Abstract
Among a collection of 74 aprt mutations induced by treatment of plateau phase Chinese hamster ovary CHO cells with the radiomimetic DNA double-strand cleaving agent neocarzinostatin, nine were large-scale rearrangements. Molecular analysis indicated that all nine were highly conservative, non-homologous reciprocal exchanges, most of which were intrachromosomal as determined by fluorescence in situ hybridization. All but one of the parental sequences contained potential double-strand cleavage sites positioned such that the observed rearrangements could be explained by drug-induced double-strand breakage followed by trimming, templated patching and ligation of the exchanged ends. Predicted non-complementary 3′ overhangs were often preserved in the newly formed junctions, suggesting alignment-based fill-in of the overhangs. Banding of metaphase spreads of these mutants, and of a number of mutants induced by the functionally similar compound bleomycin, revealed that bleomycin-induced reciprocal exchange mutants had multiple additional chromosome alterations and considerable chromosomal heterogeneity within each mutant line. In contrast, neocarzinostatin-induced reciprocal exchange mutants, as well as bleomycin-induced base substitution and single base deletion mutants, retained stable pseudodiploid karyotypes similar to that of the parent line. Thus, some reciprocal exchanges arising from misjoining of double-strand breaks were associated with global chromosomal instability, while other ostensibly similar events were not.
INTRODUCTION
In mammalian cells, radiation and other DNA-cleaving agents induce a variety of both stable and unstable chromosome aberrations, at least some of which probably result from errors in repair of double-strand breaks (DSBs). In particular, misjoining of the exchanged ends of two such breaks on different chromosomes may account for many if not most of the reciprocal translocations and dicentric chromosomes induced by these agents (1–4). However, it has also been shown that, following treatment with radiation or radiomimetic drugs, a subpopulation of the progeny cells acquire a persistent global chromosomal instability and continue to accumulate additional chromosome alterations many generations after treatment (2,5,6). Although the mechanism of this delayed effect is not known, it is generally agreed that the effect cannot be due to residual radiation-induced DNA damage, because such damage, even if not repaired, would be diluted out as the cells proliferated. Thus, the persistence of these presumably indirect genomic effects raises the question of whether even the early chromosome alterations that occur in the first few generations after treatment might be due to an induced global chromosome instability, rather than to the misrepair of DSBs.
In an attempt to distinguish between these possibilities, we previously analyzed aprt mutations induced by treatment of CHO-D422 cells with bleomycin in plateau phase. The mutations included several large-scale rearrangements (7), and by a combination of ligation-mediated PCR, DNA sequencing and fluorescence in situ hybridization, we showed that these rearrangements were highly conservative reciprocal translocations between aprt and unrelated sequences on different chromosomes (the one exception being a qualitatively similar three-way exchange; 8). Moreover, each translocation breakpoint corresponded to an expected double-strand cleavage site, strongly suggesting that the translocations did indeed result directly from the misjoining of the exchanged ends of two DSBs. In some cases, it was possible to reconstruct the breakage and repair events at single nucleotide resolution.
Neocarzinostatin (zinostatin, NCS) is a member of the enediyne family of radiomimetic natural products. Like bleomycin, zinostatin induces free radical-mediated DSBs, but the breaks differ in chemistry, geometry and sequence specificity from those induced by bleomycin (9,10). In an attempt to determine in detail whether and how these breaks are misjoined, zinostatin-induced aprt rearrangements were likewise analyzed at the chromosome and DNA sequence levels.
MATERIALS AND METHODS
Cell lines with rearranged aprt genes were from a mutant collection generated previously by treatment of confluence-arrested CHO-D422 cells (hemizygous for aprt) for 2 days with low concentrations of zinostatin (11). DNA for molecular analysis was prepared from each mutant approximately 30 cell divisions after isolation. Cytogenetic analysis was performed between 30 and 40 cell divisions after isolation.
Following PCR-based mapping (12), each rearrangement breakpoint was amplified by nested ligation-mediated PCR, using as a template MboI- or Tsp509I-cleaved mutant genomic DNA ligated to BamHI- or EcoRI-cut pUC19 (13). A third inner nested aprt primer was end labeled and used to sequence the resulting PCR product across the breakpoint. To determine whether the two sequences which had become linked to aprt in each mutant were themselves linked in non-mutant cells (as in a reciprocal exchange), PCR was performed using DNA from the CHO-D422 parent line and a primer from each of the two non-aprt genomic sequences, with the primers directed toward each junction with aprt. The products were sized on low melting point agarose gels, in some cases isolated as single bands, and sequenced using one of the PCR primers or an internal primer. (A complete list of the more than 50 genomic primers used is available from the authors on request.)
For cytogenetic analysis and fluorescence in situ hybridization, metaphase spreads were prepared from coded cultures of each mutant cell line and Giemsa/trypsin/Giemsa (GTG) banded by standard procedures (14). The λDM2 and λB21 probes (kindly provided by Mark Meuth, University of Utah) (15) were directly labeled with the fluorophores Spectrum Green and Spectrum Orange, respectively, using a nick translation kit (Vysis). A mixture consisting of 500 ng of each labeled probe plus 10 µg of repetitive C0t-1 DNA (16) from CHO-D422 cells (to suppress non-specific hybridization) was precipitated, resuspended in hybridization buffer (Vysis), denatured at 70°C for 5 min and suppression-hybridized for 1 h at 37°C. In situ hybridization was performed as described (14), except that the slides were first denatured at 70°C for 70 s and then hybridized with the probes at 37°C for 16 h. Twenty metaphases were analyzed per cell line.
