Abstract
MelR is an Escherichia coli transcription factor belonging to the AraC family. It activates expression of the melAB operon in response to melibiose. Full-length MelR (MelR303) binds to two pairs of sites upstream of the melAB transcription start site, denoted sites 1′ and 1 and sites 2 and 2′, and to a fifth site, R, which overlaps the divergent melR promoter. The C-terminal domain of MelR (MelR173) does not activate transcription. Here we show that, like MelR303, when MelR173 binds to sites 1 and 2 it recruits CRP to bind between these sites. Hence, the C-terminal domain is involved in heterologous interactions. MelR173 binds to the R site, which has 11 of 18 bp identical to sites 1 and 2 but, surprisingly, does not bind to site 1′, which has 12 of 18 bp identical, nor to site 2′. Using electrophoretic mobility shift assays, we show that the binding of MelR303 to sites 1′ and 2′ is due to cooperative binding with the adjacent site. This homologous cooperativity requires the N-terminal domain of the protein. Activation of the melAB promoter requires MelR to occupy site 2′, which overlaps the –35 hexamer. Hence, both domains of MelR are required for transcription activation.
INTRODUCTION
The MelR protein of Escherichia coli is a transcription activator required for the metabolism of the disaccharide melibiose (1). MelR activates expression of the melA and melB genes, encoding an α-galactosidase and a melibiose permease, respectively. These genes are co-transcribed from a single promoter, pmelAB, which is 234 bp from the divergent melR promoter, pmelR (Fig. 1). Transcription from pmelAB is totally dependent on MelR and the presence of melibiose in the medium (2), while transcription from pmelR depends on the global transcription activator, the cyclic AMP receptor protein (CRP) (3).
Figure 1.
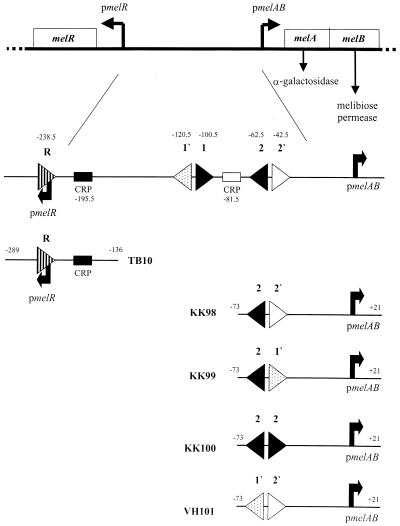
Schematic diagram of the melR and melAB intergenic region and some of the fragments used in this paper. The top part of the figure illustrates the divergent melAB and melR promoters and their transcripts; horizontal arrows indicate the corresponding transcription start sites. DNA sequences are numbered with respect to the melAB transcript start as +1. The locations of the DNA sites for MelR are indicated by horizontal triangles. Each triangle indicates an 18 bp sequence and its orientation, and the position of the centre of each site is denoted. The horizontal shaded boxes denote 22 bp DNA sites for CRP. One site centred at position –195.5 is responsible for activation of the melR promoter, whilst the other, centred at position –81.5 is responsible for cooperative binding of CRP with MelR (21). The bottom part of the figure illustrates the different promoter fragments made for this study. These are shown schematically with the same shading for each 18 bp sequence as in the upper diagram.
MelR is a member of the AraC family of transcription activators. Members of this family are found in a large number of different bacterial species (reviewed in 4,5). There are more than 100 proteins in this group, including MarA, Rob, SoxS, RhaR, XylS and the virulence protein VirF. Many of the proteins in this family contain ∼300 amino acids, with a C-terminal, DNA-binding domain of ∼120 amino acids and an N-terminal domain that binds a small ligand activator. The DNA-binding domains of these proteins show substantial similarity and are presumed to share a common tertiary structure, however, the N-terminal domains vary considerably. The structure of MarA, a 129 amino acid activator, which only contains the C-terminal homologous region of the protein, has been determined bound to DNA (6). It contains seven helices with two helix–turn–helix motifs bound to two adjacent major grooves of 16 bp DNA, bending it by 27°. The structure of Rob, another family member where the DNA-binding domain is in the N-terminal part of the protein, has also been determined bound to a DNA target (7). The structures of the DNA-binding domains of Rob and MarA are superimposable with a root mean squared deviation of only 0.9 Å for all main chain atoms. However, in Rob only one of the two helix–turn–helix motifs binds to the major groove of the DNA, the other is free in solution. The structure of the N-terminal domain of AraC has also been determined (8). It contains a β-barrel that binds the sugar and three α-helices, two of which form an antiparallel four-helix bundle with a second domain, giving a dimer.
Transcription activation by AraC has been studied intensively (reviewed in 9,10) and serves as a model for activation by other proteins of the family. AraC is the activator of the ara regulon that is essential for arabinose transport and metabolism. In the absence of the sugar arabinose, it binds to two 16 bp sites (denoted O2 and I1) 200 bp apart at the araBAD promoter, forming a repression loop. In the presence of arabinose, AraC binds to I2, which is adjacent to I1, rather than to O2. This breaks the repression loop and the presence of the activator at I2, next to the RNA polymerase, activates transcription. AraC-dependent transcription initiation at the araBAD promoter is increased by CRP, which binds to a single DNA site upstream of I1 and I2 (11).
In previous work, we have shown that MelR binds to two identical 18 bp sites upstream of pmelAB (named sites 1 and 2) in both the presence and absence of melibiose (12,13). Site 1 is centred between base pairs 100 and 101 upstream of the melAB transcription start site (i.e. at position –100.5), while site 2 is centered at position –63.5. These sites form a perfect inverted repeat, separated by 20 bp. We also showed that a C-terminal fragment of MelR (MelR173) binds to the same two sites (14). Both full-length MelR (MelR303) and MelR173 bend DNA at sites 1 and 2 to similar extents (15). However, in contrast to the C-terminal domains of AraC (16) and XylS (17) or the small MarA and SoxS proteins (18), MelR173 does not activate transcription (14).
