Abstract
Background
The aim of this study was to establish a primary mouse gastric carcinoma cell line.
Methods
Gastric adenocarcinoma in the body region was induced in immunocompetent BALB/c mice using N-Methyl-N-nitrosourea and a 2% NaCl solution. Fresh gastric cancer tissue samples were cultured in 1640 medium supplemented with 10% fetal bovine serum for primary culture and subculture. Cellular morphology was assessed via light microscopy, and a cell growth curve was established. Genomic and proteomic analyses were conducted to characterize the molecular features of the cell lines. This cell line demonstrated a 100% success rate in forming subcutaneous tumors in BALB/c mice. By integrating proteomic profiles from clinical gastric cancer patients and the murine subcutaneous tumor model, several molecular targets suitable for preclinical investigation were identified. Trametinib, a MEK inhibitor, was employed as a model compound in our preclinical study.
Results
A novel gastric carcinoma cell line, designated MCC, was established from BALB/c mice. This cell line exhibited a doubling time of approximately 33 h. Genomic and proteomic analyses identified mutations frequently observed in clinical gastric cancer patients, such as Kras, Egfr, and Ccnd3. Additionally, MCC overexpresses proteins, including SLC1A5, MCM6, and ITGA2, which are significantly upregulated in gastric cancer tissues compared to adjacent non-cancerous tissues. The MCC cell line demonstrated stable tumorigenicity in immunocompetent BALB/c mice, forming subcutaneous tumors that closely resemble the proteomic profile of clinical gastric cancer samples. This high concordance facilitated the identification of several potential therapeutic targets for gastric cancer. Preclinical studies with trametinib revealed that treatment effectively inhibited gastric cancer growth, likely mediated through the activation of immune cells, particularly neutrophils and T cells.
Conclusions
The MCC cell line serves as an indispensable model for gastric cancer research, offering a robust platform for investigating tumor development and progression. Its exceptional tumorigenic capacity and strong concordance with clinical proteomic profiles underscore its significance in translational research, facilitating the discovery of novel therapeutic targets and elucidation of molecular pathways critical for developing effective treatment strategies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-024-03633-6.
Keywords: Gastric carcinoma, MCC, Genomic, Proteomic, Trametinib
Background
In 2022, approximately 970,000 new cases of gastric cancer were reported worldwide, ranking it as the fifth most common and the fifth deadliest malignancy globally. The incidence rate in males is twice that of females [1]. Gastric cancer exhibits a high prevalence in East Asia, with China alone accounting for about 360,000 new cases annually, representing nearly 37% of the global incidence [2]. Furthermore, gastric cancer is characterized by significant heterogeneity [3]. Most patients are asymptomatic in the early stages, resulting in late-stage diagnosis when the prognosis is poor, with a 5-year relative survival rate of only 6% [4]. The current clinical treatment options for gastric cancer are limited, primarily involving surgical resection combined with chemotherapy, which has a low efficacy rate. In the realm of targeted therapy, only trastuzumab, targeting ERBB2/HER2, and ramucirumab, targeting VEGFR2, have received FDA approval for the treatment of gastric cancer, which is less than 10% of the gastric cancer patient population [5]. In recent years, the emergence of immunotherapy has shown promise, with pembrolizumab demonstrating exceptional efficacy in gastric cancer patients with high PD-L1 expression. Nevertheless, the proportion of patients with this target remains low [6]. Effective biomarkers and therapeutic targets for the clinical treatment of gastric cancer are still lacking.
With the rise of precision medicine, the identification of biomarkers has facilitated the stratification of patients, thereby enabling the adoption of tailored diagnostic and therapeutic strategies [7]. Compared to the traditional pathology-based Lauren classification system for gastric cancer [8], molecular subtyping through omics approaches such as genomics [9], transcriptomics [10], proteomics [11, 12], and single-cell transcriptomics [13, 14] offers potential therapeutic targets by delineating the characteristics of different patient subtypes. The collection and analysis of large-scale clinical data have established a robust foundation for the application of big data in clinical treatment, while also presenting further challenges. These potential biological targets necessitate extensive preclinical research. To validate these targets and their biological functions, there is an urgent need to develop high-throughput, stable, and clinically relevant preclinical tumor models.
The tumor microenvironment, comprising tumor cells, stromal cells, infiltrating immune cells, blood vessels, and non-cellular components of the extracellular matrix, plays a pivotal role in tumor growth and progression through intricate interactions [15]. Studies have demonstrated that the tumor microenvironment can serve both as a predictive marker for patient treatment and prognosis, and as a therapeutic target to overcome tumor heterogeneity and genetic instability [16]. This is particularly pertinent in immunotherapy, where immune cells within the tumor microenvironment provide significant insights for the precision treatment of patients [17]. Consequently, to bridge the gap between omics-based big data and clinical application, it is essential to establish high-throughput subcutaneous models in immunocompetent mice for preclinical evaluation of potential therapeutic targets.
Approximately 90% of clinical gastric cancer cases are adenocarcinomas [18]. However, the available mouse gastric cancer cell line commonly used for modeling in immunocompetent mice is limited to the mouse forestomach carcinoma (MFC) cell line, which originates from squamous cell carcinoma of the mouse forestomach—a region not present in humans—thus lacking clinical relevance [19]. To address this limitation, we have established a gastric adenocarcinoma cell line derived from the mouse gastric body to develop syngeneic mouse models of gastric cancer, thereby providing a valuable tool for preclinical anti-tumor research. Through genomic and proteomic analyses of this cell line and the resultant subcutaneous tumors, we have characterized their molecular features, establishing a foundation for their pre-clinical applications. Utilizing this model, we conducted preclinical studies on trametinib, an inhibitor of MEK targeting Map2k1, a target overexpressed in gastric tumors compared to adjacent non-tumorous tissues. Our results demonstrate that trametinib effectively inhibits gastric cancer growth by activating immune responses within the tumor without causing adverse effects. Notably, neutrophil degranulation proteins, may play a crucial role in this anti-tumor activity.
