Abstract
Background
Colorectal cancer (CRC) exhibits a high incidence globally, with the liver being the most common site of distant metastasis. At the time of diagnosis, 20–30% of CRC patients already present with liver metastases. Colorectal liver metastasis (CRLM) is a major cause of mortality among CRC patients. The pathogenesis of CRLM involves complex molecular mechanisms and the hepatic immune microenvironment, but current clinical prevention and treatment are significantly limited. Recent studies have revealed that the major facilitator superfamily domain containing protein-2a (Mfsd2a) plays a pivotal role in the development and metastasis of various cancers. For instance, Mfsd2a inhibits gastric cancer initiation and progression and may impact angiogenesis. However, the mechanisms by which Mfsd2a influences CRC progression and liver metastasis remain unclear.
Methods
In this study, we conducted a survival analysis of Mfsd2a in colorectal cancer using data from the GEPIA and GEO databases, and examined the expression differences between primary tumor (PT) and liver metastasis (LM). We further assessed the clinical significance and prognostic relevance of Mfsd2a through immunohistochemical analysis of tissue samples from 70 CRLM patients. Moreover, Kaplan-Meier analysis was used to perform survival analysis on these patients. The biological function of Mfsd2a in CRLM was confirmed by a series of experiments conducted both in vitro and in vivo. Additionally, we investigated downstream molecular pathways using western blot, Co-immunoprecipitation, immunofluorescence, and mass spectrometry techniques.
Results
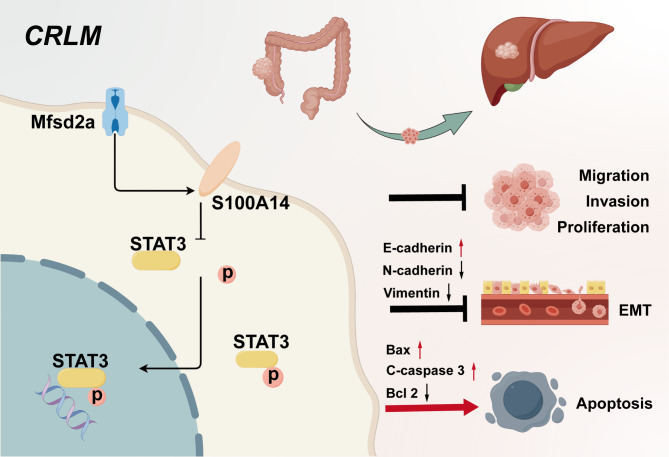
We observed that Mfsd2a expression is reduced in LM compared to PT, and higher Mfsd2a levels are associated with better prognosis in CRLM patients. Furthermore, function assays demonstrated that Mfsd2a suppresses CRC cells proliferation, migration, invasion, and EMT in vitro, while also delaying tumor growth and liver metastasis in vivo. Mechanistically, Mfsd2a interacts with S100A14, enhancing its expression and inhibiting phosphorylation of STAT3. In addition, the STAT3 activator colivelin partially reversed the inhibitory effect of Mfsd2a overexpression on the progression of colorectal cancer and liver metastasis.
Conclusion
In summary, Mfsd2a inhibits colorectal cancer progression and liver metastasis by interacting with S100A14, thereby suppressing the phosphorylation of STAT3. Mfsd2a functions as a tumor suppressor in CRLM and could be a promising therapeutic target for treating CRC patients with liver metastasis.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05994-y.
Keywords: CRC, CRLM, Mfsd2a, EMT, S100A14, STAT3
Background
Colorectal cancer (CRC) ranks among the top three gastrointestinal malignancies in terms of incidence and mortality [1]. The liver is the most common site of distant metastasis for CRC, with approximately 15–25% of patients presenting with liver metastasis at initial diagnosis, and over half of CRC patients ultimately developing liver metastasis [2]. Compared to patients without liver metastasis, those with colorectal liver metastasis (CRLM) have a significantly lower 5-year survival rate, typically below 20% [3, 4]. Liver metastasis is a major cause of mortality among CRC patients [5]. CRLM is emerging as a significant global health challenge [6]. Extensive research highlights the critical role of the interaction between tumor biology and the microenvironment in CRLM development. However, effective therapeutic strategies or targeted drugs remain unavailable, leading to suboptimal clinical outcomes [7]. Therefore, it is crucial to identify potential biomarkers for CRLM and to explore the potential mechanisms of colorectal cancer metastasizes to the liver.
Major facilitator superfamily domain containing protein-2a (Mfsd2a) is a lipid metabolism-associated membrane protein that transports a wide range of substrates such as sugars, polyols, drugs, and neurotransmitters and plays a key role in maintaining the blood-brain barrier [8–10]. Recent studies have emphasized its involvement in various cancers. Mfsd2a acts as a tumor suppressor in lung cancer by regulating tumor cell cycle and matrix adhesion [11], and it can prevent metastatic brain tumors by restoring intracellular docosahexaenoic acid transport [12]. Additionally, expression level of Mfsd2a is positively correlated with improved patient survival in hepatocellular carcinoma [13]. Mfsd2a inhibits gastric cancer initiation and progression and may impact angiogenesis [14], with Mfsd2a potentially serving as a predictive biomarker for anti-PD-1 immunotherapy response in advanced gastric cancer [15]. In the colon, Mfsd2a promotes endothelial generation of lipid mediators that resolve inflammation and reduces colitis in mice [16]. However, its role in CRC progression and liver metastasis remains unexplored.
This study aims to investigate the regulatory mechanisms of Mfsd2a in CRLM. Analysis of public datasets and clinical outcomes indicated that the expression level of Mfsd2a in liver metastasis (LM) was positively correlated with the prognosis of CRLM patients. Experiments in vivo and in vitro demonstrated that Mfsd2a inhibited the proliferation, migration, and epithelial-to-mesenchymal transition (EMT) of CRC cells, confirming that Mfsd2a served as a tumor suppressor restraining CRC progression and liver metastasis. Moreover, we found that Mfsd2a regulated the expression and phosphorylation of the signal transducer and activator of transcription 3(STAT3) by interacting with its downstream protein S100 calcium-binding protein A14 (S100A14), thereby inhibiting the development of CRLM. These findings suggest that Mfsd2a holds promise as a therapeutic target and prognostic biomarker for CRLM.
Materials and methods
Patients and samples collection
All tissue samples were obtained from patients with colorectal liver metastasis (CRLM) who underwent synchronous resection of primary colorectal tumor and liver metastasis at the First Affiliated Hospital of Nanjing Medical University, without concurrent primary or metastatic malignancies at other sites. Informed consent was obtained from all patients before the collection of clinical samples, and consent forms were signed. This study was approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (Approval No: 2024-SR-526).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissue or cells using an RNA extraction kit (esunbio, Shanghai, China) according to the manufacturer’s instructions, and reverse transcribed into cDNA. qRT-PCR was performed using SYBR Green fluorescent dye, and target gene expression levels were normalized to GAPDH expression. Primer sequences used for amplification are detailed in Table S1.
Western blot
Tissues and cells were lysed using RIPA buffer (Beyotime, Shanghai, China) supplemented with PMSF (Beyotime, Shanghai, China). Proteins were separated by SDS-PAGE (Epizyme, Shanghai, China) and then transferred to PVDF membranes (Merck Millipore, Burlington, MA, USA). Membranes were incubated with primary and secondary antibodies, and protein bands were visualized using an enhanced chemiluminescence detection reagent (NcmECL High, Suzhou, China). Details of all antibodies are listed in Table S2.
Hematoxylin and eosin(H&E), immunohistochemistry(IHC), and immunofluorescence staining
The specimens were fixed in 4% neutral buffered formalin and then embedded in paraffin. Tissue sections were stained with hematoxylin and eosin. Standard immunohistochemical protocols were employed for tissue assessment.
For immunofluorescence staining, fixed tissue sections were placed in an antigen retrieval solution containing EDTA (pH = 9.0) from a retrieval kit (Servicebio, China) and heated in a microwave for antigen retrieval. After blocking with 1% BSA for 30 min, primary antibodies were applied and incubated overnight at 4 °C. After PBS washes, secondary antibodies specific to the respective species were applied for 50 min, and nuclei were stained with DAPI. The slides were examined and recorded using an inverted confocal fluorescence microscope (NIKON ECLIPSE C1).
