Abstract
DNMT3A encodes a DNA methyltransferase involved in development, cell differentiation, and gene transcription, which is mutated and aberrant-expressed in cancers. Here, we revealed that loss of DNMT3A promotes malignant phenotypes in lung cancer. Based on the epigenetic inhibitor library synthetic lethal screening, we found that small-molecule HDAC6 inhibitors selectively killed DNMT3A-defective NSCLC cells. Knockdown of HDAC6 by siRNAs reduced cell growth and induced apoptosis in DNMT3A-defective NSCLC cells. However, sensitive cells became resistant when DNMT3A was rescued. Furthermore, the selectivity to HDAC6 inhibition was recapitulated in mice, where an HDAC6 inhibitor retarded tumor growth established from DNMT3A-defective but not DNMT3A parental NSCLC cells. Mechanistically, DNMT3A loss resulted in the upregulation of HDAC6 through decreasing its promoter CpG methylation and enhancing transcription factor RUNX1 binding. Notably, our results indicated that HIF-1 pathway was activated in DNMT3A-defective cells whereas inactivated by HDAC6 inhibition. Knockout of HIF-1 contributed to the elimination of synthetic lethality between DNMT3A and HDAC6. Interestingly, HIF-1 pathway inhibitors could mimic the selective efficacy of HDAC6 inhibition in DNMT3A-defective cells. These results demonstrated HDAC6 as a HIF-1-dependent vulnerability of DNMT3A-defective cancers. Together, our findings identify HDAC6 as a potential HIF-1-dependent therapeutic target for the treatment of DNMT3A-defective cancers like NSCLC.
Key words: DNMT3A, NSCLC, Synthetic lethal, HDAC6, HIF-1
Graphical abstract
This study reveals a novel synthetic lethality approach to combat tumor-based epigenetic enzyme interaction.
1. Introduction
Cancer is considered as a genetic disease1. Gene mutations, including oncogenes and tumor suppressor genes (TSGs), play essential roles in the development and progression of cancer2. Driven mutation in oncogenes, such as KRAS and EGFR, would contribute to the malignant phenotype3,4. Similarly, the mutation in TSGs, such as TP53 and VHL, usually results in the loss of their function, which would lead to the imbalance between oncogenes and TSGs, and further promote tumorigenesis5,6. In parallel with gene mutation, epigenetic alteration, through affecting gene expression, also plays a crucial role in cancer7,8. Interestingly, recent studies disclose that mutation in epigenetic enzymes is relatively frequent9,10. Thus, how to understand and take advantage of the frequent genetic mutation in epigenetic enzymes would be a crucial issue for the development of a novel anticancer approach.
DNA methyltransferase (DNMT), a kind of earlier discovered epigenetic enzyme, participates in DNA methylation to regulate various biological functions, including embryo development, cell differentiation, and gene transcription, and is associated with tumorigenesis11, 12, 13. Among DNMT family members, DNMT1, called maintenance DNMT, and DNMT3A and DNMT3B, called de novo DNMT, display the key role in the regulation of DNA methylation14. Although the inhibitors targeting DNMT, such as 5-azacytidine (5-AzaC) and 5-aza-2′-deoxycytidine (Decitabine)15,16, have been applied in the treatment of leukemia17,18, the impact of DNMTs on tumors and their therapeutic value are still controversial. A recent study discovered that the mutation rate of DNMT3A is the highest in the DNMT family, and mutation in DNMT3A was frequent in various tumors19,20, which mostly resulted in loss of function (LOF) of DNMT3A21. However, the biological function and clinical value of DNMT3A LOF mutation in cancer need to be further explored.
With the introduction of the first DNA damage repair (DDR) based synthetic lethal drug (PARP inhibitor olaparib in cancer with BRCA1 LOF mutation), research in DDR synthetic lethal has been an ideal strategy to target cancers with a mutation in certain DDR-related TSGs22, 23, 24. Previous studies revealed some potential DDR synthetic lethal gene pairs, such as PTEN/CHD125, NOXA/RUNX126, and SLFN11/ATR27. Similar to DDR gene pairs, epigenetic regulation enzymes also display interactive and complementary characteristics. Recently, some epigenetic synthetic lethal pairs were identified28, including ARID1A/ARID1B and ARID1A/EZH229, 30, 31. However, the epigenetic synthetic lethal gene pairs have not been studied extensively.
In this study, we revealed the TSG role of DNMT3A in lung cancer. In addition, histone deacetylase 6 (HDAC6), as a synthetic lethal partner of DNMT3A loss, was identified by utilizing an epigenetic inhibitor library. Interactively, DNMT3A loss would result in the upregulation of HDAC6 by affecting its promoter cytosine-phosphate-guanine (CpG) methylation. Mechanistically, the hypoxia-inducible factor (HIF-1) pathway was found to be involved in this epigenetic synthetic lethal partner. These findings clarify the underlying mechanisms driving the malignant phenotype of DNMT3A-deficient lung cancer and broaden the field for the development of a novel epigenetic synthetic lethal strategy in cancer therapy.
2. Materials and methods
2.1. Cell culture
The human non-small cell lung cancer (NSCLC) cell lines A549, NCI-H460, NCI-H1299, and acute myeloid leukemia cell lines HL-60 and NB-4 were obtained from the American Type Culture Collection, and DNMT3AKO cells were constructed by our laboratory. Cell lines were cultured in RPMI 1640 Medium (Gibco, C11875500BT) supplemented with 10% fetal bovine serum (Gibco, 10270106) and 1% penicillin/streptomycin (Gibco,15140122).
2.2. Analysis of clinical specimens
Paraffin-embedded clinical tissue specimens from 80 lung adenocarcinoma patients were obtained from Shanghai Outdo Biotech. Clinical specimens used in this study were approved by the Committee for Ethical Review of Research Involving Human Subjects at Shanghai Biochip National Engineering Research Center (Ethical number: YB M-05-02). Written informed consent was obtained from the patients or their legal guardians before tissue sampling. The research design and methods were carried out in accordance with the Declaration of Helsinki's Guidelines and all applicable laws regarding the use of human study subjects. Among the 80 patients, 30 were determined as stage III, 47 were stage II, and 3 patients were stage I. The immunohistochemical staining of specimens was scored according to the intensity of dye color, the percentage of positive cells, and the calculated average score in studied cases. The scores above the average value were designated as high expression, and scores below the average were designated as low expression.
2.3. Epigenetics compound library screening
For screening, cells were seeded in 96-well plates at 2000–3000 cells/well and cultured overnight. The compounds (Med Chem Express, HY-L005) were added to the plates according to the concentration. After 72 h of drug treatment, viable cells were quantified with Cell-Counting-Kit-8 (Med Chem Express, HY-K0301) according to the manufacturer's instructions at indicated time points. All experiments were performed in triplicate.
2.4. Cell viability assay
For cell viability, cells were seeded in 96-well plates at 2000–3000 cells/well and cultured overnight. The agents were added to the plates according to the concentration. After drug treatment, viable cells were quantified with Cell-Counting-Kit-8 (Med Chem Express, HY-K0301) according to the manufacturer's instructions at indicated time points. All experiments were performed in triplicate.
2.5. Colony formation assay
Cells were seeded in plates and incubated for 3 days, the agents were added to the plates with various concentrations. Every 2–3 days, fresh medium with agents was added to the plates. After culturing for 7–10 days, the cells were fixed with methanol for 15 min and stained with 0.1% crystal violet for half an hour, and the colonies were imaged and counted.
2.6. Apoptosis assay
For the assay of apoptotic cells, 0.2 million cells were seeded into 6-well plates and cultured overnight. After 72 h of drug treatment, cells were harvested and examined using an Annexin V–PI Apoptosis Detection Kit (BD, 556547) according to the manufacturer's protocol and analyzed by flow cytometry.
2.7. siRNA transfection
Cells were seeded into 6-well plates and cultured overnight. Cells were transfected with specific small interfering RNAs (siRNAs) according to the manufacturer's instruction (Thermo Fisher Scientific, L3000015) for 48–72 h. All siRNAs were obtained from BIONEER. The following siRNA sequences were used:
siDNMT3A#1: GCGUCACACAGAAGCAUAU
siDNMT3A#2: CCUCAGAGCUAUUACCCAA
siHDAC6#1: GAGGUAAAGAAGAAAGGCA
siHDAC6#2: CACUUCGAAGCGAAAUAUU
siVHL: UAUCACACUGCCAGUGUAUAC
2.8. Western blot analysis
Whole-cell lysates were isolated in RIPA buffer (Cell Signaling Technology, 9806S) supplemented with the protease inhibitor (Med Chem Express, HY-K0010) and the phosphatase inhibitor (Med Chem Express, HY-K0022). Total protein was separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and then transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% nonfat dried milk, and incubated with the indicated primary antibody and secondary antibodies. The protein bands were visualized with ECL detection reagents (PE, NEL105001EA). The primary antibodies used were anti-DNMT3A (Cell Signaling Technology, 32578), anti-HDAC6 (Cell Signaling Technology, 7558), anti-PARP (Santa Cruz, sc-7150), anti-HIF-1α (Proteintech, 20960-1-AP), anti-VHL (Proteintech, 24756-1-AP), anti-β-actin (anti-ACTB, Santa Cruz, sc-8432).