RESULTS
Zinostatin-induced aprt rearrangements are reciprocal exchanges
Of 74 independent zinostatin-induced aprt mutations analyzed, nine were large-scale rearrangements. As previously reported (12), PCR-based mapping indicated that in each rearrangement most if not all of the aprt gene (17) was still present, but there was a small region across which no PCR products could be generated, consistent with either a large insertion or the separation of the two halves of aprt to distant genomic sites. The breakpoints of each rearrangement were then recovered by ligation-mediated PCR and ∼50–200 bp of non-aprt sequence beyond each breakpoint were determined. To recover the original non-aprt parental sequences, PCR was performed with DNA from non-mutant cells and primers from the two sequences that had been linked to aprt. In every case a prominent PCR product was obtained with a length approximately equal to the sum of the distances from each primer to the junction with aprt, and the sequences of these products were identical to the non-aprt sequences initially recovered by ligation-mediated PCR.
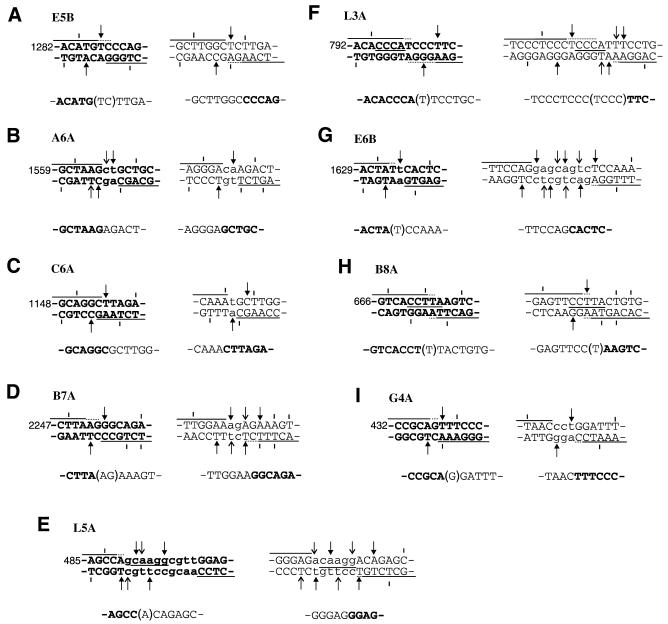
Thus, each rearrangement was a reciprocal exchange between aprt and an unrelated sequence (Fig. 1). Like bleomycin-induced exchanges (7), zinostatin-induced exchanges were highly conservative; for 9 of the 18 parental sequences, every base pair of sequence was retained in one or the other newly formed junction, and only four sequences unambiguously lost >2 bp. The maximum loss was 10 bp, from the aprt parental sequence of mutant L5A.
Figure 1.
Sequence analysis of zinostatin-induced reciprocal translocations. The top line shows the aprt (bold) and non-aprt parental sequences. Arrows indicate specifically positioned DSBs that could have accounted for the observed translocations and short vertical bars indicate DSBs that could not; different arrow styles show pairing of cleavage sites in opposite strands, on a two base 3′ stagger. Lines above and below indicate the extent of sequences from each parent that were unequivocally retained in each repair joint; dotted portions indicate sequences that were present in the joints but could have come from either parent. Lines between the strands indicate microhomologies shared by the two parental sequences. Lower case letters indicate nucleotides that were deleted, i.e. not found in either of the final repair joints. The bottom line shows joint sequences (one strand only), with sequences that could have come from either parent shown in parentheses. Numbering of aprt sequence positions follows the convention of Phear et al. (17). Longer segments of the non-aprt sequences (110–348 bp) have been deposited in GenBank (accession nos AF452455–AF452463).
BLASTN searches of GenBank, for sequences homologous to each non-aprt sequence, revealed that the breakpoint region of mutant E6B was nearly identical to a 50 bp sequence near the 3′ end of intron 14 of the hamster 3-hydroxy-3-methylglutaryl coenzyme A gene (one mismatch; P < 10–27). The other sequences showed less extensive homology to various coding and non-coding human and rodent sequences. However, none of the sequences were identified as containing any of the common rodent interspersed repeat elements.
Junction sequences are consistent with joining of exchanged DSB ends
The most straightforward mechanism by which DNA-cleaving agents could induce non-homologous reciprocal exchanges would be misjoining of the exchanged ends of two DSBs. By comparing the known potential sites of zinostatin-induced double-strand cleavage in each parental sequence with the bases retained in each junction sequence, it is possible to assess whether any given exchange is consistent with such a mechanism. Zinostatin induces bistranded damage on a two base 3′ stagger, at the trinucleotide sequences AGT·ACT, TGT·ACA, AGC·GCT, AGA·TCT, TGC·GCA, AGG·CCT, AAT·ATT and AAA·TTT (all sequences are 5′→3′·5′→3′ and are listed in order of decreasing cleavage frequency; underlining indicates the nucleotides attacked) (12,18,19). In one strand, typically the C-containing strand of NGN·NCN sequences, the target nucleotide is oxidized at the C5′ position, leaving a non-ligatable nick with 5′-aldehyde and 3′-phosphate termini. In the opposite strand, attack can be either at the C-1′ position (as at AGC sequences), resulting in a C-1′-oxidized abasic site that can break down to form a one base gap with 3′- and 5′-phosphate termini, or at C-4′ (as at AGT·ACT sequences), resulting in a one base gap with 3′-phosphoglycolate (-PO4CH2COOH) and 5′-phosphate termini (9,10).