Cell Culture, transfection, and establishment of stable cell lines
The HEK 293T and the colorectal cancer cell lines(RKO, CT26)used in this study were obtained from the Cell Bank of the Chinese Academy of Sciences (CASCB, Shanghai, China). The RKO cell line was cultured in DMEM (GIBCO, NY, USA) supplemented with 10% fetal bovine serum (GIBCO) and 1% penicillin-streptomycin (GIBCO). The HEK 293T and CT26 cell lines were cultured in RPMI-1640 (GIBCO) medium supplemented with 10% FBS and 1% penicillin-streptomycin. All cell lines were maintained in a 5% CO2 humidified incubator (Thermo Fisher Scientific, MA, USA) at 37 °C.
To establish stable cell lines, the lentiviral vectors for Mfsd2a overexpression and S100A14 knockdown plasmids were designed by GenePharma (Shanghai, China). Target cells were transfected with these lentiviral vectors, and stable transfectants were selected using puromycin. Verification was performed using qRT-PCR and Western Blotting. Detailed sequences are listed in Table S3.
Subcutaneous tumor and CRLM models in mouse
Animal experiments for this study were approved by the Institutional Animal Care and Use Committee (IACUC-2306050) of the First Affiliated Hospital of Nanjing Medical University. 6–8 weeks old BALB/c male mice were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and housed under standard laboratory conditions.
BALB/C mice were grouped according to experimental requirements, with six mice per group. A total of 5 × 10^6 lentivirus-transduced CT26 cells were injected into the right axillary region of the mice. Once subcutaneous tumors were successfully established, tumor size was measured every three days for 21 days. After euthanasia, subcutaneous tumors were excised for weighing and immunohistochemical staining.
For the CRLM model, after successful anesthesia, the abdomen was opened, and the spleen was isolated and exposed. Then, 1 × 106 lentivirus-transduced CT26/Luc cells in 100 µl PBS were injected into the spleen. After withdrawing the needle, local pressure was applied to the injection site for hemostasis, and the spleen was removed 15 min later. Two weeks post-surgery, mice were euthanized, and liver tissues were harvested for weighing, imaging, and subsequent tissue embedding.
CCK-8 assay
Cells were seeded in 96-well plates at a density of 2,000 cells per well, with 100 µl of complete medium per well. At 0, 24, 48, 72, 96, and 120 h, cells were incubated with the CCK-8 reagent (Biosharp, Hefei, China) for 2 h. Absorbance at 450 nm was measured using a spectrophotometer (Thermo Fisher Scientific, Pittsburgh, PA, USA).
Colony formation assay
Cells were seeded in 6-well plates at a density of 1,000 cells per well and cultured for 14 days. After 14 days, cells were fixed with 4% paraformaldehyde for 30 min and then stained with 1% crystal violet for 20 min. The number of colonies in each well was recorded.
EdU proliferation assay
Logarithmically growing tumor cells were seeded in 24-well plates and cultured for 12 h. EdU staining was performed according to the instructions provided with the EdU Assay Kit (BeyoClick™ EdU-555) to assess cell proliferation. Images were captured using a fluorescence microscope (Zeiss, Jena, Germany).
Transwell assay
Invasion and migration assays were conducted using Transwell chambers (Corning, USA), with or without Matrigel (BD Biosciences, USA) coating. RKO and CT26 cells (2 × 10^4) were seeded in the upper chamber and incubated in 200 µl of serum-free medium, while the lower chamber contained 600 µl of complete medium with 10% FBS. After 24 h of incubation, invasive cells on the lower surface of the membrane were fixed with 4% paraformaldehyde and stained with crystal violet. Images were captured and recorded using an inverted microscope.
Wound healing assay
Cells were seeded in 6-well plates at an appropriate density and grown to near confluence. After 24 h of incubation in serum-free medium, a cross-shaped wound was created on the bottom of the wells using a 200 µl pipette tip. Cell migration into the wound area was observed and photographed at 0, 24, and 48 h using an inverted microscope. The width of the wound was measured and recorded to determine the wound healing rate.
Cell apoptosis
Cell apoptosis was assessed using the APC Annexin V Apoptosis Detection Kit (640932, BioLegend). After collection, cells were stained with Annexin V-APC and PI at room temperature for 15 min, following the manufacturer’s instructions. The cells were resuspended in buffer for flow cytometric analysis (CytoFLEX S, Beckman). All data were analyzed using FlowJo (V10.8.1) and CytoExpert (V2.4.0.28).
Immunoprecipitation (IP), silver staining, and mass spectrometry
Cells were lysed with NP-40 (Beyotime, Shanghai, China) to extract proteins. Antibodies were incubated with Protein A/G magnetic beads (Beyotime, Shanghai, China) to capture antigen-antibody complexes. The protein complexes were separated by SDS-PAGE and detected by Western blotting using appropriate antibodies. Silver staining was performed according to the manufacturer’s instructions (Beyotime, Shanghai, China). Candidate proteins that differed significantly from the gel background were excised and sent to a commercial facility for mass spectrometry analysis.
Immunofluorescence (IF)
Cells at the appropriate density were fixed with 4% paraformaldehyde at 37 °C for 30 min and permeabilized with 0.3% Triton X-100. After blocking for 30 min, cells were incubated overnight at 4 °C with specific primary antibodies. The next day, cells were incubated with fluorescent secondary antibodies (Beyotime, Shanghai, China) for 2 h and stained with DAPI (Beyotime, Shanghai, China) to visualize the nuclei. Fluorescent images were captured using a confocal fluorescence microscope.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 9.0 software. Comparisons between two groups were conducted using two-tailed Student’s t-test, while one-way ANOVA was used for multiple comparisons. The X2 test was used to analyze the correlation between Mfsd2a expression and clinicopathological variables. Overall survival (OS) and progression-free survival (PFS) were analyzed using the Kaplan-Meier analysis method. The relevant experiments presented in this study were performed at least three times independently, and data are presented as mean ± SD. Statistical significance was considered at p < 0.05 (ns. = not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Result
Mfsd2a expression is downregulated in CRLM and associated with poor prognosis
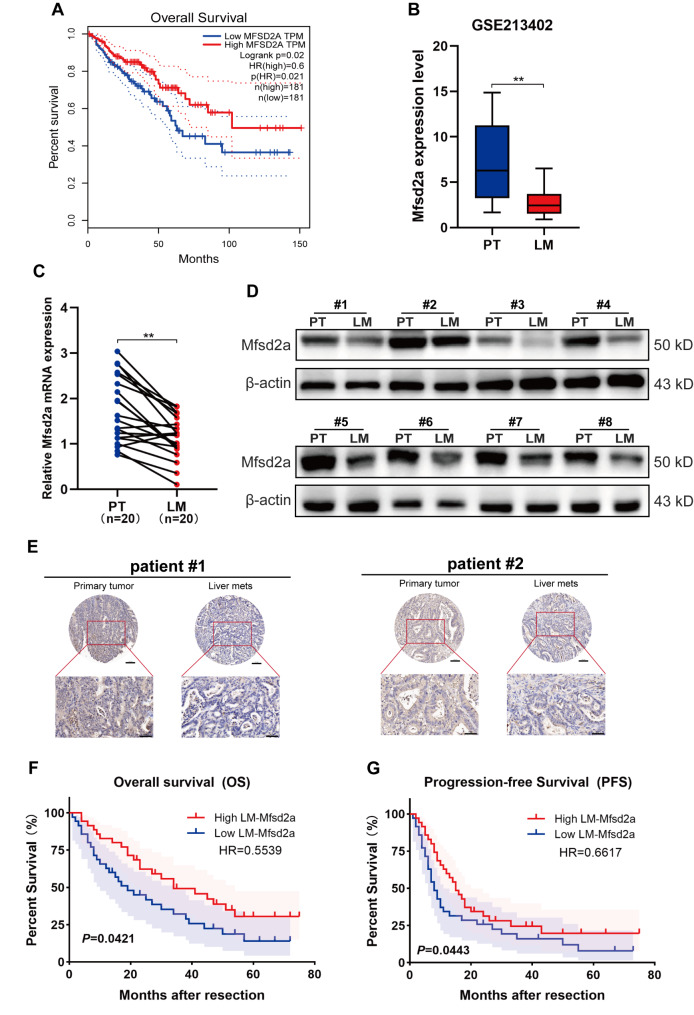
Mfsd2a has been confirmed to act as a tumor suppressor in several cancers, including lung cancer [11], gastric cancer [14], and hepatocellular carcinoma [13]. However, its role and mechanisms in colorectal cancer (CRC) and colorectal liver metastasis (CRLM) remain unclear. Analysis using the TIMER2.0 database(http://timer.cistrome.org/) revealed no statistically significant difference in Mfsd2a expression between CRC tumor tissues and normal tissues (Fig.S1A). Nevertheless, Survival analysis indicated that CRC patients with higher Mfsd2a expression had a longer life expectancy (Fig. 1A). Recent research has shown that CRLM is associated with low survival rates and mortality in CRC patients [5, 17]. Therefore, we hypothesized that Mfsd2a is involved in CRLM. Analysis of the GEO dataset (GSE213402) revealed that Mfsd2a transcript levels were downregulated in liver metastasis (LM) compared to primary tumor (PT) (Fig. 1B). Subsequent validation by qRT-PCR and Western blot confirmed that Mfsd2a expression was significantly reduced in LM compared to PT (Fig. 1C, D). Additionally, immunohistochemical analysis further confirmed decreased Mfsd2a protein expression in LM (Fig. 1E). We classified 70 CRLM patients into two different groups with high or low Mfsd2a expression based on the immunohistochemical analysis of Mfsd2a in LM tissues. The results showed that low expression of Mfsd2a was associated with an increased number of metastases(p = 0.022), poorer degree of differentiation (p = 0.025), higher postoperative recurrence rates (P = 0.031), and elevated clinical risk scores (CRS) (P = 0.030) by analyzing the relationship between Mfsd2a and the clinical-pathological features of CRLM patients (Table S4). Kaplan-Meier survival analysis indicated that CRLM patients with low Mfsd2a expression had significantly shorter overall survival (OS) (p = 0.0421, HR = 0.5539) and progression-free survival (PFS) (p = 0.0443, HR = 0.6617) after liver resection (Fig. 1F, G). Overall, our results suggest that reduced Mfsd2a expression in LM correlates with poor prognosis in CRLM patients.