Recently we have shown that there are three additional binding sites for MelR in the intergenic region between pmelAB and pmelR. At the pmelR promoter, MelR binds at site R, centred just downstream of the transcription start point, and represses its own transcription (19). Upstream of pmelAB, in addition to sites 1 and 2, there are two further MelR-binding sites; site 1′, centred at –120.5, and site 2′, centred at –42.5 (20). Sites 1 and 1′ and sites 2 and 2′ each form 18–2–l8 bp imperfect inverted repeats that bind MelR303 (Fig. 1). We have also found that CRP binds cooperatively with MelR303, between sites 1 and 2, helping to fill site 2′ and activate transcription (20,21). To further examine the mode of transcription activation by MelR, in this study we have explored the binding of MelR173 to these additional sites and the effect of CRP and compared this to the binding of full-length MelR (MelR303).
MATERIALS AND METHODS
Bacterial strains, plasmids and oligonucleotides
Standard methods for recombinant DNA manipulations were used throughout this work (22). The bacterial strains, plasmids and DNA fragments used are listed in Table 1.
Table 1. Bacterial strains, DNA fragments and plasmids used in this work.
| Reference | ||
|---|---|---|
| Escherichia coli strains | ||
| BL21 (λDE3) | T7RNApol+ F– ompT rB– mB– | 23 |
| WAM131 | ΔlacU169 melR+ | 20 |
| WAM132 | ΔlacU169 ΔmelR | 20 |
| DNA fragments | ||
| KK39 | DNA fragment containing pmelAB sequences bounded by an EcoRI site at –188 and a SalI–PstI–HindIII linker at +21 | 20 |
| KK43 | DNA fragment containing pmelAB sequences bounded by an EcoRI site at –136 and a SalI–PstI–HindIII linker at +21 The KK43 fragment carries a GC→AT transversion at –73, creating a unique BglII site between MelR-binding sites 1 and 2 | 12 |
| KK81 | DNA fragment carrying the intergenic region between pmelAB and pmelR bounded by an EcoRI site at position –312 and a SalI–PstI–HindIII linker at position +21 with respect to the melAB transcription start site | 20 |
| KK98 | DNA fragment containing pmelAB sequences from –73 to +21, bounded by an EcoRI site and a SalI–HindIII linker. The fragment contains a 2 bp mutation at –52 and –53 to give an NsiI site between sites 2 and 2′ | This work |
| KK99 | DNA fragment based on KK98, with further mutations in site 2′ to give a fragment containing sites 2 and 1′ in an 18–2–18 bp repeat | This work |
| KK100 | DNA fragment based on KK98, with further mutations in site 2′ to give a fragment containing two copies of site 2 in an inverted repeat, separated by 2 bp | This work |
| TB10 | DNA fragment carrying pmelR sequences bounded by an EcoRI site at position –289 and HindIII site at position –136 with respect to the melAB transcription start site | This work |
| VH101 | DNA fragment based on KK98, with further mutations in site 2 to give a fragment containing sites 1′and 2′ in an 18–2–18 bp repeat | This work |
| Plasmids | ||
| pAA121 | pBR322 derivative for cloning EcoRI–HindIII fragments | 35 |
| pCM117–173 | pET9d derivative encoding MelR173 on an NcoI–BamHI fragment under the control of a T7 promoter | 14 |
| pCM117–303 | pET9d derivative encoding full-length MelR on an NcoI–BamHI fragment under the control of a T7 promoter | 14 |
| pCM118–173 | pBR322 derivative encoding MelR173 under the control of the CRP-independent galP2 promoter | 14 |
| pCM118–314 | pBR322 derivative encoding full-length MelR with a N-terminal 11 amino acid fusion under the control of the CRP-independent galP2 promoter | 14 |
| pJW15 | pAA121 derivative carrying the melR gene expressed from its own promoter | 30 |
| pLysS | pACYC184 derivative which supplies low levels of T7 lysozyme | 23 |
| pRW50 | Low copy number lac expression vector, also encoding resistance to tetracycline | 28 |
| pVH-173 | pET9d derivative encoding MelR173 with an additional N-terminal His6 tag on an NsiI–BamHI fragment under the control of a T7 promoter | This work |
Construction of KK98, KK99, KK100 and VH101
A series of DNA fragments were constructed containing site 2 of pmelAB and a second DNA binding site for MelR, increasingly like site 2, in an 18–2–18 bp arrangement (Figs 1 and 2). The starting DNA was KK43, which contains sites 1′, 1, 2 and 2′ of pmelAB on an EcoRI–HindIII fragment (12). It also contains two PstI sites, one at –23 of pmelAB and the second as part of a SalI–PstI–HindIII linker (from pUC9). The site in the linker was destroyed by PCR mutagenesis, using the oligodeoxynucleotide primer D14314 and a primer upstream of the EcoRI site. D14314 covers the HindIII site and replaces CTGCAG of the PstI site with ATGCAG. This leaves a unique PstI site at position –23 of pmelAB.
Figure 2.
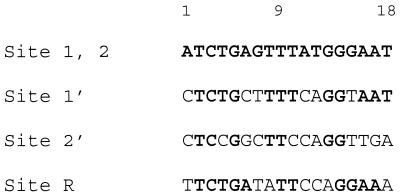
Sequences of the MelR-binding sites in the melR and melAB intergenic regions. The sequences are written in the orientation of site 2. Bases identical to sites 1 and 2 are in bold.
HindIII SalI melA
D14314: 5′-GCAAAGCTTGGATGCAGGTCGACGGATCTC-3′
Initially, pUC9 containing the modified EcoRI–HindIII KK43 insert was cut with EcoRI and PstI to remove pmelAB sequences upstream of –23. The two 18 bp sites in pmelAB were then constructed in two halves using a series of oligodeoxynucleotides on NsiI–PstI fragments for the downstream site or on EcoRI–NsiI fragments for the upstream half.
For the downstream site of pmelAB, pairs of complementary oligodeoxynucleotides containing the following sequences were synthesized, annealed and cloned into the pUC9 vector. This gave constructs containing different 18 bp sequences upstream of position –23 of pmelAB flanked upstream by an EcoRI–BglII–NsiI linker. These sequences corresponded to the 2′ site sequence in KK98, the 1′ site sequence in KK99 and the 1 (or identical 2) site sequence in KK100 (underlined).