2.9. RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Invitrogen, 15596026) according to the manufacturer's protocol. First-strand cDNA synthesis was set up using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, K1622). Quantitative real-time PCR analysis was performed by using Top Green qPCR SuperMix (Transgene, AQ601-04) by the manufacturer's protocols. GAPDH was used as the endogenous control. All qRT-PCRs were set up in triplicates using three biological replicates for each sample.
2.10. Immunohistochemical staining
Tumor samples acquired from the mice bearing H460/H1299 cells were fixed in 4% neutral buffered formalin and embedded in paraffin. Samples were dewaxed, and hydrated with xylene and ethanol, and citric acid buffer was used for antigen repair. Tumor samples were incubated with primary antibody and secondary antibody. DAB and hematoxylin were added for staining and observed under a microscope. The primary antibodies used were anti-DNMT3A (Proteintech, 20954-1-AP), anti-HDAC6 (Proteintech, 12834-1-AP), anti-Ki67 (Abcam, ab15580), anti-HIF-1α (Proteintech, 20960-1-AP), anti-VHL (Proteintech, 24756-1-AP).
2.11. In vivo tumor xenograft animal model
BALB/c nude mice (body weight 18–22 g) were maintained in a specific-pathogen-free (SPF) facility. NCI-H460/H1299 cells (1 × 106–2 × 106 cells in 0.2 mL phosphate-buffered saline) were subcutaneously injected into the right flank of BALB/c nude mice. The drugs were administered to mice by intraperitoneal injection for 21 consecutive days. After 21 days, the mice were sacrificed, the tumors were photographed and the tumor volumes were analyzed. Tumor volumes were measured using calipers every 2 days and calculated by Eq. (1):
| Tumor volume = 1/2 × (Long diameter) × (Short diameter)2 | (1) |
2.12. Statistical analysis
Statistical analyses were performed using SPSS V 26.0 software. Data in all graphs were represented as mean ± standard error of mean (SEM) of biological triplicates by using GraphPad Prism version 8.0. Statistical significance was determined by Student's t-test (independent samples t-test), one-way analysis of variance (ANOVA) followed by Tukey's or Dunnett's T3 multiple comparisons test. P values < 0.05 were considered statistically significant.
3. Results
3.1. The aberrant alteration and the role of DNMT3A in cancer
Due to the paradoxical role of DNMT3A in cancer, we analyzed the expression and clinical significance of DNMT3A in various cancers according to the Kaplan–Meier Plotter database. As shown in Fig. 1A, the median survival of patients with low expression of DNMT3A was 56 months compared to 99 months for patients with high DNMT3A expression. The patients with the decreased expression of DNMT3A predict a poor prognosis, suggesting the TSG role of DNMT3A in cancer. The inactivation of TSGs is mostly caused by mutation. Therefore, we assessed the clinical value of DNMT3A mutation using the ICGC database. Our analyzed data indicated that there was a 12.0% frequency of DNMT3A mutation in cancer. Importantly, mutation of DNMT3A was associated with poor prognosis (Fig. 1B), which further confirmed its possible TSG role in cancer. According to the mutation rate of DNMT3A in various tumors, lung cancer is located in the front ranks (Fig. 1C and Supporting Information Fig. S1). Thus, we next choose lung cancer as a study example. TCGA database revealed that, compared with normal tissues, lung cancer tissues expressed DNMT3A at a lower level (Fig. 1D). Furthermore, we measured the expression level of DNMT3A in 80 patients of lung cancer. In line with the database data, patients with DNMT3A low expression predicted a bad survival rate in lung cancer (Fig. 1E). To explore the function of DNMT3A in lung cancer, we established the DNMT3A knockout (DNMT3AKO) model using the CRISPR-Cas9 approach. As expected, knockout of DNMT3A resulted in an enhanced ability in proliferation and self-renewal in H460 and A549 cell lines (Fig. 1F). On the contrary, overexpression of DNMT3A would lead to the reduction of proliferation and self-renewal in H460 and A549 cell lines (Fig. 1F). Taken together, the above results indicate that the aberrant alteration of DNMT3A is frequent and DNMT3A may act as a tumor suppressor gene in lung cancer, which suggests the crucial role of DNMT3A in cancer.
Figure 1.
Effects of DNMT3A deletion on the biological characteristics of non-small cell lung cancer. (A) The correlation between DNMT3A expression and survival probability is shown. Log-rank (Mantel–Cox) test, P < 0.001. (B) The overall survival of lung cancer patients with DNMT3A mutation (n = 779) and no DNMT3A mutation (n = 5695). Log-rank (Mantel–Cox) test, P < 0.001. (C) The incidence of DNMT3A mutations in 32 types of cancer (10,967 samples, lung adenocarcinoma 4.24% and lung squamous cell carcinoma 1.64%). The majority of DNMT3A mutations are loss of function or likely loss of function. (D) The expression of DNMT3A in normal tissues (n = 50) and tumor tissues (n = 51) in the TCGA database. Log-rank (Mantel–Cox) test, ∗∗∗P < 0.001. (E) The correlation between DNMT3A protein expression level and overall survival in lung cancer is shown. The sections of tumor tissues from two lung cancer patients showed the immunohistochemical detection of DNMT3A protein. Scale bars, 50 μm. Log-rank (Mantel–Cox) test, ∗P < 0.05. (F) Expression of DNMT3A in DNMT3A knockout and overexpression cells, and effect of DNMT3A expression on cell proliferation and self-renewal ability. Data are presented as mean ± SEM (n = 3). ∗P < 0.05, ∗∗P < 0.01. P values were determined using Student's t-test (independent samples t-test).
3.2. Knockout of DNMT3A induces tumor growth and drug resistance
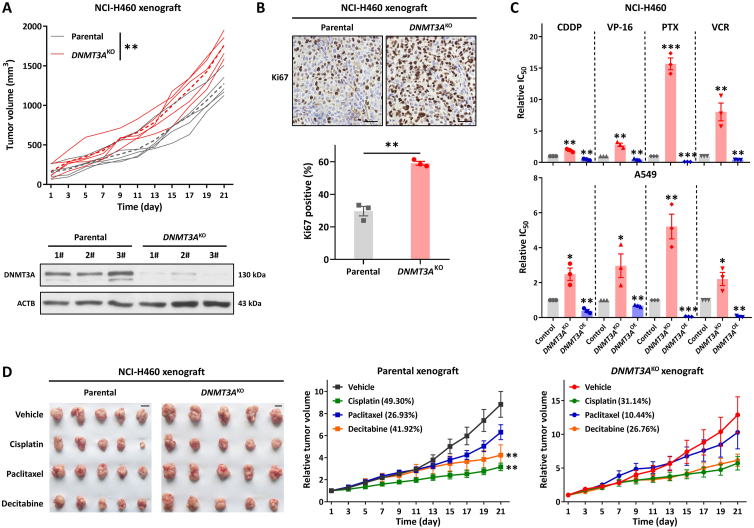
To further explore the role of DNMT3A in lung cancer, we established subcutaneous xenograft models with NCI-H460 parental and DNMT3AKO cells. As expected, knockout of DNMT3A resulted in rapid tumor growth in a mouse model. Compared with the parental H460 xenograft model, the H460-DNMT3AKO xenograft displayed an increasing tumorigenesis capability (Fig. 2A). In addition, the proliferation biomarker, Ki67, was increased in H460-DNMT3AKO tumor tissues as compared with parental H460 tumor tissues (Fig. 2B), which confirmed the TSGs role of DNMT3A in lung cancer. Previous studies indicated that epigenetic enzyme mutation would contribute to drug resistance32, therefore, we next assessed the effects of gene manipulation of DNMT3A on drug sensitivities in lung cancer. As shown in Fig. 2C, knockout of DNMT3A resulted in the decrease of sensitivities of H460 and A549 cell lines to frequently-used chemotherapy agents, including cisplatin (CDDP), etoposide (VP-16), paclitaxel (PTX) and vincristine (VCR), whereas overexpression of DNMT3A could increase the sensitivities of H460 and A549 cell lines to chemotherapy agents. Consistent with the in vitro results, the H460-DNMT3AKO xenograft mouse model also showed a reduction of sensitivities to chemotherapy agents, including cisplatin, paclitaxel, and DNMT inhibitor decitabine, as compared with the parental xenograft model (Fig. 2D and Supporting Information Fig. S2A). Moreover, the weight of mice did not decrease significantly (Fig. S2B). The above results demonstrate that DNMT3A exhibits a TSG role in lung cancer.