Fig. 1.
Mfsd2a is downregulated in CRLM and associated with poor prognosis. (A) Kaplan-Meier survival analysis of Mfsd2a expression in CRC patients from the GEPIA2 database. (B) Mfsd2a expression levels in paired PT and LM from the GEO dataset (GSE213402). (C,D) Relative mRNA levels (n = 20) and protein expression levels (n = 8) of Mfsd2a in PT and LM tissues from CRLM patients. (E) Representative IHC images of Mfsd2a expression in PT and paired LM from CRLM patients. (F,G) Kaplan-Meier survival curves for overall survival (OS) and progression-free survival (PFS) in 70 CRLM patients, stratified by median Mfsd2a expression levels in LM. (*P < 0.05, **P < 0.01, ***P < 0.001)
Mfsd2a inhibits CRC cell proliferation and tumor growth
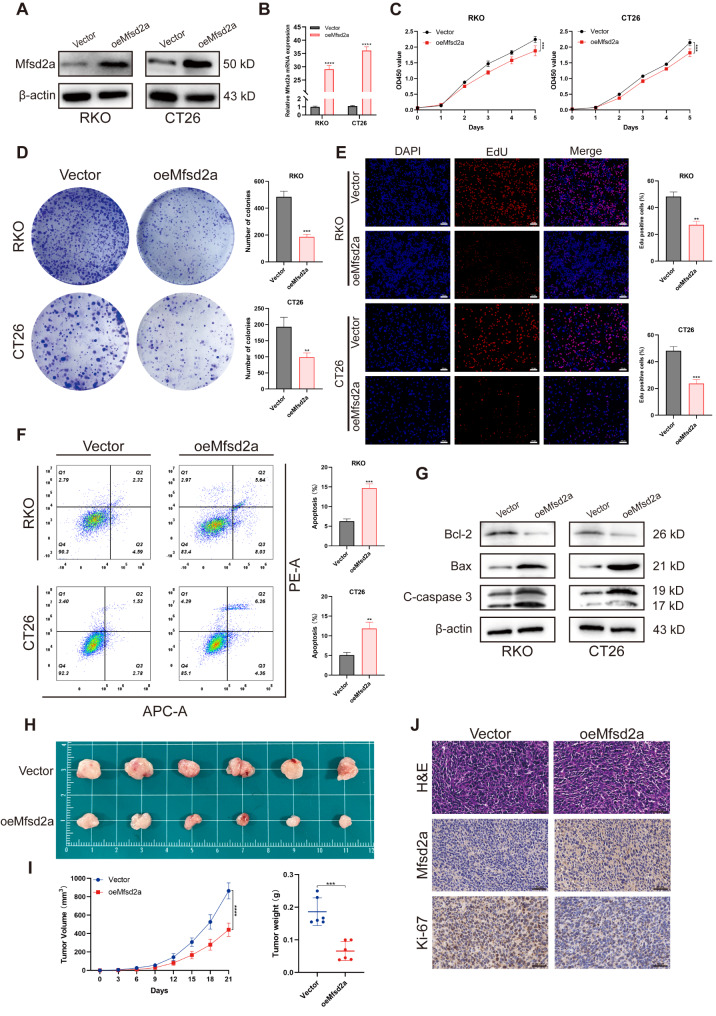
Cell proliferation provides the foundation for tumor metastasis and expansion in distant organs, serving as a key driving of tumor growth and metastasis [7]. To investigate the biological function of Mfsd2a in the progression of CRLM, we used the highly metastatic CRC cell lines RKO and CT26 for in vitro and in vivo experiments. Stable Mfsd2a-overexpressing (oeMfsd2a) cell lines were generated through lentiviral transduction (Fig. 2A, B). Overexpression of Mfsd2a significantly inhibited the proliferation and colony formation of RKO and CT26 cells (Fig. 2C, D). Consistently, EdU staining further confirmed that overexpression of Mfsd2a inhibited cell proliferation (Fig. 2E). Furthermore, we observed that overexpression of Mfsd2a promoted apoptosis in RKO and CT26 cells (Fig. 2F, G).
Fig. 2.
Mfsd2a inhibits CRC cell proliferation in vitro and in vivo. (A, B) Overexpression efficiency of Mfsd2a in RKO and CT26 cells were validated by Western blot and qRT-PCR. (C) CCK-8 assay was performed on oeMfsd2a RKO and CT26 cells. (D,E) EdU and colony formation assays were used to evaluate the proliferative capacity of oeMfsd2a RKO and CT26 cells. (F) Apoptosis was detected using the APC Annexin V Apoptosis Detection Kit with PI by flow cytometry. (G) Western blot analysis of apoptosis-related protein expression. (H) Subcutaneous tumors were established by injecting oeMfsd2a CT26 cells in BALB/c mice. (I) Volume and weight of subcutaneous tumors. (J) H&E staining, Mfsd2a, and Ki-67 staining of subcutaneous tumors. (*P < 0.05, **P < 0.01, ***P < 0.001,****P < 0.0001)
To determine the effects of Mfsd2a on proliferative capacity in vivo, we established a subcutaneous tumor model in BALB/c mice. Compared to the control group, the oeMfsd2a group showed a significant reduction in both tumor volume and weight (Fig. 2H, I). In addition, immunohistochemistry (IHC) staining revealed reduced Ki-67 expression in the oeMfsd2a group (Fig. 2J). Overall, both in vitro and in vivo experiments demonstrated that Mfsd2a inhibits the proliferation of CRC cells.