KK98
EcoRI NsiI Site 2′ PstI
(G)AATTCAGATCTATGCATAACCTGGAAGCCGGAGGTTTTCTGCA(G)
KK99
EcoRI NsiI Site 1′ PstI
(G)AATTCAGATCTATGCATTACCTGAAAAGCAGAGGTTTTCTGCA(G)
KK100
EcoRI NsiI Site 2 PstI
(G)AATTCAGATCTATGCATTCCCATAAACTCAGATGTTTTCTGCA(G)
The upstream DNA binding site for MelR in the KK98, KK99 and KK100 fragments, site 2, was amplified as an EcoRI–NsiI fragment using the following primers with KK43 as template:
EcoRI Site 2
Upstream: 5′-GCAGAATTCCCGGGGATCTGAGTTTAT
Downstream: 5′-GCAAAGCTTATGCATTCCCATAAACTCAG
NsiI Site 2
This corresponds to the sequence –73 to –51 of pmelAB between two linkers. This fragment was inserted between the EcoRI and NsiI sites flanking the downstream half-sites to give KK98, KK99 and KK100, containing two DNA binding sites for MelR organised in an 18–2–18 bp inverted repeat with a central NsiI site.
The VH101 DNA was constructed by replacing the EcoRI–NsiI fragment in KK98 with two complementary oligonucleotides containing the sequence of site 1′.
EcoRI Site 1′ NsiI
5′-(G)AATTCCCGGGGCTCTGCTTTTCAGGTAATGCA(T)
Construction of TB10
This was done by PCR mutagenesis using KK81 (20) cloned in pAA121 as a template and the following primers.
HindIII primer: 5′-AGCAAGCTTCGCTGCTGCACATAAA-3′
EcoRI primer: 5′-GCAGAATTCCCTCATGATACTCGGA-3′
This resulted in an EcoRI–HindIII fragment carrying pmelR with the EcoRI site upstream of the pmelR DNA site for CRP and the HindIII site downstream of the melR translation initiation codon (–289 to –136 with respect to the pmelAB initiation start site).
Construction of pVH-173
The plasmid pVH-l73, encoding MelR173 with a His6 tag at the N-terminus, was constructed from pCM118–173 (14), a pET9d derivative containing DNA encoding the last 173 amino acids of MelR between NcoI and BamHI sites. To do this, complementary oligodeoxynucleotides containing the sequence
NsiI NcoI
5′-CATGCATCACCACCACCACCACTC-3′
were annealed. These form a linker containing a NsiI site and a NcoI site flanking DNA encoding a hexahistidine linker (underlined). Plasmid pCM118–173 was restricted with NcoI and ligated to the linker. The resulting DNA was transformed into E.coli and the resulting colonies tested for the presence of the additional NsiI site in the plasmid. The orientation of the linker was determined by examining the length of the small XhoI–NsiI fragment in the plasmid. Finally, the insert was sequenced to confirm the orientation of the linker and the initiation codon of MelR173.
Preparation of pure proteins
MelR303 was overexpressed in E.coli strain BL2I(λDE3) [pLysS] (23) transformed with the T7 expression plasmid pCM117–303 (14), as described (20). Traces of nuclease left after the heparin–agarose column were removed by passing the protein through an additional Q-Sepharose column equilibrated in 20 mM Tris–HCl pH 7.0, 0.1 mM EDTA, 20% (v/v) glycerol, 10 mg/l phenylmethylsulphonyl fluoride (PMSF), 0.1 mM dithiothreitol (DTT), 300 mM NaCl.
MelRl73 was overexpressed in E.coli strain BL21(λDE3) [pLysS] transformed with pVH-l73. The cells were grown in luria broth. Protein expression was induced by addition of 1 mM isopropyl β-d-thiogalactoside, at a cell density giving an A560 of ∼0.5, and the cells were harvested by centrifugation 4 h after induction. The expressed protein was found to be in inclusion bodies. It was purified on a nickel–agarose column, after denaturation in guanidine hydrochloride, as described for MarA (24). Fractions containing MelR173 were diluted 4-fold to reduce the NaCl concentration, loaded onto a heparin–agarose column and eluted with a 250 mM–1 M NaCl gradient in 50 mM sodium phosphate buffer, pH 7.6, 20% glycerol, 0.5 mM EDTA, 10 mg/l PMSF. The purified MelR173 protein eluted at ∼700 mM NaCl. It was stored at –20°C. Comparison of electrophoretic mobility shift assay (EMSA) and DNase I footprints, using crude cell extracts containing MelR173 expressed from pCM117–173 (without the His tag and without denaturation) with protein purified as above, confirm that the DNA binding specificity is not affected by the His tag or the purification method. This is in agreement with similar experiments on full-length MelR303 (25).
Protein concentrations were determined either from Bradford assays, calibrated with bovine serum albumin (BSA) (26) or from the absorbance at 280 nm using the equation (27):
ɛ280 = (no. of Trp × 5500) + (no. of Tyr × 1490).
For MelR303 ɛ280 = 42 400 M–1 cm–1, while for MelR173 ɛ280 = 21 430 M–1 cm–1.
Measurement of promoter activities in vivo
DNA fragments containing the melAB or melR promoters were cloned into pRW50, a low copy number vector (28), to give promoter::lac fusions. The activity of the enzyme β-galactosidase in cells carrying these recombinants was measured by the Miller method (29). Cells were grown in minimal media with fructose as the carbon source, either with or without melibiose, as in our previous work (1). Assays were performed in isogenic melR+ (WAM131) or ΔmelR (WAM132) strains (20).
DNase I footprints
DNase I footprinting experiments were performed as in our previous work (20). Incubations contained 4–10 nM purified KK39 EcoRI–HindIII fragment (20) that had been specifically labelled at the HindIII end with [γ-32P]ATP and polynucleotide kinase. Incubations contained 10 mM melibiose, 1.2 µM MelR303, 11 µM MelR173 and 75 nM CRP as indicated. After DNase I treatment, footprint patterns were analysed on polyacrylamide sequencing gels that were calibrated with Maxam–Gilbert sequence ladders.