Figure 2.
Knockout of DNMT3A promotes tumor growth and drug resistance in vivo. (A) Tumor growth in BALB/c-nu mice xenografted with NCI-H460-DNMT3AKO and parental cells (n = 5). The expression level of DNMT3A in mouse tumor tissues (B) Ki67 staining of tumor tissue cells in xenograft mice. Scale bars, 25 μm. (C) Effect of DNMT3A expression on the sensitivity of chemotherapeutic drugs (CDDP, VP-16, PTX, and VCR). (D) CDX-bearing mice (n = 5 mice per group) were treated with vehicle (three times per week), CDDP (4 mg/kg, twice per week), PTX (5 mg/kg, twice per week), or decitabine (1 mg/kg, three times per week) through intraperitoneal administration. The graph shows the tumor image and relative tumor volume, with the tumor growth inhibition value. Scale bars, 1 cm. Data are presented as means ± SEM (n = 3–5). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. In (A) and (B), P values were determined using Student's t-test (independent samples t-test). In (C) and (D), P values were determined using one-way ANOVA with Tukey's or Dunnett's T3 multiple comparisons test.
3.3. Identification of a synthetic lethal interaction of DNMT3A and HDAC6
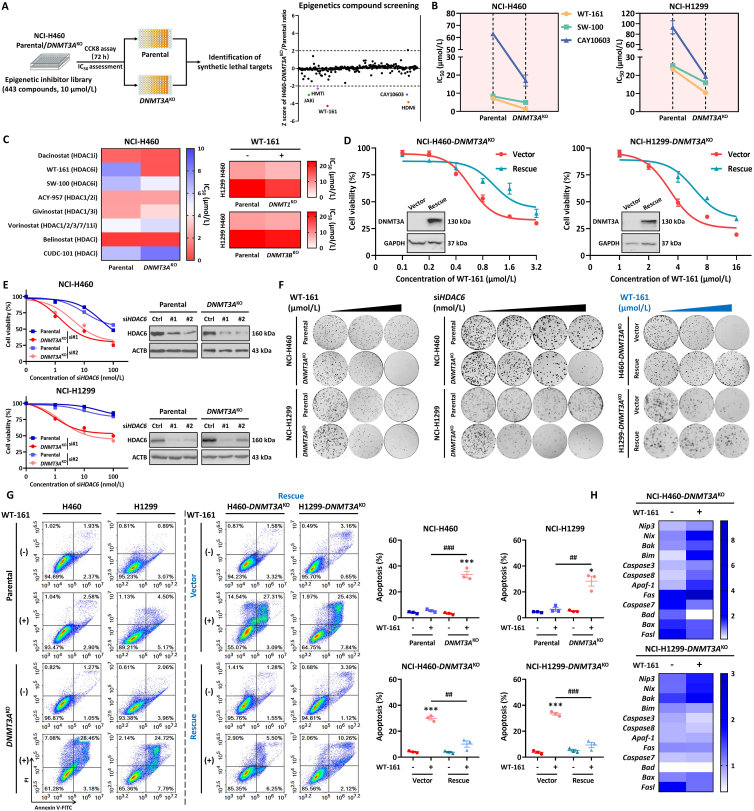
Based on the above finding, we next performed a drug screening with a total of 443 epigenetic enzyme inhibitors in DNMT3A proficient (parental) and DNMT3AKO cells to identify an epigenetic synthetic lethal partner with TSG DNMT3A. Drug testing and viability assay were conducted with a single concentration of 10 μmol/L. To exclude the effect of high selectivity but with low efficacy, we used the Z score to assess the selectivity of compounds. Out of 443 compounds, we identified 5 inhibitors that showed higher efficiency in H460-DNMT3AKO cells compared with parental cells. Notably, within the identified hits, we found JAKi, which is in line with previous reports (Fig. 3A and Supporting Information Fig. S3A)33. Among the 5 identified inhibitors, two inhibitors, namely WT-161 and CAY10603, belong to HDAC6-specific inhibitors. The selective action of HDAC6 inhibition was confirmed by various inhibitors targeting HDAC6 in both H460-DNMT3AKO and H1299-DNMT3AKO cell lines (Fig. 3B). To clarify the specific action of HDAC6 inhibition, we also detected the cell viability of parental and DNMT3AKO cell lines after being treated with additional HDAC inhibitors. The results indicated that only HDAC6-specific inhibitors displayed a selectivity inhibitory action to DNMT3AKO cell lines (Fig. 3C). Furthermore, we also evaluated the HDAC6 inhibition in other DNMT isoforms (DNMT1 and DNMT3B) knockout cells. As shown in Fig. 3C and Fig. S3B, treatment with HDAC6 inhibitor WT-161 exhibited a similar inhibitory action in both parental and DNMT1KO or DNMT3BKO cell lines. The above results indicate synthetic lethal action between DNMT3A and HDAC6 is a specific and unique interaction. To confirm that the selective action of HDAC6 inhibition on NSCLC cell growth was due to the on-target effect of DNMT3A deficiency, we restored DNMT3A expression in DNMT3AKO cell lines and found that DNMT3A expression reduced the sensitivity of cells to WT-161 (Fig. 3D), demonstrating the on-target effect of the DNMT3A construct. Together these data elucidated that HDAC6 inhibition selectively reduces the growth of DNMT3A-deficient NSCLC cells.
Figure 3.
Identification and validation of synthetic lethal target. (A) Flow chart for synthetic lethality screening of epigenetics compound library. After NCI-H460 parental and DNMT3AKO cell lines were treated with epigenetics compound library (10 μmol/L) for 72 h, cell viability was determined by CCK8 assay. Z-score scatterplot of relative lethality for epigenetics compound library screening. Most compounds have a Z-score index between 1 and –1, and compounds with a Z-score below −2 were deemed “hit”. In (A), 256 compounds are shown (the remaining 187 compounds are shown in Supporting Information). (B) NCI-H460/NCI-H1299 parental and DNMT3AKO cells were treated with HDAC6 selective inhibitors (WT-161, SW-100, CAY10603) for 72 h, and the cell viability was detected by CCK8 assay. (C) A heatmap showing the IC50 values of the selected compound on NCI-H460 parental and DNMT3AKO cells (left), and the efficacy of WT-161 in NCI-H460/NCI-H1299 parental or DNMT1KO/DNMT3BKO cells (right). (D) Effect of DNMT3A re-expression on the synthetic lethality of the HDAC6 inhibitor in NCI-H460-DNMT3AKO and NCI-H1299-DNMT3AKO cells. (E) The cell viability of parental and DNMT3AKO cells after gene intervention of HDAC6 expression. (F) Effects of HDAC6 inhibition (0, 0.16, 0.32 μmol/L for H460 and 0, 2, 4 μmol/L for H1299) or knockdown (0, 20, 40, 60 nmol/L for H460 and H1299) on the colony formation ability of lung cancer cells with different DNMT3A expression levels. (G) The apoptosis induction by HDAC6 inhibitor WT-161 (0, 1 μmol/L for H460 and 0, 5 μmol/L for H1299) in lung cancer cells with different DNMT3A expression levels. (H) The heatmap showed the changes of apoptosis-related target genes in NCI-H460-DNMT3AKO and NCI-H1299-DNMT3AKO under WT-161 treatment. Data are presented as means ± SEM (n = 3). ∗P < 0.05, ∗∗∗P < 0.001, compared with control group. ##P < 0.01, ###P < 0.001, compared with the indicated group. P values were determined using Student's t-test (independent samples t-test).