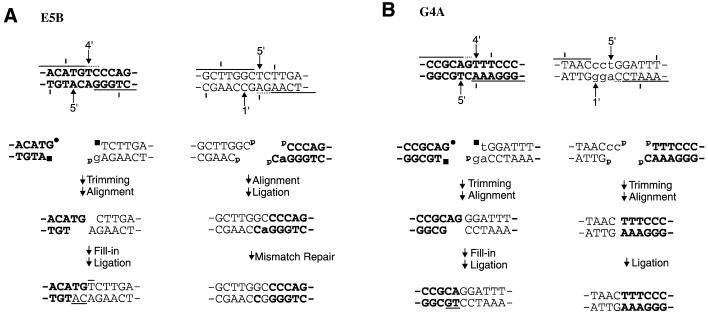
Comparison of translocation breakpoints with predicted DSB sites revealed that there was not a significant excess of DSB sites at the breakpoints, as compared with the gene as a whole, and only one of the exchanges (G4A, Fig. 2B) had a breakpoint that was unambiguously associated with an AGT·ACT sequence, which are the most prominent sites of zinostatin-induced DSBs (12,18). Nevertheless, with the single exception of the aprt sequence of mutant B8A, every parental sequence contained one or more potential DSB sites that could account for the observed exchanges, assuming that they arose by joining of the exchanged DSB ends (Fig. 1). In a few cases, there was only one potential DSB site in each parental sequence that could have produced the observed junctions, and it was thus possible to reconstruct the specific nucleotide removal and fill-in events that would have been required, as shown for two of the mutants in Figure 2. Besides the highly conservative nature of the end-processing, a striking feature of the inferred processing is the apparent preservation of 3′ overhangs in the final repair joints. Such preservation would require fill-in of the 3′ overhangs prior to ligation, presumably using the aligned 3′ terminus of the other end of the break as the primer for fill-in (as in the lower strand of the left-hand repair joint in Fig. 2B). Such fill-in has been reproduced in human, Xenopus and hamster whole-cell extracts and shown to be strictly dependent on alignment of the two ends by the DNA end-binding protein Ku (20–22). Of the 11 parental sequences with uniquely determined DSB sites, eight showed apparent fill-in of a 3′ overhang on at least one end of the DSB.
Figure 2.
Possible intermediates in formation of reciprocal changes, based on the known chemistry and specificity of zinostatin-induced DNA cleavage. The top line shows the parental sequences and potential DSB sites, with notation as in Figure 1. Numbers indicate the specific deoxyribose carbon attacked in each nucleotide. The second line shows the predicted DNA ends, with 5′- or 3′-phosphate (p), 3′-phosphoglycolate (circle) and 5′-aldehyde (square) termini indicated; lower case letters indicate nucleotides that were unequivocally removed from the final joint. The third line shows trimming and/or other intermediate processing, including removal of 3′-phosphate and 3′-phosphoglycolate groups; 5′-aldehyde nucleotides are also assumed to be removed, since they cannot be ligated. The last line shows the final repair joints, with overlines and underlines indicating nucleotides that must have been filled in by de novo synthesis.
In many systems, crossover points for illegitimate recombination events often occur at microhomologies (usually only a few base pairs) common to the two sequences, with one copy of the microhomology being retained at the newly formed junction (23–26). Although three of the zinostatin-induced exchanges had 4–5 bp microhomologies between aprt and non-aprt parental sequences at the respective breakpoints, only one of the six resulting junctions (Fig. 1F, right-hand junction) was consistent with microhomology-based splicing of the two sequences. Thus, models involving end joining by single-strand exposure and annealing (27,28) are apparently not applicable to these exchanges. There were no non-templated nucleotide additions at any of the junctions.
