Abstract
Background
Pituitary neuroendocrine tumors, PitNETs, are often aggressive and precipitate in distant metastases that are refractory to current therapies. However, the molecular mechanism in PitNETs’ aggressiveness is not well understood. Developmental pluripotency-associated 4 (DPPA4) is known as a stem cell regulatory gene and overexpressed in certain cancers, but its function in the context of PitNETs’ aggressiveness is not known.
Methods
We employed both rat and human models of PitNETs. In the rat pituitary tumor model, we used prenatal-alcohol-exposed (PAE) female Fischer rats which developed aggressive PitNETs following estrogen treatment, while in the human pituitary tumor model, we used aggressively proliferative cells from pituitary tumors of patients undergone surgery. Various molecular, cellular, and epigenetic techniques were used to determine the role of DPPA4 in PitNETs’ aggressiveness.
Results
We show that DPPA4 is overexpressed in association with increased cell stemness factors in aggressive PitNETs of PAE rats and of human patients. Gene-editing experiments demonstrate that DPPA4 increases the expression of cell stemness and tumor aggressiveness genes and promotes proliferation, colonization, migration, and tumorigenic potential of PitNET cells. ChIP assays and receptor antagonism studies reveal that DPPA4 binds to canonical WINTs promoters and increases directly or indirectly the WNT/β-CATENIN control of cell stemness, tumor growth, and aggressiveness of PitNETs. Epigenetic studies show the involvement of histone methyltransferase in alcohol activation of DPPA4.
Conclusions
These findings support a role of DPPA4 in tumor stemness and aggressiveness and provide a preclinical rationale for modulating this stemness regulator for the treatment of PitNETs.
Keywords: DPPA4, histone methyltransferase, pituitary neuroendocrine tumors, prenatal alcohol, WNT/β-CATENIN
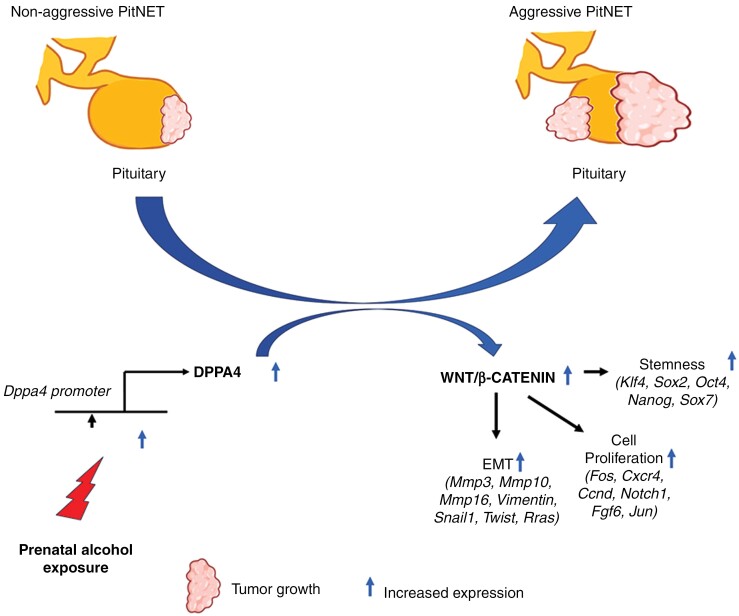
Graphical Abstract
Graphical Abstract.
Key Points.
DPPA4 gene is overexpressed in adult aggressive pituitary neuroendocrine tumors.
Expanded stem cell niche within tumor microenvironment promotes aggressiveness.
Increased cell stemness is dependent on WNT/β-CATENIN signaling.
Importance of the Study.
Pituitary tumor cells are reprogrammed to an aggressive state following prenatal alcohol exposure through the overexpression of DPPA4 via the histone H4 methylation. DPPA4 promotes stem cell niche within the tumor microenvironment through WNT/β-CATENIN signaling that enhances tumor cell proliferation, colonization, and migration. Targeting DPPA4 modifiers is a potential treatment for aggressive tumors.
Pituitary neuroendocrine tumors (PitNETs) are common intracranial tumors that can be hormone-secreting and are an important cause of hypogonadism and infertility in humans.1 Some of these tumors show a lack of sensitivity to therapy or recur during follow-ups and exhibit aggressive behavior, characterized by gross invasion of the surrounding tissues or distance metastasis.2,3 Pangenomic classification of PitNETs has recently been described.4 Surgery is usually deployed as second-line therapy given the well-established sensitivity of these masses to dopamine agonist therapy, though aggressive prolactinomas respond poorly, with a high chance of recurrence.5,6 Chemotherapy employing temozolomide (TMZ) is used to control aggressive prolactinomas.1 However, a significant number of cases of aggressive prolactinomas and prolactin-secreting pituitary carcinomas continue to progress despite TMZ therapy.3 Immunotherapy, based on the use of immune checkpoint inhibitors, has also been considered in cases resistant to TMZ but shows variable outcomes.7–9 Management of aggressive PitNETs is a challenge because of the unexplained etiology of this disease.10
Stem cell involvement has recently been suggested in the development of aggressiveness and drug-resistant behaviors of PitNETs. It has been shown that an activated phenotype of the pituitary-resident stem cells is present in tumorigenic glands and tumors in humans.11,12 Also, pituitary stem-like cells are shown to be resistant to dopamine agonist treatment because of the low dopamine D2 receptor (D2R) in these cells.13 In the prolactinomas of estrogen-treated PAE female rats, tumor cells formed spheres in the ultra-low attachment plate and expressed stem cell marker genes. These pituitary tumorspheres retained their stemness during passaging and induced solid tumors in immunodeficient mice,14 identifying important stem cell characteristics.15 The Ki67 labeling index in pituitary tumors of PAE rats is shown to be >4%,14 which is in the range that indicates pituitary invasiveness.16 Thus, the PAE female rat model appears to be useful in identifying important molecular pathways regulating stemness and tumor aggressiveness in the pituitary.
DPPA4 is one of the embryonic stem cell transcription factors known to regulate the genetic machinery of embryonic development.17 DPPA4 was traced as the predominant binding site for the core pluripotency factors OCT4, SOX2, and NANOG in human embryonic stem cells (hESCs) and pluripotent germ cell tumors18,19 and has been shown to generate oncogenic foci in in vitro assays.20 DPPA4 is also overexpressed in the PAE rat’s placenta and in hESCs.21 In this study, we determined that DPPA4 expresses and regulates the stemness and aggressiveness of tumors in pituitaries of estrogen-treated PAE female rats. We also verified these novel roles of DPPA4 in aggressive PitNETs of adult human patients.
Methods
Animal Model of PAE
The PAE animal model was adopted from a previously published study.14 Fisher-344 rats were obtained from Harlan Laboratories and housed in pairs in open-type shoebox cages with Bedcob bedding and maintained in a room under the controlled condition of a 12-hour light–dark cycle and a constant temperature of 22°C. The animals were bred and on gestation day (GD) 7, they were fed ad libitum with rat chow (AD), a liquid diet containing ethanol (AF), or pair-fed (PF) an isocaloric control liquid diet (Bio-Serv). Alcohol-fed animals were first acclimatized to ethanol by feeding them with liquid diets containing increasing concentrations of ethanol (from 1.7% to 5.0% v/v) from GD7 to GD10, and then fed 6.7% v/v ethanol from GD11 to GD21. Both AF and PF offspring were cross-fostered on postnatal day 2 (PND2) to untreated lactating dams to avoid any compromised nurturing by their mothers. Furthermore, we maintained a litter size of 8 pups/dam to limit the nurturing effect on overall body growth. We weaned pups on PND21 and housed them by sex. A single female offspring was utilized from each experimental group of rat litters. At the age of 60 days, each female rat underwent a bilateral ovariectomy using 2% isoflurane anesthesia and 2.5% bupivacaine subcutaneously (sc) to induce local analgesia and was sc implanted with an estradiol-17β (Sigma-Aldrich) filled 1-cm silastic capsule (Dow Corning). Analgesic drug treatment was continued for 3 days after surgery for prevention of pain. After 120 days of estradiol implants, some rats were euthanized and their pituitary tumor tissues were collected and used for experimentation. Other animals were perfused transcranially and their pituitaries were removed and fixed with 10% formalin for histology. Animal surgery and care were performed in accordance with institutional guidelines and the Rutgers Institutional Animal Care and Use Committee (IACUC) approved protocol (#999900286).
Rat Pituitary Tumor Cell Cultures
Primary cultures of rat pituitary tumor (RPT) cells were prepared using a published method with minor modifications14,22 (see Supplementary Methods for details).
Human Pituitary Tumor Tissue Cell Culture
Human pituitary tumor (HPT) tissue samples were obtained from patients who underwent surgical treatment at Rutgers Cancer Institute of New Jersey. HPT tissue fragments were immediately processed for primary cell cultures according to previously described protocols with minor modifications23,24 (Supplementary Methods).
Human Pituitary Tumor Cell Immortalization
Immortalization of HPT cells was done by using the SV40 T Antigen and hTERT Antigen Cell Immortalization Kit (ALSTEM#CILV01/CILV02) and followed the previously published protocols.25 After transfection, cells were subsequently selected and maintained in a medium containing 1 μg/mL puromycin for 2 weeks (Supplementary Methods).
CRISPR Knockdown of Dppa4
Primers containing the target DNA clustered regularly interspaced short palindromic repeats (CRISPR) sequences of the Dppa4 gene were used to generate the gRNA by in vitro transcription (IVT) using the GeneArt Precision gRNA Synthesis Kit (Invitrogen). To target the gene, 3 gRNA sequences were used for AF pituitary cells, and 4 gRNA sequences were used for human pituitary cells (Supplementary Tables 1 and 2). A total of 2400 ng of gRNA was used to knockdown the Dppa4 gene using CRISPRMAX lipofectamine26 (see Supplementary Methods for details).