EMSA
For the titration experiments, the EcoRI–HindIII fragments carrying MelR-binding sites were purified, end-labelled using [α-32P]dATP and the Klenow fragment of DNA polymerase and used in gel retardation assays as in our previous work (30). Binding assays were done in 30 µl of buffer containing 40 mM Tris–HCl pH 8.0, 100 mM KCl, 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 0.1 mg/ml BSA, 5 µg/ml herring sperm DNA. No difference was observed for gels run in the presence or absence of melibiose. For quantitative studies on fragments containing two DNA binding sites, eight or 16 samples with 0–2 µM protein were run in parallel on separate lanes. Up to three bands of radioactivity were observed in each lane, corresponding to the free DNA, a retarded band and a second, more retarded band. The amount of radioactivity in each band was determined using a phosphorimager plate, measuring the intensity using ImageQuant 3.3 (Molecular Dynamics). The fraction of the total radioactivity found in each band was fitted to the equations below (31) using non-linear regression in Sigmaplot (SPSS Inc.).
Ffree = 1/Z
F1 = K1 [P]/Z
F2 = K2 [P]2/Z
Z = 1 + K1 [P] + K2 [P]2
where Ffree is the fraction of DNA species in the free band, F1 is the fraction of DNA species in the first retarded band and F2 is the fraction of DNA species in the second, more retarded band. [P] is the protein concentration in subunits (we have assumed that a monomer binds to each site; the degree of cooperativity is not affected by this assumption). K1 is the macroscopic association constant for a single protein molecule binding to the DNA. K2 is the macroscopic association constant for two protein molecules binding simultaneously to free DNA.
The (macroscopic) constants K1 and K2 are related to the (microscopic) association constants for binding to two individual sites A and B on the DNA as below.
K1 = kA + kB
K2 = kA kB kAB
where kA is the association constant for site A, kB is the association constant for site B and kAB is the cooperativity factor between site A and site B, i.e. the degree to which binding to both sites is influenced by the neighbouring site.
When different protein preparations were used, the relative affinities of the pairs of fragments remained the same, as did the ratios of K1/K2, but the absolute values differed, presumably due to different specific activities of the preparations. For these reasons it is not possible to compare binding constants for a given DNA sequence between proteins or between preparations, just the relative affinities of different DNA sequences for the same protein and the cooperativity values.
RESULTS
In vivo transcription
To examine the effect of the C-terminal domain of MelR on transcription in vivo, we used derivatives of the lac expression vector pRW50 (28) carrying pmelAB::lac (12) and pmelR::lac fusions (Tables 2 and 3). Expression of β-galactosidase from the plasmids was measured in the WAM132 ΔmelRΔlac strain of E.coli (20) grown in the presence or absence of melibiose. Either MelR303 or MelR173 was expressed in this strain from a second plasmid in trans. For pmelAB, expression was also tested in the parent WAM131 melR+Δlac strain (20).
Table 2. In vivo activities of pmelAB::lac fusions in pRW50 in E.coli strains WAM131 (ΔlacU169) and WAM132 (ΔlacU169 ΔmelR).
| Plasmid | β-Galactosidase activity of KK43 [Miller units (29)] | |||||
|---|---|---|---|---|---|---|
| WAM132 (ΔmelR host) | WAM131 (melR+ host) | |||||
| –melibiose | +melibiose | Fold induction | –melibiose | +melibiose | Fold induction | |
| pAA121 | 4 | 1.8 | 0.5 | 2.7 | 188 | 70 |
| pJW15 | 9 | 1581 | 175 | 30 | 1431 | 47 |
| pCM118–314 | 6 | 1377 | 229 | 6 | 1367 | 227 |
| pCM118–173 | 2.5 | 2.8 | 1.0 | 3 | 2 | 0.7 |
β-Galactosidase levels in two strains of E.coli containing pmelAB::lac fusions in pRW50 are shown. The promoter fragment used was KK43 containing sites 1′, 1, 2 and 2′. Cells were grown ± melibiose Cells also contained multicopy plasmids expressing full-length MelR (pJW15, pCM118–314), N-terminally truncated MelR173 (pCM118–173) or a control (pAA121). Assays were performed as in Webster et al. (1). Assays vary by <10% when repeated independently.
Table 3. In vivo activities of pmelR::lac fusions in pRW50 in E.coli strain WAM132 (ΔlacU169 ΔmelR) (19).
| Plasmid | β-Galactosidase activity of TB10 [Miller units (29)] | |||
|---|---|---|---|---|
| –melibiose | Relative activity | +melibiose | Relative activity | |
| pAA121 | 235 | 100% | 243 | 100% |
| pJW15 | 149 | 63% | 107 | 44% |
| pCM118–314 | 74 | 32% | 61 | 25% |
| pCM118–173 | 57 | 24% | 54 | 22% |
β-Galactosidase levels in E.coli containing pmelR::lac fusions in pRW50 are shown. The promoter fragment used was TB10. Cells were grown ± melibiose. Cells also contained multicopy plasmids expressing full-length MelR (pJW15, pCM118–314), N-terminally truncated MelR173 (pCM118–173) or a control (pAA121). Assays were performed as in Webster et al. (1). Assays vary by <10% when repeated independently.
For pmelAB in the absence of melibiose, very little expression of lacZ is observed in either the melR+ or ΔmelR strain (Table 2). In the ΔmelR strain, in the presence of melibiose, β-galactosidase is expressed from pmelAB in the presence of pJWl5 and pCM118–314, carrying melR (see Table 1), but not in the presence of pCM118–173, carrying melR173. In the melR+ strain, expression of β-galactosidase from pmelAB is induced by melibiose and this is increased by the presence of multicopy plasmids pJW15 and pCM118–314, carrying melR. However, the presence of pCM118–173, carrying melR173, severely reduces expression from pmelAB. This is in accordance with our previous studies, in a different strain of E.coli, showing that transcription of pmelAB requires both MelR303 and melibiose and that MelR173 does not activate the promoter but acts as a trans-dominant repressor (2,14).
At the pmelR promoter, expression is dependent on CRP (3), so lacZ cloned under pmelR is expressed in the ΔmelR strain in the absence of MelR303 and MelR173 (Table 3). However, less expression is observed in the presence of plasmids carrying either full-length melR or melR173, i.e. both proteins repress expression from pmelR. This repression occurs both in the presence and absence of melibiose.