Most but not all zinostatin-induced exchanges are intrachromosomal
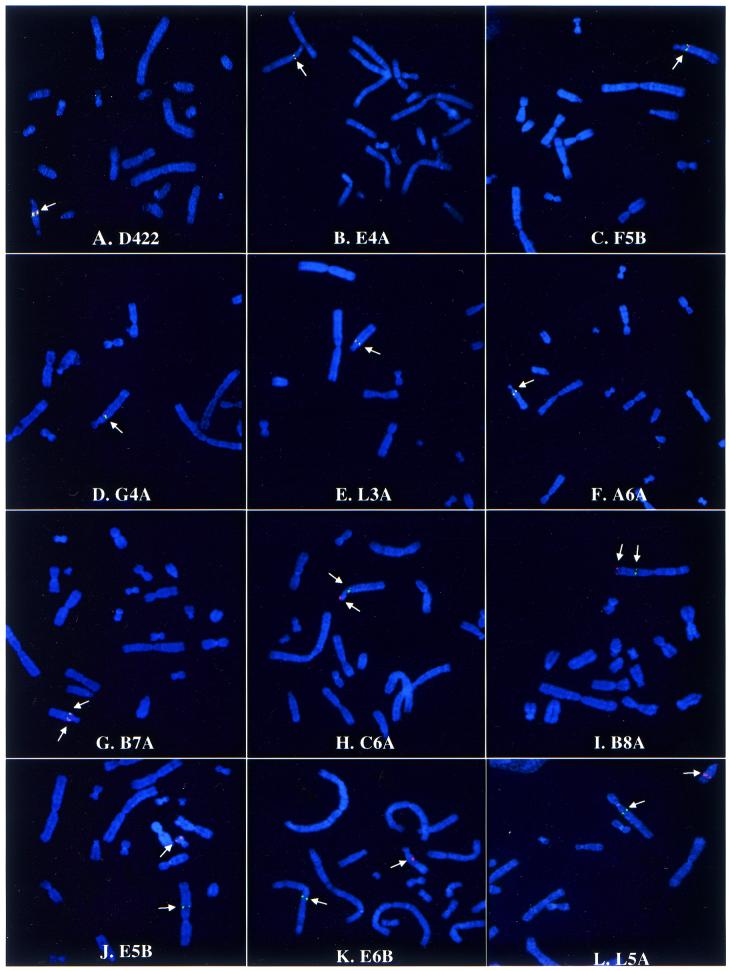
To determine whether the aprt and non-aprt sequences involved in each rearrangement were on the same chromosome, metaphase spreads of each mutant cell line were hybridized with probes corresponding to sequences 50 kb upstream (λDM2, green fluorescence) and 10 kb downstream (λB21, red fluorescence) from aprt (15). As reported previously (29), in the original CHO-K1 parental line these probes produced two yellow (red + green) signals as expected, corresponding to the two aprt loci on chromosome Z4 (which is a pericentric inversion of the normal chromosome 3) and chromosome Z7 (30). In the CHO-D422 cell line, an aprt hemizygote from which all the zinostatin-induced mutants were generated, only the signal on Z4 was retained and, as expected, the same single coincident (or nearly coincident) signal on Z4 was seen in zinostatin-induced base substitution (E4A) and small deletion (F5B) aprt mutants (Fig. 3A–C). In the zinostatin-induced reciprocal exchange mutants, four different patterns were detected. Mutants G4A, L3A and A6A were indistinguishable from the CHO-D422 parental line, with a single coincident yellow signal on an apparently unaltered Z4 chromosome (Fig. 3D–F), suggesting that these rearrangements were intrachromosomal and confined to the immediate vicinity of aprt. Mutant B7A had a normal appearing Z4 with a slight but consistent separation of the red and green signals (Fig. 3G), suggesting an inversion or other rearrangement encompassing a slightly larger region of the chromosome (the separation could be more clearly seen in interphase nuclei; see cover of this issue). Mutant C6A showed a Z4 of slightly altered morphology (Fig. 3H), in which the red signal was translocated from the long to the short arm, consistent with an inversion encompassing the centromere. Mutant B8A showed red and green signals well separated on a new chromosome about twice the size of Z4 (Fig. 3I), suggesting a complex rearrangement involving at least one other chromosome. Finally, mutants E5B, E6B and L5A showed red and green signals separated to different chromosomes (Fig. 3J–L), consistent with an interchromosomal reciprocal translocation with one breakpoint within aprt. Thus, five of nine zinostatin-induced exchanges were intrachromosomal, as opposed to only one of seven bleomycin-induced translocations (P ≈ 0.08) (7). The interchromosomal exchanges did not have any consistent characteristics at the sequence level that distinguished them from the intrachromosomal exchanges.
Figure 3.
Fluorescence in situ hybridization analysis of reciprocal exchanges. Green and red fluorescence signals correspond to sequences immediately upstream and downstream of aprt, respectively. (A–F) In the parental strain D422, as well as in a base substitution mutant (E4A), a small deletion mutant (F5B) and three reciprocal exchange mutants (G4A, L3A and A6A), the signals were coincident on an apparently unaltered Z4 chromosome. (G) In mutant B7A the signals were nearly coincident, but a consistent separation could be seen in most metaphases, and more distinctly in interphase nuclei (not shown). (H) In mutant C6A the red signal was transposed from the long to the short arm of Z4, suggesting an inversion encompassing the centromere. (I) In mutant B8A, the signals were well separated on a chromosome twice the size of Z4, suggesting a complex interchromosomal event. (J–L) In mutants E5B, E6B and L5A the signals were separated to different chromosomes, indicating an interchromosomal reciprocal translocation at the aprt locus. The very slight displacements of red and green signals in mutants F5B and G4A are within the normal range seen with parental cells and thus do not in themselves suggest large-scale rearrangements; note that these displacements were largely transverse to the axis of the chromosome, unlike the longitudinal separation consistently seen on both chromatids in mutant B7A.
Zinostatin-induced reciprocal exchanges are not accompanied by global chromosomal instability
A previous study of aprt rearrangements induced by the radiomimetic antibiotic bleomycin showed that all but one of them were highly conservative reciprocal exchanges between unrelated sequences (7), similar to those described herein. Cytogenetic analysis of these mutants revealed that all but one of the exchanges were interchromosomal and that most were accompanied by several additional chromosomal alterations, with considerable heterogeneity among individual cells of each mutant clone. These results suggested a possible phenomenological link between bleomycin-induced reciprocal translocations and global chromosomal instability.