3.4. HDAC6 inhibition reduces the growth of DNMT3A-deficient NSCLC cells by inducing cell apoptosis
Given that pharmacology inhibition of HDAC6 by compounds might be mediated by off-target action, we next assayed whether specific siRNA for HDAC6 produced a selective action in DNMT3A-deficient NSCLC cells. As shown in Fig. 3E, knockdown of HDAC6 by two siRNAs could result in an obvious concentration-dependent reduction of cell growth in both DNMT3AKO cell lines, but just produced a weak decrease in parental cell lines, which mimicked the action of HDAC6 inhibitors. To further assess the synthetic lethal action between DNMT3A and HDAC6, we also performed colony formation analysis. Our results revealed that consistent with cell viability data, inhibition of HDAC6 by both pharmacology inhibitor and gene manipulation led to selective reduction of colony formation number in H460-DNMT3AKO and H1299-DNMT3AKO cell lines, whereas restoration of DNMT3A in DNMT3AKO cell lines alleviate their inhibitory action (Fig. 3F). Next, to explore the underlying mechanism, we detected the cell apoptosis status, which is considered as the main death pathway of synthetic lethal, after being treated with pharmacology inhibitors or given gene manipulation. The apoptosis measurement data indicated that HDAC6 inhibitor WT-161 treatment resulted in an obvious increase of apoptosis rate (Annexin V positive cell), from ∼2% to ∼31%, in H460-DNMT3AKO cells, compared that, from ∼4% to ∼5%, in H460 parental cells. Similar results were obtained from H1299 parental and DNMT3AKO cell lines (Fig. 3G). Consistent with pharmacology inhibitor treatment, the addition of specific HDAC6 siRNA also led to selectively increasing cell apoptosis in DNMT3AKO cells (Fig. S3C). The DNMT3A-deficient-dependent apoptosis induction of HDAC6 inhibition was further confirmed by the restoration of DNMT3A (Fig. 3G). Rescue of DNMT3A significantly alleviated the apoptosis rate, from ∼30% to ∼10%, in H460/H1299-DNMT3AKO cell lines. To further validate the synthetic lethal effect of DNMT3A and HDAC6, we found that the hotspot mutation DNMT3A R882H in acute myeloid leukemia (AML) is a loss-of-function mutation34. We introduced DNMT3A R882H into lung cancer cell lines H460 and H1299 to simulate the real DNMT3A LOF mutation in the clinic as much as possible and then investigated the effect of the HDAC6 inhibitor WT-161 on cells expressing the R882H mutation. The results showed that similar to the effect of WT-161 on DNMT3A knockout cells, WT-161 selectively inhibited the growth of cells expressing R882H. This indicated that HDAC6 inhibitor selectively killed cells with DNMT3A functional defects (Supporting Information Fig. S4A and S4B). Moreover, we selected two AML cell lines HL-60 and NB-4, treated cells with siRNAs to silence DNMT3A, and assessed the effect of WT-161 on AML cells. The results also confirmed the synthetic lethality of DNMT3A/HDAC6 in AML cells (Fig. S4C and S4D). In addition, we also found that some apoptosis-related genes were up-regulated in DNMT3AKO cells treated with WT-161 (Fig. 3H). Taken together, the above results demonstrated that inhibition of HDAC6 by both pharmacology inhibitor and gene manipulation could selectively reduce cell viability in DNMT3AKO cells by inducing cell apoptosis.
3.5. HDAC6 inhibition by WT-161 is effective in DNMT3A-deficient NSCLC mouse model
To validate whether HDAC6 inhibition could be effective in vivo, we evaluated the efficacy of HDAC6 inhibitor WT-161 in mice carrying H460 and H1299 NSCLC xenografts (Fig. 4A). WT-161 treatment resulted in a significant reduction in tumor volume in the DNMT3AKO xenograft model, whereas the parental xenograft model did not change in tumor volume. Similarly, WT-161 administration also led to a dose-dependent decrease in tumor volume (Fig. 4A), with a maximal inhibition rate (IR) of 69.63%, and positive expression of Ki67 (Fig. 4B). Importantly, the administration of WT-161 also significantly induced apoptosis measured by cleaved PARP in H460-DNMT3AKO xenograft tumors compared to that in parental xenograft tumors (Fig. 4C). To further explore the role of the DNMT3A in the anti-tumor efficacy of HDAC6 inhibition in vivo, we applied the Tet-on model to animal experiments. The results showed that in the absence of doxycycline (dox) induction, after treatment with WT-161, the tumor growth in H460-DNMT3AKO xenograft mice was significantly decreased, with IR 39.53% (Fig. 4D). After the addition of the inducer to rescue DNMT3A expression, the tumor inhibitory efficacy of WT-161 in mice receiving H460-DNMT3AKO cells was significantly weaker than that in the vector-construct group (IR: 25.29% vs. 65.65%), supporting DNMT3A deficiency as an essential contributor of WT-161 efficacy (Fig. 4D). The body weight and viscera index of the H460-DNMT3AKO xenograft model were not significantly altered during drug administration (Supporting Information Fig. S5). Taken together, these data indicate that HDAC6 inhibition by WT-161 is effective in DNMT3A-deficient NSCLC mouse models by suppressing cell proliferation and inducing cell apoptosis.
Figure 4.
HDAC6 inhibition by WT-161 is effective in DNMT3A-deficient NSCLC mouse model. (A) Representative images of NCI-H460/NCI-H1299 parental and DNMT3AKO xenograft tumors and statistical analysis of the relative growth inhibition (n = 5). Scale bars, 1 cm. (B) Evaluation of Ki67 expression in the indicated xenograft tumors by IHC assay. Scale bars, 25 μm. The statistics of Ki67 positive cell rate in the indicated xenograft tumors are shown at the bottom. (C) Western blot analysis of DNMT3A and PARP expression in NCI-H460 parental and DNMT3AKO xenograft tumors. (D) Image of a tumor xenograft model with DNMT3A expression induced by doxycycline, 1–10 days without doxycycline induction and 11–20 days with doxycycline induction. The tumor growth curve is shown on the right (n = 5). Data are presented as means ± SEM (n = 3–5). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, compared with control group. ##P < 0.01, ###P < 0.001, compared with the indicated group. P values were determined using Student's t-test (independent samples t-test) and one-way ANOVA with Tukey's or Dunnett's T3 multiple comparisons test.
3.6. DNMT3A inhibition induces HDAC6 through reducing promoter methylation status
Understanding the mode of interaction of synthetic lethal partners is critical for the formulation of clinical therapeutic strategy. To address this issue, we next explored how knockout of DNMT3A affects HDAC6 expression. As shown in Fig. 5A, the protein expression and activity of HDAC6 were obviously upregulated in DNMT3A-deficient NSCLC cell lines as compared with parental cell lines. On the contrary, the other HDACs' isoforms were not significantly changed (Supporting Information Fig. S6). Additionally, the restoration of DNMT3A would lead to the down-regulation of HDAC6 protein in DNMT3A-deficient NSCLC cell lines (Fig. 5A). This expression pattern was mimicked at the mRNA level (Fig. 5B), supporting HDAC6 induction by DNMT3A inhibition at transcription regulation. Notably, knockdown of DNMT3A by siRNA also resulted in the upregulation of HDAC6 (Fig. 5C), which further confirmed the regulation relationship between DNMT3A and HDAC6. In view of the role of DNMT3A in DNA methylation, we next detected the expression of HDAC6 after being treated with DNMT inhibitor 5-AzaC in NSCLC cell lines. As shown in Fig. 5D, treatment with 5-AzaC would induce the upregulation of HDAC6 in both mRNA and protein levels, demonstrating that the expression of HDAC6 might be regulated by DNA methylation. Usually, DNA methylation in the promoter region could block transcription factor's binding and contribute to transcription suppression35. Therefore, we next analyzed the transcription factor binding site in the HDAC6 promoter region using the JASPAR prediction system. The analyzed results indicated that transcription factors RUNX1 and TFAP2A own a relatively higher matrix score (Fig. 5E). Next, we investigated whether the transcription factors could regulate the transcription activation of HDAC6. Our results indicated that the reporter gene of HDAC6 promoter could be activated by overexpression of RUNX1 and TFAP2A in H460 cells, and their activation can be enhanced by treatment with 5-AzaC (Fig. 5F), suggesting the status of promoter methylation of HDAC6 might affect its transcription activation by transcription factors. In addition, the transcription level of HDAC6 was also enhanced by RUNX1 overexpression, but not TFAP2A, in the DNMT3AKO cells and 5-AzaC treated cells (Fig. 5F), which indicated that RUNX1 played a more important role in methylation-dependent HDAC6 regulation. To deeply explore the interaction between DNA methylation and transcription factor in the HDAC6 promoter region, we performed bisulfite sequencing analysis. Our data revealed that, as compared with parental cells, there were 3 sequencing regions, containing transcription factor binding sites, in the HDAC6 promoter that showed a decrease of CpG methylation in DNMT3AKO cell lines (Fig. 5E). Consistent with this result, the treatment of 5-AzaC also contributed to a reduction of CpG methylation in HDAC6 promoter regions (Fig. 5E). Importantly, ChIP results demonstrated that knockout of DNMT3A could result in an enrichment of RUNX1 on the promoter region of HDAC6 in both NCI-H460 and NCI-H1299 cell lines (Fig. 5G). Consistent with the above data, the GEPIA database analyzed results demonstrated that the expression of RUNX1 was positively related to the HDAC6 level, which indicated that RUNX1 might be a transcription activator of HDAC6 (Fig. 5H). Next, we performed immunohistochemistry to assess the relationship between DNMT3A and HDAC6 in NSCLC patients' tumor tissue. Pearson correlation analysis showed a negative correlation between DNMT3A and HDAC6 in the studied 80 tumor tissues (Fig. 5I), which was consistent with in vitro results. Taken together, our results elucidate that inhibition of DNMT3A would induce upregulation of HDAC6 through reducing promoter DNA methylation status.