CRISPR Knockin of Dppa4
Eleven target gRNA sequences and 1 control gRNA sequence were cloned into pCC_05-hU6-BsmBI-sgRNA(E + F)-barcode-EFS-dCas9-NLS-VPR-2A-Puro-WPRE. (The plasmid was taken from Addgene plasmid RRID: Addgene_139090.) The gRNA sequences were chosen from −580 to −54 upstream of the rat Dppa4 transcription start site. The 5 gRNA sequences (without PAM) were used for knockin of Dppa4 (Supplementary Tables 1 and 2) using lipofectamine 300027 (Supplementary Methods).
RNA Isolation and Sequencing
RNA was isolated using an RNeasy kit from QIAGEN. RNA quantity was assessed using the Nanodrop ND-100 (Thermo Scientific). RNA-Seq library preparation and sequencing were carried out from Genewiz of Azenta Life Science. Using DESeq2, a comparison of gene expression between groups of samples was performed. The Wald test was used to generate P values and log2 fold changes. Genes with an adjusted P value < .05 and absolute log2 fold change >1 were designated as differential expression of genes (DEGs) for each comparison28 (Supplementary Methods).
IPA Analysis
The list of genes with log2 fold change value for different experimental groups compared to AD was uploaded to Ingenuity Pathway Analysis (IPA; www.qiagen.com/ingenuity; Qiagen) and was used to integrate genes and biological pathways. The entire analysis in IPA was done with constant parameters and the cutoff was set from −1 to +1. All the datasets were analyzed under the same parameters, and we only included the experimentally observed results with minimal prediction levels. The overrepresented cancer-related canonical pathways compared to AD were identified in all experimental groups, and molecules involved in those pathways were detected (Supplementary Methods).
RT PCR (qPCR)
The qPCR was done according to the previous protocol26 with minor modifications (Supplementary Table 3).
Knockdown/Inhibition of β-Catenin
The β-Catenin was knocked down through the transfection of shRNA plasmid (SC-270011-SH) containing puromycin resistance gene. One microgram of plasmid was mixed with Santa Cruz Biotechnology’s shRNA plasmid transfection reagent (sc-108061) at different ratios (1:1 to 1:6), and then transfection was done using shRNA plasmid transfection medium (sc-108062). The scrambled plasmid was used as a control.29 For the inhibition of the β-CATENIN protein, AF and HPT cells were treated for 48 hours with the inhibitor of IWR-1-endo (1 to 20 µM in 0.5% DMSO) which promotes the β-CATENIN protein degradation through AXIN2 protein stabilization.30 In the in vivo study, IWR-1 was administered intratumorally (5 mg/kg) every 2 days for 2 weeks30 in the animal with tumor xenografts. Control animals were treated with vehicle following a similar administration schedule and euthanized at the end of the treatment schedule (see Supplementary Methods for details).
Subcutaneous Xenograft Experiments
NOD/SCID mice or NSG mice for HPT (Charles River Laboratories) were used for xenograft studies as previously described.31 Briefly, 1 × 106 rat pituitary tumor cells or HPT cells were subcutaneously injected with an equal volume of Matrigel (354248, Corning) in the right flank of each animal. Tumors were measured using electronic calipers and tumor volumes were calculated as V = LxW2 × 0.5 (Supplementary Methods).
In Vitro Treatment of MM102
To confirm the role of H3K4me3 in the activation of Dppa4 promoter, AD and AF cells were treated with three different concentrations of H3K4me3 blocker MM102 (25, 50, and 75 µM), according to the previous protocol32 (Supplementary Methods).
Cell Proliferation Assay, Transwell Migration Assay, Colony Formation Assay
The cell proliferation rate was determined using an MTT assay. Transwell migration and colony formation assays were performed according to the previous protocol26 (Supplementary Methods).
Immunocytochemistry, Immunoblotting, and Immunohistochemistry Staining
Immunocytochemical analysis was performed according to the method described by Teotia et al., 2019.33 The immunoblot was performed following the previous protocol from our lab,31 and immunohistochemistry was performed as described previously34 (Supplementary Table 1).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were conducted using a ChIP assay kit (Sigma-Aldrich) following the manufacturer’s instructions35 (Supplementary Methods)
TCF/LEF Reporter Assay
A TCF/LEF Reporter kit (Wnt Signaling Pathway, 60500) was purchased from BPS Bioscience, and the reporter assay was carried out according to the manufacturer’s protocol using lipofectamine 2000 with some modifications (see the supplementary file for more details).
Rescue Assay
In the rescue assay, Dppa4 and β-Catenin were modulated in either or both ways. Lipofectamine 2000 was used to transfect overexpression plasmid in HPT cells according to the manufacturer’s instructions, while knockdown was performed using the CRISPR protocol (see the supplementary file for more details).
Statistical Analysis
Graph Pad Prism (version 9) software was used for statistical analysis of data. Data are presented as mean ± SEM. The significance between treatment groups and controls was assessed using unpaired t-test/1-way/2-way ANOVA. Post hoc analysis for one-way ANOVA was done using the Newman–Keuls test, and for 2-way ANOVA, it was done using Dunnett’s multiple comparison test. For survival data, Kaplan–Meier curves were generated using Graph Pad Prism. P < .05 was considered significant. Additional details regarding quantitation and statistical analysis are provided in the figures and figure legends. For cell culture studies where treatments were given, the experiments were repeated three times. Western blotting micrographs shown in the figures are representative images/blots.
Results
Aggressive PitNETs Produce DPPA4 and Have Stemness-Like Characteristics
We employed PAE female Fischer rats with pituitary tumors (RPTs) and human pituitary tumors (HPTs). In the PAE animal model, three treatment groups were included (1) alcohol-fed liquid diet (AF), (2) ad libitum fed rat chow (AD), and (3) pair-fed isocaloric liquid diet (PF). The AD and PF groups served as control. Characterization of the tumors in pituitaries of estrogen-treated AF rats revealed that these tumors were highly vascularized and often penetrated to the sphenoid bone when compared to estrogen-treated AD or PF rats (Supplementary Figure 1). AF rat tumors had high levels of PRL, low concentrations of D2R, elevated levels of stemness-related proteins (OCT-4, KLF4, SOX-2, and SNAIL-1),36 and several aggressiveness tumor marker proteins (Ki67, PTTG, CD-44, MMP9)37 as determined by immunohistochemistry (Supplementary Figure 2) and Western blot measurements (Supplementary Figure 3). AF rats without the estrogen treatment showed a moderate increase in PRL and stem cell related proteins in the pituitary as compared to AD and PF rats (Supplementary Figures 4 and 5). Cells prepared from pituitary tumors of AF rats showed rapid proliferation, high colony formation, and cell migration compared to pituitary tumor cells of control AD and PF rats in cultures (Figure 1a to c), confirming that AF pituitary tumor cells are more aggressive than control cells in monolayer cultures as found previously in pituitary tumorsphere cultures.14
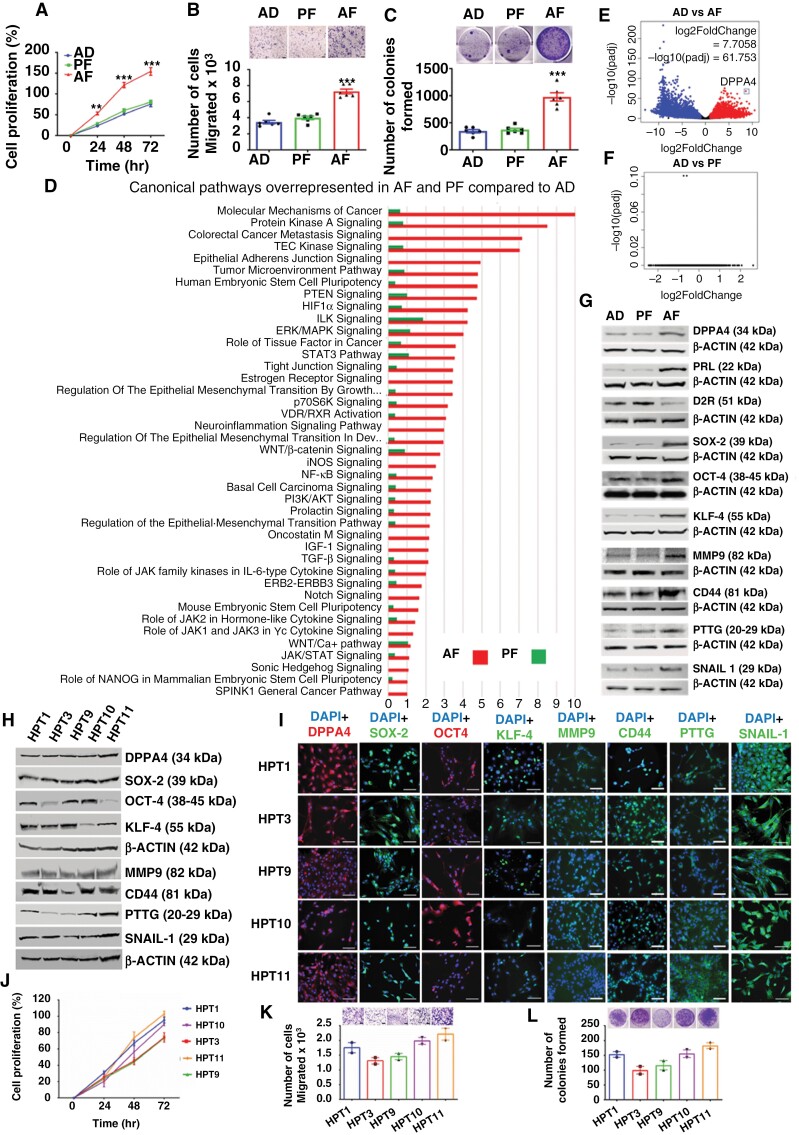
Figure 1.
