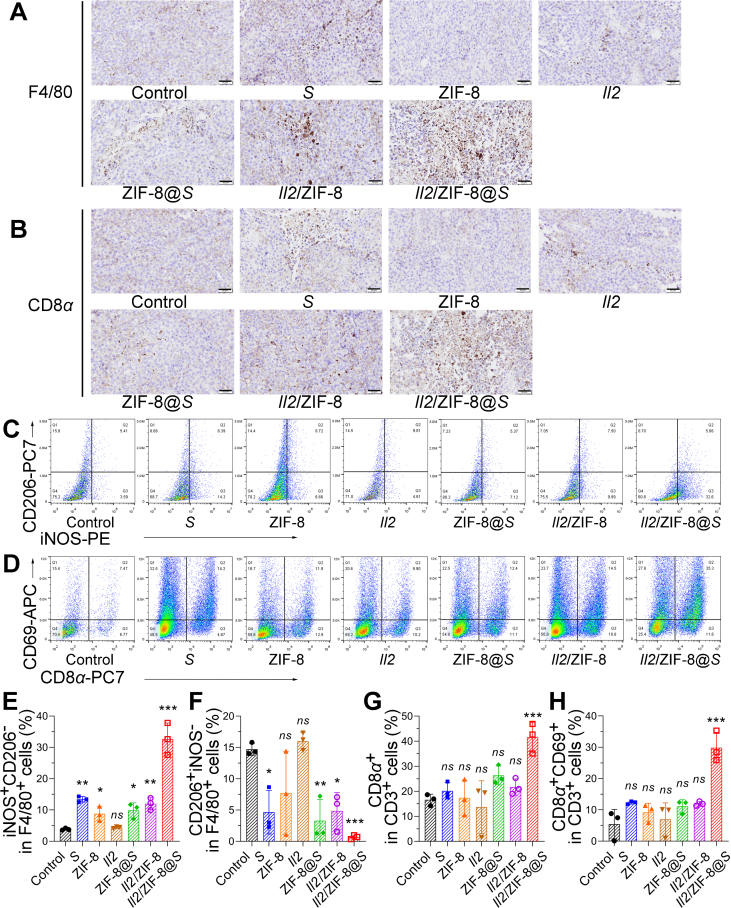
Since both Salmonella and IL2 were potent simulators for anti-tumor immune responses, the status of T cells and macrophages were analyzed by IHC and flow cytometry for mice receiving different regimes. IHC assays of F4/80, CD8α, and CD4 demonstrated a higher infiltration level for macrophages (Fig. 5A), CD8+ cytotoxic T lymphocytes (CTLs) (Fig. 5B), and CD4+ T cells (Supporting Information Fig. S5A) in melanoma treated with Il2/ZIF-8@Salmonella, compared to other treatment regimes. Meanwhile, flow cytometry assay showed an increase in iNOS+CD206- macrophages and a decrease in CD206+iNOS− macrophages, indicating that macrophages were polarized towards an anti-tumor M1 status in tumors treated with Salmonella, ZIF-8@Salmonella Salmonella, Il2/ZIF-8, and Il2/ZIF-8@Salmonella, while Il2/ZIF-8@Salmonella treatment induced the most potent effect (Fig. 5C, E, and F). Similarly, the percentages of CTL, CD69+ CTL (Fig. 5D, G, and H), CD4+ T cells, and CD69+ CD4+ T cells (Fig. S5B‒S5D) were remarkably elevated in tumors treated with Il2/ZIF-8@Salmonella. Moreover, macrophages and T cells showed a similar trend in the spleen. Specifically, iNOS+CD206- cells expanded significantly in total F4/80+ splenocytes, and the percentage of CD69+CD8α+ cells also increased in CD3+ splenocytes (Fig. S5E‒S5I). Collectively, Il2/ZIF-8@Salmonella could induce a potent activation of macrophages and T cells in favor of anti-tumor immune responses in both TME and peripheral lymphoid organs.
Figure 5.