Methods
Animals and in situ induction of mouse gastric adenocarcinoma
Eight-week-old male BALB/c mice were purchased from Beijing HFK Bioscience Co., LTD (Beijing, China) and housed under standard specific pathogen-free (SPF) conditions. Gastric adenocarcinoma was induced using N-methyl-N-nitrosourea (MNU, Toronto Research Chemicals Co., Toronto, Canada) and sodium chloride (NaCl; Beijing Chemical Works, Beijing, China). Mice received 120 ppm MNU and 2% NaCl alternately in their drinking water for one week each, over a period of 2 months. Afterward, they returned to a normal diet for 9 months. All procedures were approved by the Institutional Animal Care and Use Committee of the National Protein Science Center (Beijing Proteome Research Center).
Establishment of the MCC cell line
Gastric adenocarcinoma tissues from mice were rinsed with sterile phosphate-buffered saline (PBS) containing antibiotics, minced, and digested with 0.25% trypsin (Gibco, Thermo Fisher Scientific, USA). The resulting suspension was cultured in 1640 medium with 10% fetal bovine serum (FBS; Gibco, Grand Island, USA) under 5% CO2 at 37 °C. Monoclonal screening was performed after cell harvest, and cells were passaged over 70 generations, resulting in the mouse corpus cancer (MCC) cell line.
MCC-derived allografting mice and preclinical study
For cell-derived allograft experiments, 8-week-old male BALB/c mice were used. MCC cells (2 × 106 cells) were subcutaneously implanted into the right flank. When tumors reached approximately 100 mm3, mice were randomized into control and treatment groups (n = 5 per group). Treatment groups received either the MEK inhibitor Trametinib (1 mg/kg, p.o. daily) or anti-PDL1 antibody (200 μg/mouse, i.p. every three days), while controls received saline. Tumor volume and body weight were measured every two days. Tumor volumes were calculated as TV = (L*W2)/2. Mice were euthanized at the end of the experiment.
Collection of tumor and immune organs
On day 28 post-MCC injection, tumors from five mice were collected for analysis. On day 18 post-treatment, tumors from the anti-PD-L1 and control groups were weighed. Tumors and four immune organs (lymph nodes, spleen, thymus, bone marrow) from the trametinib and control groups were collected on day 18 post-treatment for further analysis.
H&E staining and pathological study
Tumors were fixed in formalin, embedded in paraffin, sectioned at 4 μm, and stained with hematoxylin and eosin. Slides were examined using an Olympus BX53 fluorescence microscope (Tokyo, Japan). Two experienced pathologists reviewed the sections.
Cell growth assay
MCC cells (3000 cells/well) were cultured in 96-well plates. CCK-8 solution (10 μL; Dojindo Molecular Technologies, USA) was added at various time points, and absorbance was measured at 450 nm using a microplate reader (Biorad).
Whole exome sequencing (WES)
Genomic DNA from MCC cells was extracted, processed, and sequenced on the Illumina HiSeq 2000 platform. Raw reads were filtered to remove low-quality data.
Extraction and digestion of proteome
Cells and tumors were subjected to protein extraction in lysis buffer (1% sodium deoxycholate, 10 mM Tris(2-carboxyethyl) phosphine, 40 mM 2-chloroacetamide, and 100 mM Tris–HCl pH 8.8). After heating at 95 °C for 5 min and sonicating for 5 min (3 s on and 3 s off, amplitude 30%), the tissue lysates were centrifuged at 16,000 g for 10 min at 4 °C, and the supernatants were collected as whole tissue extract (WTE). One hundred micrograms of protein (the protein concentration determined by Thermo Nanodrop One) was digested overnight with trypsin (Promega, USA) at 37 °C, and the digestion was stopped by formic acid at a final concentration of 1%. Precipitated sodium deoxycholate was removed by centrifugation at 4 °C with 16,000 g for 10 min. The supernatants were collected, desalted, vacuum-dried, and stored at − 80 °C until subsequent liquid chromatography tandem mass spectrometry (LC–MS/MS) analysis.
LC–MS/MS analysis
Dried peptide samples were redissolved in Solvent A (0.1% formic acid in water) and loaded onto a trap column (100 μm × 2 cm, homemade; particle size, 1.9 μm; pore size, 120 Å; SunChrom, USA) with a maximum pressure of 280 bar using Solvent A, then separated on a home-made 150 μm × 30 cm silica microcolumn (particle size, 1.9 μm; pore size, 120 Å; SunChrom, USA) with a gradient of 5–35% mobile phase B (80% acetonitrile and 0.1% formic acid) at a flow rate of 600 nL/min for 120 min, followed by a wash for 10 min using 95% mobile phase B (0.1% formic acid in acetonitrile). The eluted peptides were ionized under 2 kV. MS was operated under a data-dependent acquisition (DDA) mode. For detection with Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific, USA). The most intense ions selected under top-speed mode were isolated in the Quadrupole with a 1.6 m/z window and fragmented by higher energy collisional dissociation (HCD) with normalized collision energy of 35%, then measured in the linear ion trap using the rapid ion trap scan rate. Automatic gain control targets were 5 × 105 ions with a maximum injection time of 50 ms for full scans and 5 × 103 with 35 ms for MS/MS scans. Dynamic exclusion time was set at 18 s.
MS data processing
MS data were processed using Firmiana [20]. Data were searched against the NCBI RefSeq mouse proteome database using Mascot 2.3, with a false discovery rate (FDR) of 1%. Protein abundance was represented as the fraction of total (FOT), and missing values were substituted with zeros.
Bioinformatics and statistical analysis
Principal component analysis (PCA) and Student's t-test identified differentially expressed proteins (DEPs). Pathway enrichment analysis was conducted using STRING, with results considered significant at FDR < 0.05. Immune cell analysis was performed using CIBERSORT. Data visualization was done using R packages such as “ggplot2”, “dplyr”, and “pheatmap”.
Data availability
The proteomics data, the spectral library, and details regarding all identified proteins have been submitted to the ProteomeXchange Consortium (https://www.iprox.cn/).