DPPA4 expression is elevated in aggressive PitNETs. (a to c) The aggressiveness of rat pituitary tumor (RPT) cells obtained from prenatal alcohol-fed (AF), pair-fed, (PF), ad libitum-fed (AD) rats was determined by cell proliferation (a), cell migration (b), and colony formation (c); n = 6. (d to f) RNA-seq data of AF, PF, and AD cells are analyzed by IPA and overrepresented canonical pathways are shown by histograms (d) and the expression difference between AD vs AF and AD vs PF are shown by volcano plots (e and f). (g) Western blot data of DPPA4, PRL, D2R, stem cell marker, and tumor aggressiveness marker proteins in AF, PF, and AD cells; n = 6. (h to i) DPPA4 and stem cell factors are expressed in aggressive patient-derived PitNET (HPT) cells. Expression of DPPA4 and various stem cell regulatory proteins were detected by Western blot (h) and immunofluorescence (i) in 5 different human pituitary tumor cells (HPT1-5) prepared from patient-derived tumor tissues that expresses more DPPA4 and stem cell regulatory proteins than those tumor tissues produced non-viable cells (Supplementary Figure 7). (j to l) Cell proliferation (j), cell migration (k), and colony formation (l) profiles of HPT1-5 cells (n = 2). Scale bar represents 100 μM in immunofluorescence figures. Data shown in histograms are mean ± SEM and were analyzed using 1-way or 2-way analysis of variance (ANOVA) with the Newman Keuls post hoc test or Dunnett’s multiple comparison test *P < .05, **P < .01, and ***P < .001 between AF and controls (AD, PF).
To investigate the key tumor regulatory genes, we first employed a next-generation sequencing approach. The transcriptional changes induced by PAE in pituitary tumor cells of AF, AD, and PF animals were examined using RNA-seq. Differential expression of genes compared to AD was calculated for individual genes in AF and PF. The fold change value was converted to log2fold change and the datasets were uploaded in IPA to perform core analysis. IPA clusters the molecules in different biological pathways depending on their functions. This clustering of molecules in IPA identified 41 overrepresented (−log(P-value) > 1) canonical pathways associated with cancer in the AF group compared to the AD group (Figure 1d). Out of 41 overrepresented canonical pathways, 39 canonical pathways in the PF group showed −log(P-value) < 1 which identified a similarity between PF and AD groups. Therefore, only AF cells showed significant alterations in cancer-related canonical pathways compared to AD cells. Next, we calculated log2(fold change) and −log10(padj) from the P value adjusted as obtained by RNA-seq analysis. The log2(fold change) vs −log10(padj) was plotted to generate a volcano plot to show the comparison between both AD vs AF (Figure 1e) and AD vs PF (Figure 1f). Among all the upregulated genes, we found one gene Dppa4 which showed a marked difference [log2(fold change) value of 7.7058 and a −log10(padj) value of 61.753] compared to the other upregulated cancer-related genes. RNA-seq data demonstrating Dppa4 gene overexpression in AF cells, compared to AD and PF cells, is confirmed by Western blots (Figure 1g; Supplementary Figure 6m) and immunofluorescence analyses (Supplementary Figure 6c). In addition, a high level of DPPA4 protein was detected in pituitary tumor tissues in AF rats compared to those in AD and PF rats by both immunocytochemistry (Supplementary Figure 2c) and by Western blot (Supplementary Figure 3c). AF rat pituitary cells also produced high levels of PRL, low D2R, and elevated levels of stem cell-related proteins SOX-2, OCT-4, KLF-4, aggressive tumor marker proteins MMP9, CD44, PTTG, and EMT protein SNAIL-1 (Supplementary Figure 6). Together these data suggest that AF rat pituitary tissues and cells express elevated levels of DPPA4 and proteins related to stemness, tumor aggressiveness, and EMT, together with high cell proliferation, migration, and colony formation characteristics.
Pituitary tumor samples obtained from 10 patients (age range: 18 to 73 years; 5 males and 5 females; Supplementary Table 1) undergoing surgery were also utilized in this study. We found 5 of these tumor tissues and/or cells (HPT1, HPT3, HPT9, HPT10, and HPT11) expressed elevated mRNA levels and protein levels of DPPA4 (Supplementary Figure 7a,b), WNT/FZD (Supplementary Figure 7c,d) stem cell markers (KLF4, SOX-2, SNAIL-1, OCT-4, and NESTIN;Supplementary Figure 7e to i), aggressive tumor markers (MMP-9, MMP16;Supplementary Figure 7j,k), cell proliferation and cancer progression marker [PTTG, CD44, EGF, FGFR, EGFR;Supplementary Figure 7l to p],38 and EMT activator (N-CADHERIN; Supplementary Figure 7q) compared to HPT2, HPT5, HPT6, HPT7 and HPT8 tissues. We also found that cells expressing higher levels of DPPA4, stemness, and tumor aggressiveness markers grew well in cell culture conditions (HPT1, HPT3, HPT9, HPT10, and HPT11). Subcultures of these cells remain positive for DPPA4, stem cell marker, and aggressive tumor marker proteins and produce multiple hormones (Figure 1h,i; Supplementary Figure 7s,t). These HPT cells showed rapid proliferation (Figure 1j) and high colony formation (Figure 1k) and cell migration (Figure 1l). These data suggest that HPT cells produce high levels of aggressive tumor markers and express elevated levels of DPPA4 and stemness-related proteins together with high cell proliferation, colony formation, and migration characteristics.
DPPA4 Is Involved in Increasing Stemness and Aggressiveness of PitNET Cells
CRISPR-mediated gene knockdown and knockin approaches were first used to investigate the role of DPPA4 using RPT cells. After a 48-hour treatment, Western blot assays were conducted and determined that the knockdown was successful (Figure 2a). The cell growth response to DPPA4 knockdown revealed that this gene knockdown inhibited the growth rate of AF cells (Figure 2b). The ability of cells to migrate following DPPA4 knockdown was found to be decreased (Supplementary Figure 8a). Also, the cells with DPPA4 knockdown showed a lesser ability to form colonies compared to controls (Supplementary Figure 8b). In addition, AF/KD cells showed reduced tumorigenic potential after inoculation into immunodeficient mice (Figure 2c). Tumor size and tumor wet weight (Supplementary Figure 8c), and the tumor volume (Figure 2c) were reduced in mice with AF/KD cells compared to the control cells. The survival analysis of AF cell xenografts shows that the mean life span of animals injected with AF/KD cell xenografts was significantly increased (Figure 2d).
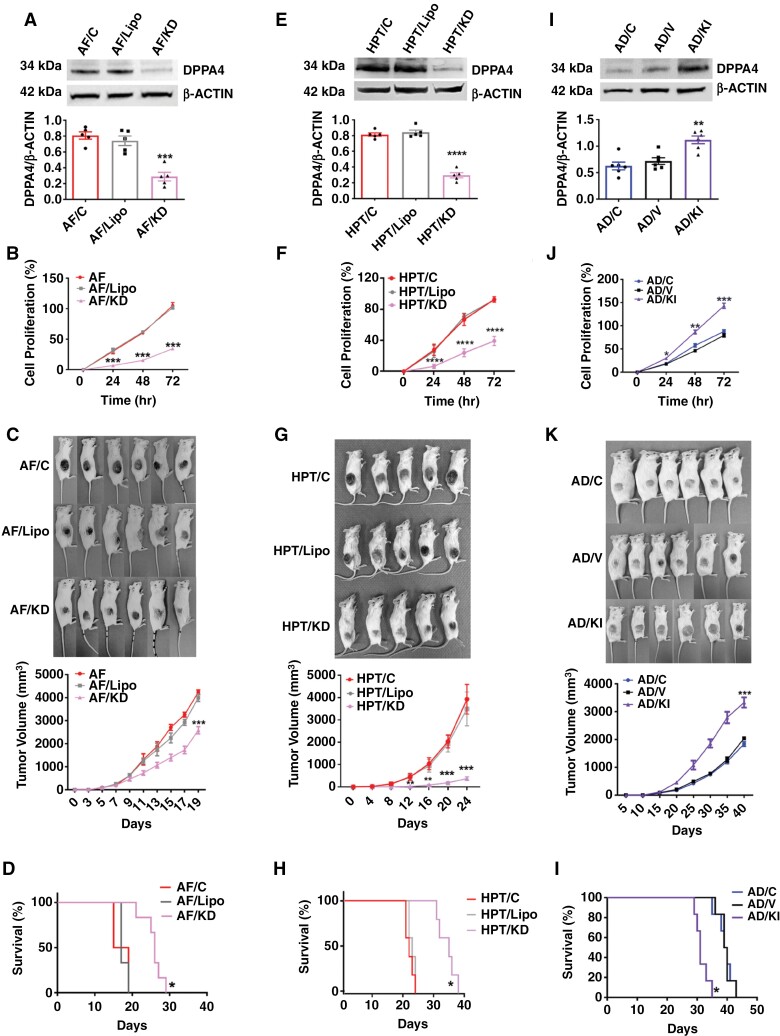
Figure 2.
Effect of CRISPR knockdown and knockin of Dppa4 in the aggressiveness of PitNETs. (a to d) Effects of DPPA knockdown on RPT cells—AF cells untreated (AF/C), lipofectamine (AF/Lipo), or lipofectamine with gRNA (AF/KD). DPPA4 protein levels by Western blots (a). Cell proliferation rate (b). Pictures of animals with tumors in each group and tumor volume changes (c). Effects on survival time (d); n = 6. (e to h) Effects of DPPA knockdown on HPT cells—HPT cells untreated (HPT/C), lipofectamine (HPT/Lipo), or lipofectamine with gRNA (HPT/KD). DPPA4 protein levels by Western blots (e). Cell proliferation rate (f). Pictures of animals with tumors in each group and tumor volume changes (g). Effects on survival time (h); n = 5. (i to l) Effects of DPPA4 knockin on RPT cells—AD cells untreated (AD/C), lipofectamine (AD/V), or lipofectamine with gRNA (AD/KI). DPPA4 protein levels by Western blots (i). Cell proliferation rate (j). Pictures of animals with tumors in each group with tumor volume changes (h). Effects on survival time (l); n = 6. Data shown are mean ± SEM and were analyzed using 1-way or 2-way analysis of variance (ANOVA) with the Newman Keuls post hoc test or Dunnett’s multiple comparison test. ***P < .001, ****P < .0001, AF/KD vs AF/C and AF/Lipo or HPT/KD vs HPT/C and HPT/Lipo. Kaplan–Meier survival analysis was used to test significant differences between survival curves and mean survival time for mice from each group (d, h, and i). n = 6, *P < .05, AF/KD vs AF/C and AF/Lipo; n = 5, *P < .05, HPT/KD vs HPT/C and HPT/Lipo; n = 6, *P < .05, AD/KI vs AD/C and AD/V.