The analysis of anti-tumor immune responses induced by Il2/ZIF-8@Salmonella in murine melanoma model. (A) The representative IHC results showing the F4/80 in melanomas treated with indicated regimes. Scale bars = 50 μm. (B) The representative IHC results showing the CD8α in melanomas treated with indicated regimes. Scale bars = 50 μm. (C) Representative images for the flow cytometry analysis of CD206 and iNOS expression in CD45+F4/80+ cells in melanomas. (D) Representative images for the flow cytometry analysis of CD8α and CD69 expression in CD45+CD3+ intratumoral lymphocytes. (E) The histogram showing the percentage of iNOS+CD206- in F4/80+ cells in melanomas treated with indicated regimes. n = 3. Data were expressed as mean ± SD. Student’ t test was carried out using PBS-treated group as control for each treatment regime. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant. (F) The histogram showing the percentage of CD206+ iNOS-in F4/80+ cells in melanomas treated with indicated regimes. n = 3. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant. (G) The histogram showing the percentage of CD8α+ in CD3+ lymphocytes in melanomas treated with indicated regimes. n = 3. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗∗∗P < 0.001; ns, not significant. (H) The histogram showing the percentage of CD8α+CD69+ in CD3+ lymphocytes in melanomas treated with indicated regimes. n = 3. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗∗∗P < 0.001; ns, not significant.
3.5. In vivo analysis of therapeutic efficacy of Il2/ZIF-8@Salmonella in murine orthotopic HCC model and melanoma pulmonary metastasis model
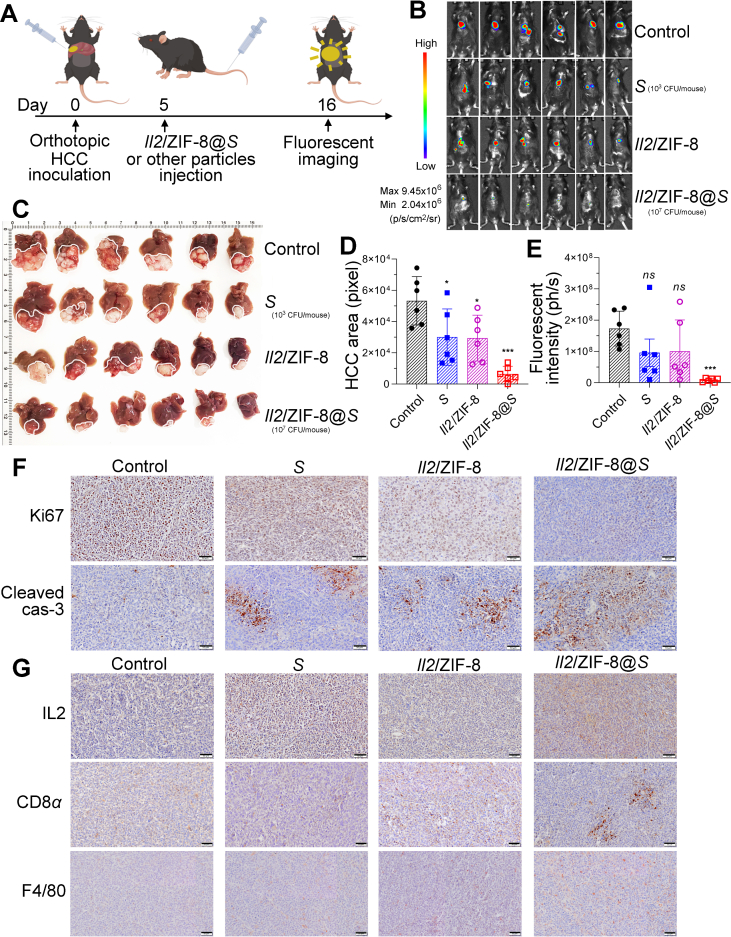
In addition to murine melanoma model, the therapeutic efficacy of Salmonella, Il2/ZIF-8 particles, and Il2/ZIF-8@Salmonella were compared in orthotopic HCC model. A single dose of various regimes was administrated intravenously on Day 5 after the inoculation of luciferase-labeled HCC cells in liver lobes, and in situ fluorescent signals were imaged on Day 16 (Fig. 6A). The fluorescent imaging detected obvious signals in untreated mice and mice receiving Salmonella or Il2/ZIF-8 particles, while the signals were minimal in mice treated with Il2/ZIF-8@Salmonella (Fig. 6B and E), in consistency with the images of resected liver tissues (Fig. 6C and D). The IHC results also demonstrated significantly suppressed cell proliferation and augmented apoptosis in HCC treated with Il2/ZIF-8@Salmonella, as showed by weakened Ki67 staining and stronger signal of cleaved Caspase-3 (Fig. 6F, Supporting Information Fig. S6). Similar to the phenomenon observed in the melanoma model, IL2 expression level and the infiltration of CTLs and macrophages were significantly elevated in HCC treated with Il2/ZIF-8@Salmonella, compared to other treatment regimes (Fig. 6G).
Figure 6.