DNase I footprinting
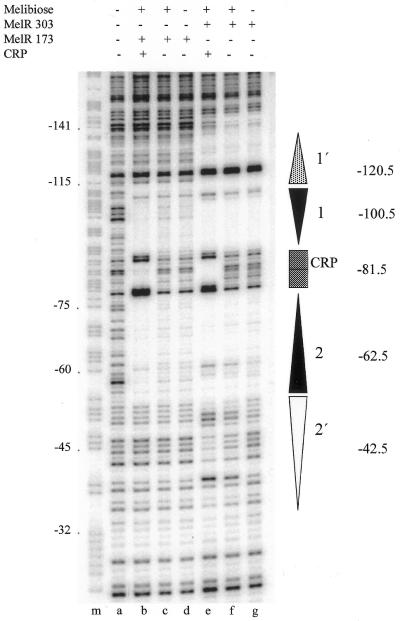
DNase I footprinting experiments were used to examine the binding of full-length MelR303 and MelR173 at pmelAB. Figure 3 shows that, as expected, MelR303 protects sites 1′, 1 and 2 from DNase I cleavage, both in the absence (lane g) and presence (lane f) of melibiose. In the presence of melibiose (Fig. 3, lane f), additional weak protection is observed at site 2′. When CRP is added (Fig. 3, lane e), the protection at site 2′ is more pronounced, and strong additional protection is observed between sites 1 and 2. This is in agreement with our previous results showing that CRP binds cooperatively with MelR between sites 1 and 2, enhancing occupation of site 2′ (20,21). MelR173 protects sites 1 and 2 from DNase I cleavage (Fig. 3, lanes c and d) but, in contrast to MelR303, there is no protection of sites 1′ or 2′ either in the presence or absence of melibiose. Addition of CRP in the presence of MelR173 causes protection of DNA between sites 1 and 2 and increases the protection at sites 1 and 2, but does not extend the footprint at site 1′ or 2′ (Fig. 3, lane b). Parallel DNase I footprinting experiments at pmelR show that MelR303 and MelR173 each protect site R from cleavage (data not shown).
Figure 3.
DNase I footprint analysis of MelR and CRP binding to pmelAB DNA. The figure shows an autoradiogram of a polyacrylamide sequencing gel on which DNA cleavage due to attack by DNase I was analysed. The DNA was the KK39 EcoRI–HindIII fragment containing the melAB promoter specifically labelled on the promoter template strand at the HindIII end. Prior to DNase I treatment, the labelled DNA was preincubated with melibiose, MelR303, MelRl73, CRP and cAMP as indicated. The gel is calibrated with a Maxam–Gilbert G+A reaction (lane m) with the melAB transcript start point as ±1. The locations of the different MelR- and CRP-binding sites are indicated as vertical arrows or boxes as in Figure 1.
EMSA
The above results show that, while MelR303 can bind to all five previously identified sites in the pmelR–pmelAB intergenic region, MelR173 only binds to the two identical sites, sites 1 and 2, and to site R. MelR173 is unable to occupy site 1′ or 2′ at pmelAB. One explanation for this could be that the differences in the sequences between sites 1′ and 2′ and sites 1 and 2 (Fig. 2) prevent MeR173 from binding to the former sites. Alternatively, the small 2 bp spacing between sites 1′ and 1 (and 2′ and 2) may prevent two molecules of MelR173 from binding simultaneously to the 18–2–18 bp repeat. To distinguish between these possibilities, we constructed a series of DNA fragments, KK98, KK99 and KK100, each containing two MelR-binding sites; a constant site with the sequence of site 2 and a second site, 2 bp away, increasingly close in sequence to site 2 (Fig. 1). The KK98 fragment contains the sequences of site 2 and site 2′, as for the pmelAB promoter but with a 2 bp change to give a NsiI restriction site between sites 2 and 2′. This gives a 2 bp change at the 3′ end of site 2′ (as oriented in Fig. 2), so it is closer to the sequence of site 2. The KK99 fragment contains site 2 and the sequence of site 1′ instead of site 2′ in KK98, and so is similar to sites 1 and 1′. The KK100 fragment contains two site 2 sequences in an inverted repeat. We then examined binding of purified MelR303 and MelR173 to each of these fragments by EMSA.
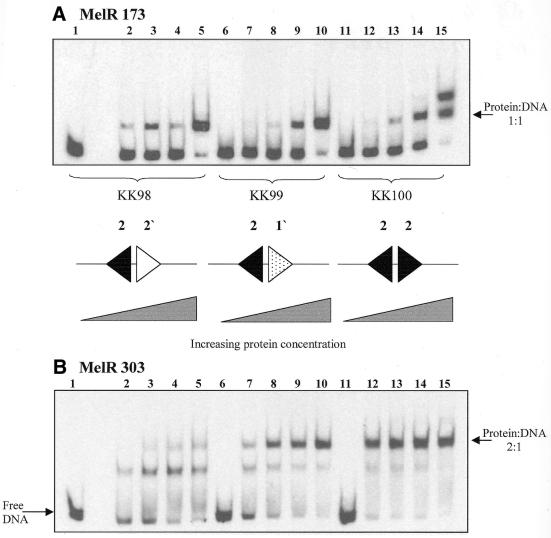
Figure 4A shows that with the DNA fragments KK98 and KK99 MelR173 gives only one band of lower mobility than free DNA, suggesting that a 1:1 protein:DNA complex is formed. In contrast, with the KK100 DNA fragment, containing two copies of site 2, two bands of lower mobility than the free DNA are observed, most likely corresponding to 1:1 and 2:1 protein:DNA complexes, respectively. To compare the relative association constants of the protein with different fragments, we used gels in which two different fragments of DNA were examined in parallel, incubated with eight different protein concentrations. Different combinations of pairs of fragments were used in order to compare the macroscopic association constants for all the fragments directly (Table 4). These measurements show that MelR173 binds with the same affinity, K1, to the DNA fragments KK98 and KK99. For the KK100 fragment, K1, the association constant of a single protein molecule, is approximately twice that for the other two fragments. K2, the association constant for two protein molecules binding simultaneously, is approximately equal to K12/4. These values of K1 and K2 are those expected for protein binding independently to two equivalent sites, each with a microscopic affinity constant the same as for KK98. These studies show that MelR173 can bind to two adjacent DNA sites independently, but does not bind to site 1′ or 2′.
Figure 4.