Figure 5.
DNMT3A inhibition induces HDAC6 through reducing promoter methylation status. (A) Expression level and activity of HDAC6 in NCI-H460/H1299 with different DNMT3A expression levels. (B) Relative HDAC6 mRNA level in NCI-H460/H1299 with different DNMT3A expression levels. (C) HDAC6 protein expression level in NCI-H460/H1299 cells after knockdown of DNMT3A by siRNA (50 nmol/L). (D) Changes of HDAC6 mRNA and protein levels in NCI-H460/NCI-H1299 parental cells treated with 5-azacytidine (10 μmol/L for H460 and 5 μmol/L for H1299, 24 h). (E) Predicted transcription factor binding site and bisulfite sequencing analysis in HDAC6 promoter region. Bisulfite sequencing was used to analyze the altered methylation status in the CG site of HDAC6 promoter under different DNMT3A expression levels. 5-Azacytidine treated with 5 μmol/L for 24 h. (F) Dual luciferase reporter assay and RT-qPCR were used to evaluate HDAC6 promoter activity and mRNA levels after overexpression of transcription factors in different DNMT3A expression states. (G) The binding of RUNX1 to HDAC6 promoter in parental and DNMT3AKO cells was detected by ChIP-qPCR. (H) The correlation between RUNX1 and HDAC6 expression was analyzed by the GEPIA database (lung adenocarcinoma and lung squamous cell carcinoma). (I) Immunohistochemistry combined with Pearson correlation analysis was used to analyze the correlation between the expression of DNMT3A and HDAC6 in the tumor tissues of 80 patients. N: Normal tissue, T: Tumor tissue. Data are presented as means ± SEM (n = 3). ∗P < 0.05, ∗∗P < 0.01, compared with control group. #P < 0.05, ###P < 0.001, compared with the indicated group. P values were determined using Student's t-test (independent samples t-test) and one-way ANOVA with Tukey's or Dunnett's T3 multiple comparisons test.
3.7. DNMT3A deficiency results in HDAC6–VHL–HIF-1 axis activation
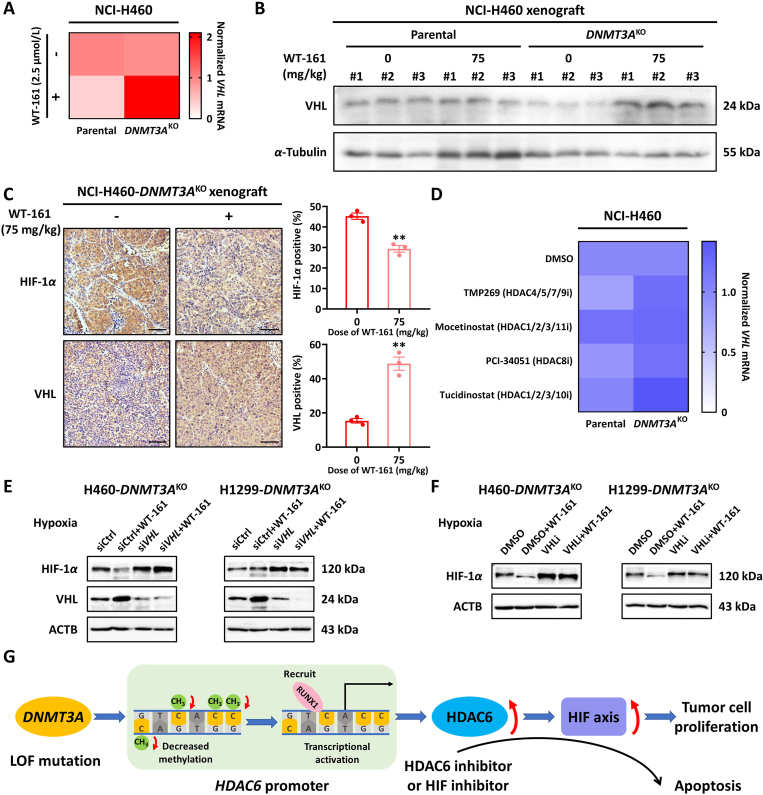
To identify HDAC6 targets in DNMT3A-deficient NSCLC cells, we performed RNA sequencing analysis in H460 parental cells, H460-DNMT3AKO cells, and WT-161 treated DNMT3AKO cells. Since DNMT3A is a DNA methyltransferase and possible gene silencing function36, we first focused our analysis on the upregulated genes in DNMT3A-deficient cells. Our results showed that there were 1165 upregulated genes in H460-DNMT3AKO cells relative to parental cells. Next, in view of the selectivity anti-growth action of HDAC6 inhibition, we analyzed the down-regulated genes in HDAC6 inhibitor WT-161 treated DNMT3AKO cells as compared with vehicle control. There were 1966 down-regulated genes in WT-161 treated cells. Notably, there were 465 converged genes in both groups, suggesting their crucial role in this synthetic lethal interaction (Fig. 6A). Integrated KEGG pathway and Gene set enrichment analysis (GSEA) revealed that the converged genes were significantly enriched in pathways related to HIF-1, a well-known oncogenic pathway (Fig. 6B). To further explore the relationship between HDAC6 inhibition and the HIF-1 pathway, we next detected the expression levels of some HIF-1 target genes, in the WT-161 treated DNMT3AKO cell lines (Supporting Information Fig. S7A and S7B). Besides, we also detected the expression of HIF-1α and VHL in cells under normoxia or hypoxia and analyzed the expression of HIF-1α and VHL in patients through the cBioPortal database. The results showed that DNMT3A deletion or loss of function resulted in high expression of HIF-1α and low expression of VHL (Fig. S7C–S7E). The role of HIF-1 in DNMT3A/HDAC6 synthetic lethal was confirmed by cell viability assay. Our results indicated that knockdown of DNMT3A by specific siRNA could enhance the inhibitory action of WT-161 in H460 cell lines, which was consistent with our previous results. In contrast to H460 parental cells, the enhanced inhibitory action between WT-161 and DNMT3A siRNA disappeared in HIF-1α knockout H460 cell lines (Fig. 6C). Similar results were obtained in colony formation assays (Fig. 6D). The above results suggest that the synthetic lethal action of DNMT3A/HDAC6 is dependent on the HIF-1 pathway. According to GSEA data, the HIF-1 pathway was inhibited by WT-161 treatment, thus we next investigated whether inhibition of the HIF-1 pathway could mimic the WT-161 function. The results revealed that treatment with various HIF-1 inhibitors, including PX-478 and LW6, could obviously reduce colony formation in DNMT3AKO NSCLC cell lines as compared with those in parental NSCLC cell lines (Fig. 6E). Additionally, FACS analysis also demonstrated that HIF-1 inhibitors could selectively induce cell apoptosis in DNMT3AKO NSCLC cells (Fig. 6F). HIF-1 pathway exhibits its function through regulating the tumor microenvironment, therefore we next investigated the role of the HIF-1 pathway in DNMT3A/HDAC6 synthetic lethal interaction using a xenograft mouse model. Consistent with our previous results, HIF-1 inhibitor PX-478 treatment would result in an obvious reduction of tumor growth in the DNMT3AKO xenograft model, with an inhibition rate of 86.35% and no gross toxicity (Supporting Information Fig. S8). However, the anti-tumor efficacy of PX-478 was significantly attenuated, with an inhibition rate of 29.74%, in the DNMT3A parental xenograft model (Fig. 6G). Consistent with tumor inhibitory efficacy, treatment with PX-478 also significantly induces cleavage of PARP, an apoptosis biomarker of tumor tissues in the DNMT3AKO xenograft model (Fig. 6G). These data clarified that the block of the HIF-1 pathway owned a similar biology function with HDAC6 inhibition in DNMT3A-deficient cells.
Figure 6.