To determine whether zinostatin-induced reciprocal exchanges were associated with a similar chromosomal instability, mutants harboring such exchanges were subjected to GTG banding (Table 1). In stark contrast to the bleomycin-induced reciprocal exchange mutants, the zinostatin-induced reciprocal exchange mutants, including those showing translocations at the chromosomal level, had much more stable karyotypes (1.0 ± 0.3 versus 4.4 ± 0.7 chromosome alterations per cell, P < 0.001). Although some individual lines were analyzed 5–10 passages later than others, average ages of the zinostatin- and bleomycin-induced cultures at harvest were similar and there was no apparent tendency for later passage cells to harbor either more or fewer chromosomal alterations.
Table 1. Chromosome alterations in zinostatin-induced and bleomycin-induced aprt mutants.
| Mutant | No. of chromosomes | Altered chromosomesa | Cytogenetic heterogeneityb | |
|---|---|---|---|---|
| Mode | Range | |||
| Zinostatin-induced reciprocal exchange mutants | ||||
| G4A | 21 | 18–22 | 0 | – |
| L3A | 20 | 19–20 | 1–2 | – |
| A6A | 21 | 20–22 | 0–2 | ± |
| B7A | 21 | 20–21 | 0 | – |
| C6A | 21 | 20–21 | 1–2 | – |
| B8A | 21 | 20–21 | 1–4 | + |
| E5B | 21 | 20–22 | 0–1 | ± |
| E6B | 21 | 20–21 | 0–2 | ± |
| L5A | 21 | 20–21 | 1–2 | ± |
| Bleomycin-induced reciprocal exchange mutants | ||||
| L4C | 21 | 20–21 | 6–7 | + |
| J5A | 21 | 19–21 | 5–8 | ± |
| K6B | 21 | 20–22 | 4–7 | + |
| P7B | 21 | 20–42 | 2–8 | + |
| L3A | 20 | 19–21 | 2–5 | + |
| K3A | 21 | 19–23 | 0–3 | ± |
| P5C | 21 | 19–42 | 1–4 | + |
| Bleomycin-induced base substitution mutants | ||||
| J7A | 21 | 20–21 | 0–1 | – |
| K8C | 40 | 38–42 | 2–4 | ± |
| F5A | 21 | 19–21 | 1–3 | ± |
| M7B | 21 | 20–21 | 0 | – |
| K5A | 21 | 20–21 | 1–3 | ± |
| Bleomycin-induced –1 deletion mutants | ||||
| F3 | 21 | 20–21 | 0–2 | – |
| K3B | 21 | 21 | 0 | – |
| P5A | 21 | 20–21 | 0 | – |
| I4A | 22 | 21 | 0–2 | ± |
| I6A | 21 | 21 | 1–4 | ± |
| K7B | 21 | 21 | 0 | – |
| P8A | 21 | 21 | 0–1 | – |
| H8A | 20 | 19–20 | 1–3 | ± |
| Parental | ||||
| D422 | 21 | 21 | 0 | ± |
aExcluding chromosome Z4, site of the aprt locus.
bCytogenetic heterogeneity was assessed according to the fraction of metaphases that had at least one altered chromosome not present in the dominant karyotype. Those with >40% were scored positive (+); those with 20–40% were scored marginal (±) and those with <20% were scored negative (–).
To determine whether chromosomal instability in the bleomycin-induced mutants might have arisen by some effect of bleomycin treatment distinct from the reciprocal translocation itself, mutants harboring bleomycin-induced base substitutions and single base (–1) deletions, derived from the same treated cultures as the translocation mutants, were also subjected to GTG banding. All of these point mutants had stable karyotypes similar to those of parental D422 cells and of the zinostatin-induced rearrangement mutants (Table 1). Thus, chromosomal instability appeared to be specifically associated with bleomycin-induced translocation events, rather than with bleomycin treatment as such. However, zinostatin-induced reciprocal exchanges, even though they were very similar to bleomycin-induced exchanges at the molecular level, were not associated with chromosomal instability.
DISCUSSION
While chromosomal reciprocal translocations induced by ionizing radiation and radiomimetic compounds have been studied exhaustively, very few of these events have been examined at the DNA sequence level. For the purposes of elucidating the detailed processing involved in such events, radiomimetic drugs have the distinct advantage of inducing breaks that have a defined geometry and chemistry and are to some extent sequence specific. Thus, in the case of bleomycin-induced translocations in CHO aprt, we were able to establish a statistically significant correspondence between translocation breakpoints and potential sites of drug-induced DSBs, strongly suggesting that the translocations were indeed formed by misjoining of exchanged DSB ends (7). However, all of these mutants also harbored additional chromosome rearrangements that were not present in the parental line and appeared to vary among individual cells in the culture. This result suggests that the mutants had acquired some degree of global chromosomal instability, raising the possibility that the observed translocations could have been a result of this instability, rather than a direct result of misjoining of bleomycin-induced DSBs.
The large-scale aprt rearrangements induced by zinostatin were very similar to those induced by bleomycin under the same conditions; they constituted a similar proportion of the total mutants, they were all reciprocal exchanges, they rarely involved loss of >1–2 bp from the parental sequences and they showed a conspicuous absence of even very short homologies at the breakpoints. Curiously, however, the zinostatin-induced mutants showed little if any evidence of chromosomal instability; rather, except for the chromosomes directly involved in the aprt translocation, the pseudodiploid karyotype of the CHO parental line was preserved. This result strengthens the argument that these translocations result directly from misjoining of zinostatin-induced DSBs rather than from some indirect effect on overall chromosomal stability. Unfortunately, the two base stagger of the zinostatin-induced breaks undermines attempts to establish a correlation between such sites and translocation breakpoints, because it increases the probability of fortuitous correspondence between cleavage sites and breakpoints. Thus, although all but one of the translocations were consistent with misjoining of exchanged DSB ends, the conclusion that the exchanges probably resulted from DSB misjoining is based more on a lack of plausible alternative mechanisms than on the sequence specificity of these events.