Results
Establishment of a new mouse gastric cancer cell line (MCC)
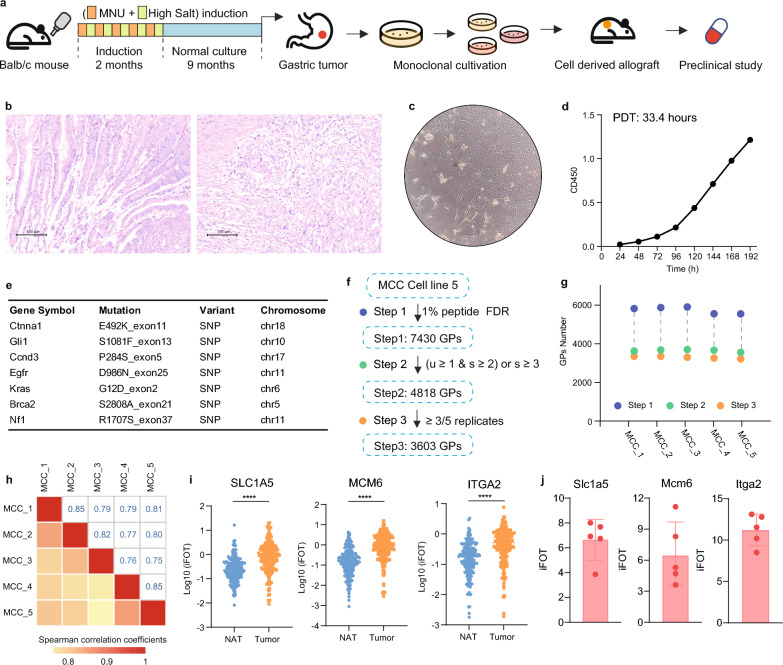
We induced gastric corpus adenocarcinoma in BALB/c mice, which are characterized by their intact immune systems, through the implementation of a protocol that involved alternating administration of 120 ppm N-Methyl-N-nitrosourea (MNU), an N-nitroso compound and a direct-acting alkylating agent, and 2% NaCl solution in the drinking water over a period of 2 months, with each solution given continuously for one week, followed by a maintenance phase of 9 months with a standard diet (Fig. 1a). The tumors in the gastric corpus were independently evaluated by two pathologists and diagnosed as adenocarcinomas (Fig. 1b). To facilitate scalable preclinical studies, we collected gastric adenocarcinoma tissue blocks and cultured them in a 10% fetal bovine serum (FBS) 1640 medium after digestion with 0.25% trypsin. Subsequently, we performed monoclonal screening and propagated the cells for more than 70 generations, resulting in the establishment of a cell line derived from mouse corpus adenocarcinoma, named MCC (mouse corpus carcinoma). Microscopic observation revealed that the majority of MCC cells exhibited spindle and elliptical shapes, with a minority displaying a polygonal morphology, and grew as adherent monolayers with epithelial characteristics (Fig. 1c). The population doubling time (PDT) was determined to be 33.4 h, indicating that the in vitro growth capacity of MCC cells can be sustained (Fig. 1d).
Fig. 1.
Establishment of a new mouse carcinoma gastric cancer cell line. a Schematic representation of the establishment process for the mouse corpus carcinoma (MCC) cell line. b Pathological H&E staining of normal corpus tissue compared to orthotopic gastric corpus adenocarcinoma specimens. c Morphological characteristics of the MCC cell line. d Growth curves demonstrating the proliferation rate of MCC cells. e Gene mutations identified in the MCC cell line. f Proteomic data sets of the MCC cell line using various filtering criteria. g Total number of proteins identified in each biological replicate of MCC cell lines. h Correlation analysis across five biological replicates of MCC cell lines. i Protein expression levels of SLC1A5, MCM6, and ITGA2 in gastric cancer tissues (tumor) and adjacent normal tissues (NAT) from patients with gastric adenocarcinoma. j The relative abundance (iFOT) of Slc1a5, Mcm6, and Itga2 in MCC cell lines
Genomic analysis of MCC was conducted, incorporating data from The Cancer Genome Atlas (TCGA)9, cBio cancer genomics portal (cBioPortal) [21], and Catalogue of Somatic Mutations in Cancer (COSMIC) [22] databases to analyze mutation profiles in clinical gastric cancer patients. We found that MCC harbors various mutations present in clinical gastric cancer patients, including Kras, Egfr, and Ccnd3 (Fig. 1d, supplementary tables S1). To better understand the biological property of the MCC cell line, we further performed proteomic analysis on five biologically parallel MCC cell lines. A total of 7430 proteins were detected, and after rigorous quality control, 4818 high-confidence proteins were selected, with 3603 proteins detectable in at least three out of five samples (Fig. 1f, supplementary tables S2). Each sample detected approximately 3300 proteins (Fig. 1g), and the Spearman correlation coefficients among the five samples were around 0.8, indicating stability among the biologically parallel samples of the MCC cell line (Fig. 1h). Subsequent bioinformatics analysis revealed that proteins such as SLC1A5 (with a mean T/NAT ratio of approximately 308 and a p-value of 2.5E-11), MCM6 (with a mean T/NAT ratio of approximately 372 and a p-value of 8.3E-14), and ITGA2 (with a mean T/NAT ratio of approximately 42 and a p-value of 1.1E-7) exhibited significantly higher expression in tumor tissues compared with adjacent normal tissues in human gastric cancer samples (Fig. 1i) [12]. These proteins were also detectable in the MCC cell line (Fig. 1j), indicating that the MCC cell line exhibits characteristics reminiscent of gastric adenocarcinoma and holds promise for clinical applications.