DNMT3A deficiency results in HDAC6–VHL–HIF-1 axis activation. (A) The Venn diagram shows the differentially expressed genes (DEG) between DNMT3AKOvs. parental and DNMT3AKO + WT-161 (2.5 μmol/L, 48 h) vs. DNMT3AKO. (B) The bubble map and GSEA analysis showed the most obvious signaling pathway with different conditions. (C) DNMT3A was knocked down by siRNA (80 nmol/L) in NCI-H460 parental and HIF-1αKO cells and cell activity was detected by CCK8 after treatment with WT-161 for 48 h. (D) A colony formation assay was used to investigate the changes in the proliferation ability of NCI-H460 parental and HIF-1αKO cells after DNMT3A knockdown. (E) HIF-1α inhibitors (PX-478 and LW6) were treated on NCI-H460/NCI-H1299 parental and DNMT3AKO cells, and the proliferation ability of the cells was investigated by colony formation assay. (F) Annexin V-PI staining showed changes in apoptotic ability of NCI-H460/NCI-H1299 parental and DNMT3AKO cells after treatment with PX-478 (25 μmol/L for H460, 6 μmol/L for H1299) or LW6 (15 μmol/L for H460 and H1299). (G) A mouse xenograft model was established using NCI-H460 parental and DNMT3AKO cells (n = 5). The column chart showed the relative growth inhibition after treatment with PX-478 (15 mg/kg, five times per week). The expression of DNMT3A and cleaved PARP in xenografts was analyzed by Western blot. Data are presented as means ± SEM (n = 3–5). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, compared with control group. #P < 0.05, ###P < 0.001, compared with the indicated group. P values were determined using Student's t-test (independent samples t-test).
Von Hippel-Lindau (VHL) is a tumor suppressor gene, that is highly expressed in normal tissues and benign tumors but decreased in a variety of malignant tumors37. Lots of studies have shown that the deficiency of VHL tumor suppressor gene can inhibit the degradation of HIF-1α, thereby increasing the expression of HIF-1α and causing a series of changes in cells38. Based on this, we wondered whether the down-regulation of HIF-1 expression caused by HDAC6 inhibitor in DNMT3AKO cells was related to the change of VHL expression. We detected the mRNA and protein expression of VHL in H460/H1299 cells and H460 tumor tissues after WT-161 treatments, and the results showed that the mRNA level of VHL increased after WT-161 treated DNMT3AKO cells (Fig. 7A). In addition, we detected the expression of HIF-1α and HIF-1β in cells under hypoxia. The results showed that the expression of HIF-1α in DNMT3A knockout cells increased, while the expression of HIF-1α decreased with the addition of WT-161, and the expression of HIF-1β did not change significantly (Supporting Information Fig. S9). Similarly, the level of VHL protein in DNMT3AKO tumor tissues after WT-161 treatment also increased, and HIF-1α expression was downregulated (Fig. 7B and C). In addition, we also found that inhibitors of other subtypes of HDAC had no significant effect on the mRNA expression level of VHL in DNMT3AKO cells (Fig. 7D). These results indicated that HDAC6 inhibitor WT-161 induced the down-regulation of HIF-1 by upregulating the expression of VHL in DNMT3AKO cells. In order to explore whether the HIF-1α regulation by HDAC6 inhibition is dependent on VHL. We treated the cells with VHL siRNA to investigate whether HDAC6 inhibitors can still affect the expression of HIF-1α under the condition of silencing VHL. The results showed that the expression of HIF-1α was not affected by the addition of WT-161 after transient silence of VHL under hypoxia (Fig. 7E). We also used a VHL inhibitor and observed the same phenomenon (Fig. 7F). Together, the above results demonstrate that the synthetic lethal action of DNMT3A/HDAC6 is mediated by the VHL–HIF-1 pathway.
Figure 7.
Increased expression level of VHL after treatment with WT-161. (A) Changes of VHL mRNA expression in NCI-H460 parental and DNMT3AKO cells after WT-161 treatment (2.5 μmol/L, 48 h). (B) Detection of VHL protein expression in parental and DNMT3AKO tumor tissues treated by WT-161 (75 mg/kg, 21 days) by Western blot. (C) Expression of HIF-1α and VHL in tumor tissue of xenograft mice by IHC assay. Scale bars, 50 μm. (D) Effects of inhibitors of other HDAC subtypes (HDAC 4/5/7/9 inhibitor: TMP269, HDAC 1/2/3/11 inhibitor: Mocetinostat, HDAC 8 inhibitor: PCI-34051, HDAC 1/2/3/10 inhibitor: Tucidinostat, 2.5 μmol/L, 48 h) on VHL mRNA expression levels. (E, F) The effect of HDAC6 inhibitor WT-161 (2.5 μmol/L, 48 h) on HIF-1α expression after VHL siRNA (100 nmol/L, 48 h) or VHL inhibitor VH-298 (40 μmol/L, 48 h) treatment under hypoxia. (G) Schematic diagram showing the increase of HDAC6 induced by DNMT3A loss, which leads to HIF axis activation. Data are presented as means ± SEM (n = 3). ∗∗P < 0.01. P values were determined using Student's t-test (independent samples t-test).
4. Discussion
There has been a growing interest in using synthetic lethality to identify personalized treatment strategies for cancers with specific genetic defects39,40. DNMT3A, a DNA methyltransferase enzyme, was reported to own a relatively high mutation rate in various malignant tumors20. Patients with DNMT3A mutations, mostly loss-of-function mutation, usually predict a bad prognosis41. However, the role and therapeutic value of DNMT3A mutation were still not well known. Here, we demonstrated that DNMT3A owned a tumor suppressor function in lung cancer. Using the epigenetic inhibitors library, a synthetic lethality interaction between DNMT3A and HDAC6 in lung cancer was identified. Mechanistically, DNMT3A loss results in the upregulation of HDAC6 by decreasing its promoter methylation level and enhancing its transcription activation. As a result, the oncogenic HIF-1 pathway is activated in DNMT3A-deficient cell lines. Treatment with HDAC6 inhibitor could retard tumor growth in vitro and in vivo in DNMT3A-deficient NSCLCs via blocking HIF-1 activation (Fig. 7G).
DNMT3A is originally recognized as a pro-oncogenic enzyme, whose overexpression or activation would result in the loss of tumor suppressor genes, such as p1642, 43, 44, CDH1, and RASSF1A45, through enhancing their promoter CpG methylation. Deng et al.46 found that silencing of DNMT3A with RNA interference inhibited melanoma growth and metastasis. In addition, several reports revealed that overexpression of DNMT3A might be associated with malignant characteristics such as high invasiveness and recurrence in melanoma, vulvar squamous cell carcinomas47, and pituitary adenomas45. Paradoxically, several studies have shown that deletion of DNMT3A will contribute to tumor progression in various tumors21. Here, we verified that knockout of DNMT3A in NSCLC cell lines led to malignant phenotypes, including proliferation, self-renewal, and multiple drug resistance. Furthermore, clinical study data showed that NSCLC patients with loss of DNMT3A might have a bad prognosis. Taken together, our results demonstrate that DNMT3A displays a tumor suppressor role in NSCLC, which suggests the function of DNMT3A is highly specific, depending on the tissue and cell type.
Tumor suppressor genes are usually considered undruggable. Recently, the success of synthetic lethality provides a promising approach to target tumor suppressor genes. It is reported that DNMT3A executes its function dependent on interaction and regulation by the other epigenetic enzymes, such as EZH248 and DNMT111. Therefore, we identified a synthetic lethal interaction partner of DNMT3A with a chemical epigenetic inhibitors screen. Our finding disclosed that pharmacology inhibition of HDAC6 selectively reduced the growth and survival of DNMT3A-deficient NSCLC cells and mouse models. Although we found that multiple HDAC6 inhibitors (WT-161, SW-100, CAY10603) differed in the strength of their effects on DNMT3A (Fig. 3B), all three inhibitors specifically target HDAC6, so differential effects due to target inconsistency are essentially ruled out. We speculate that the reason for the difference in the effects of HDAC6-specific inhibitors may be related to the potency, efficacy, and distribution of the drugs in vivo. Some researchers have demonstrated the anticancer activity of WT-161 in human multiple myeloma cell xenograft mouse model49. It has also been reported that WT-161 and the BET inhibitor have synergistic anti-tumor efficacy against osteosarcoma xenografts, and their combined therapy may be a potential therapeutic strategy50. It shows that WT-161 has the strongest effect in vivo, which suggests that WT-161 has a better druggability.
Integrated analysis of transcription, protein interaction, and bisulfite sequencing revealed that DNMT3A knockout decreased promoter methylation of HDAC6 and increased its transcription activation of RUNX1. As a result, the upregulation of HDAC6 induced the malignant phenotype of NSCLCs. Thus, inhibition of HDAC6 would lead to selective death in DNMT3A-deficient cells. In fact, the epigenetic-based synthetic lethality of DNMT3A/HDAC6 can be extended to other epigenetic pairs31, such as EZH2/ARIDA30, SMARCB/HDAC51, CREBBP/p30052, and H3K27me/IDH53. Our results elucidate that the synthetic lethal-based DNA methylation and histone acetylation regulation might be a novel potential strategy. Therefore, deeply digging into the epigenetic synthetic lethality interaction would pave a new pathway for the precision treatment of cancer.