Mfsd2a suppresses CRC cell migration, invasion, EMT and CRLM formation
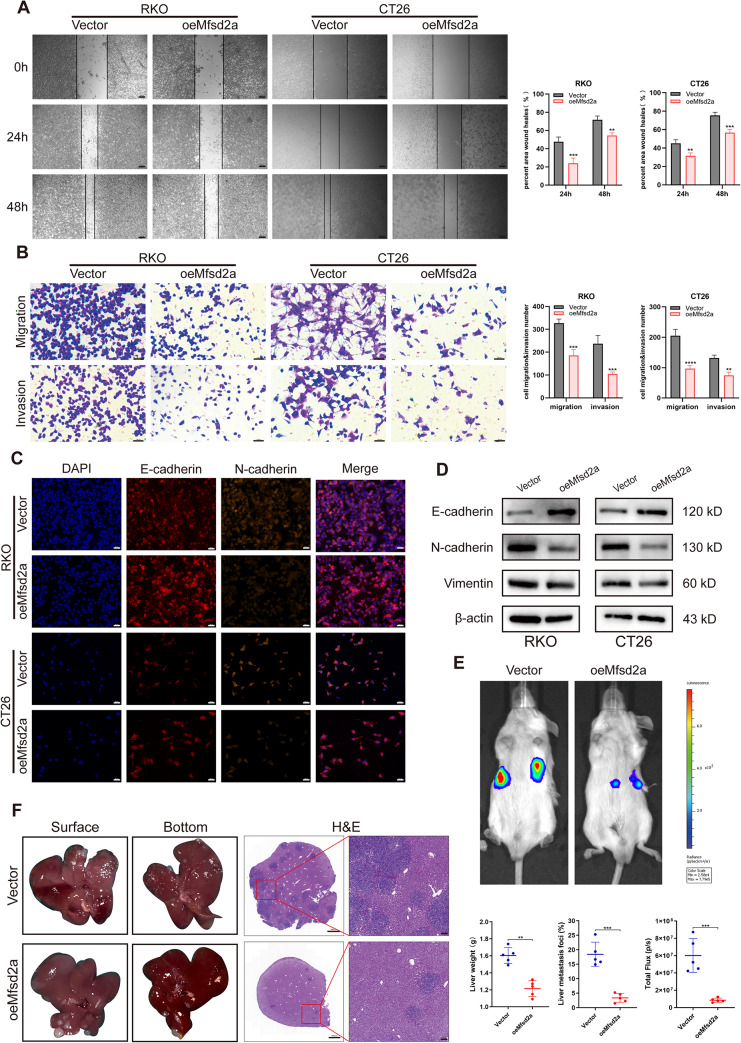
Previous studies have demonstrated that the migration and invasion enable tumor cells to overcome barriers and spread by penetrating blood vessels and tissues, playing a crucial role in the progression of CRLM [18, 19]. In order to explore the role of Mfsd2a in CRLM, we conducted follow-up studies. In wound healing and Transwell assays, oeMfsd2a RKO and CT26 cells exhibited significantly reduced migration and invasion compared to control groups (Fig. 3A, B). Given that epithelial-to-mesenchymal transition (EMT) has been critical for cancer cell migration, invasion, and metastasis [20, 21], we investigated EMT-related markers in oeMfsd2a CRC cells. Immunofluorescence (Fig. 3C) and western blot (Fig. 3D) analyses demonstrated that Mfsd2a overexpression led to increased E-cadherin and decreased N-cadherin and vimentin expression. These results indicate that Mfsd2a effectively inhibits CRC cell migration, invasion and EMT.
Fig. 3.
Mfsd2a inhibits CRC cell migration, invasion, EMT, and CRLM formation. (A,B) Wound healing assays and Transwell assays were used to evaluate the effects of Mfsd2a overexpression on RKO and CT26 cell migration and invasion. (C,D) Immunofluorescence and Western blot analyses were performed to assess the expression of EMT-related proteins. (E) Representative bioluminescence images from the CRLM model. (F) Representative images of CRLM formation and H&E staining of liver metastasis sections in the CRLM model with oeMfsd2a CT26 cells, including statistical results for liver weight and percentage of liver metastatic lesions. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
We then established a highly standardized liver metastasis mouse model using oeMfsd2a CT26/Luc cells. Compared to the control group, tumors with overexpression of Mfsd2a exhibited reduced bioluminescence intensity (Fig. 3E), as well as significantly fewer metastatic nodules and decreased liver weight (Fig. 3F). Furthermore, histological analysis of metastatic liver tissues, following fixation and H&E staining, revealed fewer and smaller metastatic lesions in the oeMfsd2a group. Accordingly, Mfsd2a inhibits the progression of CRLM in vivo.
Mfsd2a directly interacts with downstream protein S100A14
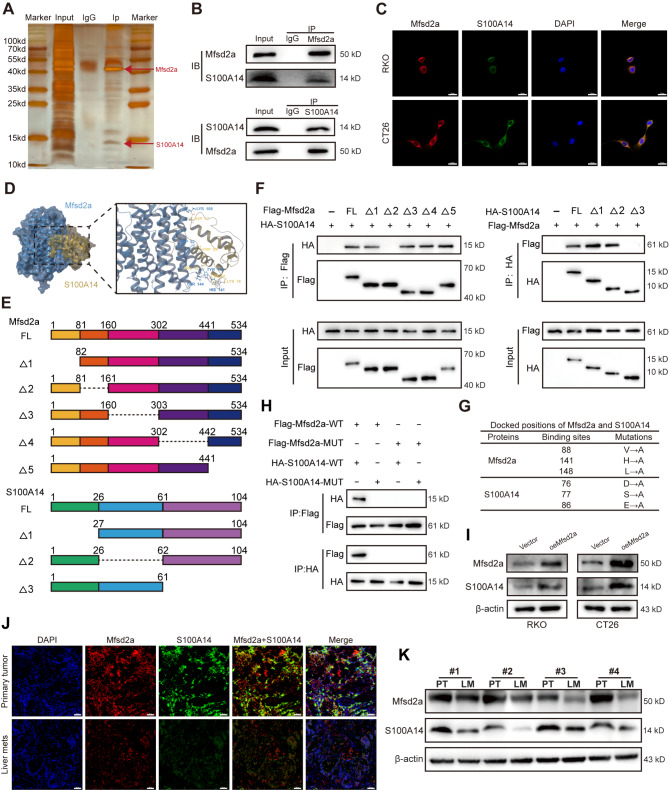
To explore how Mfsd2a inhibits CRC progression and liver metastasis, we performed silver staining and immunoprecipitation coupled with mass spectrometry (IP/MS) to identify proteins that interact with Mfsd2a in CRC cells (Fig. 4A). We identified S100 calcium-binding protein A14 (S100A14) (Fig.S2A), an S100 calcium-binding protein that inhibits tumor metastasis in various malignancies including prostate cancer, nasopharyngeal carcinoma, and gastric cancer, and is associated with EMT [22–24]. Next, Co-immunoprecipitation (Co-IP) and immunofluorescence staining confirmed the direct interaction between Mfsd2a and S100A14 (Fig. 4B, C). We used Schrodinger Software for Protein-Protein Docking analysis, and the results showed that Mfsd2a and S100A14 could bind together stably (Fig. 4D). In pursuit of determining the specific regions involved in this interaction, we first overexpressed both FLAG-tagged Mfsd2a and HA-tagged S100a14 in 293T cells, and then designed a series of truncation mutants guided by protein structure (Fig. 4E). Co-IP analysis showed that residues 82–160(△2) of Mfsd2a and residues 62–104(△3) of S100A14 were essential for Mfsd2a-S100A14 interaction (Fig. 4F). We then mutated the predicted binding sites between Mfsd2a and S100A14 (Fig. 4G). Co-IP assays suggested that the interaction between Mfsd2a and S100A14 was effectively impeded by mutations in the Mfsd2a-binding site (FLAG-Mfsd2a-MUT) or S100A14-binding site (HA-S100A14-MUT), whether introduced individually or in conjunction (Fig. 4H). In addition, the protein expression of S100A14 was significantly increased in oeMfsd2a CRC cells (Fig. 4I). However, the mRNA expression of S100A14 in CRC cells remained unchanged regardless of Mfsd2a expression (Fig.S2B). Furthermore, Western blot and immunofluorescence staining in CRLM patients demonstrated that S100A14 expression was reduced in LM compared to PT and was positively correlated with Mfsd2a expression (Fig. 4J, K). Overall, these results suggest that Mfsd2a has a positive correlation and direct interaction with S100A14.
Fig. 4.