EMSA of MelR173 and MelR303 binding to fragments containing different pairs of MelR-binding sites. The figure shows autoradiograms of electrophoretic mobility shift assays performed with labelled KK98, KK99 and KK100 DNA fragments as indicated, with either MelR173 (A) or MelR303 (B). MelR173 concentrations: lanes 1, 6 and 11, 0 nM; lanes 2, 7 and 12, 7.6 nM; lanes 3, 8 and 13, 11.4 nM; lanes 4, 9 and 14, 38 nM; lanes 5, 10 and 15, 150 nM. MelR303 concentrations: lanes 1, 6 and 11, 0 nM; lanes 2, 7 and 12, 1.5 nM; lanes 3, 8 and 13, 4.4 nM; lanes 4, 9 and 14, 8.9 nM; lanes 5, 10 and 15, 29 nM. The sequences of the MelR-binding sites in the fragments are shown schematically as in Figure 1.
Table 4. Relative asociation constants from EMSA of MelR173 with different DNA fragments.
| DNA fragment | K1 (106 M) | K2 (1012 M2) | kA (106 M) | kB (106 M) | kAB |
|---|---|---|---|---|---|
| KK98 | 7 ± 0.73 | 0 | 7 | 0 | 0 |
| KK99 | 8.2 ± 0.8 | 0 | 8.2 | 0 | 0 |
| KK100 | 17 ± 1.8 | 65 ± 15 | 8.5 | 8.5 | 0.9 |
The K1 and K2 values are the macroscopic association constants from several gels with the data fitted simultaneously in Sigmaplot. From these K1 and K2 values, the microscopic association constants for binding to individual sites, kA and kB, and the cooperativity constant, kAB, have been estimated for the KK100 DNA fragment with two equivalent sites, assuming that kA and kB are identical. For the KK98 and KK99 fragments, kA is assumed to be equal to K1. The errors shown are those from the fit, which are similar to those obtained from fitting data from each gel individually for a given protein preparation. When different protein preparations were used the relative affinities of the pairs of fragments remained the same, as did the ratios of K1/K2, but the absolute values differed, presumably due to different specific activities of the preparations.
Figure 4B shows the equivalent experiment with MelR303. With the KK98 DNA fragment, MelR303 gives a single band of lower mobility than free DNA, with a second band of weak intensity only observed at high concentrations of protein. With the KK99 fragment, two bands of lower mobility than free DNA are observed, the second one being more intense than the lower one at medium protein concentrations. With the KK100 DNA fragment, the most retarded band is much more intense than the lower retarded band even at the lowest protein concentration. The relative macroscopic association constants to pairs of fragments were measured as described for MelR173 (Table 5). These show that K1 is similar for the KK98 and KK99 DNA fragments and about half that for the KK100 fragment, suggesting that this again is approximately equal to the affinity of binding to site 2. For the KK100 fragment, K2 is much larger than expected by independent binding to two sites, giving about a 30-fold cooperativity for binding to both sites (Table 5).
Table 5. Relative association constants from EMSA of MelR303 with different DNA fragments.
| DNA fragment | K1 (106 M) | K2 (1012 M2) | kA (106 M) | kB (106 M) | kAB |
|---|---|---|---|---|---|
| KK98 | 1.3 ± 0.2 | 2.8 ± 0.6 | 1.25 | n.d. | n.d. |
| KK99 | 1.2 ± 0.3 | 19 ± 2.2 | 1.25 | n.d. | n.d. |
| KK100 | 2.5 ± 0.3 | 46 ± 9 | 1.25 | 1.25 | 31 |
The K1 and K2 values are the macroscopic association constants from several gels with the data fitted simultaneously in Sigmaplot. From these K1 and K2 values, the microscopic association constants for binding to individual sites, kA and kB, and the cooperativity constant, kAB, have been estimated for the KK100 DNA fragment with two equivalent sites, assuming that kA and kB are identical. For the KK98 and KK99 fragments, kA is assumed to be the same as for KK100 with MelR303. The errors shown are those from the fit, which are similar to those obtained from fitting data from each gel individually for a given protein preparation. When different protein preparations were used the relative affinities of the pairs of fragments remained the same, as did the ratios of K1/K2, but the absolute values differed, presumably due to different specific activities of the preparations. The values of the association constants for this preparation are much lower than for the one used in Figures 4 and 5. n.d., not determined.
To examine the cooperativity of MelR303 binding further, a DNA fragment, VH101, containing the site 1′ sequence next to site 2′, was constructed by replacing the consensus site 2 sequence in KK98 with site 1′. No binding of MelR303 to this fragment was observed even at the highest protein concentration tested, 6.4 µM (data not shown). This shows that binding of MelR303 to site 1′ or 2′ alone is extremely weak. The binding of MelR to these sites in the KK98 and KK99 DNA fragments must therefore be due primarily to cooperative binding with MelR303 bound at the adjacent consensus sites. Independent experiments show that if site 2 is mutated no binding is seen in a fragment containing sites 2 and 2′ (32).
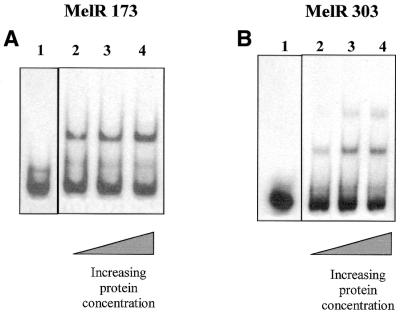
We also examined binding of both MelR303 and MelR173 to the R site, in fragment TB10 (Fig. 5). Both proteins bind to this DNA fragment in a similar way to their binding to the KK98 fragment; MelR303 gives two bands of lower mobility than the free DNA, with the second band filling at high protein concentrations, but MelRl73 gives only a single band. Comparison of the binding of MelR to the KK98 and TB10 DNA fragments on the same gel gives similar values of K1 and K2 for MelR303 for both fragments, showing that they have similar affinity, while MelR173 binds slightly less tightly to the TB10 fragment.
Figure 5.
EMSA of MelR173 and MelR303 binding to the TB10 fragment containing site R. The figure shows autoradiograms of EMSAs performed with labelled TB10 DNA fragments carrying the R site, with either MelR173 (A) or MelR303 (B). MelR173 concentrations: lane1, 0 nM; lane 2, 22 nM; lane 3, 44 nM; lane 4, 88 nM. MelR 303 concentrations: lane 1, 0 nM; lane 2, 6.7 nM; lane 3, 20 nM; lane 4, 34 nM.