An adventitious feature of the two base stagger of zinostatin-induced DNA cleavage is that analysis of the resulting exchange events can in theory provide insight into the fate of 3′ overhangs on DSBs. From such analysis, it could be inferred that 3′ overhangs were often preserved during end joining. Preservation of 3′ overhangs has also been demonstrated for end joining in cell extracts and appears to involve fill-in of the 3′ overhang primed from the 3′ terminus of the opposite end of the break (Fig. 2), with the two ends being aligned by Ku protein (20,22,31). It would thus be predicted that 3′ overhangs would not be preserved during end joining in Ku-deficient cells, but the lack of a Ku-deficient, aprt-hemizygous CHO strain has precluded a direct test of this hypothesis. Our own attempts to generate such a strain have thus far been unsuccessful, but it might be possible to perform similar studies using some other locus, such as the much larger hprt gene, as a target.
Although there are significant differences between zinostatin and bleomycin in both the chemistry and the geometry of the induced DSBs, it is not immediately clear how such differences could account for the specific observed differences in the resulting rearrangements or in genomic stability. In particular, unlike zinostatin-induced DSBs, bleomycin-induced DSBs are predominantly either blunt ended or have single base 5′ overhangs and always have 5′-phosphate termini (9,32). A previous study in CHO cells (33) suggested that the structure of DSB termini may be important determinants of their potential to trigger chromosomal instability but, curiously, in this case bleomycin and zinostatin both induced instability while restriction endonucleases did not. Another difference is that zinostatin, unlike bleomycin, is highly unstable in dilute solution (34), so that even though it was replenished every 12 h, it is likely that most breakage occurred within the first few hours after each replenishment. Thus, zinostatin-treated cells may have sustained several waves of DSBs, while bleomycin-treated cells would have been subjected to a more constant level of breakage. Bleomycin is also noted for inducing large cell-to-cell variations in the level of DNA damage (35), so it is conceivable that the chromosomally unstable bleomycin-induced reciprocal translocation mutants were derived from a subpopulation of heavily damaged cells, although such cells would presumably be much less likely to survive. Finally, bleomycin is very poorly taken up by mammalian cells (36), so that the extracellular concentrations required to induce comparable DNA damage and cytotoxicity are ∼1000-fold greater for bleomycin (11) than for zinostatin (12). Thus, in light of recent evidence for involvement of non-DNA targets in radiation-induced genomic instability (37), one could speculate that perhaps chromosomal instability was somehow triggered by a combination of a DSB misjoining event and bleomycin-induced free radical damage to the cell membrane. This proposal could account for the fact that chromosomal instability was seen in bleomycin-induced rearrangement mutants, but not in either zinostatin-induced rearrangement mutants or bleomycin-induced base substitution and small deletion mutants.
Only a few other reciprocal translocations induced by DNA double-strand-cleaving agents have been sequenced. Chemotherapy patients treated with DNA topoisomerase II inhibitors often develop secondary leukemias harboring t(4,11) or t(11,15) reciprocal translocations that could arise by misjoining of drug-stimulated, topoisomerase-mediated DSBs. Molecular analysis of translocations in three such leukemias (38,39) revealed highly conservative reciprocal exchanges, similar to those presented here. Intriguingly, t(4,11) translocations in primary leukemias were much more complex, with deletions and duplications of hundreds of base pairs, supporting the view that the simpler chemotherapy-associated translocations were in fact formed by misjoining of drug-induced DSBs.
In contrast, however, reciprocal translocations in mouse embryonic stem cells, resulting from site-specific enzymatic cleavage at two I-SceI sites on two different chromosomes, showed much larger terminal deletions at the junctions, ranging from 13 bp to >2 kb (40). In this model system, each I-SceI site was positioned in a defective bacterial neo (neomycin resistance) gene introduced into the genome by homologous recombination. Upon expression of I-SceI, breakage at the two sites would result in a 538 bp direct repeat at two of the resulting DNA ends, and detection of a translocation depended on decatenation of this repeat to a single copy at the newly formed junction (producing a functional neo gene). This unusual requirement may have somehow promoted deletions at the second junction as well. A second difference is that these experiments, unlike those of the present study, were performed in log phase cultures; thus, it is possible that end-joining events tend to be more conservative in confluence-arrested G0 phase cells than in log phase cells. A third possibility is that embryonic stem cells, which seem to depend more heavily on the homologous recombination pathway for DSB repair (41), may have a less well-developed, more error-prone non-homologous end-joining pathway(s) than adult somatic cells. A fourth possibility is that the modified termini of free radical-mediated or topoisomerase-mediated DSBs may protect the termini from exonucleolytic degradation until they are sequestered by repair complexes; thus, enzymatically induced DSBs may be more susceptible to degradation. Finally, it is possible that artificially introduced sequences may have an altered chromatin conformation or some other unusual structural property that tends to reduce the fidelity of end joining.
Thus, because of the difficulties of the experimental systems, relatively little is known of the factors that control the quite variable extent of terminal deletions during end-joining repair. Analysis of many more misrepair events, involving various types of DSBs, in a variety of normal and repair-deficient cells will be required in order to achieve even a rudimentary understanding of the mechanisms that enforce the fidelity of end joining, and of why these mechanisms sometimes fail.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants CA40615 and RD1HD33527 from the National Institutes of Health.