Mfsd2a directly interacts with downstream protein S100A14. (A) Sensitive silver staining was used to identify interacting proteins. (B) Co-IP analysis demonstrated the interaction between Mfsd2a and S100A14. (C) Immunofluorescence staining showed colocalization of Mfsd2a and S100A14 in CRC cells. (D) Predicted binding model of Mfsd2a and S100A14. (E) Schematic of Mfsd2a and S100A14 in full length and truncations. (F) Immunoprecipitation and Western blot analyses illustrating interactions between FLAG-tagged truncated Mfsd2a and HA-tagged truncated S100A14 proteins in 293T cells. (G) Docked positions of Mfsd2a and S100A14 and mutations of Mfsd2a and S100A14 interaction sites. (H) Immunoprecipitation and Western blot analyses showing interactions between FLAG-tagged mutated Mfsd2a and HA-tagged mutated S100A14 in 293T cells. (I) S100A14 protein expression in oeMfsd2a CRC cells. (J,K) Immunofluorescence and Western blot analyses showed a positive correlation between Mfsd2a and S100A14 protein levels in CRLM patients
Mfsd2a inhibits malignant progression in CRC cells by targeting S100A14
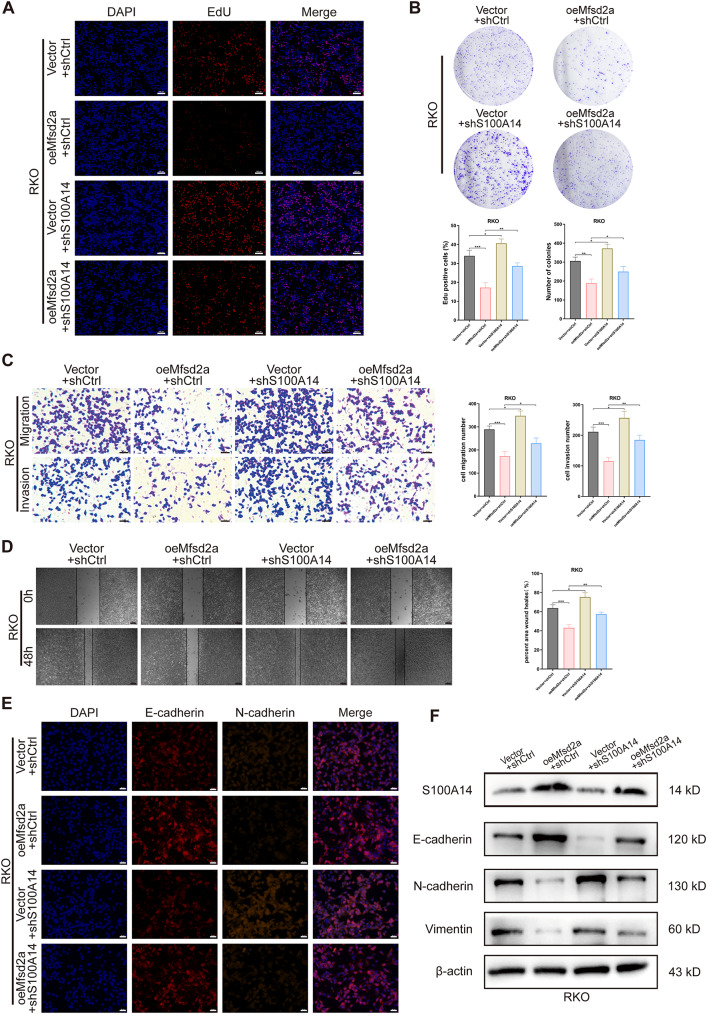
To confirm that S100A14 is a downstream target of Mfsd2a, we performed rescue experiments in CRC cells. First, we knocked down S100A14 in oeMfsd2a CRC cells. EdU and colony formation assay showed that S100A14 knockdown partially restored the proliferative capacity in oeMfsd2a CRC cells (Fig.S3A, B;Fig. 5A, B). Meanwhile, knockdown of S100A14 antagonized the pro-apoptotic effect of oeMfsd2a CRC cells (Fig.S3C, D). In addition, the results of wound healing, Transwell and EMT experiments showed that Mfsd2a overexpression inhibited CRC cells invasion and migration and impaired the EMT process, but knockdown of S100A14 partially reversed these results (Fig.S3E-H, Fig. 5C-F). Collectively, these results suggest that Mfsd2a inhibits CRC cells progression in vitro by upregulating S100A14.
Fig. 5.
S100A14 knockdown rescues the inhibitory effects of oeMfsd2a RKO cells proliferation, invasion and EMT. (A,B) EdU and colony formation assays assessed changes in cell proliferation following S100A14 knockdown in oeMfsd2a RKO cells. (C,D) Transwell and wound healing assays validated that S100A14 knockdown partially antagonized the effects of Mfsd2a overexpression on RKO cells migration and invasion. (E,F) Immunofluorescence and Western blot analyses measured the expression of S100A14 and EMT-related proteins in different groups. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
Mfsd2a inhibition of tumor growth and CRLM progression in vivo is dependent on S100A14
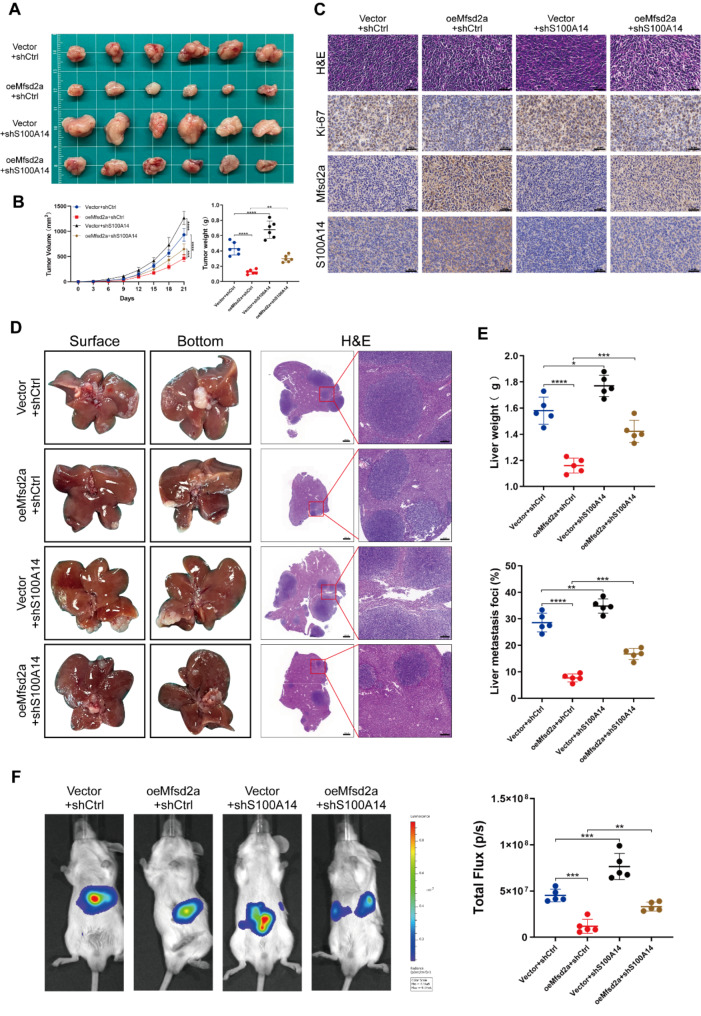
To further validate the regulatory effect of Mfsd2a on S100A14, we conducted experiments in vivo. In subcutaneous tumor models, Mfsd2a suppressed tumor growth, but this suppression was reversed by S100A14 knockdown (Fig. 6A, B). IHC staining results showed a higher expression of Ki-67 in the oeMfsd2a + shS100A14 group compared to the oeMfsd2a + shCtrl group (Fig. 6C). Accordingly, in liver metastasis models, knockdown of S100A14 partially reversed the effect of oeMfsd2a on liver metastatic tumor formation in mice (Fig. 6D-F). These results suggest that Mfsd2a suppresses tumor growth and CRLM progression in vivo through S100A14.
Fig. 6.