In vivo analysis of therapeutic efficacy of Il2/ZIF-8@Salmonella in murine orthotopic HCC model. (A) The schematic illustration for the schedule of the establishment, treatment and imaging for the orthotopic HCC model. (B) The fluorescent imaging of mice carrying murine orthotopic HCC for mice receiving various regimes on Day 16 after the inoculation of HCC cells. (C) The image of the resected liver tissues. (D) The histogram showing the areas of fluorescent signals for mice receiving various regimes. n = 6. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗P < 0.05; ∗∗P < 0.01. (E) The histogram showing the total fluorescent intensity of each mouse after the administration of various regimes. n = 6. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗∗∗P < 0.001; ns, not significant. (F) The representative IHC results showing the protein level of Ki67 and cleaved caspase-3 in HCC treated with indicated regimes. Scale bars = 50 μm. (G) The representative IHC results showing the expression of IL2, CD8α and F4/80 in HCC treated with indicated regimes. Scale bars = 50 μm.
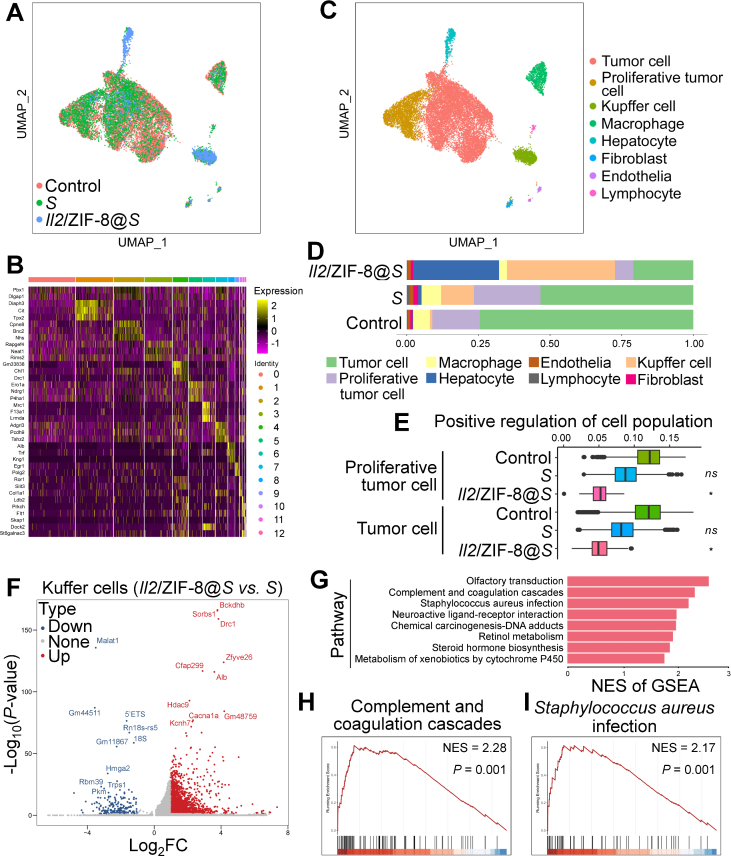
Meanwhile, the alteration of cell components intratumorally by the administration of Salmonella or Il2/ZIF-8@Salmonella was analyzed by single-cell RNA sequencing analysis of cryopreserved HCC tissues (Fig. 7A). Cell identities were determined according to the expression pattern of signature genes (Fig. 7B). Tumor cells, Kupffer cells (liver-resident macrophages), macrophages and hepatocytes are the major intratumoral cell types in murine HCC model (Fig. 7C). Interestingly, even though Salmonella monotherapy induced remarkable expansion of Kupffer cell population in TME, the proportion of proliferative tumor cells did not exhibit dramatic shrinkage (Fig. 7D). On the contrary, the combinatorial Il2/ZIF-8@Salmonella simultaneously resulted in a potent expansion of intratumoral Kupffer cell population and a dramatic shrinkage of tumor cell population (Fig. 7D), which was in consistence with the expression indexes for signature genes associated with positive regulation of cell population in tumor cells in TME (Fig. 7E). Moreover, we identified the differentially expressed genes in Kupffer cells for HCC tissues treated with Salmonella and Il2/ZIF-8@Salmonella, and performed Gene Set Enrichment Analysis (GSEA) analysis (Fig. 7F). Among the 8 pathways significantly enriched in the Kupffer cells of Il2/ZIF-8@Salmonella group, two of them were closely related with pro-inflammatory immune responses, namely “Complement and coagulation cascades” and “Staphylococcus aureus infection”, indicating that the Kupffer cells underwent a transformation towards immune-activating status (Fig. 7G‒I, Supporting Information Fig. S7A‒S7F).
Figure 7.