CRISPR-mediated gene knockdown approach was also used in HPT cells. In the knockdown experiment, 4 gRNAs were effective in knockdown of the DPPA4 gene (HPT/KD) compared to no treatment controls (Figure 2e). Also, HPT/KD cells had a decreased cell proliferation rate (Figure 2f), cell migration rate (Supplementary Figure 9a), and number of colonies formed compared to those in control groups (Supplementary Figure 9b). In addition, HPT/KD cells showed reduced tumorigenic potential in in vivo when tumor growth was studied after inoculation into immunodeficient mice. Tumor size (Figure 2g), tumor wet weight at the completion of study, and the tumor volume (Supplementary Figure 9c) were reduced in mice with HPT/KD cells compared to control cells. The survival analysis of HPT cell xenografts showed that the life span of animals injected with HPT/KD cell xenografts was significantly increased (Figure 2h).
DPPA4 knockin experiments were also conducted in AD cells which had low expression of DPPA4. (All HPT cells had high expression of DPPA4 and were not included in the knockin study). The DPPA4 gene was knocked in using CRISPR by cloning 5 gRNAs into AD cells (AD/KI) and confirming overexpression of the DPPA4 protein by Western blot (Figure 2i). We observed an increased cell proliferation rate (Figure 2j), cell migration number (Supplementary Figure 10a), and cell colony formation number (Supplementary Figure 10b) in AD/KI cells compared to those in control cells. In addition, AD/KI cells showed increased tumorigenic potential following inoculation into immunodeficient mice (Figure 2k). Tumor size and wet weight (Supplementary Figure 10c), and the tumor volume was increased in mice with AD/KI cells compared to the control cells (Figure 2k). Furthermore, the mean life span of animals with AD/KI cell xenografts was reduced compared to those with control cell xenografts (Figure 2l).
Together these data support a role of DPPA4 in tumor aggressiveness in both RPT and HPT cells.
RNA-seq Analysis Identifies WNT and β-CATENINE as the Critical Genes Involved in DPPA4 Actions in PitNET Cells
RNA-seq analysis was first conducted to identify the critical genes involved in the DPPA4 mechanism of action. RNA-seq data were obtained as the differential expression of genes (DEGs) for all 7 experimental groups (AD, PF, AF, AF/Lipo, AF/KD, AD/V, and AD/KI). Log2fold change values (with respect to AD) were calculated for individual genes in all experimental groups and the core analysis was performed in IPA. The effects were monitored on the same 41 canonical pathways (obtained from AD vs AF RNA-seq data; Figure 1d). Seventeen pathways were found to be involved with DPPA4 (Figure 3a) as DPPA4 knockdown affected those cancer-associated canonical pathways while DPPA4 knockin reversed this condition (Figure 3a and b). Thus, it can be assumed that the DPPA4 protein is the key factor associated with these canonical pathways and cancer aggressiveness. The −log(P-value) values of the 17 canonical pathways were overlapped among AD/V vs AD/KI and AF vs AF/Lipo (Figure 3b and c), which suggest their association with DPPA4. We used a Venn diagram to show the common and exclusive molecules among AF, AF/Lipo, and AF/KD (Figure 3d); between AD vs AD/KI (Figure 3e); and among AF, AF/KD, and AD/KI (Figure 3f). The Venn diagram showed 111 molecules that were common in AF and AD/KI and 17 molecules that were common in AF, AD/KI, and AF/KD. The AF, AD/KI, and AF/KD groups showed some unique molecules also, but these were not included in the study as they were not associated with DPPA4 expression. This finding indicated that the knockin of DPPA4 in AD restored the expression of 128 genes among 489 (341 + 111 + 17 + 20) genes in AF cells. Now, these 128 (111 + 17) genes/molecules were classified into different signaling pathways based on their function and used to generate a gene ontology plot (Figure 3g). The plot showed the proportion of major cancer-associated pathways (Stemness, WNT, proliferation, and EMT) in the whole event as obtained from different canonical pathways and Venn diagrams. Using Rstudio, a heatmap was generated to show differential expression of these 128 molecules in AF, AF/KD, and AD/KI groups (Figure 3h). These data identified activation of Wnt/β-catenin, Wnt/Ca2+, NF-κB, Notch, PI3K-Akt, P-38-Mapk, EMT, Stat3, and other signaling pathways related to cancer in the AF group that were selectively downregulated in the AF/KD group. Also, the overexpression of Dppa4 in AD cells (AD/KI) upregulated those signaling pathways that are overexpressed in AF cells (AF/C) (Figure 3e). A qPCR array confirmed the alteration of these pathways which showed increased expression of various Wnt signaling molecules (Wnt5, Wnt10B, Wnt7B, Fzd1, β-catenin, N-Cadherin)38 and reduced expression of E-Cadherin; increased stem cell factors (Klf4, Sox2, Nanog, Oct4, Sox7;Figure 3i to j)39 and EMT factors (Mmp3, Mmp10, Vimentin. Mmp16, Snail1, Twist, Rras;Figure 3k)37,40 and cell proliferation regulators (Fos, Cxcr4, Ccnd, Notch1, Fgf6, Jun;Figure 3l)41 in AD/KI cells compared to AF/KD. The RNA-seq analysis in HPT cells showed similar observations. The volcano plot showed a significant difference between HPT/C vs HPT/KD but not for HPT/C vs HPT/Lipo (Supplementary Figure 11 a,b). The knockdown of DPPA4 in HPT cells showed a significant decrease in overrepresented cancer canonical pathways compared to the control cells (Supplementary Figure 11c). The common molecules among the HPT/C, HPT/Lipo, and HPT/KD cells were identified through Venn diagram (Supplementary Figure 11d) and gene ontology plot classified them according to their function (Supplementary Figure 11e). The heatmap analysis showed the differential expression of those common molecules and additional heatmap analysis showed the changes in DPPA4, Wnt, and EMT related molecules (Supplementary Figure 11f,g).
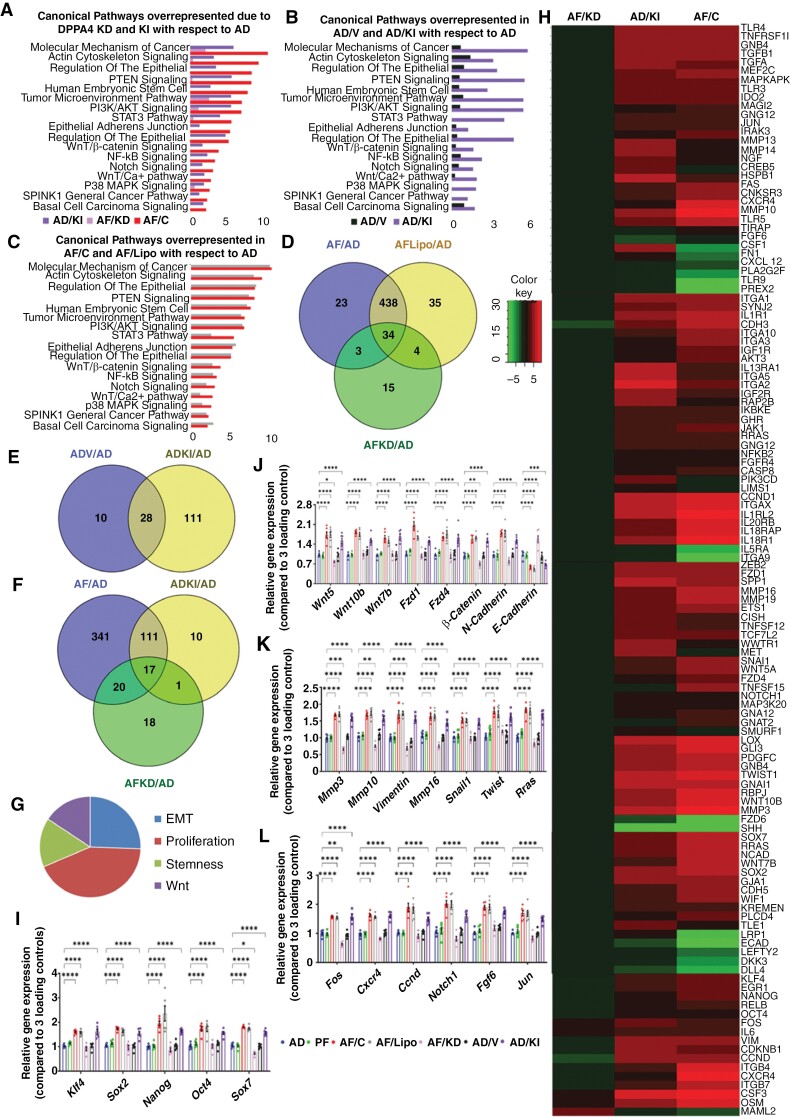
Figure 3.
Gene expression profile changes following Dppa4 knockdown and knockin implicate WNT/β-CATENIN signaling involvement. (a to g) RNA-seq analysis of the effects of Dppa4 knockdown (AF/KD vs AF/C) and knockin (AD/KI vs AD) in the cancer-related canonical pathways. IPA analysis identified canonical pathways overrepresented in AF/KD and AD/KI vs AF/C (a), AD/V vs AD/KI (b), and AF/Lipo vs AF/C (c). Venn diagrams show the common and differentially expressed molecules among the AF, AF/Lipo, and AF/KD groups (d); AD/V and AD/KI groups (e); and AF, AD/KI, and AF/KD groups (f). Gene ontology plot shows changes in biological functions (g). Heatmap analysis of genes observed in AF/KD, AD/KI, AF/C (h); n = 3. (i to l) Validation of RNA-seq data using qPCR analysis. Relative quantification of Klf4, Sox2, Nanog, Oct4, Sox7 (i); Wnt5, Wnt10b, Wnt7b, Fzd1, Fzd4, β-Catenin, N-Cad, E-Cad (j); Mmp3, Mmp10, Vimentin, Mmp16, Snail, Twist, Rras (k); and Fos, Jun, Cxcr4, Ccnd, Notch1, fgf6 (l) in AF/C, AF/Lipo, AF/KD, AD/V, and AD/KI cells. Data shown are mean ± SEM (n = 6) and were analyzed using 1-way analysis of variance (ANOVA) with the Newman Keuls post hoc test. *P < .05, **P < .01, and ***P < .001 vs AD, PF, AF/KD, and AD/V.