Establishment of a cell-derived allograft (CDA) model
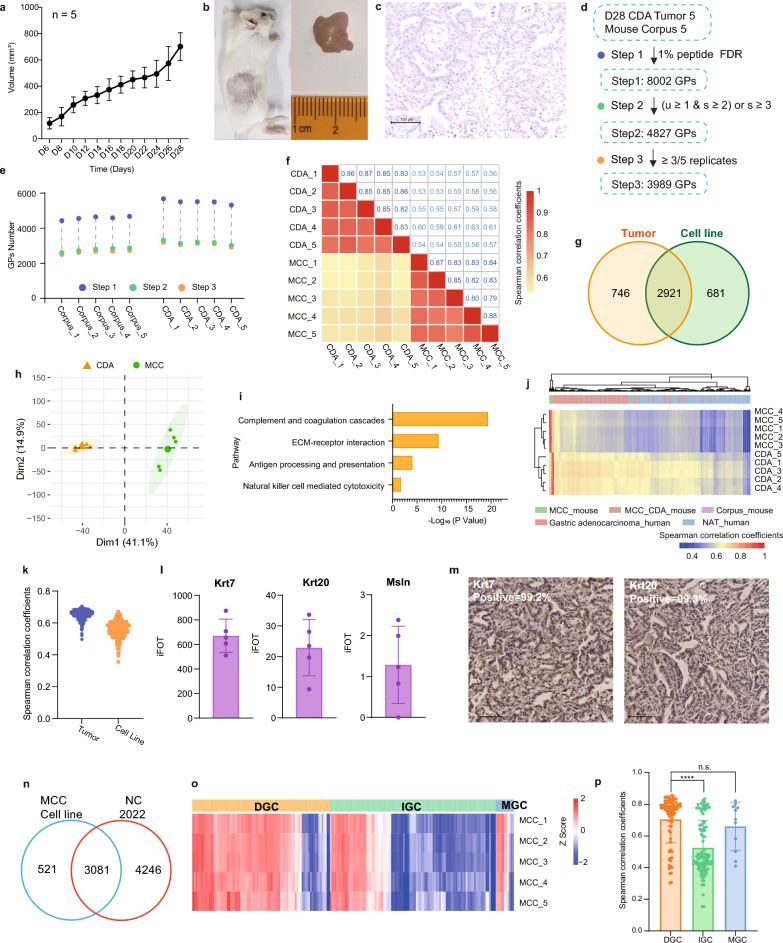
In our experimental design, we inoculated 106 MCC cells into the subcutaneous tissue of syngeneic BALB/c mice at 8 weeks of age to generate an allograft model of gastric adenocarcinoma with intact murine immune responses. Approximately 6 days subsequent to the inoculation, palpable subcutaneous tumors with a volume of 100 mm3 were observed, achieving a tumorigenesis of 100%. Approximately 24 days post-inoculation, the tumor volume had escalated to approximately 500 mm3 (Fig. 2a). At day 28, when the tumors had reached an average volume of 700 mm3, they were harvested for analysis (Fig. 2b). Histopathological assessment of the MCC-CDA tumors by two independent pathologists confirmed the adenocarcinomatous nature of the tumors (Fig. 2c). Proteomic profiling was conducted on corpus and subcutaneous tumor tissues from five mice, yielding a total of 8002 proteins. After stringent quality control measures, 4827 proteins were deemed as high-confidence, with 3989 of these proteins being detectable in a minimum of three samples from either the corpus or MCC-CDA tumors (Fig. 2d, supplementary tables S3). The number of detected proteins in each corpus sample was approximately 2600, while the subcutaneous MCC-CDA tumor samples exhibited around 3100 detected proteins (Fig. 2e).
Fig. 2.
MCC cell-derived allograft model in BALB/c mouse. a Growth curve of MCC cell-derived allografts (CDA). b Representative images of a tumor-bearing mouse (left) and subcutaneous MCC-CDA tumor tissue (right). c Pathological H&E staining of subcutaneous MCC-CDA tumor specimens. d Proteomic data sets of mouse normal corpus and MCC-CDA tumors using different filtering criteria. e Total number of proteins identified in each biological replicate of mouse normal corpus and MCC-CDA tumors. f Correlation analysis between biological replicates of MCC cell lines and MCC-CDA tumors. g Venn diagram showing proteins identified and quantified in MCC cell lines and MCC-CDA tumors. h Principal component analysis (PCA) of proteomic data differentiating MCC cell lines and MCC-CDA tumors. i KEGG pathway enrichment in MCC-CDA tumors. j Hierarchical clustering and heatmap of Spearman correlation coefficients comparing MCC cell lines and MCC-CDA tumors to clinical gastric cancer tumors and NAT. k Distribution of Spearman correlation coefficients for MCC cell lines and MCC-CDA tumors relative to clinical gastric cancer tumors. l The relative abundance (iFOT) of classic markers of clinical gastric adenocarcinoma (Krt7, Krt20, and Msln) in MCC-CDA tumors. m IHC staining results of Krt7 and Krt20 in MCC CDA tumors. n Venn diagram comparing the MCC cell line proteome with the clinical gastric cancer patient proteome. o Correlation heat map of 5 MCC cell line bioparallel samples and clinical gastric cancer patients. p Bar plot showing Pearson correlation between the MCC cell line and clinical gastric cancer patients of different Lauren histological subtypes
A comparative proteomic analysis of the MCC-CDA tumors and the original MCC cell line revealed a Spearman correlation coefficient of 0.57 (Fig. 2f), and principal component analysis (PCA) highlighted significant divergence between the two datasets (Fig. 2h). The MCC-CDA tumors displayed 746 proteins that were specifically expressed (Fig. 2g), which were enriched in pathways associated with the tumor microenvironment, such as complement and coagulation cascades, ECM-receptor interaction, antigen processing and presentation, and natural killer cell mediated cytotoxicity (Fig. 2i), suggesting that the subcutaneous tumors in immunocompetent mice are influenced by the local immune microenvironment. The unsupervised hierarchical clustering (HC) analysis of the Spearman correlation coefficient from the MCC cell line and MCC-CDA tumors with clinical gastric cancer patient proteomic data demonstrated a strong correlation between MCC cell line and MCC-CDA tumors with clinical gastric cancer tumor tissues (Gastric adenocarcinoma_human) (Fig. 2j). The Spearman correlation coefficients for the MCC cell line in relation to clinical gastric cancer tumors and for MCC-CDA tumors in relation to clinical gastric cancer tumors were 0.58 and 0.65, respectively (Fig. 2k). Proteins commonly utilized as markers for clinical gastric adenocarcinoma, including Krt7, Krt20, and Msln, were prominently expressed in CDA tumors (Fig. 2l), with IHC staining confirming positivity for Krt7 and Krt20 in over 99% of tumor cells (Fig. 2m).