Single-cell RNA sequencing analysis of cryopreserved HCC tissues after the treatment of Il2/ZIF-8@Salmonella. (A) The preliminary Uniform Manifold Approximation and Projection (UMAP) spectrum after harmony balancing batch effects showing the degree of fit between HCC samples collected from mice receiving indicated treatment. (B) Heat map showing the typical gene expression patterns of different cell populations after preliminary dimensionality reduction clustering for total cells from all samples. (C) UMAP colored by cell types for total cells from all samples. (D) Bar plot showing the cell type proportion for each phenotype group in HCC samples receiving indicated treatment. ∗P < 0.05; ns, not significant. (E) Box plot showing the scores of “positive regulation of cell population proliferation” pathway for proliferative tumor cells and tumor cells in HCC samples receiving indicated treatment. (F) Volcano plot showing differentially expressed genes between Kupffer cells in Il2/ZIF-8@Salmonella-treated HCC and Salmonella-treated HCC. (G) Bar plot showing significantly up-regulated gene clusters in GESA analysis for Kupffer cells in Il2/ZIF-8@Salmonella-treated HCC compared to Salmonella-treated HCC. (H–I) GSEA plot of “Complement and coagulation cascades” gene cluster (H) and “Staphylococcus aureus infection” gene cluster (I) by comparing gene expression patterns between Kupffer cells in Il2/ZIF-8@Salmonella-treated HCC and Salmonella-treated HCC.
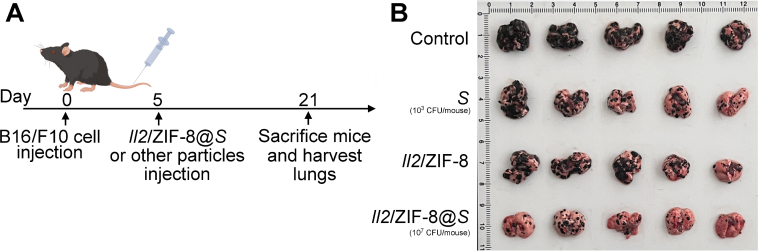
Apart form orthotopic HCC model, melanoma pulmonary metastasis model was also used to evaluate the anti-cancer efficacy of Il2/ZIF-8@Salmonella. In brief, mice bearing melanoma pulmonary metastasis loci received various treatments, and they were sacrificed on Day 16 post-treatment (Fig. 8A). The images of resected lungs showed that the lungs of untreated group were mostly covered by black metastasis loci, while the lungs of Il2/ZIF-8@Salmonella-treated group exhibited relatively healthy morphology with significantly reduced metastasis loci, demonstrating that Il2/ZIF-8@ Salmonella could potently suppress the formation of pulmonary metastasis in vivo (Fig. 8B).
Figure 8.
In vivo analysis of therapeutic efficacy of Il2/ZIF-8@Salmonella in murine melanoma pulmonary metastasis model. (A) The schematic illustration for the schedule of the establishment, treatment and imaging for the melanoma pulmonary metastasis model. (B) The image of resected lungs.
Taken together, Il2/ZIF-8@Salmonella thoroughly re-shaped TIME by inducing the expansion and pro-inflammatory transformation of Kupffer cells, thus achieving a potent anti-cancer effect in orthotopic HCC model and melanoma pulmonary metastasis model.
3.6. In vivo evaluation of safety profile for Il2/ZIF-8@Salmonella
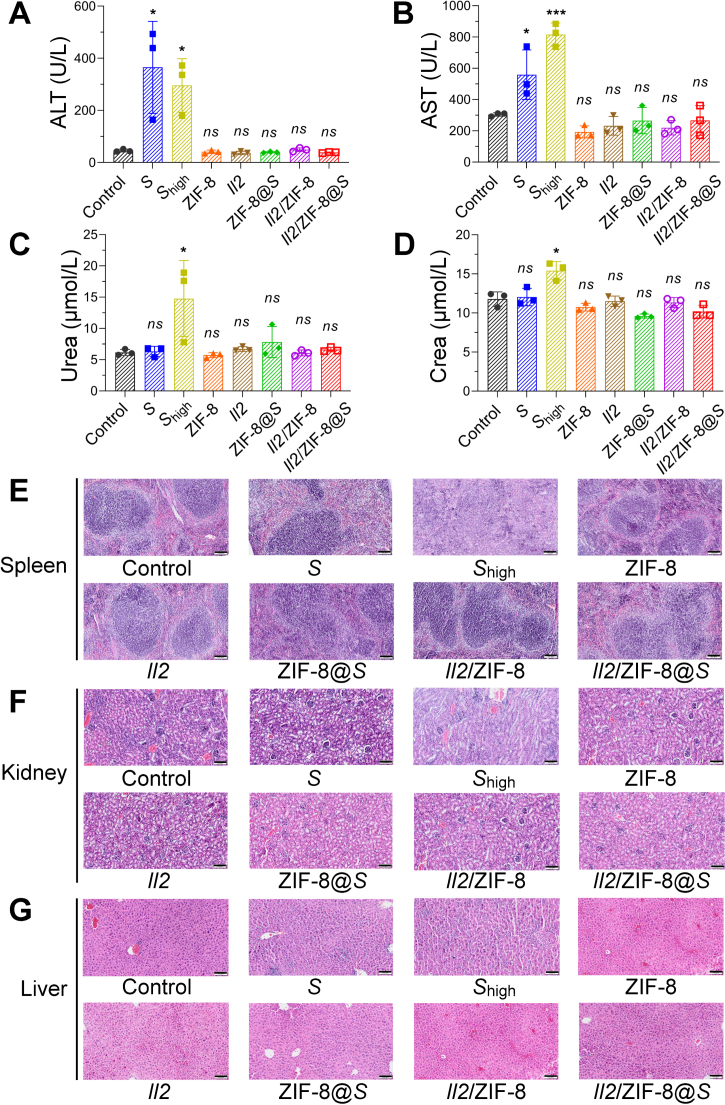