DISCUSSION
To better understand the molecular mechanisms of transcription activation at pmelAB, we have compared binding of the full-length MelR transcription activator, MelR303, to that of its C-terminal domain, MelR173, which does not activate transcription, using DNase I footprinting and EMSA. The footprinting studies in this paper show that the C-terminal domain of MelR, MelR173, binds to sites 1, 2 and R in the same way as the full-length MelR303, protecting the same base pairs. Our previous studies showed that both proteins bend the DNA at site 1 or 2 to a similar extent (15). This suggests that the interactions of the two proteins with DNA are similar and so require solely the C-terminal domain of MelR. The footprinting study also shows that MelR173, when bound at sites 1 and 2, is sufficient to recruit CRP to pmelAB, between sites 1 and 2. This site contains only a few bases in common with a consensus CRP site. We have recently shown that binding of CRP and MelR are cooperative, involving protein–protein contacts (21). These heterologous contacts must occur with the C-terminal domain of MelR.
The footprinting studies on the KK39 DNA fragment and the EMSAs show that MelR173 cannot bind to site 2′ or 1′. MelR173 binds independently to the two identical sites in the KK100 fragment, showing that it can bind to two adjacent sites, but it does not bind to sites 1′ and 2′ in the KK98 or KK99 fragments. In contrast, MelR303 binds the adjacent sites in the KK100 DNA fragment with high cooperativity. It binds site 2′ or 1′ in the KK98 and KK99 fragments, where there is an adjacent site 2, but it does not bind to the VH101 fragment with site 1′ adjacent to site 2′, i.e. in the absence of site 2. This shows that binding of MelR303 at sites 1′ and 2′ in KK98 and KK99 is due solely to the cooperativity of binding with the adjacent site 2 and that this cooperativity requires the N-terminal domain of MelR. The difference in cooperativity between MelR303 and MelR173 is also seen at the R site in TB10. The DNA sequence adjacent to the R site bears little resemblance to site 2, yet MelR303 gives two retarded bands in EMSA, while MelR173 gives only one, with similar affinity to the KK98 fragment. Our interpretation of the more retarded band is that MelR303, once bound at a target site, binds weakly to the adjacent DNA due to its highly cooperative DNA binding. As the protein–DNA interactions appear to be identical in both MelR173 and MelR303, cooperativity of binding to the adjacent DNA sites is probably due to protein–protein interactions involving the N-terminal domain of MelR. These findings have important implications with respect both to the DNA specificity of MelR and its mode of transcription activation.
The binding of MelR173 and MelR303 to site R in the TB10 fragment but not to an isolated site 1′ in VH101 suggests that the R site is a stronger binding site than site 1′. However, the R site only contains 11 of 18 bp in common with site 2, while site 1′ contains 12 of 18 bp identical (Fig. 2). This suggests that the bases in site R that are the same as in site 2 but not conserved in site 1′, i.e. A6 and G15, are more important for binding MelR than those changed at site R, i.e. T8, G13 and T18. Mutational studies of the DNA-binding site of AraC (33) showed that two groups of bases, 8 bp apart, termed the A and the B boxes, are important for protein binding. The crystal structure of MarA with its cognate sequence (6) shows direct hydrogen bond contacts to two groups of bases 9 bp apart, the equivalent of the A and B boxes. One group of 3 bp contact one helix–turn–helix and the other group of 2 bp contact the other helix–turn–helix motif. Our results suggest that A6 and G15 may be part of the A box and B box elements for MelR while T8, G13 and T18 may lie outside these contact sites. Further studies to define the DNA binding site for MelR are underway.
The current studies support our previous suggestion that transcription activation at pmelAB requires the binding of MelR to site 2′, which overlaps the –35 sequence of the promoter (20). The requirement of cooperative binding of MelR at sites 2 and 2′ explains the trans-dominant negative effect of MelR173 at pmelAB, despite the inability of MelR173 to bind to the critical site 2′. MelR173 competes with MelR303 binding to site 2 and, hence, prevents the binding of MelR303 to site 2′. The inability of MelR173 to bind to site 2′ explains in part why MelR173 cannot activate transcription. The N-terminal domain is required for cooperativity while the C-terminal domain is required for DNA binding, hence, both domains are required for transcription activation of pmelAB.
Surprisingly, the optimal MelR-binding sites are not optimal for transcription activation. Base pairs –34 and –37 are critical for transcription, yet differ from site 2 (34). When these bases are retained, transcription activation by MelR303 is increased if site 2′ is changed to resemble the site 1/site 2 sequence, but is much more markedly increased with the sequence of site 1′. In both cases activation is still dependent on melibiose (20) and neither of these improved promoters is activated by MelR173 (data not shown). Hence, DNA binding is necessary but not sufficient for transcription activation.
The fact that a poor binding site at –35 enhances transcription activation by MelR emphasises the importance of the cooperative interactions between MelR proteins at adjacent sites in pmelAB. In addition to these interactions, cooperative binding of MelR with CRP also enhances transcription activation (20,21). Our studies show that both domains of MelR are involved in cooperative interactions; the C-terminal domain in interactions with CRP, while the N-terminal domain is required for interactions between MelR proteins. Cooperative binding of one or more regulatory proteins to a series of DNA binding sites at a promoter is found in many genomes. In such cases, as at pmelAB, the critical site for activation must be the weakest site, which is filled last as a result of the cooperative interactions.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Rosemary A. Parslow for skilful purification of MelR and Christine L. Webster for promoter assays in vivo. This study was funded with a BBRSC project grant and a BBSRC PhD studentship to V.J.H.