To further establish clinical relevance, proteomic samples from 194 clinical gastric cancer (GC) patients were analyzed using stringent quality control criteria: protein identifications were accepted at a peptide- and protein-level false discovery rate (FDR) of 1%, and proteins were considered valid if they met one of the following conditions: at least one unique strict peptide and two strict peptides, or more than three strict peptides. After applying these filters, 3,081 proteins were commonly identified in both the clinical samples and MCC cell line (Fig. 2n). Pearson correlation analysis revealed that the MCC cell line demonstrated the highest correlation with diffuse gastric cancer (DGC) (r = 0.71), followed by mixed-type gastric cancer (MGC, r = 0.66) and intestinal-type gastric cancer (IGC, r = 0.53) (Fig. 2o, p). These findings indicate that the MCC cell line more closely resembles DGC at the proteomic level than IGC or MGC. This analysis positions the MCC cell line as a highly relevant model for DGC research. The cell line's characteristics also make it an ideal system for preclinical drug screening, particularly for targeted therapies and immune interventions. Given these advantages, we believe the MCC cell line will significantly contribute to the development of more effective treatments for DGC.
MCC-CDA model of gastric adenocarcinoma for preclinical studies
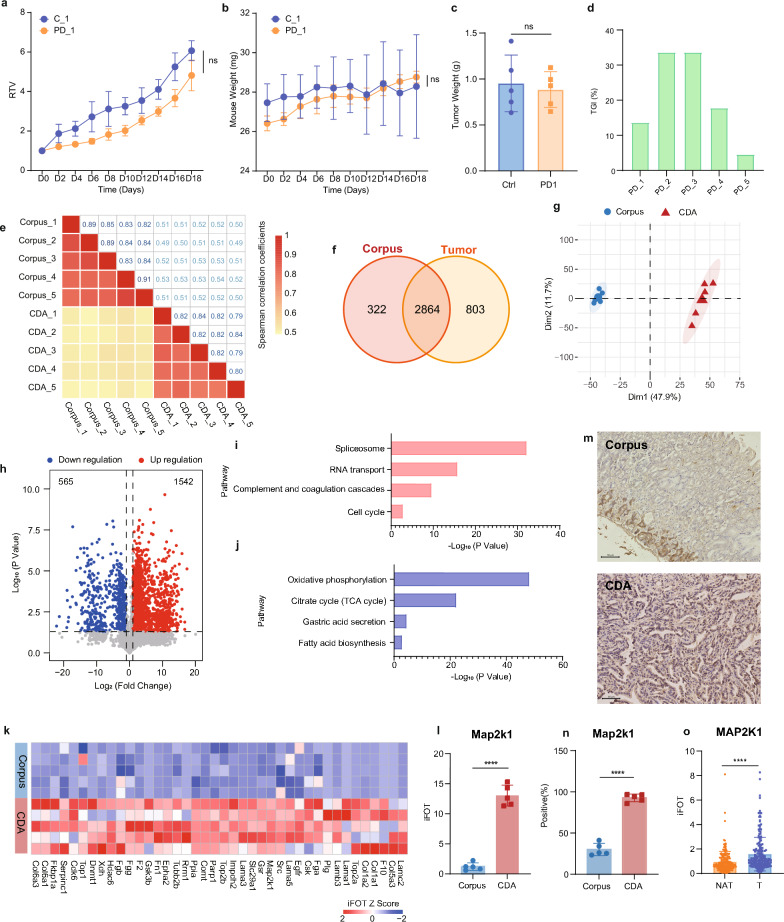
The immunocompetent murine allograft model of gastric adenocarcinoma may serve as a convenient platform for investigating the modulation of antitumor immunity within the tumor microenvironment or for developing novel immunotherapeutic strategies in vivo. We further explored the potential of this model for anti-PD-L1 (200 μg/mouse, i.p. every three days) therapy, a treatment modality that has demonstrated clinical benefits for a subset of patients with gastric adenocarcinoma [23]. The experimental results revealed that anti-PD-L1 treatment did not significantly inhibit the growth of gastric cancer in mice compared to control treatment (Fig. 3a, supplementary tables S4) and did not impact mouse body weight (Fig. 3b). The lack of significant difference in tumor weight between the treatment and control groups on day 18 post-administration (Fig. 3c) and the mean tumor growth inhibition (TGI) rate of only approximately 20% (Fig. 3d) suggest that this model is refractory to anti-PD-L1 therapy, mirroring the response of the majority of clinical gastric cancer patients to such treatment. Consequently, this model could be utilized to inform clinical treatment strategies for gastric cancer patients who do not respond to anti-PD-L1 therapy.
Fig. 3.