Mfsd2a inhibition of tumor growth and liver metastasis in vivo depends on S100A14. (A,B) Subcutaneous tumor models established with CT26 cells from different groups reveal the rescuing effect of S100A14 on Mfsd2a in vivo. (C) H&E, Ki-67, Mfsd2a, and S100A14 staining of subcutaneous tumors. (D,E) Representative images of the constructed CRLM model and H&E staining of liver metastasis sections from CT26 cells in different groups, including corresponding liver weights and percentage of metastatic lesions. (F) Representative bioluminescence images of CRLM models from different groups. (*P < 0.05, **P < 0.01, ***P < 0.001)
Mfsd2a inhibits CRLM through the S100A14/STAT3 axis
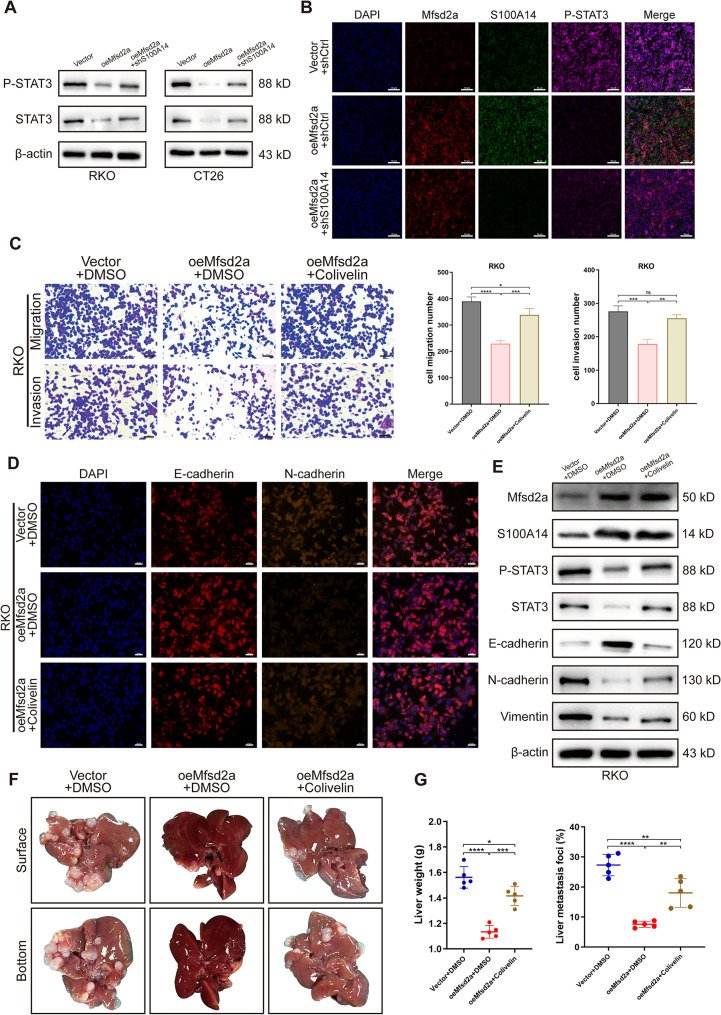
As widely recognized, the signal transducer and activator of transcription 3 (STAT3) is closely associated with the progression, metastasis, and EMT of various cancers, including CRC [25–28]. Recent research has revealed that STAT3 overexpression drives the initiation, progression, metastasis, and recurrence of CRC, with elevated phosphorylated STAT3 (P-STAT3) levels strongly correlated with poor prognosis in patients [29, 30]. Additionally, our previous studies revealed that activation of the IL-6/STAT3 pathway significantly contributes to postoperative recurrence in CRLM patients [31]. Moreover, it has been shown that S100A14 induces STAT3 degradation and inhibits its phosphorylation level in CRC cells [32]. Consequently, we examined the changes in STAT3 and P-STAT3 protein expression levels in CRC cells. Surprisingly, we found that the expression of both STAT3 and P-STAT3 was significantly reduced in oeMfsd2a CRC cells compared to the control group, while knockdown of S100A14 led to increased expression levels of both (Fig. 7A). Furthermore, immunofluorescence staining of liver tissues from CRLM mouse model showed similar results. (Fig. 7B).
Fig. 7.
Mfsd2a inhibits CRLM via the S100A14/STAT3 axis. (A) Protein expression levels of STAT3 and P-STAT3 were assessed in CRC cells co-transfected with Mfsd2a overexpression and/or S100A14 knockdown. (B) Immunofluorescence intensity of Mfsd2a, S100A14, and P-STAT3 was evaluated in CRLM models established with CT26 cells. (C) Transwell assays demonstrated that colivelin rescued the inhibitory effects of Mfsd2a overexpression on CRC cells migration and invasion. (D) Immunofluorescence analysis was performed to examine the expression of EMT-related proteins in CRC cells. (E) Western blot analysis was conducted to determine the expression levels of Mfsd2a, S100A14, and EMT-related proteins in CRC cells. (F,G) CRLM models showed that colivelin partially counteracted the inhibitory effect of Mfsd2a on liver metastasis formation. Mice were intraperitoneally injected with 1 mg/kg colivelin or DMSO every other day for 2 weeks. (*P < 0.05, **P < 0.01, ***P < 0.001,****P < 0.0001)
We further validated the role of STAT3 in regulating Mfsd2a-mediated CRC cell metastasis by using the STAT3 activator colivelin. In vitro experiments demonstrated that colivelin (10 µM) partially reversed the inhibitory effects of Mfsd2a overexpression on CRC cell migration, invasion, and EMT progression (Fig. 7C-E). Moreover, intraperitoneal injection of colivelin (1 mg/kg) partially antagonized the inhibitory effect of Mfsd2a overexpression on CRLM formation (Fig. 7F, G). These results reveal that Mfsd2a inhibits CRLM through the S100A14/STAT3 axis.
Discussion
Colorectal cancer (CRC) is the third most common malignant tumor worldwide [33, 34], with the liver being the most frequent site of metastasis and a major cause of mortality [2, 35]. Therefore, identifying new targets of colorectal cancer metastasis (CRLM) and understanding the specific mechanisms of CRLM progression are crucial for the treatment of CRLM. In our study, we found that Mfsd2a was downregulated in CRLM and strongly correlated with postoperative prognosis in CRLM patients. Experiments in vitro and vivo showed that Mfsd2a inhibits progression of CRC and CRLM. Then, we identified S100 calcium-binding protein A14 (S100A14) as an interacting downstream target of Mfsd2a. Mechanistic investigations indicated that Mfsd2a modulates the phenotype of CRC cells and CRLM formation by inhibiting STAT3 expression and phosphorylation through upregulation of S100A14. Moreover, a STAT3 activator colivelin partially neutralizes the inhibitory effects of Mfsd2a. These results suggest that Mfsd2a plays a pivotal role in the development of CRLM and may serve as a potential prognostic marker for CRLM.
The Major Facilitator Superfamily (MFS) is one of the largest transporter families, comprising over 70 members [36]. The major facilitator superfamily domain containing protein-2a (Mfsd2a), located on chromosome 1p34.2, is a member of the MFS. It shares structural similarities with bacterial N/melibiose transporters and features a 12-transmembrane α-helix domain structure [37]. While previous studies have emphasized its critical role in lipid transport and maintenance of the blood-brain barrier [8, 38], emerging research has highlighted that Mfsd2a plays diverse roles in various cancers. Mfsd2a exhibits tumor-suppressive functions by blocking the cell cycle of lung cancer cells and impairing their adhesion and migration properties [11]. It also reprograms the tumor microenvironment to activate T cell responses, thereby enhancing the efficacy of anti-PD-1 immunotherapy in gastric cancer [15]. High expression of Mfsd2a predicts favorable prognosis in both gastric cancer and hepatocellular carcinoma [13, 14]. Interestingly, despite the TIMER2.0 database revealed no statistically significant difference of Mfsd2a expression in CRC tissues, prognosis data indicate that expression level of Mfsd2a positively correlates with prognosis of CRC patients. Given the significant impact of CRLM on patient survival, we further investigated the differential expression of Mfsd2a in CRLM. Our results revealed that Mfsd2a expression is markedly lower in liver metastases compared to primary CRC tissues. We observed that Mfsd2a inhibits CRC cell proliferation, migration, invasion, and EMT in vitro, and suppresses CRLM development in mice. These findings suggest that Mfsd2a may act as a metastasis suppressor in CRLM.