REFERENCES
- 1.Webster C.L. Kempsell,K., Booth,I. and Busby,S.J.W (1987) Organisation of the regulatory region of the Escherichia coli melibiose operon. Gene, 59, 253–263. [DOI] [PubMed] [Google Scholar]
- 2.Webster C.L., Gardner,L. and Busby,S.J.W. (1989) The E. coli melR gene encodes a DNA binding protein with affinity for specific sequences located in the melibiose-operon regulatory region. Gene, 83, 207–213. [DOI] [PubMed] [Google Scholar]
- 3.Webster C.L., Gaston,K. and Busby,S.J.W. (1988) Transcription from the E. coli melR promoter is dependent on the cyclic AMP receptor protein. Gene, 68, 297–305. [DOI] [PubMed] [Google Scholar]
- 4.Gallegos M.-T., Schleif,R., Bairoch,A., Hofmann,K. and Ramos,J.-L. (1997) AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev., 61, 393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin R.G. and Rosner,J.L. (2000) The AraC transcriptional activators. Curr. Opin. Microbiol., 4, 132–137. [DOI] [PubMed] [Google Scholar]
- 6.Rhee S., Martin,R., Rosner,J. and Davies,D. (1998) A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl Acad. Sci. USA, 95, 10413–10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon H.J., Bennik,M.H.J., Demple,B. and Ellenberger,T. (2000) Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nature Struct. Biol., 7, 424–430. [DOI] [PubMed] [Google Scholar]
- 8.Soisson S.M., MacDougall-Shackleton,B., Schlief,R. and Wolberger,C. (1997) Structural basis for ligand-related oligomerization of AraC. Science, 276, 412–425. [DOI] [PubMed] [Google Scholar]
- 9.Schleif R. (2000) Regulation of the l-arabinose operon of Escherichia coli. Trends Genet., 16, 559–565. [DOI] [PubMed] [Google Scholar]
- 10.Schleif R. (1996) Two positively regulated systems, ara and mal. In Neidhardt,F. (ed.), Escherichia coli and Salmonella. ASM Press, Washington, DC, Vol. 1, pp. 1300–1309.
- 11.Zhang X. and Schleif,R. (1998) Catabolite activator protein mutations affecting the activity of the araBAD promoter. J. Bacteriol., 180, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caswell R., Webster,C.L. and Busby,S.J.W. (1992) Studies on the binding of the E. coli MelR transcription activator protein to operator sequences at the melAB promoter. Biochem. J., 287, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams J., Michán,C., Webster,C.L. and Busby,S.J.W. (1994) Interactions between the E coli MelR transcription activator protein and operator sequences at the melAB promoter. Biochem. J., 300, 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michán C., Busby,S.J.W. and Hyde,E.I. (1995) The Escherichia coli MelR transcription activator: production of a stable fragment containing the DNA-binding domain. Nucleic Acids Res., 23, 1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourgerie S., Michán,C., Thomas,M., Busby,S.J.W. and Hyde,E.I. (1997) DNA binding and DNA bending by the MelR transcription activator protein. Nucleic Acids Res., 25, 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bustos S.A. and Schleif,R. (1993) Functional domains of the AraC protein. Proc. Natl Acad. Sci. USA, 90, 5638–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaldalu N., Toots,U., de Lorenzo,V. and Ustav,M. (2000) Functional domains of the TOL plasmid transcription factor XylS. J. Bacteriol., 182, 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen S.P., McMurry,L.M. and Levy,S.B. (1993) Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol., 175, 1484–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wade J.T., Belyaeva,T.A., Hyde,E.I. and Busby,S.J.W. (2000) Repression of the Escherichia coli melR promoter by MelR: evidence that full repression requires the formation of a repression loop. Mol. Microbiol., 36, 223–229. [DOI] [PubMed] [Google Scholar]
- 20.Belyaeva T.A., Wade,J.T., Webster,C., Howard,V.J., Thomas,M., Hyde,E.I. and Busby,S.J.W. (2000) Transcription activation at the Escherichia coli melAB promoter: the role of MelR and the cyclic AMP receptor protein. Mol. Microbiol., 36, 211–222. [DOI] [PubMed] [Google Scholar]
- 21.Wade J.T., Belyaeva,T.A., Hyde,E.I. and Busby,S.J.W. (2001) A simple mechanism for co-dependence on two activators at an Escherichia coli promoter. EMBO J., 20, 7149–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniatis T., Fritsch,E. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Studier F.W. (1991) Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol., 219, 37–44. [DOI] [PubMed] [Google Scholar]
- 24.Jair K.-W., Martin,R.G., Rosner,J.L., Nobuyuki,F., Ishihama,A. and Wolf,R.E. (1995) Purification and regulatory properties of MarA, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance properties. J. Bacteriol., 177, 7100–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamai E., Belyaeva,T.A., Busby,S.J.W. and Tsuchiya,T. (2000) Mutations that increase the activity of the promoter of the Escherichia coli melibiose operon improve the binding of MelR, a transcription activator triggered by melibiose. J. Biol. Chem., 275, 17058–17063. [DOI] [PubMed] [Google Scholar]
- 26.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 27.Pace C.N., Vajdos,F., Fee,L., Grimsley,G. and Gray,T. (1995) How to measure and predict the molar absorption-coefficient of a protein. Protein Sci., 4, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodge J., Fear,J., Busby,S.J.W., Gunasekaran,P. and Kamini,N. (1992) Broad host-range plasmids carrying the E. coli lactose and galactose operons. FEMS Lett., 95, 271–276. [DOI] [PubMed] [Google Scholar]
- 29.Miller J. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Caswell R., Williams,J., Lyddiatt,A. and Busby,S.J.W. (1992) Overexpression, purification and characterisation of the E coli MelR transcription activator protein. Biochem. J., 287, 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fried M. and Crothers,D.M. (1981) Equilibria and kinetics of lac repressor operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res., 9, 6505–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wade J.T. (2001) Transcriptional regulation in the Escherichia coli melibiose operon: interations of MelR and CRP. PhD thesis, University of Birmingham, Birmingham, UK.
- 33.Niland P. and Muller-Hill,B. (1996) How AraC interacts specifically with its target DNAs. J. Mol. Biol., 264, 667–674. [DOI] [PubMed] [Google Scholar]
- 34.Keen J., Williams,J. and Busby,S.J.W. (1996) Location of essential sequence elements at the E. coli melAB promoter. Biochem. J., 318, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelsall A., Evans,C. and Busby,S.J.W. (1985) A plasmid vector that allows fusion of the E. coli galactokinase gene to the translation start point of other genes. FEBS Lett., 180, 155–159. [Google Scholar]