MCC-CDA gastric adenocarcinoma model for preclinical studies. a Relative tumor volume (RTV) growth curves of subcutaneous MCC-CDA tumors in BALB/c mice, measured every two days. b Changes in body weight of mice in control (ctrl) and anti-PD-L1 groups. c Tumor weight at day 18 in control and anti-PD-L1 groups. d Tumor growth inhibition (TGI) at day 18 in control and anti-PD-L1 groups. e Correlation analysis across biological replicates of mouse normal corpus and MCC-CDA tumors. f Venn diagram showing proteins identified and quantified in both mouse normal corpus and MCC-CDA tumors. g Principal component analysis (PCA) of proteomic data differentiating mouse normal corpus and MCC-CDA tumors. h Volcano plot showing differential protein expression between normal corpus and MCC-CDA tumors. i KEGG pathway enrichment of proteins upregulated in MCC-CDA tumors. j KEGG pathway enrichment of proteins downregulated in MCC-CDA tumors. k Comparison of drug target proteins between normal corpus and MCC-CDA tumors. l The relative abundance (iFOT) of ap2k1 in MCC-CDA tumors. m IHC staining results of Map2k1 in mouse normal corpus(up) and MCC-CDA tumors(down). n Proportion of Map2k1-positive cells in mouse normal corpus and MCC-CDA tumors. o Protein expression of MAP2K1 in tumor tissues and NAT from patients with gastric adenocarcinoma
To further explore the clinical translational potential of this model, we compared the proteomes of the tumor and normal gastric tissue from mice. The Spearman correlation coefficients for biological replicates were approximately 0.86 and 0.82 for the corpus and MCC-CDA tumors, respectively, whereas the coefficient for comparison between corpus and MCC-CDA tumors was 0.52 (Fig. 3e). Among the identified proteins, 2864 were common to both corpus and MCC-CDA tumors, 322 were unique to the corpus, and 803 were exclusive to the MCC-CDA tumors (Fig. 3f). Principal component analysis (PCA) demonstrated significant differences between the proteomes of normal gastric tissue and MCC-CDA tumors (Fig. 3g). Differential protein expression analysis (Student's t-test, p < 0.05, ratio of treatment to control group > 2 or < 0.5) revealed that 1542 proteins were significantly upregulated in the MCC-CDA tumors (Fig. 3h), with enrichment in tumor-associated pathways such as spliceosome, RNA transport, complement and coagulation cascades, and cell cycle (Fig. 3i). Additionally, 562 proteins were significantly downregulated in the MCC-CDA tumors (Fig. 3h), predominantly involved in gastric function-related pathways including oxidative phosphorylation, citrate cycle (TCA cycle), gastric acid secretion, and fatty acid biosynthesis (Fig. 3j).
By integrating our findings with the FDA drug database, we identified potential preclinical study targets for our immunocompetent murine gastric CDA model (Fig. 2k). Notably, Map2k1 exhibited high expression in the CDA gastric cancer model (Fig. 2l), Immunohistochemical analysis further confirmed a marked upregulation of MAP2K1 in MCC tumors compared to normal gastric tissue (Fig. 2m, n). Specifically, MAP2K1 positivity was observed in approximately 29% of cells in normal gastric tissue, whereas the proportion increased to approximately 93% in MCC tumors, reflecting a more than threefold elevation. Consistent with these findings, MAP2K1 was significantly upregulated in clinical gastric cancer tissues relative to adjacent non-cancerous tissues (Fig. 2o). These observations highlight the potential of MAP2K1 as a therapeutic target, prompting the selection of its inhibitor, trametinib, for preclinical evaluation.
MEKi therapy represses gastric adenocarcinoma via immune activation
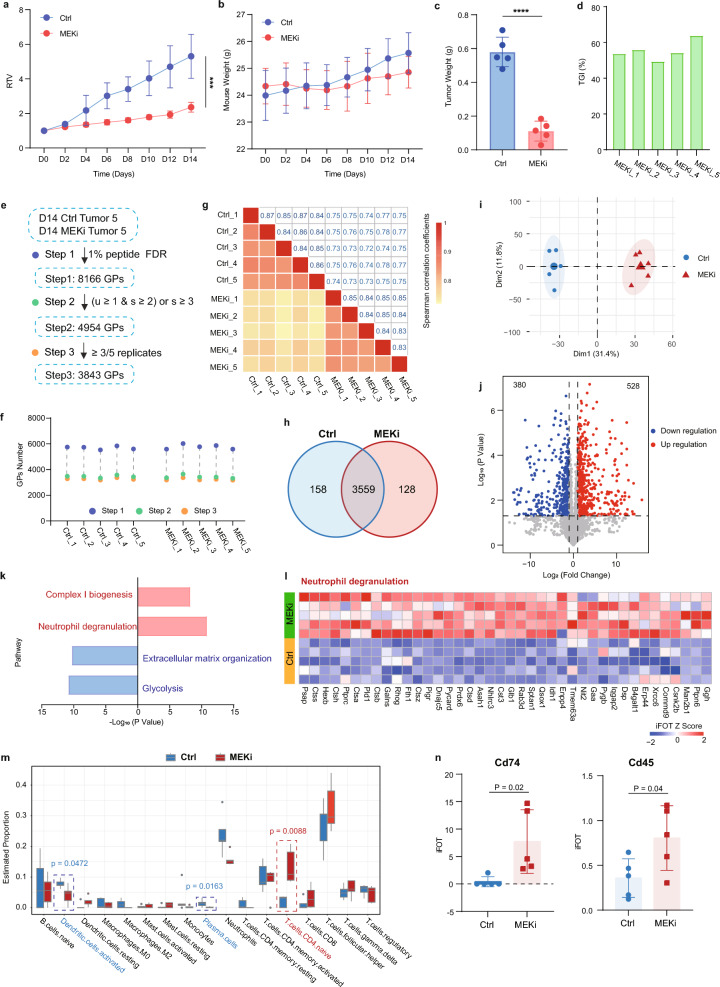
To investigate the impact of trametinib on gastric cancer progression, we conducted a preclinical study using the MCC-CDA gastric adenocarcinoma model in BALB/c mice. Mice bearing tumors were administered trametinib (1 mg/kg, orally, MEKi) or saline solution daily. The mice were weighed every two days, and tumor size was measured over 18 days. Trametinib effectively slowed the growth trend of subcutaneous gastric cancer tumors (Fig. 4a, supplementary tables S5). Additionally, tumors were harvested and weighed 18 days post-treatment. The analysis of tumor weight correlated with tumor volume (Fig. 4c) and tumor growth inhibition (TGI) (Fig. 4d). There was no significant difference in body weight between mice treated with saline solution and those treated with trametinib (Fig. 4b), indicating the safety and efficacy of trametinib in the treatment of gastric cancer.
Fig. 4.