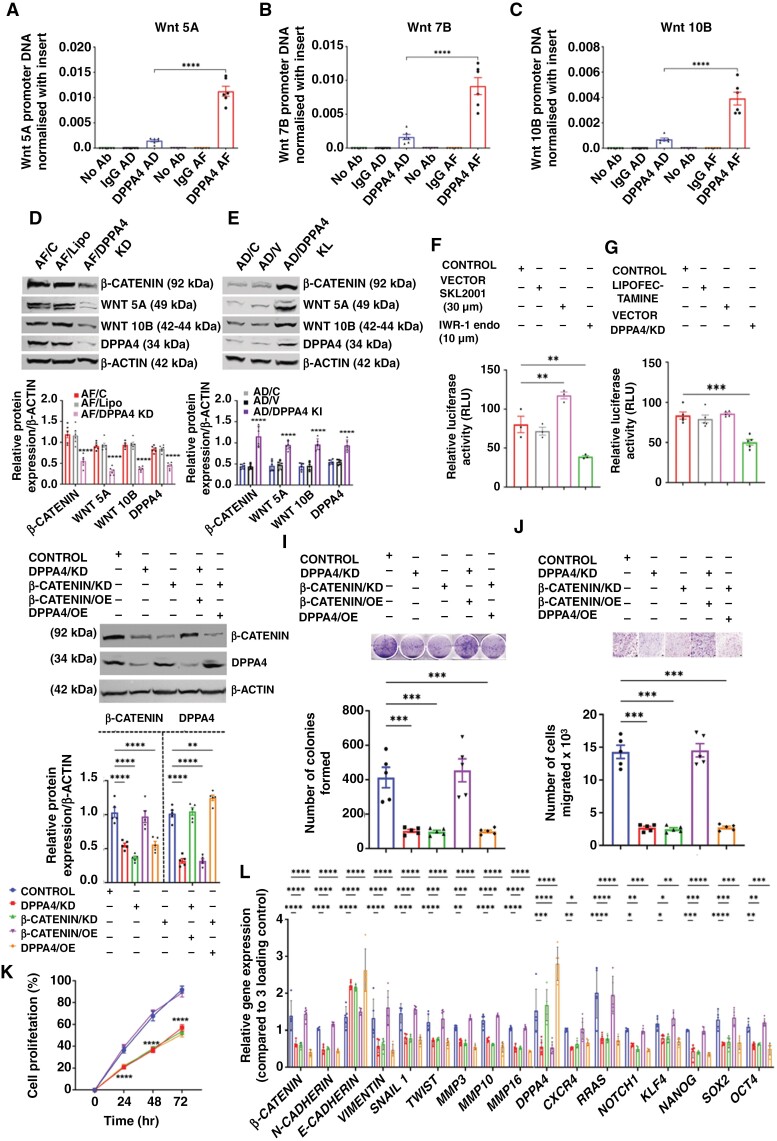
Previously it has been shown that WNT signaling has a regulatory role in cancer cell stemness and growth.42,43Wnt was found to be as major in DPPA4-activated genes in the RNA-seq experiment among the other target genes. Therefore, the WNT pathway may participate in DPPA4-activated cancer stemness and growth. To determine this, the DPPA4 protein was pooled down, and qPCR was performed against Wnt 5A, Wnt 10B, and Wnt 7B DNA as these specific Wnts were upregulated in the RNA-seq datasets. The AF group showed significantly higher levels of all three Wnt promoter DNAs interacting with the DPPA4 protein compared to AD (Figure 4a to c). This result suggests that the DPPA4 protein has the ability to bind to the promoter of canonical Wnts to alter their transcriptions. The knockdown of Dppa4 in AF cells reduced the expression of β-CATENIN, WNT 5A, and WNT 10B, while knockin of Dppa4 in AD cells increased their levels compared to the control cells (Figure 4d and e). TCF/LEF β-Catenin promoter activity was measured in terms of dual luciferase activity. Application of SKL2001 (30 µM) β-Catenin agonist after 24 hours of reporter plasmid transfection increased the relative luciferase activity after 24 hours of treatment which, on the other hand, was reduced significantly by 10 µM of IWR-1-endo treatment (Figure 4f, Supplementary Figure 12). After establishing the promoter assay model, Dppa4 was knocked down in transfected cells after 24 hours. After 36 hours of knockdown, a significant reduction was observed in relative luciferase activity compared to control cells (Figure 4g). Next, we performed a rescue assay, where we modulated Dppa4 and β-Catenin in either or both ways. The knockdown of Dppa4 reduced the levels of both DPPA4 and β-CATENIN, while the CRISPR knockdown of β-Catenin didn’t alter DPPA4 expression, which established DPPA4 as upstream of the WNT–β-CATENIN pathway (Figure 4h). Both of these knockdowns reduced cell migration, colony formation, cell proliferation, and β-Catenin-related gene expression (Figure 4i to l). The overexpression of β-CATENIN (β-Catenin overexpression plasmid) in Dppa4 knockdown cells increased the β-CATENIN protein level and also rescued the cell migration, colony formation, cell proliferation, and β-Catenin-related gene expression (Figure 4i to l). However, there was no improvement observed in cell migration, colony formation, cell proliferation, and β-Catenin-related gene expression when DPPA4 was overexpressed in β-Catenin knockdown cells (Figure 4i to l). Lipofectamine 2000 was used to transfect overexpression plasmid which did not show any change with respect to the control (Supplementary Figure 13). Therefore, it can be assumed that DPPA4 increased the aggressiveness in PitNETs through the activation of the WNT–β-CATENIN pathway.
Figure 4.
Dppa4 increased tumor aggressiveness through the activation of the Wnt pathway in PitNETs. (a to c) ChIP assay data show the interaction of DPPA4 protein with 3 different Wnt promoters—Wnt 5A (a), Wnt 7B (b), Wnt 10B (c). (d and e) Effect of DPPA4 knockdown (d) and knockin (e) in protein levels of β-CATENIN, WNT 5A, WNT 10B and DPPA4. (f and g) Luciferase reporter assay to measure TCF/LEF (β-Catenin) promoter activity in HPT cells after treatment with β-CATENIN agonist or antagonist treatments (f) or knockidown of Dppa4 (g). Lipofectamine 2000 (vector) was used to transfect the reporter plasmid in HPT cells. After 24 hours of transfection, the cells were treated with SKL2001 (β-CATENIN agonist) and IWR-1-endo (β-CATENIN antagonist) separately. After 24 hours of treatment, relative luciferase activity was measured (firefly/renilla) and expressed as percentage in Control (HPT/C), vector (lipofectamine 2000), SKL2001 (HPT treated with SKL2001), IWR-1-endo (HPT treated with IWR-1-endo). Lipofectamine represents the CRISPRMAX lipofectamine used to transfect gRNA Cas9 complex. Control (HPT/C), lipofectamine (CRISPRMAX), vector (lipofectamine 2000), DPPA4/KD (HPT/KD). (h–k) Rescue assay showed the requirement of β-CATENIN for the downstream activity of DPPA4. Control (HPT/C), DPPA4/KD (Dppa4 knockdown HPT), β-CATENIN/KD (β-Catenin knockdown HPT cells), β-CATENIN/OE (HPT cells transfected with β-Catenin overexpression plasmid using lipofectamine 2000), DPPA4/OE (HPT cells transfected with Dppa4 overexpression plasmid using lipofectamine 2000). The immunoblot data shows the level of β-CATENIN and DPPA4 where both were manipulated in either way (h). Colony formation assay (i), transwell migration assay (j), cell proliferation assay (k). (l) Expression changes of genes that are associated with β-Catenin pathways (l), n = 5. Data shown are mean ± SEM and were analyzed using 1-way or 2-way ANOVA with the Newman Keuls post hoc test or Dunnett’s multiple comparison test. *P < .05, **P < .01, and ***P < .001, ****P < .0001 vs Control (HPT/C).