DDBJ/EMBL/GenBank accession nos AF452455–AF452463
REFERENCES
- 1.Bender M.A., Griggs,H.G. and Bedford,J.S. (1974) Mechanisms of chromosomal aberration production 3. Chemicals and ionizing radiation. Mutat. Res., 23, 197–212. [DOI] [PubMed] [Google Scholar]
- 2.Galloway S.M. (1994) Chromosome aberrations induced in vitro: mechanisms, delayed expression and intriguing questions. Environ. Mol. Mutagen., 23 (suppl. 24), 44–53. [DOI] [PubMed] [Google Scholar]
- 3.Preston R.J. (1990) Mechanisms of induction of specific chromosomal alterations. Basic Life Sci., 53, 329–336. [DOI] [PubMed] [Google Scholar]
- 4.Savage J.R. (1989) The production of chromosome structural changes by radiation: an update of Lea (1946), Chapter VI. Br. J. Radiol., 62, 507–520. [DOI] [PubMed] [Google Scholar]
- 5.Marder B.A. and Morgan,W.F. (1993) Delayed chromosomal instability induced by DNA damage. Mol. Cell. Biol., 13, 6667–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadhim M.A., Lorimore,S.A., Townsend,K.M., Goodhead,D.T., Buckle,V.J. and Wright,E.G. (1995) Radiation-induced genomic instability: delayed cytogenetic aberrations and apoptosis in primary human bone marrow cells. Int. J. Radiat. Biol., 67, 287–293. [DOI] [PubMed] [Google Scholar]
- 7.Wang P., Zhou,R.-H., Zou,Y., Jackson-Cook,C.K. and Povirk,L.F. (1997) Highly conservative reciprocal translocations formed by apparent joining of exchanged DNA double-strand break ends. Proc. Natl Acad. Sci. USA, 94, 12018–12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Povirk L.F. (1998) A highly conservative, cyclically permuted, non-homologous exchange among three unrelated DNA sequences in bleomycin-treated CHO cells. Int. J. Radiat. Biol., 74, 561–564. [DOI] [PubMed] [Google Scholar]
- 9.Dedon P.C. and Goldberg,I.H. (1992) Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin and calicheamicin. Chem. Res. Toxicol., 5, 311–332. [DOI] [PubMed] [Google Scholar]
- 10.Povirk L.F. (1996) DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat. Res., 355, 71–89. [DOI] [PubMed] [Google Scholar]
- 11.Povirk L.F., Bennett,R.A.O., Wang,P., Swerdlow,P.S. and Austin,M.J.F. (1994) Single base-pair deletions induced by bleomycin at potential double-strand cleavage sites in the aprt gene of stationary phase Chinese hamster ovary D422 cells. J. Mol. Biol., 243, 216–226. [DOI] [PubMed] [Google Scholar]
- 12.Wang P. and Povirk,L.F. (1997) Targeted base substitutions and small deletions induced by neocarzinostatin at the APRT locus in plateau-phase CHO cells. Mutat. Res., 373, 17–29. [DOI] [PubMed] [Google Scholar]
- 13.Mizobuchi M. and Frohman,L.A. (1993) Rapid amplification of genomic DNA ends. Biotechniques, 15, 214–216. [PubMed] [Google Scholar]
- 14.Jackson-Cook C., Bae,V., Edelman,W., Brothman,A. and Ware,J. (1997) Cytogenetic characterization of the human prostate cancer cell line P69SV40T and its novel tumorigenic sublines M2128 and M15. Cancer Genet. Cytogenet., 87, 14–23. [DOI] [PubMed] [Google Scholar]
- 15.Davis R. and Meuth,M. (1994) Molecular characterization of multilocus deletions at a diploid locus in CHO cells: association with an intracisternal-A particle gene. Somat. Cell Mol. Genet., 20, 287–300. [DOI] [PubMed] [Google Scholar]
- 16.Landegent J.E., in de Wal,N.J., Dirks,R.W., Baas,F. and van der Ploeg,M. (1987) Use of whole cosmid cloned genomic sequences for chromosomal localization by non-radioactive in situ hybridization. Hum. Genet., 77, 366–370. [DOI] [PubMed] [Google Scholar]
- 17.Phear G., Armstrong,W. and Meuth,M. (1989) Molecular basis of spontaneous mutation at the aprt locus of hamster cells. J. Mol. Biol., 209, 577–582. [DOI] [PubMed] [Google Scholar]
- 18.Dedon P.C. and Goldberg,I.H. (1990) Sequence-specific double-strand breakage of DNA by neocarzinostatin involves different chemical mechanisms with a staggered cleavage site. J. Biol. Chem., 265, 14713–14716. [PubMed] [Google Scholar]
- 19.Povirk L.F., Houlgrave,C.W. and Han,Y.-H. (1988) Neocarzinostatin-induced DNA base release accompanied by staggered oxidative cleavage of the complementary strand. J. Biol. Chem., 263, 19263–19266. [PubMed] [Google Scholar]
- 20.Chen S., Inamdar,K.V., Pfeiffer,P., Feldmann,E., Hannah,M.F., Yu,Y., Lee,J.W., Zhou,T., Lees-Miller,S.P. and Povirk,L.F. (2001) Accurate in vitro end-joining of a DNA double-strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem., 276, 24323–24330. [DOI] [PubMed] [Google Scholar]
- 21.Pfeiffer P. and Vielmetter,W. (1988) Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res., 16, 907–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldmann E., Schmiemann,V., Goedecke,W., Reichenberger,S. and Pfeiffer,P. (2000) DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res., 28, 2585–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth D.B. and Wilson,J.H. (1988) Illegitimate recombination in mammalian cells. In Kucherlapati,R. and Smith,G.R. (eds), Genetic Recombination. American Society for Microbiology, Washington, DC, pp. 621–653.