As shown by experiments mentioned above, Il2/ZIF-8@Salmonella potently activated anti-tumor immune responses without intolerable negative influences on body weights and spleen volumes in mice bearing melanoma (Fig. S4F‒S4H). Here in this section, the toxicity to liver, kidney, spleen, and hematopoietic system was systematically evaluated for Salmonella, ZIF-8 particles, Il2 plasmids, and various combinatorial formulations. The serum concentrations of Alanine Aminotransferase (ALT), Aspartate Aminotransferase (AST), Crea and Urea showed that liver functions were severely impaired by free Salmonella, regardless of dosage, while kidney damage was mostly observed in mice receiving a high dose of Salmonella. In parallel, these serum indicators remained stable in mice receiving Il2/ZIF-8@Salmonella treatment, indicating this formulation did not cause obvious tissue damage for the liver or kidney (Fig. 9A‒D). Similarly, peripheral blood cell indexes showed that ZIF-8 particles, Salmonella, and the combinatorial formulations would lead to a moderate decrease in lymphocyte counts, but none of these treatment groups reached a critical value (Supporting Information Fig. S8A‒S8H). Furthermore, the histology of the spleen, kidney and liver were evaluated by H&E staining. In consistence with previous observations, H&E images showed that a high dose of free Salmonella resulted in obvious shrinkage of lymphatic germinal center in the spleen, glomerular swelling in the kidney, and hepatocyte necrosis in the liver, while mice treated with Il2/ZIF-8@Salmonella exhibited normal histological features in these major organs (Fig. 9E‒G).
Figure 9.
The analysis for the toxicity of Il2/ZIF-8@Salmonella to the liver, kidney, and spleen on Day 8 after treatment. (A–D) The histograms showing serum concentrations of ALT (A), AST (B), Crea (C) and Urea (D) in mice treated with indicated regimes. Shigh indicated 107 CFU Salmonella/mouse in this figure and successive figures. n = 3. Data were expressed as mean ± SD. Students' t test was carried out using PBS-treated group as control for each treatment regime. ∗P < 0.05; ∗∗∗P < 0.001; ns, not significant. (E–G) Representative images of H&E staining of spleens (E), kidneys (F), and livers (G) in mice treated with indicated regimes. Scale bar = 100 μm.
4. Discussions and conclusions
Here, we reported a TME-targeted gene-delivery system based on live bacteria carrying MOF-DNA complex, named plasmid/ZIF-8@Salmonella, which would selectively accumulate in tumor and phagocytosed by macrophages, thus leading to the expression of therapeutic proteins. In this work, we employed a plasmid carrying Il2 gene as an example. We demonstrated that IL2 could be expressed and secreted by TAMs via plasmid/ZIF-8@Salmonella, leading to the re-activation of TIME as well as the suppression of tumor growth (Fig. 10).
Figure 10.
The schematic illustration showing that Il2/ZIF-8@Salmonella actively delivers Il2 gene to TAM to induce intratumoral IL2 production and modulates the immune-suppressive TIME towards an active status.