DPPA4 Uses WNT/β-CATENINE Signaling to Increase Stemness and Aggressiveness of PitNET Cells
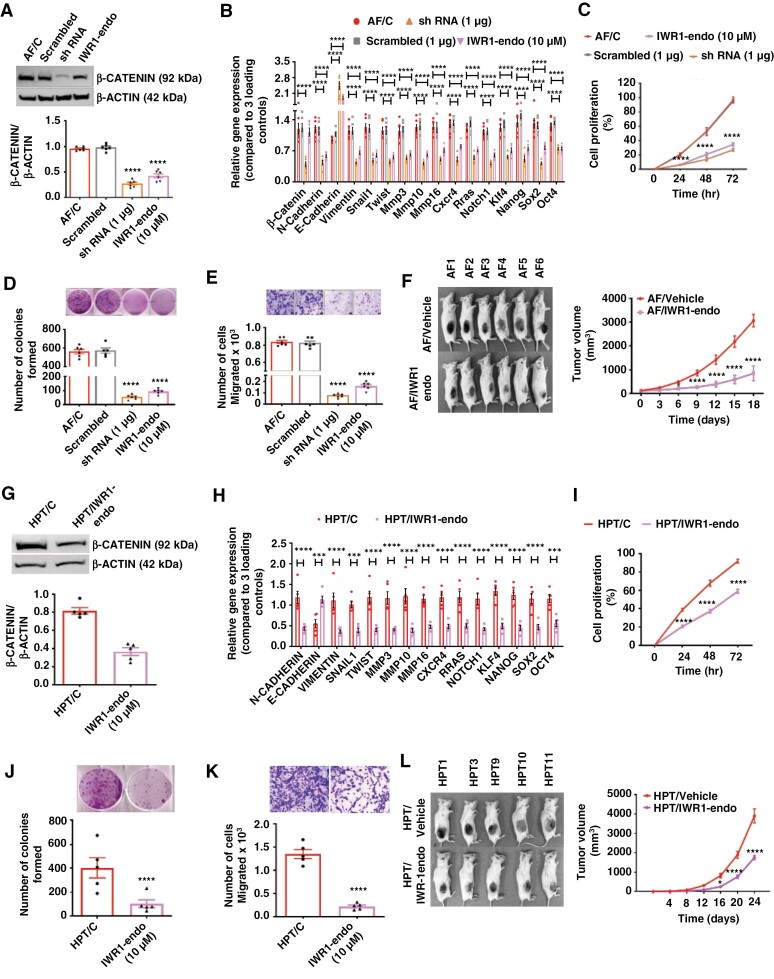
As DPPA4 uses WNT-β-CATENIN signaling to control pituitary tumor cell stemness, growth, and metastasis, we targeted β-CATENIN. We knocked down β-catenin with shRNA plasmid and IWR-1-endo, a potent inhibitor of the WNT/β-CATENIN pathway44 (Figure 5a; Supplementary Figure 14a to d). Relative quantification of gene expression levels following β-catenin shRNA or IWR-1-endo treatment in AF cells revealed that these treatments were able to alter the gene levels of EMT factors (increasing E-cadherin levels while reducing N-cadherin and other mesenchymal transition regulator [Vimentin, Snail1, Twist, Mmp3; Mmp10, Mmp16] levels), stemness regulators (Klf4, Nanog, Sox 2, and Oct4), and cell proliferation regulators (Cxcr4, Rras, and Notch1) (Figure 5b). β-catenin shRNA or IWR-1-endo treatment suppressed the rate of cell proliferation (Figure 5c), colony formation (Figure 5d), and cell migration (Figure 5e) of AF cells. In addition, IWR-1-endo reduced the tumorigenic potential of AF cells following inoculation into immunodeficient mice (Figure 5f). The tumor size (Supplementary Figure 15a), the tumor growth as determined by weekly measurements of tumor volume (Figure 5f), and the tumor weight (Supplementary Figure 15b) at the end point were decreased in mice treated with IWR-1-endo compared to those treated with vehicle. We also tested the effects of IWR-1-endo in HPT cells. Application of IWR-1-endo in HPT cells showed a significant reduction in the level of β-CATENIN (Figure 5g; Supplementary Figure 12); increased mRNA levels of E-cadherin mRNA; reduced levels of N-cadherin, Vimentin, Snail1, Twist, Mmp3; Mmp10, and Mmp16 mRNA, reduced stemness regulators Klf4, Nanog, Sox 2, and Oct4 and cell proliferation regulators Cxcr4, Rras, and Notch1 (Figure 5h); suppressed the rate of cell proliferation (Figure 5i); reduced colony formation (Figure 5j); minimized cell migration (Figure 5k); and minimized the tumorigenic potential of HPT cells following inoculation into immunodeficient mice. The tumor size (Supplementary Figure 16a), the tumor growth as determined by weekly measurements of tumor volume (Figure 5i), and the tumor weight (Supplementary Figure 16b) at the end point were decreased in mice treated with IWR-1-endo compared to those treated with vehicle. These data support a role for WNT/β-CATENIN in the mechanism through which DPPA4 increased the cancerous growth.
Figure 5.
Knockdown and inhibition of β-Catenin through shRNA and IWR-1-endo treatment implicate WNT/β-CATENIN signaling. (a to f) Effects on RPT cells—treated with scrambled plasmid, shRNA plasmid for β-Catenin (1 µg concentration was determined by a dose-response study; Supplementary Figure 11a) or IWR-1-endo (10 µM concentration was determined by a dose–response study; Supplementary Figure 11b to d). β-CATENIN protein levels determined by Western blots (a). Expression changes of genes that are associated with β-Catenin pathways (b). Effects on cell proliferation rate (c). Colony-forming abilities (d). Changes in cell migration (e). Pictures of animals with tumors in each group and tumor volume changes (f), n = 6. (i to l) Effects on HPT cells treated with IWR-1-endo (concentration was determined by a dose–response study; Supplementary Figure 13). Western blot analysis of β-CATENIN (g). Expression changes of genes that are associated with β-Catenin pathways (h). Effects on cell proliferation rate (i). Colony-forming abilities (j). Changes in cell migration (k). Pictures of animals with tumors in each group with tumor volume changes (l), n = 5. Data shown are mean ± SEM and were analyzed using 1-way or 2-way ANOVA with the Newman Keuls post hoc test or Dunnett’s multiple comparison test. *P < .05, **P < .01, and ***P < .001, ****P < .0001 AF/IWR-1-endo vs AF/C, AF/IWR-1-endo, ShRNA vs AF/C, Scrambled and AF/vehicle or HPT/IWR-1-endo vs HPT/C or HPT/vehicle.
An Epigenetic Mechanism is Involved in DPPA4 Overexpression by PAE in the Pituitary
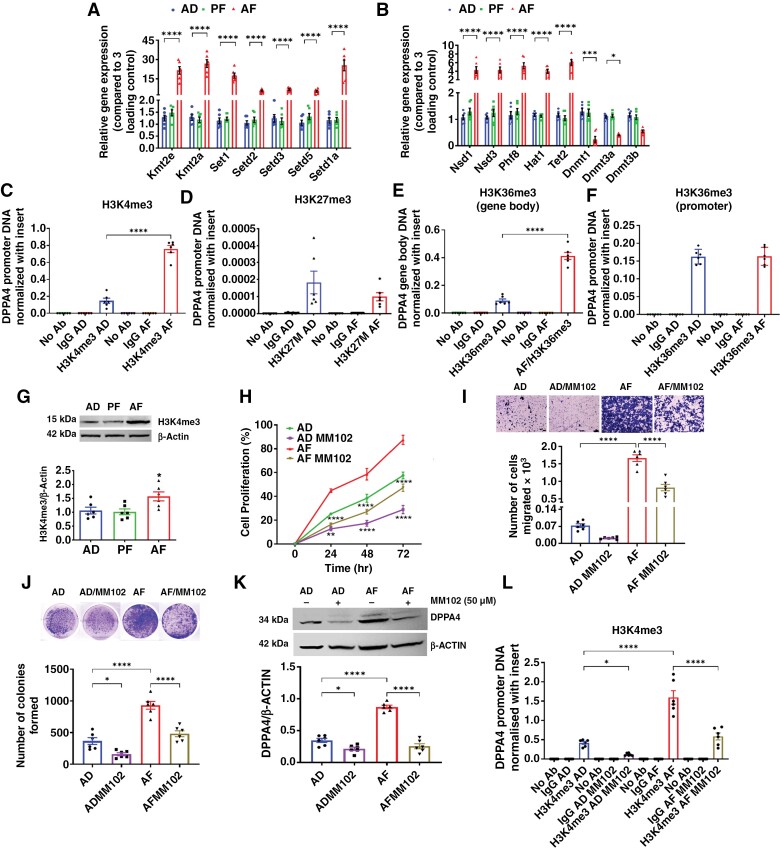
Owing to the function of DPPA4 in early embryonic development,45 PAE could have modified chromatin structure and affected DNA methylation and expression of DPPA4 in RPT cells. We investigated the epigenetic mechanisms involved in PAE effects on Dppa4 expression. Analysis of the RNA-seq data obtained from AD and AF cells identified changes in transcriptomes related to epigenetic modification by PAE (Supplementary Table 4). A qPCR array was performed to check the expression of these epigenetic regulators. Results show that the regulatory genes of H3K4me3, H3K9ac, H3K36me3, and DNA demethylation (TETs) were overexpressed in AF cells, while genes involved in H3K27me3 and DNA methylation (DNMTs) were downregulated in AF cells compared to those in AD cells (Figure 6a and b). It is noteworthy that the genes linked with H3K4me3 (Kmt2e, Kmt2a, and Set1) and H3K36me3 (Setd1a) showed the maximum fold changes among the other genes. The ChIP assay confirmed the changes in H3K4me3, H3K36me3, and H3K27me3 after PAE (Figure 6c to f). A significant fold enrichment in H3K4me3 was seen at the Dppa4 promoter region. Though H3K36me3 showed an insignificant change in the promoter region of Dppa4, the level was significantly higher in the gene body. However, there were no significant differences observed in H3K27me3 (Figure 6f). These data suggest H3K4me3 marks in the Dppa4 promoter primarily drive the overexpression of this protein in AF cells. An elevated level of the H3K4me3 mark was also observed in AF pituitary tumor cells (Figure 6g; Supplementary Figure 6l) and tissues by Western blotting (Supplementary Figure 3k) and immunohistochemistry (Supplementary Figure 2i). Finally, to confirm the role of H3K4me3 in the expression of Dppa4, 50 µM of MM-102 blocker specific to the MLL1 catalytic subunit of the H3K4 methyltransferase enzyme was applied, which suppressed the rate of cell proliferation (Figure 6h, Supplementary Figure 17), minimized cell migration (Figure 6i), and reduced colony formation (Figure 6j). Treatment with MM-102 also reduced the protein levels of DPPA4 markedly in AF cells and minimally in AD cells (Figure 6k). Additionally, MM-102 decreased the level of H3K4me3 at the promoter (Figure 6l). These data suggest that PAE epigenetically modifying the Dppa4 promoter to increase its expression promotes pathogenesis in the pituitary gland.
Figure 6.