To further investigate the molecular mechanisms by which Mfsd2a inhibits CRLM progression, we performed IP/MS and identified S100A14 as a downstream binding partner of Mfsd2a. Molecular docking analysis predicted the interaction mode between Mfsd2a and S100A14. S100A14, a member of the S100 family, is involved in various intracellular functions, including cell proliferation, apoptosis, migration, and invasion [39]. Previous research has shown that S100A14 inhibits proliferation and invasion in oral cancer [40, 41], metastasis in nasopharyngeal and gastric cancers [23, 24], and cell growth and EMT in prostate cancer through the Hippo signaling pathway [22]. Additionally, increased S100A14 expression is associated with poor clinical outcomes in ovarian, breast, and cervical cancers [42], whereas decreased expression correlates with adverse prognosis and disease progression in colorectal, small intestine, gastric, and esophageal cancers [42–45]. In CRC, S100A14 plays a critical role involved in cell motility, adhesion, and growth changes [46], and its expression level is inversely correlated with CRC metastatic potential [44]. Our research reveals that Mfsd2a interacts with S100A14 in CRC cells and upregulates of S100A14. Ectopic knockdown of S100A14 partially reverses the effect of oeMfsd2a on inhibiting proliferation, migration, invasion, EMT, and CRLM progression. These findings suggest that Mfsd2a exerts its inhibitory effects on CRC progression and liver metastasis through S100A14.
The STAT3 signaling pathway is widely recognized as a key oncogenic driver associated with the onset and progression of CRC [47]. Recent research has shown that overexpression of STAT3 initiates CRC and promotes its progression, metastasis and recurrence, with elevated levels of P-STAT3 are closely associated with poor prognosis in CRC patients [29, 30]. Additionally, our previous studies found that activation of the IL-6/STAT3 pathway promotes postoperative recurrence in CRLM patients [31]. STAT3 activation is typically regulated through proteasomal degradation and upstream signaling molecules [48], and targeting STAT3 has shown to inhibit tumor growth and metastasis in vitro and in vivo [49]. Previous studies have also indicated that S100A14 induces STAT3 degradation and inhibits its phosphorylation in CRC cell lines [32]. Accordingly, our experiments reveal that oeMfsd2a inhibits the levels of STAT3 and P-STAT3 in CRC cell lines, and P-STAT3 expression is significantly reduced in the oeMfsd2a group of the CRLM mouse compared to the control group. Furthermore, treatment with the STAT3 activator colivelin partially antagonized the inhibitory effects of Mfsd2a on CRC cells both in vitro and in vivo.
Conclusion
In summary, our findings provide the novel evidence that Mfsd2a acts as a potent inhibitor of colorectal liver metastasis (CRLM) and functions as a biomarker for favorable prognosis in CRLM patients. Mechanistically, Mfsd2a suppresses the initiation and progression of CRLM by interacting with S100A14 to inhibit STAT3 phosphorylation. Our findings may provide evidences to explore novel therapeutic strategy targeting Mfsd2a for CRLM patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Jiangsu Province Hospital Core Facility Center (http://yqgx.jsph.org.cn) for the technical support and Jiangsu Provincial Medical Innovation Center, Jiangsu Provincial Medical Key Laboratory. Some cartoon components were from FigDraw (www.figdraw.com) for model drawing.
Abbreviations
- CRC
Colorectal cancer
- CRLM
Colorectal liver metastasis
- Mfsd2a
Major facilitator superfamily domain containing protein-2a
- S100A14
S100 calcium-binding protein A14
- STAT3
Signal transducer and activator of transcription 3
- EMT
Epithelial-to-mesenchymal transition
- OS
Overall survival
- PFS
Progression-free survival
- CRS
Clinical risk scores
- H&E
Hematoxylin and eosin
- IHC
Immunohistochemistry
- IF
Immunofluorescence
- Co-IP
Co-immunoprecipitation
- IP/MS
Immunoprecipitation coupled with mass spectrometry
- FBS
Fetal bovine serum
- DMSO
Dimethyl sulfoxide
Author contributions
Linfeng Sun: Investigation, Formal analysis, Data curation, Conceptualization, Writing – original draft. Xiangdong Li: Investigation, Formal analysis, Data curation, Conceptualization, Writing – original draft. Yuhao Xiao: Investigation, Formal analysis, Data curation. Wenjie Yu: Data curation, Formal analysis, Investigation. Xuyang Chen: Investigation, Formal analysis, Data curation. Ziyi Wang: Investigation, Formal analysis, Data curation. Nan Xia: Investigation, Formal analysis, Data curation. Xuejiao Chen: Investigation, Formal analysis, Data curation. Minhao Chen: Data curation. Haoliang Zhu: Data curation. Jie Li: Data curation. Jie Wei: Data curation. Sheng Han: Resources, Writing – original draft. Liyong Pu: Writing – review & editing, Writing – original draft, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Funding
This project was supported by grants from the National Natural Science Foundation of China (NSFC) (Grant Nos. 81870443 and 81273261) and Jiangsu Provincial Medical Innovation Center (ZDXYS202201), Jiangsu Provincial Medical Key Laboratory (CXZX202203).
Data availability
The GSE213402 dataset was downloaded from the GEO datasets of human patients with CRLM from the National Center for Biotechnology Information (NCBI) database. All data, analytic methods, and study materials will be made available to other researchers upon reasonable request.
Declarations
Ethics approval and consent to participate
This study was conducted under the approval and supervision of the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University in accordance with the Declaration of Helsinki (2024-SR-526). Written consent was obtained from all participants. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University (IACUC-2306050).
Consent for publication
All authors agreed on the manuscript.
Competing interests
The authors have no competing interests to declare.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Linfeng Sun, Xiangdong Li, Yuhao Xiao and Wenjie Yu are first authors and contributed equally to this work.
Contributor Information
Sheng Han, Email: hanshengss@163.com.
Liyong Pu, Email: puliyong@njmu.edu.cn.