Compared with other drug delivery systems, the most prominent features of live bacteria are their capacity of active targeting towards the poorly vesiculated region within tumors, as well as their sustained therapeutic effect originated from in situ proliferation intratumorally. Therefore, bacteria are referred as “smart motors” to deliver active compounds to positions which are difficult to reach for traditional vehicles, such as glioma and pancreatic cancer24,25. However, along with these advantages, the accumulated risk of sever systemic infection after heavy or repeated dosages of live bacteria constantly haunts the fundamental and translational research processes of BCTs like the sword of Damocles. Therefore, the delivered bioactive compounds should possess a high potency ratio and long half-life to solve the inherent contradiction of effects and toxicity, such as therapeutic genes and antibodies. Our study is an example of combining BCT with a cytokine gene. In addition, Hu et al.26 reported that an E. coli strain expressing TNFα-binding antibodies fused with immunotoxins exhibited a synergistic anti-tumor effect. Recently, Liu et al.27 reported that a VNP20009 strain secreting a nanobody against TNFα robustly transformed “cold” tumor with immune suppression to “hot” tumor with anti-tumor immune activation.
Apart from live bacteria, hollow cell walls of dead Gram-negative bacteria, also called bacteria ghosts (BGs), are often used as vehicles for targeted drug delivery28. BGs are prepared by the plasmid-driven expression of gene E cloned from bacteriophage ϕX174, which forms numerous transmembrane tunnels and leads to the leakage of all cytoplasmic content29. Thus, BGs have a large loading capacity as vehicles for bioactive compounds, while maintaining all surface PAMPs for the induction of natural immune responses without the risk of systemic infections related with excessive bacterial proliferation. By contrast, BGs have the inherent disadvantages of impaired passive/active TME-targeting activity due to large particle size and loss of motility, as well as short-term therapeutic effects due to loss of self-renewal. Therefore, BGs are intensively investigated for the applications in vaccination (including cancer vaccines) and systemic immune boosters for treating infectious diseases30, 31, 32. Recently, emerging reports have shown that bacterial outer membrane vesicles (OMVs), which are much smaller compared to BGs, are able to accumulate in TME via EPR effects. Therefore, OMVs have been investigated as immune-activating nano-vehicles for chemotherapies and gene therapies against tumors33,34. Compared to live bacteria, BGs and OMVs obviously have the advantage in safety profile, but their impaired tissue penetration capacity required further rational optimization for wider applications in cancer therapy.
According to our research, macrophages endocytose and express the cargo gene carried by plasmid/ZIF-8@Salmonella at a significantly higher efficiency compared to tumor cells. However, macrophages are far outnumbered by tumor cells in TME, so the ideal genes to be delivered by this system should possess extensive anti-tumor effects via a paracrine action and low toxicity to macrophages. Moreover, the form of delivered nucleic acids is also an optimizable factor. Apart from plasmids, viruses, mRNA, and other forms of nucleic acids could be utilized to produce secretory proteins. Moreover, siRNA, circRNA, and even sgRNA for gene editing could also be delivered to modulate biological behaviors of TAMs. In this study, plasmid, virus and mRNA could be used to mediate the production of IL2, and plasmid was chosen due to the advantage of high safety and low cost. However, the fast loss rate during cell proliferation limits the performance of plasmids. It would be interesting to evaluated whether virus could further improve the therapeutic effects of BCT-MOF-DNA therapies.
Nowadays, the importance of microbiome in cancer therapy has received intense attention, and the research community of BCT grows rapidly, marked by emerging mechanistic insights, engineered bacteria strains, as well as innovative combination strategies. With a deeper understanding of the crosstalk between bacteria-host immune responses and anti-tumor immune responses, bacteria would become an active player in the new era of multidisciplinary synthetic cancer therapy.
Author Contributions
Yiting Qiao: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Miao Luo: Data curation, Formal analysis, Investigation. Yufei Wang: Data curation, Formal analysis, Project administration, Validation. Haoxiang Qi: Data curation, Formal analysis, Software. Menglan Wang: Methodology. Yunxin Pei: Visualization. Mengqing Sun: Methodology. Zhengguo Zhang: Methodology. Jiacheng Huang: Project administration, Software. Pengyu Gong: Methodology. Shusen Zheng: Funding acquisition, Resources. Jianxiang Chen: Conceptualization, Funding acquisition, Project administration, Resources, Supervision.
Conflicts of interest
The authors have no conflicts of interest to declare.
Acknowledgments
This study was supported by “Pioneer” and “Leading Goose” R&D Program of Zhejiang Province (2022C03004 to Jianxiang Chen, China), Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022029C to Yiting Qiao, China), National Natural Science Foundation of China (82373888 to Yiting Qiao, 82072646, 82372664 to Jianxiang Chen), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2023-PT320-02 to Yiting Qiao, China). The joint foundation of National Administration of Traditional Chinese Medicine & Zhejiang Province - major project (GZY-ZJ-KJ-24045 to Jianxiang Chen, China). Research Unit Project of Chinese Academy of Medical Sciences (2019‒I2M‒5‒030 to Shusen Zheng, China). Postgraduate research innovation project of Hangzhou Normal University (2022HSDYJ SKY244 to Yunxin Pei, 2022HSDYJ SKY015 to Menglan Wang, China).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2024.07.020.