Proteomics links suppression of tumor to immune activation by MEKi therapy. a Relative tumor volume (RTV) growth curves of subcutaneous MCC-CDA tumors in BALB/c mice, measured every two days. b Changes in body weight of mice in control (ctrl) and MEKi groups. c Tumor weight at day 14 in control and MEKi groups. d Tumor growth inhibition (TGI) at day 14 in control and MEKi groups. e Proteomic data sets of MCC-CDA tumors in control and MEKi groups with different filtering criteria. f Total number of proteins identified in each biological replicate of control and MEKi tumors. g Correlation analysis of biological replicates in control and MEKi tumor groups. h Venn diagram of proteins identified and quantified in control and MEKi tumor groups. i Principal component analysis (PCA) of proteomic data differentiating control and MEKi tumor groups. j Volcano plot showing differential protein expression between control and MEKi tumor groups. k KEGG pathway enrichment for proteins upregulated (red) and downregulated (blue) in MEKi tumors. l Heatmap of proteins related to the neutrophil degranulation pathway in control and MEKi tumor groups. m Boxplot illustrating mean CIBERSORT scores in control and MEKi tumor groups, with p-values from two-sided chi-square tests. n Protein expression of Cd74 and Cd45 in control and MEKi tumor groups
Building on these findings, we explored the mechanism of action of trametinib in the treatment of gastric cancer through proteomic analysis. A total of 8166 proteins were detected in both the control and MEKi treatment groups, with 4954 high-confidence proteins identified after stringent quality control. Among these, 3843 proteins were detected in at least three samples of gastric body or MCC-CDA tumor (Fig. 4e, supplementary tables S6). The number of detected proteins in each tumor sample from both the control and MEKi groups was approximately 3200 (Fig. 4f), with Spearman correlation coefficients of approximately 0.85 and 0.84 between biological replicates, respectively. The correlation coefficient between tumors from the control and MEKi groups was 0.75 (Fig. 4g).
The Venn diagram of the protein intersection between the control and MEKi groups revealed that 3559 proteins were detected in both control and MEKi treatment groups, 158 proteins were only detected in the control group, and 128 proteins were only detected in the MEKi treatment group (Fig. 4h). Additionally, principal component analysis (PCA) was generated to elucidate the discernible changes in protein expression profiles between the control and MEKi groups (Fig. 4i). Differentially expressed proteins (Student’s t-test, p < 0.05, ratio of treatment to control group > 2 or < 0.5) were identified, and pathway enrichment analysis was conducted. Trametinib specifically upregulated 528 proteins and downregulated 380 proteins in the MEKi group (Fig. 4j). Pathway enrichment analysis revealed that trametinib primarily upregulated immune-related signaling pathways, including complex I biogenesis and neutrophil degranulation. Conversely, extracellular matrix organization and glycolysis pathways were the most significantly enriched in the downregulated protein group (Fig. 4k, l). Further CIBERSORT analysis [24] found that the score of CD4 naive T cells significantly increased after drug treatment (Fig. 4m), and the expression of key molecules in T cell signaling, Cd74 and Cd45, also significantly increased (Fig. 4n). This suggests that the inhibitory effect of trametinib treatment on gastric cancer CDA tumors may be mediated by modulating the tumor microenvironment, particularly affecting neutrophils and T cells.
Discussion
In recent years, there has been growing recognition of the critical role played by the immune microenvironment and systemic immunity in tumor progression, driving the demand for preclinical models that accurately reflect the complexities of gastric cancer (GC) within an immunocompetent framework. In this study, we established a novel murine gastric adenocarcinoma cell line, designated MCC, derived from primary tumors in the gastric corpus of mice. The MCC cell line exhibits robust proliferative capacity and reliably forms syngeneic allograft tumors with a 100% engraftment rate in immunocompetent mice, providing a robust platform for preclinical research. Through comprehensive proteomic analyses of MCC-derived tumors, normal gastric corpus tissues, and MCC cell lines, we demonstrated the translational potential of this model in elucidating GC pathogenesis and identifying therapeutic targets. These findings underscore the clinical relevance of the MCC cell line and its utility for evaluating therapeutic strategies in a physiologically relevant context.
The MCC cell line is particularly valuable for modeling diffuse gastric cancer (DGC), a clinically challenging subtype of GC. Proteomic comparisons revealed a high degree of similarity between MCC-derived tumors and DGC, with a correlation coefficient of 0.71, exceeding correlations with other GC subtypes such as mixed-type (MGC) or intestinal gastric cancer (IGC). This strong alignment highlights MCC’s utility in investigating the molecular features and therapeutic challenges associated with DGC, a subtype characterized by aggressive behavior, poor prognosis, and resistance to conventional treatments. The capacity of MCC cells to establish tumors in immunocompetent mice further enhances their preclinical research potential, enabling the study of therapeutic strategies within an immune-competent microenvironment. Moreover, the proteomic resemblance of MCC to clinical DGC supports its application in identifying therapeutic targets and optimizing treatment approaches tailored to this aggressive cancer subtype. These findings position the MCC cell line as a powerful model for preclinical studies, particularly in the context of DGC, providing a robust platform for understanding the molecular basis of this subtype and for testing novel therapeutic strategies. This model holds promise for advancing precision medicine and improving clinical outcomes for patients with this challenging form of GC.
While our proteomic analysis identified immune-related pathways, we acknowledge the complexity of immune-tumor interactions and the limitations inherent to the current dataset. Further studies are necessary to validate these findings and to delve deeper into the role of immune pathways in shaping the tumor microenvironment. Such research could illuminate mechanisms of tumor immune evasion and guide the development of targeted immunotherapies for DGC. Additionally, integrating multi-omics approaches may refine the molecular characterization of MCC tumors and uncover novel regulatory mechanisms, such as post-translational modifications, that could play pivotal roles in GC pathogenesis. Collectively, the MCC cell line offers significant promise as a preclinical tool, facilitating the advancement of therapeutic strategies and a deeper understanding of GC biology.
Conclusions
In conclusion, leveraging our novel mouse gastric cancer cell line (MCC), we have successfully developed a syngeneic tumor model. The observed enhancement of anti-tumor immune responses with trametinib treatment further validates the applicability and reliability of this mouse model. Our study provides an effective platform for elucidating the mechanisms of gastric cancer progression and identifying promising drug targets.
Supplementary Information
Supplementary Material 1: Table S1 MCC Cell Lines whole exome sequencing.
Supplementary Material 2: Table S2 MCC Cell Lines normalized proteome.
Supplementary Material 3: Table S3 Mouse corpus and MCC-CDA tumor normalized proteome.
Supplementary Material 4: Table S4 Anti-PD-L1 therapy in MCC-CDA model.