References
- 1.Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal Cancer: a review. JAMA. 2021;325:669–85. 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Nordlinger B, Cervantes A, Group EG. W. Advanced colorectal cancer: ESMO Clinical Practice guidelines for treatment. Ann Oncol. 2010;21(5):v93–97. 10.1093/annonc/mdq222. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Jones RP, et al. Colorectal liver metastases: a critical review of state of the art. Liver Cancer. 2016;6:66–71. 10.1159/000449348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsilimigras DI, et al. Liver metastases. Nat Rev Dis Primers. 2021;7:27. 10.1038/s41572-021-00261-6. [DOI] [PubMed] [Google Scholar]
- 6.Reboux N, et al. Incidence and survival in Synchronous and Metachronous Liver metastases from Colorectal Cancer. JAMA Netw Open. 2022;5:e2236666. 10.1001/jamanetworkopen.2022.36666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Liver metastasis from colorectal cancer: pathogenetic development, immune landscape of the tumour microenvironment and therapeutic approaches. J Exp Clin Cancer Res. 2023;42:177. 10.1186/s13046-023-02729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Zvi A, et al. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 2014;509:507–11. 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger JH, Charron MJ, Silver DL. Major facilitator superfamily domain-containing protein 2a (MFSD2A) has roles in body growth, motor function, and lipid metabolism. PLoS ONE. 2012;7:e50629. 10.1371/journal.pone.0050629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spinola M, et al. MFSD2A is a novel lung tumor suppressor gene modulating cell cycle and matrix attachment. Mol Cancer. 2010;9:62. 10.1186/1476-4598-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwary S, et al. Metastatic brain tumors disrupt the blood-brain barrier and alter lipid metabolism by inhibiting expression of the endothelial cell fatty acid transporter Mfsd2a. Sci Rep. 2018;8:8267. 10.1038/s41598-018-26636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing S, et al. The prognostic value of major facilitator superfamily domain-containing protein 2A in patients with hepatocellular carcinoma. Aging. 2019;11:8474–83. 10.18632/aging.102333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi X, Huang Y, Wang H, Zheng W, Chen S. MFSD2A expression predicts better prognosis in gastric cancer. Biochem Biophys Res Commun. 2018;505:699–704. 10.1016/j.bbrc.2018.09.156. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, et al. MFSD2A potentiates gastric cancer response to anti-PD-1 immunotherapy by reprogramming the tumor microenvironment to activate T cell response. Cancer Commun (Lond). 2023;43:1097–116. 10.1002/cac2.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ungaro F et al. MFSD2A Promotes Endothelial Generation of Inflammation-Resolving Lipid Mediators and Reduces Colitis in Mice. Gastroenterology 153, 1363–1377 e1366, 10.1053/j.gastro.2017.07.048 (2017). [DOI] [PubMed]
- 17.Liu QL, Zhou H, Zhou ZG, Chen H. N. colorectal cancer liver metastasis: genomic evolution and crosstalk with the liver microenvironment. Cancer Metastasis Rev. 2023;42:575–87. 10.1007/s10555-023-10107-0. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, et al. Th17 cells secrete TWEAK to trigger epithelial-mesenchymal transition and promote colorectal Cancer Liver Metastasis. Cancer Res. 2024;84:1352–71. 10.1158/0008-5472.CAN-23-2123. [DOI] [PubMed] [Google Scholar]
- 19.Zarour LR, et al. Colorectal Cancer Liver Metastasis: evolving paradigms and future directions. Cell Mol Gastroenterol Hepatol. 2017;3:163–73. 10.1016/j.jcmgh.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavert N. Ben-Ze’ev, A. Epithelial-Mesenchymal transition and the invasive potential of tumors. Trends Mol Med. 2008;14:199–209. 10.1016/j.molmed.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Fan M, et al. MicroRNA-30b-5p functions as a metastasis suppressor in colorectal cancer by targeting Rap1b. Cancer Lett. 2020;477:144–56. 10.1016/j.canlet.2020.02.021. [DOI] [PubMed] [Google Scholar]
- 22.Jiang S, et al. S100A14 inhibits cell growth and epithelial-mesenchymal transition (EMT) in prostate cancer through FAT1-mediated Hippo signaling pathway. Hum Cell. 2021;34:1215–26. 10.1007/s13577-021-00538-8. [DOI] [PubMed] [Google Scholar]
- 23.Meng DF, et al. S100A14 suppresses metastasis of nasopharyngeal carcinoma by inhibition of NF-kB signaling through degradation of IRAK1. Oncogene. 2020;39:5307–22. 10.1038/s41388-020-1363-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhu M, et al. Calcium-binding protein S100A14 induces differentiation and suppresses metastasis in gastric cancer. Cell Death Dis. 2017;8:e2938. 10.1038/cddis.2017.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G, Hou S, Li S, Wang Y, Cui W. Role of STAT3 in cancer cell epithelial–mesenchymal transition (review). Int J Oncol. 2024;64. 10.3892/ijo.2024.5636. [DOI] [PMC free article] [PubMed]
- 26.He T, Cui J, Wu Y, Sun X, Chen N. Knockdown of TRIM66 inhibits cell proliferation, migration and invasion in colorectal cancer through JAK2/STAT3 pathway. Life Sci. 2019;235:116799. 10.1016/j.lfs.2019.116799. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Chu D, Kawamura T, Tanaka K, He S. GRIM-19 repressed hypoxia-induced invasion and EMT of colorectal cancer by repressing autophagy through inactivation of STAT3/HIF-1alpha signaling axis. J Cell Physiol. 2019;234:12800–8. 10.1002/jcp.27914. [DOI] [PubMed] [Google Scholar]
- 28.Zhou K, et al. RAC1-GTP promotes epithelial-mesenchymal transition and invasion of colorectal cancer by activation of STAT3. Lab Invest. 2018;98:989–98. 10.1038/s41374-018-0071-2. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Luo X, Li H, Deng L, Wang Y. Genome-wide uncovering of STAT3-mediated miRNA expression profiles in colorectal cancer cell lines. Biomed Res Int. 2014;2014(187105). 10.1155/2014/187105. [DOI] [PMC free article] [PubMed]
- 30.Qin Y, et al. Laminin 521 enhances self-renewal via STAT3 activation and promotes tumor progression in colorectal cancer. Cancer Lett. 2020;476:161–9. 10.1016/j.canlet.2020.02.026. [DOI] [PubMed] [Google Scholar]
- 31.Li X, et al. Hepatocyte SGK1 activated by hepatic ischemia-reperfusion promotes the recurrence of liver metastasis via IL-6/STAT3. J Transl Med. 2023;21:121. 10.1186/s12967-023-03977-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min HY, et al. S100A14: a novel negative regulator of cancer stemness and immune evasion by inhibiting STAT3-mediated programmed death-ligand 1 expression in colorectal cancer. Clin Transl Med. 2022;12:e986. 10.1002/ctm2.986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu B, et al. Colorectal cancer incidence and mortality: the current status, temporal trends and their attributable risk factors in 60 countries in 2000–2019. Chin Med J (Engl). 2021;134:1941–51. 10.1097/CM9.0000000000001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegel RL, et al. Global patterns and trends in colorectal cancer incidence in young adults. Gut. 2019;68:2179–85. 10.1136/gutjnl-2019-319511. [DOI] [PubMed] [Google Scholar]
- 35.Page AJ, Cosgrove DC, Herman JM, Pawlik TM. Advances in understanding of colorectal liver metastasis and implications for the clinic. Expert Rev Gastroenterol Hepatol. 2015;9:245–59. 10.1586/17474124.2014.940897. [DOI] [PubMed] [Google Scholar]
- 36.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH. Jr. The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279:2022–35. 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angers M, Uldry M, Kong D, Gimble JM, Jetten AM. Mfsd2a encodes a novel major facilitator superfamily domain-containing protein highly induced in brown adipose tissue during fasting and adaptive thermogenesis. Biochem J. 2008;416:347–55. 10.1042/BJ20080165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen LN, et al. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature. 2014;509:503–6. 10.1038/nature13241. [DOI] [PubMed] [Google Scholar]
- 39.Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60:540–51. 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- 40.Sapkota D, et al. S100A14 inhibits proliferation of oral carcinoma derived cells through G1-arrest. Oral Oncol. 2012;48:219–25. 10.1016/j.oraloncology.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Sapkota D, et al. S100A14 regulates the invasive potential of oral squamous cell carcinoma derived cell-lines in vitro by modulating expression of matrix metalloproteinases, MMP1 and MMP9. Eur J Cancer. 2011;47:600–10. 10.1016/j.ejca.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Basnet S, Sharma S, Costea DE, Sapkota D. Expression profile and functional role of S100A14 in human cancer. Oncotarget. 2019;10:2996–3012. 10.18632/oncotarget.26861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diamantopoulou A et al. A Clinicopathological Analysis of S100A14 Expression in Colorectal Cancer. In Vivo 34, 321–330, 10.21873/invivo.11777 (2020). [DOI] [PMC free article] [PubMed]
- 44.Wang HY, et al. Expression status of S100A14 and S100A4 correlates with metastatic potential and clinical outcome in colorectal cancer after surgery. Oncol Rep. 2010;23:45–52. [PubMed] [Google Scholar]
- 45.Li X, Ding F, Wang L, Chen H, Liu Z. Disruption of enhancer-driven S100A14 expression promotes esophageal carcinogenesis. Cancer Lett. 2022;545:215833. 10.1016/j.canlet.2022.215833. [DOI] [PubMed] [Google Scholar]
- 46.Hashida H, Coffey RJ. Significance of a calcium-binding protein S100A14 expression in colon cancer progression. J Gastrointest Oncol. 2022;13:149–62. 10.21037/jgo-21-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gargalionis AN, Papavassiliou KA, Papavassiliou AG. Targeting STAT3 signaling pathway in Colorectal Cancer. Biomedicines. 2021;9. 10.3390/biomedicines9081016. [DOI] [PMC free article] [PubMed]
- 48.Swiatek-Machado K, Kaminska B. STAT signaling in Glioma cells. Adv Exp Med Biol. 2020;1202:203–22. 10.1007/978-3-030-30651-9_10. [DOI] [PubMed] [Google Scholar]
- 49.Niu G, et al. Gene therapy with dominant-negative Stat3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res. 1999;59:5059–63. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GSE213402 dataset was downloaded from the GEO datasets of human patients with CRLM from the National Center for Biotechnology Information (NCBI) database. All data, analytic methods, and study materials will be made available to other researchers upon reasonable request.