We disclose a mechanism by which HDAC6 inhibition reduces DNMT3A-deficient NSCLC growth and survival in a HIF-1-dependent manner. It is well known that the HIF-1 signaling pathway plays a crucial role in the development and progression of cancer by regulating its target genes, including angiogenesis-related genes, proliferation-related genes, and apoptosis-related genes54, 55, 56. In the present study, our finding indicates that the deletion of DNMT3A as a priming factor causes activation of the HIF-1 pathway in cells, but it is inhibited after treatment with HDAC6 inhibitor. The epigenetic regulation of HIF-1 has been reported by several groups. Wang et al.57 revealed that EZH2 could reduce transcription of HIF-1 and suppress cancer cell adaption to hypoxia. Chen et al.58 disclosed that epigenetic reader ZMYND8 interacted with HIF-1 and enhanced elongation of the global HIF-induced oncogenic genes. Recently, a study demonstrated that HDAC6 inhibitor Tubastatin A suppressed Th17 cell function via downregulating HIF-1α pathway59, which was consistent with our finding. Moreover, we found that HIF-1 is regulated by HDAC6, which may be caused by the down-regulation of VHL60. It is reported that the decrease of VHL expression or the loss of its function leads to the activation of HIF-1α6, and HDAC6, a subtype of HDAC II, is known to alter protein function by affecting the level of acetylation of histone or non-histone (the primary mode) substrates, may also be negatively correlated with VHL regulation. However, some researchers pointed out that HDAC6 directly acetylated HIF-1α to affect the protein expression61, and there is also evidence that HDAC6 affects the stability of HIF-1α protein by acetylating HSP9062. It suggests that the regulatory function and state of acetylation modification may be different in diverse cellular environments, which further illustrates the complexity of epigenetic regulation63,64. The regulation of HIF-1α by HDAC6 is mainly accomplished through the classical VHL under our research conditions, which complements another regulation mode between HDAC6 and HIF-1α. The specific in-depth mechanism of HDAC6 regulating VHL has to be investigated subsequently. Notably, our results indicate whether HDAC6 inhibitor or HIF-1 inhibitor could selectively retard the tumor growth in DNMT3A-deficient NSCLC in vitro and in vivo. Considering that several HDAC6/HIF-1 inhibitors are currently in clinical trials, our results provide a rationale for their application in DNMT3A-deficient tumors.
5. Conclusions
We have identified HDAC6 as a crucial factor in maintaining the growth of DNMT3A-deficient NSCLCs in a HIF-1-dependent manner, which offers opportunities to gain new insights on the molecular cross-talks among epigenetic regulation enzymes. Our findings introduce a potential therapy paradigm for treating DNMT3A-deficient cancers like NSCLC.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (82272725 to Chunfu Wu, 82073320 to Lihui Wang), “Xingliao Talents” Program of Liaoning Province (No. XLYC1902008 to Lihui Wang, China) and Natural Science Foundation of Shenyang (22-315-6-11 to Lihui Wang, China).
Author contributions
Jiayu Zhang: Writing – original draft, Visualization, Supervision, Methodology, Investigation, Data curation. Yingxi Zhao: Validation, Data curation. Ruijuan Liang: Visualization, Validation, Methodology, Investigation, Data curation. Xue Zhou: Writing – original draft, Visualization, Validation, Methodology, Investigation, Data curation. Zhonghua Wang: Validation, Investigation. Cheng Yang: Validation, Methodology, Investigation. Lingyue Gao: Visualization. Yonghao Zheng: Validation, Data curation. Hui Shao: Validation, Data curation. Yang Su: Supervision, Investigation. Wei Cui: Supervision, Investigation. Lina Jia: Supervision, Investigation. Jingyu Yang: Supervision. Chunfu Wu: Supervision, Funding acquisition. Lihui Wang: Writing – review & editing, Writing – original draft, Supervision, Project administration, Funding acquisition, Conceptualization.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2024.08.025.
Appendix A. Supporting information
The following is the Supporting data to this article:
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Alexandrov L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Jimenez F., Muinos F., Sentis I., Deu-Pons J., Reyes-Salazar I., Arnedo-Pac C., et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- 4.Harrison P.T., Vyse S., Huang P.H. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin Cancer Biol. 2020;61:167–179. doi: 10.1016/j.semcancer.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J., Cao J., Topatana W., Juengpanich S., Li S., Zhang B., et al. Targeting mutant p53 for cancer therapy: direct and indirect strategies. J Hematol Oncol. 2021;14:157. doi: 10.1186/s13045-021-01169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborty A.A., Nakamura E., Qi J., Creech A., Jaffe J.D., Paulk J., et al. HIF activation causes synthetic lethality between the VHL tumor suppressor and the EZH1 histone methyltransferase. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Bates S.E. Epigenetic therapies for cancer. N Engl J Med. 2020;383:650–663. doi: 10.1056/NEJMra1805035. [DOI] [PubMed] [Google Scholar]
- 9.Chen S., Zhao Y., Liu S., Zhang J., Assaraf Y.G., Cui W., et al. Epigenetic enzyme mutations as mediators of anti-cancer drug resistance. Drug Resist Updat. 2022;61 doi: 10.1016/j.drup.2022.100821. [DOI] [PubMed] [Google Scholar]
- 10.Han M., Jia L., Lv W., Wang L., Cui W. Epigenetic enzyme mutations: role in tumorigenesis and molecular inhibitors. Front Oncol. 2019;9:194. doi: 10.3389/fonc.2019.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z., Zhang Y. Role of mammalian DNA methyltransferases in development. Annu Rev Biochem. 2019;89:135–158. doi: 10.1146/annurev-biochem-103019-102815. [DOI] [PubMed] [Google Scholar]
- 13.Yu J., Qin B., Moyer A.M., Nowsheen S., Liu T., Qin S., et al. DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J Clin Invest. 2018;128:2376–2388. doi: 10.1172/JCI97924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattei A.L., Bailly N., Meissner A. DNA methylation: a historical perspective. Trends Genet. 2022;38:676–707. doi: 10.1016/j.tig.2022.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Gravina G.L., Festuccia C., Marampon F., Popov V.M., Pestell R.G., Zani B.M., et al. Biological rationale for the use of DNA methyltransferase inhibitors as new strategy for modulation of tumor response to chemotherapy and radiation. Mol Cancer. 2010;9:305. doi: 10.1186/1476-4598-9-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh M., Kumar V., Sehrawat N., Yadav M., Chaudhary M., Upadhyay S.K., et al. Current paradigms in epigenetic anticancer therapeutics and future challenges. Semin Cancer Biol. 2022;83:422–440. doi: 10.1016/j.semcancer.2021.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Senapati J., Shoukier M., Garcia-Manero G., Wang X., Patel K., Kadia T., et al. Activity of decitabine as maintenance therapy in core binding factor acute myeloid leukemia. Am J Hematol. 2022;97:574–582. doi: 10.1002/ajh.26496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan P., Frankhouser D., Murphy M., Tam H.H., Rodriguez B., Curfman J., et al. Genome-wide methylation profiling in decitabine-treated patients with acute myeloid leukemia. Blood. 2012;120:2466–2474. doi: 10.1182/blood-2012-05-429175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan X.J., Xu J., Gu Z.H., Pan C.M., Lu G., Shen Y., et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 20.Ley T.J., Ding L., Walter M.J., McLellan M.D., Lamprecht T., Larson D.E., et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., Yang C., Wu C., Cui W., Wang L. DNA Methyltransferases in cancer: biology, paradox, aberrations, and targeted therapy. Cancers (Basel) 2020;12:2123. doi: 10.3390/cancers12082123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Setton J., Zinda M., Riaz N., Durocher D., Zimmermann M., Koehler M., et al. Synthetic lethality in cancer therapeutics: the next generation. Cancer Discov. 2021;11:1626–1635. doi: 10.1158/2159-8290.CD-20-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lord C.J., Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalev P., Hyer M.L., Gross S., Konteatis Z., Chen C.C., Fletcher M., et al. MAT2A inhibition blocks the growth of MTAP-deleted cancer cells by reducing PRMT5-dependent mRNA splicing and inducing DNA damage. Cancer Cell. 2021;39:209–224. doi: 10.1016/j.ccell.2020.12.010. e11. [DOI] [PubMed] [Google Scholar]