Dppa4 promoter is epigenetically regulated by PAE. (a and b) Relative quantification of expression of different genes responsible for epigenetic modulation in the promoter region of the DNA in AD, PF, and AF. (c to f) ChIP assays data show the H3K4me3 level (c) and H3K27me3 level (d) in the Dppa4 promoter region. (e and f) ChIP assay shows the H3K36me3 level in the Dppa4 gene body (e) and promoter region (f), respectively. (g) The image shows the changes in protein levels of H3K4me3 in AD, PF, and AF cells. (h to k) Effects of H3K4me3 blocker MM102 (50 µM is the optimum effective dose of MM102; Supplementary Figure 15) on cell proliferation rates of AD and AF cells (h), transwell cell migration (i), colony formation (j), and DPPA4 protein levels (k). The ChIP assay shows the H3K4me3 level in DPPA4 promoter after 48 h of MM102 treatment (l). Data were analyzed using 1-way or 2-way ANOVA with the Newman Keuls post hoc test or Dunnett’s multiple comparison test. n = 6, **P < .01 and ***P < .001 vs AD and PF cells (a to d; i); ***P < .001 vs rest of the groups (e, g, and j), vs AD or AF (j); *P < .05 and ****P < .0001 as indicated by a line above the bar.
Discussion
The data presented here indicate that cells of AF rats and HPT expressed elevated levels of cell stemness marker genes and proteins, grew rapidly, showed high colony formation and cell migration, and successfully induced tumors when transplanted in immunodeficient mice, suggesting that the pituitary gland develops aggressive and invasive tumors, possibly due to an increase in the stem cell niche within the tumor microenvironment. DPPA4 acts as a nuclear factor to maintain embryonic stem cell proliferation and to reduce their differentiation behavior.46 The data obtained from this study also show that DPPA4 and the Wnt/β-catenin signaling genes are expressed in elevated levels in aggressive RPT cells and in HPT cells. Analysis of GSE datasets of previously published studies showed a positive association between DPPA4 and Wnt5 expression (Supplementary Figures 18 to 20). We have also found high levels of DPPA4 in breast cancer cells line (MDAMB 231) in which activation of Wnt/β-catenin signaling is known to be crucial for the growth and progression of tumors.47 DPPA4 knock-down reduces the cell growth and progression of these cells (Supplementary Figure 21). Thus, the data shown here support the view that WNT/β-CATENIN signaling participates in DPPA4 actions in the development of aggressive PitNETs. Previous studies have shown that nuclear accumulation of the mutant form of β-CATENIN leads to the development of a type of murine pituitary tumor that resembles human pituitary craniopharyngioma, occurs in children, tends to be resistant to treatment, and causes significant morbidity.36 In addition, mutant CTNNB1 nuclear accumulation in embryonic stem cells leads to the development of pituitary invasive adenoma.48 Similarly, it has been shown that Wnt signaling molecules play an essential role in driving the committed stem cells to the differentiated hormone-producing cells, and consistent activation of WNT/β-CATENIN signaling molecules leads to an increase in the proliferation of these cells.49 Interestingly, RPT cells of AF animals and HPT cells that had overexpression of WNT/β-CATENIN signaling molecules and DPPA4 were able to generate tumors in immunodeficient mice. Additionally, suppression of WNT/β-CATENIN signaling by the β-CATENIN blocker IWR-1-endo replicates the DPPA4 knockdown effects on proliferation rate, migration ability, colony number, and tumorigenic potentials in AF and HPT cells in immunodeficient mice. Furthermore, ChIP assay data show the DPPA4 protein binds to the promoter of canonical Wnts. These data support a role for the DPPA4-regulated WNT/β-CATENIN signaling pathway in the development of aggressive PitNETs, although how DPPA4 activates WNT/β-CATENIN has not been established. We have also identified interactions between DPPA4 and stem cell signaling. Because DPPA4 and stem cell regulatory genes are overexpressed in the AF and HPT cells, DPPA4 knockdown reduced the various signaling molecules for stemness in AF and HPT cells, while DPPA4 knockin increased stem cell marker genes in AD cells. These data suggest that the DPPA4-activated cell stemness contributes to the development of aggressive PitNETs.
We have provided evidence that PAE programs pituitary cells to express enhanced Dppa4 gene and the aggressiveness of cell tumors involving the epigenetic modification of H3K4me3. A significant fold enrichment in H3K4me3 at the Dppa4 promoter region was observed in AF cells. These data are consistent with the previous findings that alcohol increased the amount of active chromatin mark H3K4me2 in the regulatory regions of Dppa4 in alcohol-exposed hESCs.21 We also find a higher H3K36me3 level in the gene body in AF cells. Previous studies have shown the H3K36me3 mark in the gene body is associated with the active gene transcription.50 However, H3K36me3 happens to be at the 3ʹ end of the gene49 and thus, major importance was given to H3K4me3, which is known to happen in the promoter region. Furthermore, MM102, a blocker of MLL1 of the methyl transferase enzyme associated with H3K4me3, was able to suppress DPPA4 activity. Several recent studies have defined the roles of DPPA4 in priming the chromatin and maintaining developmental competency through regulating H3K4me3.17 Moreover, DPPA4 knockdown in embryonic stem cells leads to the enrichment of dimethylation at H3K9 in the promotor region of DPPA4, suggesting that DPPA4 plays an essential role in maintaining the active epigenetic status of embryonic stem cells.46 Thus, it appears that PAE epigenetically programs DPPA4 to be overexpressed, thereby maintaining stemness in adult pituitary tumor cells.
In conclusion, these data support an oncogenic role of DPPA4 in the pituitary and establish the involvement of this stemness regulatory factor in the mechanisms controlling the development of aggressive PitNETs. Because Dppa4 was found to be highly expressed in non-small-cell lung cancer tissues,51 this cell stemness regulator may also be involved in the development and progression of other cancers. Furthermore, study data suggest potential therapeutic uses of DPPA4 in the treatment of aggressive PitNETs.
Supplementary material
Supplementary material is available online at Neuro-Oncology (https://academic.oup.com/neuro-oncology).
Acknowledgments
We thank the Biorepository Services of Rutgers Cancer Institute of New Jersey for providing the human pituitary tumor samples, Dr. Peter J. Romanienko, and Rutgers Genome Editing Shared Resource for CRISPR gene editing supports, and Dr. Gokulapriya Govindarajalu for collection and preparation of a small number of human cell cultures used in this study, and Stacey Pontoriero for editorial assistance.
Contributor Information
Shaista Chaudhary, The Endocrine Program, Department of Animal Sciences, Rutgers, The State University of New Jersey, New Brunswick, New Jersey, USA.
Ujjal Das, The Endocrine Program, Department of Animal Sciences, Rutgers, The State University of New Jersey, New Brunswick, New Jersey, USA.
Shaima Jabbar, Endocrinology and Animal Biosciences Graduate Program, Rutgers, The State University of New Jersey, New Brunswick, New Jersey, USA; The Endocrine Program, Department of Animal Sciences, Rutgers, The State University of New Jersey, New Brunswick, New Jersey, USA.
Omkaram Gangisetty, The Endocrine Program, Department of Animal Sciences, Rutgers, The State University of New Jersey, New Brunswick, New Jersey, USA.
Bénédicte Rousseau, The Endocrine Program, Department of Animal Sciences, Rutgers, The State University of New Jersey, New Brunswick, New Jersey, USA.
Simon Hanft, Pituitary Tumor Program, Rutgers Cancer Institute of New Jersey, Rutgers-Robert Wood Johnson Medical School, New Brunswick, New Jersey, USA.
Dipak K Sarkar, Endocrinology and Animal Biosciences Graduate Program, Rutgers, The State University of New Jersey, New Brunswick, New Jersey, USA; The Endocrine Program, Department of Animal Sciences, Rutgers, The State University of New Jersey, New Brunswick, New Jersey, USA.
Funding
This work was supported by National Institute of Health grant R01 AA11591 and Hatch project grant NJ06160. S.J. was supported by the HCED Iraq program fellowship.
Conflict of interest statement
The authors declare no competing interests.
Authorship statement
S.C. contributed to designing experiments, performing research, analyzing data, and writing the methods; U.D. helped in designing experiments, performing research, analyzing data, and writing the methods; S.J. contributed to designing experiments and performing research; O.G. performed research; B.R. performed research; S.H., performed research; D.K.S. conceived the study and was in charge of overall direction and planning, obtained funding, and wrote the paper.
Data availability
All computer codes used in this study are the conventional code to do the RNA sequencing by Genewiz company. The coding used to generate the heatmap in R studio is regular coding. The R Script is available upon reasonable request. All the supporting information is available in the Supplementary Material and the other additional details are available from the lead contact (dipak.sarkar@rutgers.edu) upon request. The RNA-seq dataset used in this study is not deposited in the Gene Expression Omnibus (GEO). We are ready to submit those datasets anytime as per the journal requirements.