- 24.Meuth M. (1989) Illegitimate recombination in mammalian cells. In Berg,D. and Howe,M. (eds), Mobile DNA. American Society for Microbiology, Washington, DC, pp. 833–853.
- 25.Monnat R.J. Jr, Hackmann,A.F.M. and Chiaverotti,T.A. (1992) Nucleotide sequence analysis of human hypoxanthine phosphoribosyltransferase (HPRT) gene deletions. Genomics, 13, 777–787. [DOI] [PubMed] [Google Scholar]
- 26.Morris T. and Thacker,J. (1993) Formation of large deletions by illegitimate recombination in the HPRT gene of primary human fibroblasts. Proc. Natl Acad. Sci. USA, 90, 1392–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thacker J., Chalk,J., Ganesh,A. and North,P. (1992) A mechanism for deletion formation in DNA by human cell extracts: the involvement of short direct repeats. Nucleic Acids Res., 20, 6183–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karran P. (2000) DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev., 10, 144–150. [DOI] [PubMed] [Google Scholar]
- 29.Zhou R.-H., Wang,P., Zou,Y., Jackson-Cook,C.K. and Povirk,L.F. (1997) A precise interchromosomal reciprocal exchange between hotspots for cleavable complex formation by topoisomerase II in amsacrine-treated CHO cells. Cancer Res., 57, 4699–4702. [PubMed] [Google Scholar]
- 30.Adair G.M., Nairn,R.S., Brotherman,K.A. and Siciliano,M.J. (1989) Spontaneous CHO APRT heterozygotes reflect high-frequency, allele-specific deletion of the chromosome Z4 APRT gene. Somat. Cell Mol. Genet., 15, 535–544. [DOI] [PubMed] [Google Scholar]
- 31.Thode S., Schafer,A., Pfeiffer,P. and Vielmetter,W. (1990) A novel pathway of DNA end-to-end joining. Cell, 60, 921–928. [DOI] [PubMed] [Google Scholar]
- 32.Povirk L.F., Han,Y.-H. and Steighner,R.J. (1989) Structure of bleomycin-induced DNA double-strand breaks: predominance of blunt ends and single-base 5′ extensions. Biochemistry, 28, 8508–8514. [DOI] [PubMed] [Google Scholar]
- 33.Limoli C.L., Kaplan,M.I., Phillips,J.W., Adair,G.M. and Morgan,W.F. (1997) Differential induction of chromosomal instability by DNA strand-breaking agents. Cancer Res., 57, 4048–4056. [PubMed] [Google Scholar]
- 34.Kappen L.S. and Goldberg,I.H. (1980) Stabilization of neocarzinostatin nonprotein chromophore activity by interaction with apoprotein and with HeLa cells. Biochemistry, 19, 4786–4790. [DOI] [PubMed] [Google Scholar]
- 35.Östling O. and Johanson,K.J. (1987) Bleomycin, in contrast to ionizing radiation, induces extreme variation of DNA strand breakage from cell to cell. Int. J. Radiat. Biol., 52, 683–691. [DOI] [PubMed] [Google Scholar]
- 36.Roy S.N. and Horwitz,S.B. (1984) Characterization of the association of radiolabeled bleomycin A2 with HeLa cells. Cancer Res., 44, 1541–1546. [PubMed] [Google Scholar]
- 37.Limoli C.L., Ponnaiya,B., Corcoran,J.J., Giedzinski,E., Kaplan,M.I., Hartmann,A. and Morgan,W.F. (2000) Genomic instability induced by high and low LET ionizing radiation. Adv. Space Res., 25, 2107–2117. [DOI] [PubMed] [Google Scholar]
- 38.Ahuja H.G., Felix,C.A. and Aplan,P.D. (2000) Potential role for DNA topoisomerase II poisons in the generation of t(11;20)(p15;q11) translocations. Genes Chromosom. Cancer, 29, 96–105. [DOI] [PubMed] [Google Scholar]
- 39.Lovett B.D., Lo Nigro,L., Rappaport,E.F., Blair,I.A., Osheroff,N., Zheng,N., Megonigal,M.D., Williams,W.R., Nowell,P.C. and Felix,C.A. (2001) Near-precise interchromosomal recombination and functional DNA topoisomerase II cleavage sites at MLL and AF-4 genomic breakpoints in treatment-related acute lymphoblastic leukemia with t(4;11) translocation. Proc. Natl Acad. Sci. USA, 98, 9802–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson C. and Jasin,M. (2000) Frequent chromosomal translocations induced by DNA double-strand breaks. Nature, 405, 697–700. [DOI] [PubMed] [Google Scholar]
- 41.Essers J., van Steeg,H., de Wit,J., Swagemakers,S.M., Vermeij,M., Hoeijmakers,J.H. and Kanaar,R. (2000) Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J., 19, 1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]