Supplementary Material 5: Table S5 Trametinib therapy in MCC-CDA model.
Supplementary Material 6: Table S6 Ctrl and MEki groups tumor normalized proteome.
Acknowledgements
We gratefully acknowledge the financial support received from the National Natural Science Foundation of China (No. 32271498, No. 3227120162 and No. 32088101), State Key Laboratory of Proteomics (No. SKLPK202002), and the National Key Research and Development Program of China (No. 2022YFA1303200).
Abbreviations
- MFC
Mouse forestomach carcinoma
- MNU
N-Methyl-N-nitrosourea
- FBS
Fetal bovine serum
- MCC
Mouse corpus carcinoma
- PDT
Population doubling time
- TCGA
The Cancer Genome Atlas
- cBioPortal
CBio cancer genomics portal
- COSMIC
Catalogue of Somatic Mutations in Cancer
- CDA
Cell-derived allograft
- PCA
Principal component analysis
- HC
Hierarchical clustering
- WES
Whole exome sequencing
- WTE
Whole tissue extract
- LC–MS/MS
Liquid chromatography tandem mass spectrometry
- HCD
Higher energy collisional dissociation
- NCBI
National Center for Biotechnology Information
- FDR
False discovery rate
- FOT
Fraction of total
- DEPs
Differentially expressed proteins
- FDA
US Food and Drug Administration
- MEKi
MEK inhibitor
Author contributions
Yushen Wang designed the experiment, collected the data, analyzed the data, and wrote the manuscript. Xianju Li, Yi Wang and Jun Qin conceived, supervised, and funded the project.
Funding
This study was supported by the National Natural Science Foundation of China (No. 32271498, No.3227120162 and No. 32088101), State Key Laboratory of Proteomics (No. SKLPK202002), and the National Key Research and Development Program of China (No. 2022YFA1303200).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
The experiment was approved by the research protocol by the National Protein Science Center (Beijing Proteome Research Center). The approval number for animal studies is IACUC-20200818-30MT.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xianju Li, Email: xianju_li@163.com.
Yi Wang, Email: wangyi@ncpsb.org.cn.
Jun Qin, Email: jqin1965@126.com.
References
- 1.Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R, Chen R, Han B, et al. Analysis of malignant tumor prevalence in China in 2022. J Natl Cancer Cent. 2024;46:221–31. [Google Scholar]
- 3.Chalabi M. Stomach cancer gets a triple punch of therapy. Nature. 2021;600:608–9. [DOI] [PubMed] [Google Scholar]
- 4.Alsina M, Arrazubi V, Diez M, Tabernero J. Current developments in gastric cancer: from molecular profiling to treatment strategy. Nat Rev Gastroenterol Hepatol. 2022;20:155–70. [DOI] [PubMed] [Google Scholar]
- 5.Choi YY, Noh SH, Cheong J-H. Molecular dimensions of gastric cancer: translational and clinical perspectives. J Pathol Transl Med. 2016;50:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449–58. [DOI] [PubMed] [Google Scholar]
- 7.Ding C, Qin Z, Li Y, et al. Proteomics and precision medicine. Small Methods. 2019;3:1900075. [Google Scholar]
- 8.Lauren P. The two histological main types of gastric carcinoma diffuse and so-called intestinal-type carcinoma. Acta Pathol Microbiol Scand. 1965;64:31–49. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–56. [DOI] [PubMed] [Google Scholar]
- 11.Ge S, Xia X, Ding C, et al. A proteomic landscape of diffuse-type gastric cancer. Nat Commun. 2018;9:1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi W, Wang Y, Xu C, et al. Multilevel proteomic analyses reveal molecular diversity between diffuse-type and intestinal-type gastric cancer. Nat Commun. 2023;14:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, Dang M, Harada K, et al. Single-cell dissection of intratumoral heterogeneity and lineage diversity in metastatic gastric adenocarcinoma. Nat Med. 2021;27:141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Sun Z, Peng G, et al. Single-cell RNA sequencing reveals a pro-invasive cancer-associated fibroblast subgroup associated with poor clinical outcomes in patients with gastric cancer. Theranostics. 2022;12:620–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Visser KE, Joyce JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41:374–403. [DOI] [PubMed] [Google Scholar]
- 16.Qiu G-Z, Jin M-Z, Dai J-X, Sun W, Feng J-H, Jin W-L. Reprogramming of the tumor in the hypoxic niche: the emerging concept and associated therapeutic strategies. Trends Pharmaco Sci. 2017;38:669–86. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Zhang Q, Xing B, et al. Immune phenotypic linkage between colorectal cancer and liver metastasis. Cancer Cell. 2022;40:424-437.e425. [DOI] [PubMed] [Google Scholar]
- 18.Sitarz R, Skierucha M, Mielko J, Offerhaus J, Maciejewski R, Polkowski W. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian S, Gao J, Wang J, Liu Y, Dong H. Establishment of a mouse forestomach carcinoma cell line (MFC) with spontaneous hematogenous metastasis and preliminary study of its biological characteristics. Chin J Oncol. 1987;9:261–4. [PubMed] [Google Scholar]
- 20.Feng J, Ding C, Qiu N, et al. Firmiana: towards a one-stop proteomic cloud platform for data processing and analysis. Nat Biotechnol. 2017;35:409–12. [DOI] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang Y-K, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461–71. [DOI] [PubMed] [Google Scholar]
- 24.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Table S1 MCC Cell Lines whole exome sequencing.
Supplementary Material 2: Table S2 MCC Cell Lines normalized proteome.
Supplementary Material 3: Table S3 Mouse corpus and MCC-CDA tumor normalized proteome.
Supplementary Material 4: Table S4 Anti-PD-L1 therapy in MCC-CDA model.
Supplementary Material 5: Table S5 Trametinib therapy in MCC-CDA model.
Supplementary Material 6: Table S6 Ctrl and MEki groups tumor normalized proteome.
Data Availability Statement
The proteomics data, the spectral library, and details regarding all identified proteins have been submitted to the ProteomeXchange Consortium (https://www.iprox.cn/).
No datasets were generated or analysed during the current study.