- 25.Zhao D., Lu X., Wang G., Lan Z., Liao W., Li J., et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature. 2017;542:484–488. doi: 10.1038/nature21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doffo J., Bamopoulos S.A., Kose H., Orben F., Zang C., Pons M., et al. NOXA expression drives synthetic lethality to RUNX1 inhibition in pancreatic cancer. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2105691119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jo U., Murai Y., Chakka S., Chen L., Cheng K., Murai J., et al. SLFN11 promotes CDT1 degradation by CUL4 in response to replicative DNA damage, while its absence leads to synthetic lethality with ATR/CHK1 inhibitors. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2015654118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flemming A. Cancer: an epigenetic target for synthetic lethality. Nat Rev Drug Discov. 2015;14:236. doi: 10.1038/nrd4589. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Chen K., Jia Y., Chuang J.C., Sun X., Lin Y.H., et al. Dual ARID1A/ARID1B loss leads to rapid carcinogenesis and disruptive redistribution of BAF complexes. Nat Cancer. 2020;1:909–922. doi: 10.1038/s43018-020-00109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bitler B.G., Aird K.M., Garipov A., Li H., Amatangelo M., Kossenkov A.V., et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H., Cui W., Wang L. Epigenetic synthetic lethality approaches in cancer therapy. Clin Epigenetics. 2019;11:136. doi: 10.1186/s13148-019-0734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., Hlavka-Zhang J., Shrimp J.H., Piper C., Dupere-Richer D., Roth J.S., et al. PRC2 inhibitors overcome glucocorticoid resistance driven by NSD2 mutation in pediatric acute lymphoblastic leukemia. Cancer Discov. 2022;12:186–203. doi: 10.1158/2159-8290.CD-20-1771. [DOI] [PubMed] [Google Scholar]
- 33.Albrengues J., Bertero T., Grasset E., Bonan S., Maiel M., Bourget I., et al. Epigenetic switch drives the conversion of fibroblasts into proinvasive cancer-associated fibroblasts. Nat Commun. 2015;6 doi: 10.1038/ncomms10204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russler-Germain D.A., Spencer D.H., Young M.A., Lamprecht T.L., Miller C.A., Fulton R., et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356 doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L., Rau R., Goodell M.A. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaelin W.G., Maher E.R. The VHL tumour-suppressor gene paradigm. Trends Genet. 1998;14:423–426. doi: 10.1016/s0168-9525(98)01558-3. [DOI] [PubMed] [Google Scholar]
- 38.Kaelin W.G., Jr. Von Hippel-Lindau disease: insights into oxygen sensing, protein degradation, and cancer. J Clin Invest. 2022;132 doi: 10.1172/JCI162480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang B., Lyu J., Yang E.J., Liu Y., Wu C., Pardeshi L., et al. Class I histone deacetylase inhibition is synthetic lethal with BRCA1 deficiency in breast cancer cells. Acta Pharm Sin B. 2020;10:615–627. doi: 10.1016/j.apsb.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li K., You J., Wu Q., Meng W., He Q., Yang B., et al. Cyclin-dependent kinases-based synthetic lethality: evidence, concept, and strategy. Acta Pharm Sin B. 2021;11:2738–2748. doi: 10.1016/j.apsb.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel J.P., Gonen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui C., Gan Y., Gu L., Wilson J., Liu Z., Zhang B., et al. P16-specific DNA methylation by engineered zinc finger methyltransferase inactivates gene transcription and promotes cancer metastasis. Genome Biol. 2015;16:252. doi: 10.1186/s13059-015-0819-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito Y., Kanai Y., Sakamoto M., Saito H., Ishii H., Hirohashi S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology. 2001;33:561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 44.Lin R.K., Hsu H.S., Chang J.W., Chen C.Y., Chen J.T., Wang Y.C. Alteration of DNA methyltransferases contributes to 5′CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55:205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 45.Ma H.S., Wang E.L., Xu W.F., Yamada S., Yoshimoto K., Qian Z.R., et al. Overexpression of DNA (Cytosine-5)-methyltransferase 1 (DNMT1) and DNA (Cytosine-5)-methyltransferase 3A (DNMT3A) is associated with aggressive behavior and hypermethylation of tumor suppressor genes in human pituitary adenomas. Med Sci Monit. 2018;24:4841–4850. doi: 10.12659/MSM.910608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng T., Kuang Y., Wang L., Li J., Wang Z., Fei J. An essential role for DNA methyltransferase 3a in melanoma tumorigenesis. Biochem Biophys Res Commun. 2009;387:611–616. doi: 10.1016/j.bbrc.2009.07.093. [DOI] [PubMed] [Google Scholar]
- 47.Leonard S., Pereira M., Fox R., Gordon N., Yap J., Kehoe S., et al. Over-expression of DNMT3A predicts the risk of recurrent vulvar squamous cell carcinomas. Gynecol Oncol. 2016;143:414–420. doi: 10.1016/j.ygyno.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 49.Hideshima T., Qi J., Paranal R.M., Tang W., Greenberg E., West N., et al. Discovery of selective small-molecule HDAC6 inhibitor for overcoming proteasome inhibitor resistance in multiple myeloma. Proc Natl Acad Sci U S A. 2016;113:13162–13167. doi: 10.1073/pnas.1608067113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu B., Liu L., Cai F., Peng Y., Tang X., Zeng D., et al. The synergistic anticancer effect of the bromodomain inhibitor OTX015 and histone deacetylase 6 inhibitor WT-161 in osteosarcoma. Cancer Cell Int. 2022;22:64. doi: 10.1186/s12935-022-02443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muscat A., Popovski D., Jayasekara W.S., Rossello F.J., Ferguson M., Marini K.D., et al. Low-dose histone deacetylase inhibitor treatment leads to tumor growth arrest and multi-lineage differentiation of malignant rhabdoid tumors. Clin Cancer Res. 2016;22:3560–3570. doi: 10.1158/1078-0432.CCR-15-2260. [DOI] [PubMed] [Google Scholar]
- 52.Ogiwara H., Sasaki M., Mitachi T., Oike T., Higuchi S., Tominaga Y., et al. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality by apoptotic cell death due to abrogation of MYC expression. Cancer Discov. 2016;6:430–445. doi: 10.1158/2159-8290.CD-15-0754. [DOI] [PubMed] [Google Scholar]
- 53.Habiba U., Sugino H., Yordanova R., Ise K., Tanei Z.I., Ishida Y., et al. Loss of H3K27 trimethylation is frequent in IDH1-R132H but not in non-canonical IDH1/2 mutated and 1p/19q codeleted oligodendroglioma: a Japanese cohort study. Acta Neuropathol Commun. 2021;9:95. doi: 10.1186/s40478-021-01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chi J.T., Wang Z., Nuyten D.S., Rodriguez E.H., Schaner M.E., Salim A., et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. Plos Med. 2006;3 doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Semenza G.L. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33:207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mole D.R., Blancher C., Copley R.R., Pollard P.J., Gleadle J.M., Ragoussis J., et al. Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X., Wang Y., Li L., Xue X., Xie H., Shi H., et al. A lncRNA coordinates with Ezh2 to inhibit HIF-1α transcription and suppress cancer cell adaption to hypoxia. Oncogene. 2019;39:1860–1874. doi: 10.1038/s41388-019-1123-9. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y., Zhang B., Bao L., Jin L., Yang M., Peng Y., et al. ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J Clin Invest. 2018;128:1937–1955. doi: 10.1172/JCI95089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou W., Yang J., Saren G., Zhao H., Cao K., Fu S., et al. HDAC6-specific inhibitor suppresses Th17 cell function via the HIF-1α pathway in acute lung allograft rejection in mice. Theranostics. 2020;10:6790–6805. doi: 10.7150/thno.44961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ellis L., Hammers H., Pili R. Targeting tumor angiogenesis with histone deacetylase inhibitors. Cancer Lett. 2009;280:145–153. doi: 10.1016/j.canlet.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qian D.Z., Kachhap S.K., Collis S.J., Verheul H.M., Carducci M.A., Atadja P., et al. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res. 2006;66:8814–8821. doi: 10.1158/0008-5472.CAN-05-4598. [DOI] [PubMed] [Google Scholar]
- 62.Zhang D., Li J., Costa M., Gao J., Huang C. JNK1 mediates degradation HIF-1α by a VHL-independent mechanism that involves the chaperones Hsp90/Hsp70. Cancer Res. 2010;70:813–823. doi: 10.1158/0008-5472.CAN-09-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Narita T., Weinert B.T., Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20:156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 64.Hsieh W.C., Sutter B.M., Ruess H., Barnes S.D., Malladi V.S., Tu B.P. Glucose starvation induces a switch in the histone acetylome for activation of gluconeogenic and fat metabolism genes. Mol Cell. 2022;82:60–74.e5. doi: 10.1016/j.molcel.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.