Contributor Information
Yiting Qiao, Email: yitingqiao@zju.edu.cn.
Jianxiang Chen, Email: chenjx@hznu.edu.cn.
Appendix A. Supplementary data
The following is the Supporting Information to this article:
References
- 1.Starnes C.O. Coley's toxins in perspective. Nature. 1992;357:11–12. doi: 10.1038/357011a0. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen D.H., Chong A., Hong Y., Min J.J. Bioengineering of bacteria for cancer immunotherapy. Nat Commun. 2023;14:3553. doi: 10.1038/s41467-023-39224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Low K.B., Ittensohn M., Luo X., Zheng L.M., King I., Pawelek J.M., et al. In: Suicide gene therapy: methods and reviews. Springer C.J., editor. Humana Press Inc.; Totowa: 2004. Construction of VNP20009; pp. 47–59. [Google Scholar]
- 4.Toso J.F., Gill V.J., Hwu P., Marincola F.M., Restifo N.P., Schwartzentruber D.J., et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J Clin Oncol. 2002;20:142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou S., Gravekamp C., Bermudes D., Liu K. Tumour-targeting bacteria engineered to fight cancer. Nat Rev Cancer. 2018;18:727–743. doi: 10.1038/s41568-018-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valle E., Guiney D.G. Characterization of Salmonella-induced cell death in human macrophage-like THP-1 cells. Infect Immun. 2005;73:2835–2840. doi: 10.1128/IAI.73.5.2835-2840.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monack D.M., Navarre W.W., Falkow S. Salmonella-induced macrophage death: the role of caspase-1 in death and inflammation. Microbes Infect. 2001;3:1201–1212. doi: 10.1016/s1286-4579(01)01480-0. [DOI] [PubMed] [Google Scholar]
- 9.Chen J., Qiao Y., Chen G., Chang C., Dong H., Tang B., et al. Salmonella flagella confer anti-tumor immunological effect via activating Flagellin/TLR5 signalling within tumor microenvironment. Acta Pharm Sin B. 2021;11:3165–3177. doi: 10.1016/j.apsb.2021.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bantug G.R., Hess C. The immunometabolic ecosystem in cancer. Nat Immunol. 2023;24:2008–2020. doi: 10.1038/s41590-023-01675-y. [DOI] [PubMed] [Google Scholar]
- 11.Berraondo P., Sanmamed M.F., Ochoa M.C., Etxeberria I., Aznar M.A., Pérez-Gracia J.L., et al. Cytokines in clinical cancer immunotherapy. Br J Cancer. 2019;120:6–15. doi: 10.1038/s41416-018-0328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhry H., Helmi N., Abdulaal W.H., Zeyadi M., Zamzami M.A., Wu W., et al. Prospects of IL-2 in cancer immunotherapy. BioMed Res Int. 2018;2018 doi: 10.1155/2018/9056173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ptacin J.L., Caffaro C.E., Ma L., San Jose Gall K.M., Aerni H.R., Acuff N.V., et al. An engineered IL-2 reprogrammed for anti-tumor therapy using a semi-synthetic organism. Nat Commun. 2021;12:4785. doi: 10.1038/s41467-021-24987-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X.S., Zheng Z.S., Zheng M.F., Wang D., Zhang H.L., Zhang Z.Q., et al. IL-2-loaded polypeptide nanoparticles for enhanced anti-cancer immunotherapy. Chin J Polym Sci. 2023;41:1059–1068. [Google Scholar]
- 15.Deckers J., Anbergen T., Hokke A.M., de Dreu A., Schrijver D.P., de Bruin K., et al. Engineering cytokine therapeutics. Nat Rev Bioeng. 2023;1:286–303. doi: 10.1038/s44222-023-00030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tumas S., Meldgaard T.S., Vaaben T.H., Suarez Hernandez S., Rasmussen A.T., Vazquez-Uribe R., et al. Engineered E. coli Nissle 1917 for delivery of bioactive IL-2 for cancer immunotherapy. Sci Rep. 2023;13 doi: 10.1038/s41598-023-39365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yusuf V.F., Malek N.I., Kailasa S.K. Review on metal-organic framework classification, synthetic approaches, and influencing factors: applications in energy, drug delivery, and wastewater treatment. ACS Omega. 2022;7:44507–44531. doi: 10.1021/acsomega.2c05310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horcajada P., Gref R., Baati T., Allan P.K., Maurin G., Couvreur P., et al. Metal-organic frameworks in biomedicine. Chem Rev. 2012;112:1232–1268. doi: 10.1021/cr200256v. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Yuan Y., Cheng Y., Fu D., Chen Z., Wang Y., et al. Copper-based metal-organic framework overcomes cancer chemoresistance through systemically disrupting dynamically balanced cellular redox homeostasis. J Am Chem Soc. 2022;144:4799–4809. doi: 10.1021/jacs.1c11856. [DOI] [PubMed] [Google Scholar]