References
- 1. Colao A. Pituitary tumours: the prolactinoma. Best Pract Res Clin Endocrinol Metab. 2009;23(5):575–596. [DOI] [PubMed] [Google Scholar]
- 2. Raverot G, Burman P, McCormack A, et al. ; European Society of Endocrinology. European Society of Endocrinology Clinical Practice Guidelines for the management of aggressive pituitary tumours and carcinomas. Eur J Endocrinol. 2018;178(1):G1–G24. [DOI] [PubMed] [Google Scholar]
- 3. Das L, Rai A, Salunke P, et al. Temozolomide nonresponsiveness in aggressive Prolactinomas and carcinomas: management and outcomes. J Endocr Soc. 2021;6(2):bvab190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neou M, Villa C, Armignacco R, et al. Pangenomic classification of pituitary Neuroendocrine tumors. Cancer Cell. 2020;37(1):123–134.e5. [DOI] [PubMed] [Google Scholar]
- 5. Lasolle H, Ilie MD, Raverot G.. Aggressive prolactinomas: how to manage? Pituitary. 2020;23(1):70–77. [DOI] [PubMed] [Google Scholar]
- 6. Nakano-Tateno T, Lau KJ, Wang J, et al. Multimodal non-surgical treatments of aggressive pituitary tumors. Front Endocrinol (Lausanne). 2021;12:624686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duhamel C, Ilie MD, Salle H, et al. Immunotherapy in Corticotroph and Lactotroph aggressive tumors and carcinomas: two case reports and a review of the literature. J Pers Med. 2020;10(3):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sol B, de Filette JMK, Awada G, et al. Immune checkpoint inhibitor therapy for ACTH-secreting pituitary carcinoma: a new emerging treatment? Eur J Endocrinol. 2021;184(1):K1–K5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feola T, Carbonara F, Verrico M, et al. Immunotherapy for aggressive and metastatic pituitary neuroendocrine tumors (PitNETs): state-of-the art. Cancers (Basel). 2022;14(17):4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roelfsema F, Biermasz NR, Pereira AM.. Clinical factors involved in the recurrence of pituitary adenomas after surgical remission: a structured review and meta-analysis. Pituitary. 2012;15(1):71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nys C, Lee YL, Roose H, et al. Exploring stem cell biology in pituitary tumors and derived organoids. Endocr Relat Cancer. 2022;29(7):427–450. [DOI] [PubMed] [Google Scholar]
- 12. Mertens F, Gremeaux L, Chen J, et al. Pituitary tumors contain a side population with tumor stem cell-associated characteristics. Endocr Relat Cancer. 2015;22(4):481–504. [DOI] [PubMed] [Google Scholar]
- 13. Cai L, Chen J, Lu J, et al. Tumor stem-like cells isolated from MMQ cells resist to dopamine agonist treatment. Mol Cell Endocrinol. 2021;535:111396. [DOI] [PubMed] [Google Scholar]
- 14. Jabbar S, Reuhl K, Sarkar DK.. Prenatal alcohol exposure increases the susceptibility to develop aggressive prolactinomas in the pituitary gland. Sci Rep. 2018;8(1):7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17(3):313–319. [DOI] [PubMed] [Google Scholar]
- 16. Zhao D, Tomono Y, Nose T.. Expression of P27kip1 and Ki-67 in pituitary adenomas: an investigation of marker of adenoma invasiveness. Acta Neurochir (Wien). 1999;141(2):187–192. [DOI] [PubMed] [Google Scholar]
- 17. Klein RH, Knoepfler PS.. DPPA2, DPPA4, and other DPPA factor epigenomic functions in cell fate and cancer. Stem Cell Rep. 2021;16(12):2844–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chakravarthy H, Boer B, Desler M, et al. Identification of DPPA4 and other genes as putative Sox2:Oct-3/4 target genes using a combination of in silico analysis and transcription-based assays. J Cell Physiol. 2008;216(3):651–662. [DOI] [PubMed] [Google Scholar]
- 19. Madan B, Madan V, Weber O, et al. The pluripotency-associated gene DPPA4 is dispensable for embryonic stem cell identity and germ cell development but essential for embryogenesis. Mol Cell Biol. 2009;29(11):3186–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tung PY, Varlakhanova NV, Knoepfler PS.. Identification of DPPA4 and DPPA2 as a novel family of pluripotency-related oncogenes. Stem Cells. 2013;31(11):2330–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Auvinen P, Vehviläinen J, Marjonen H, et al. Chromatin modifier developmental pluripotency associated factor 4 (DPPA4) is a candidate gene for alcohol-induced developmental disorders. BMC Med. 2022;20(1):495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gonçalves PP, Stenovec M, Grácio L, Kreft M, Zorec R.. Calcium-dependent subquantal peptide release from single docked lawn-resident vesicles of pituitary lactotrophs. Cell Calcium. 2023;109:102687. [DOI] [PubMed] [Google Scholar]
- 23. Würth R, Pattarozzi A, Barbieri F, Florio T.. Primary cultures from human GH-secreting or clinically non-functioning pituitary adenomas. Bio Protoc. 2018;8(7):e2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aiello A, Cassarino MF, Nanni S, et al. Establishment of a protocol to extend the lifespan of human hormone-secreting pituitary adenoma cells. Endocrine. 2018;59(1):102–108. [DOI] [PubMed] [Google Scholar]
- 25. Daly AZ, Mortensen AH, Bando H, Camper SA.. Pituitary tumors and immortalized cell lines generated by cre-inducible expression of SV40 T antigen. Endocrinology. 2021;162(7):bqab073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rousseau B, Murugan S, Palagani A, Sarkar DK.. Beta 2 adrenergic receptor and mu opioid receptor interact to potentiate the aggressiveness of human breast cancer cell by activating the glycogen synthase kinase 3 signaling. Breast Cancer Res. 2022;24(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sousa DA, Gaspar R, Ferreira CJO, et al. In Vitro CRISPR/Cas9 transfection and gene-editing mediated by multivalent cationic liposome-DNA complexes. Pharmaceutics. 2022;14(5):1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Townsend EA, Kim RK, Robinson HL, et al. Opioid withdrawal produces sex-specific effects on fentanyl-vs.-food choice and mesolimbic transcription. Biol Psychiatry Glob Open Sci. 2021;1(2):112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H, Hui H, Li Z, et al. Pigment epithelium-derived factor attenuates myocardial fibrosis via inhibiting endothelial-to-mesenchymal transition in rats with acute myocardial infarction. Sci Rep. 2017;7:41932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martins-Neves SR, Paiva-Oliveira DI, Fontes-Ribeiro C, et al. IWR-1, a tankyrase inhibitor, attenuates Wnt/β-catenin signaling in cancer stem-like cells and inhibits in vivo the growth of a subcutaneous human osteosarcoma xenograft. Cancer Lett. 2018;414:1–15. [DOI] [PubMed] [Google Scholar]
- 31. Murugan S, Rousseau B, Sarkar DK.. Beta 2 adrenergic receptor antagonist propranolol and opioidergic receptor antagonist naltrexone produce synergistic effects on breast cancer growth prevention by acting on cancer cells and immune environment in a preclinical model of breast cancer. Cancers (Basel). 2021;13(19):4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karatas H, Townsend EC, Cao F, et al. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J Am Chem Soc. 2013;135(2):669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Teotia P, Van Hook MJ, Fischer D, Ahmad I.. Human retinal ganglion cell axon regeneration by recapitulating developmental mechanisms: effects of recruitment of the mTOR pathway. Development. 2019;146(13):dev178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyajima K, Takekoshi S, Itoh J, et al. Inhibitory effects of anti-VEGF antibody on the growth and angiogenesis of estrogen-induced pituitary prolactinoma in Fischer 344 rats: animal model of VEGF-targeted therapy for human endocrine tumors. Acta Histochem Cytochem. 2010;43(2):33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gangisetty O, Bekdash R, Maglakelidze G, Sarkar DK.. Fetal alcohol exposure alters proopiomelanocortin gene expression and hypothalamic-pituitary-adrenal axis function via increasing MeCP2 expression in the hypothalamus. PLoS One. 2014;9(11):e113228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang CV, Araujo RV, Cirqueira CS, et al. Differential expression of stem cell markers in human adamantinomatous craniopharyngioma and pituitary adenoma. Neuroendocrinology. 2017;104(2):183–193. [DOI] [PubMed] [Google Scholar]
- 37. Mete O, Ezzat S, Asa SL.. Biomarkers of aggressive pituitary adenomas. J Mol Endocrinol. 2012;49(2):R69–R78. [DOI] [PubMed] [Google Scholar]
- 38. Hassn Mesrati M, Syafruddin SE, Mohtar MA, Syahir A.. CD44: a multifunctional mediator of cancer progression. Biomolecules. 2021;11(12):1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang P, Yan R, Zhang X, et al. Activating Wnt/β-catenin signaling pathway for disease therapy: challenges and opportunities. Pharmacol Ther. 2019;196:79–90. [DOI] [PubMed] [Google Scholar]
- 40. Jia W, Zhu J, Martin TA, et al. Epithelial-mesenchymal transition (EMT) markers in human pituitary adenomas indicate a clinical course. Anticancer Res. 2015;35(5):2635–2643. [PubMed] [Google Scholar]
- 41. Zhang F, Zhang Q, Zhu J, et al. Integrated proteogenomic characterization across major histological types of pituitary neuroendocrine tumors. Cell Res. 2022;32(12):1047–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Najafi M, Farhood B, Mortezaee K.. Cancer Stem Cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234(6):8381–8395. [DOI] [PubMed] [Google Scholar]
- 43. Kim JH, Park SY, Jun Y, Kim J-Y, Nam J-S.. Roles of Wnt target genes in the journey of cancer stem cells. Int J Mol Sci. 2017;18(8):1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen B, Dodge ME, Tang W, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5(2):100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hernandez C, Wang Z, Ramazanov B, et al. Dppa2/4 facilitate epigenetic remodeling during reprogramming to pluripotency. Cell Stem Cell. 2018;23(3):396–411.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Masaki H, Nishida T, Kitajima S, Asahina K, Teraoka H.. Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J Biol Chem. 2007;282(45):33034–33042. (duplicate) [DOI] [PubMed] [Google Scholar]
- 47. Yang X, Cao D, Ma W, et al. Wnt signaling in triple-negative breast cancers: its roles in molecular subtyping and cancer cell stemness and its crosstalk with non-coding RNAs. Life Sci. 2022;300:120565. [DOI] [PubMed] [Google Scholar]
- 48. Gaston-Massuet C, Andoniadou CL, Signore M, et al. Increased wingless (Wnt) signaling in pituitary progenitor/stem cells gives rise to pituitary tumors in mice and humans. Proc Natl Acad Sci U S A. 2011;108(28):11482–11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pokholok DK, Harbison CT, Levine S, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122(4):517–527. [DOI] [PubMed] [Google Scholar]
- 50. Zhang T, Cooper S, Brockdorff N.. The interplay of histone modifications – writers that read. EMBO Rep. 2015;16(11):1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li L, Wang Y, Wang Q, et al. High developmental pluripotency‑associated 4 expression promotes cell proliferation and glycolysis, and predicts poor prognosis in non‑small‑cell lung cancer. Mol Med Rep. 2019;20(1):445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All computer codes used in this study are the conventional code to do the RNA sequencing by Genewiz company. The coding used to generate the heatmap in R studio is regular coding. The R Script is available upon reasonable request. All the supporting information is available in the Supplementary Material and the other additional details are available from the lead contact (dipak.sarkar@rutgers.edu) upon request. The RNA-seq dataset used in this study is not deposited in the Gene Expression Omnibus (GEO). We are ready to submit those datasets anytime as per the journal requirements.