- 20.Yan S., Zeng X., Wang Y., Liu B.F. Biomineralization of bacteria by a metal-organic framework for therapeutic delivery. Adv Healthc Mater. 2020;9 doi: 10.1002/adhm.202000046. [DOI] [PubMed] [Google Scholar]
- 21.Toda G., Yamauchi T., Kadowaki T., Ueki K. Preparation and culture of bone marrow-derived macrophages from mice for functional analysis. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2020.100246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C.Z., Kazmierczak R.A., Eisenstark A. Strains, mechanism, and perspective: Salmonella-based cancer therapy. Int J Microbiol. 2016;2016 doi: 10.1155/2016/5678702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masud S., Prajsnar T.K., Torraca V., Lamers G.E.M., Benning M., Van Der Vaart M., et al. Macrophages target Salmonella by Lc3-associated phagocytosis in a systemic infection model. Autophagy. 2019;15:796–812. doi: 10.1080/15548627.2019.1569297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J., Li T., Zhou N., He Y., Zhong J., Ma C., et al. Engineered Salmonella inhibits GPX4 expression and induces ferroptosis to suppress glioma growth in vitro and in vivo. J Neuro Oncol. 2023;163:607–622. doi: 10.1007/s11060-023-04369-5. [DOI] [PubMed] [Google Scholar]
- 25.Tan W., Duong M.T., Zuo C., Qin Y., Zhang Y., Guo Y., et al. Targeting of pancreatic cancer cells and stromal cells using engineered oncolytic Salmonella typhimurium. Mol Ther. 2022;30:662–671. doi: 10.1016/j.ymthe.2021.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu C.W., Chang Y.C., Liu C.H., Yu Y.A., Mou K.Y. Development of a TNF-α-mediated Trojan horse for bacteria-based cancer therapy. Mol Ther. 2022;30:2522–2536. doi: 10.1016/j.ymthe.2022.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L., Liu X., Xin W., Zhou L., Huang B., Han C., et al. A bacteria-based system expressing anti-TNF-α nanobody for enhanced cancer immunotherapy. Signal Transduct Target Ther. 2023;8:134. doi: 10.1038/s41392-023-01364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langemann T., Koller V.J., Muhammad A., Kudela P., Mayr U.B., Lubitz W. The bacterial ghost platform system: production and applications. Bioeng Bugs. 2010;1:326–336. doi: 10.4161/bbug.1.5.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henrich B., Lubitz W., Plapp R. Lysis of Escherichia coli by induction of cloned phi X174 genes. Mol Gen Genet. 1982;185:493–497. doi: 10.1007/BF00334146. [DOI] [PubMed] [Google Scholar]
- 30.Groza D., Gehrig S., Kudela P., Holcmann M., Pirker C., Dinhof C., et al. Bacterial ghosts as adjuvant to oxaliplatin chemotherapy in colorectal carcinomatosis. Oncoimmunology. 2018;7 doi: 10.1080/2162402X.2018.1424676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong S., Nan N., Sun Y., He Z., Li J., Chen F., et al. Protective immunity elicited by VP1 chimeric antigens of bacterial ghosts against hand-foot-and-mouth disease virus. Vaccines (Basel) 2020;8:61. doi: 10.3390/vaccines8010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim J., Koh V.H.Q., Cho S.S.L., Periaswamy B., Choi D.P.S., Vacca M., et al. Harnessing the immunomodulatory properties of bacterial ghosts to boost the anti-mycobacterial protective immunity. Front Immunol. 2019;10:2737. doi: 10.3389/fimmu.2019.02737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Q., Bai H., Wu W., Huang G., Li Y., Wu M., et al. Bioengineering bacterial vesicle-coated polymeric nanomedicine for enhanced cancer immunotherapy and metastasis prevention. Nano Lett. 2020;20:11–21. doi: 10.1021/acs.nanolett.9b02182. [DOI] [PubMed] [Google Scholar]
- 34.Gujrati V., Kim S., Kim S.H., Min J.J., Choy H.E., Kim S.C., et al. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 2014;8:1525–1537. doi: 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.