Abstract
We have determined the structures of complexes between the phage MS2 coat protein and variants of the replicase translational operator in order to explore the sequence specificity of the RNA–protein interaction. The 19-nt RNA hairpins studied have substitutions at two positions that have been shown to be important for specific binding. At one of these positions, –10, which is a bulged adenosine (A) in the stem of the wild-type operator hairpin, substitutions were made with guanosine (G), cytidine (C) and two non-native bases, 2-aminopurine (2AP) and inosine (I). At the other position, –7 in the hairpin loop, the native adenine was substituted with a cytidine. Of these, only the G-10, C-10 and C-7 variants showed interpretable density for the RNA hairpin. In spite of large differences in binding affinities, the structures of the variant complexes are very similar to the wild-type operator complex. For G-10 substitutions in hairpin variants that can form bulges at alternative places in the stem, the binding affinity is low and a partly disordered conformation is seen in the electron density maps. The affinity is similar to that of wild-type when the base pairs adjacent to the bulged nucleotide are selected to avoid alternative conformations. Both purines bind in a very similar way in a pocket in the protein. In the C-10 variant, which has very low affinity, the cytidine is partly inserted in the protein pocket rather than intercalated in the RNA stem. Substitution of the wild-type adenosine at position –7 by pyrimidines gives strongly reduced affinities, but the structure of the C-7 complex shows that the base occupies the same position as the A-7 in the wild-type RNA. It is stacked in the RNA and makes no direct contact with the protein.
INTRODUCTION
The coat protein dimer of phage MS2 binds specifically to an RNA hairpin in the viral genome, thereby repressing the translation of the viral replicase. This hairpin/translational operator (TR) corresponds to nucleotide positions –15 to +4 relative to the start of the replicase gene and is 19 nt in length. Coat protein binding also stimulates self-assembly of phage particles (1) and ensures that the viral RNA is preferentially encapsidated (2,3). Only one genome is packed in each capsid, and hence the TR occupies only one of the 90 binding sites in the capsid, but RNA is associated with all binding sites in a non-sequence-specific manner.
The MS2 phage capsid is built up of 180 chemically identical copies of the coat protein, arranged in a T = 3 icosahedral shell that surrounds the RNA genome. One copy of the A protein (maturation protein) is also present in the virus particle. The coat protein molecules are present in three slightly different conformations in the shell, referred to as A, B and C (Fig. 1). These are arranged as 90 dimers of two types, A/B or C/C. Virus-like capsids that do not contain viral RNA can be obtained by over-expressing the coat protein in Escherichia coli in the absence of phage RNA (4). These largely RNA-free capsids crystallise readily in a form that is isomorphous with wild-type phage (5). Short synthetic RNA hairpins containing the TR sequence can be soaked into such crystals, and the structure of the RNA–protein complex determined by X-ray crystallography (5). One TR hairpin binds to a dimer of MS2 coat protein and there are thus 90 TR RNA binding sites in the particles.
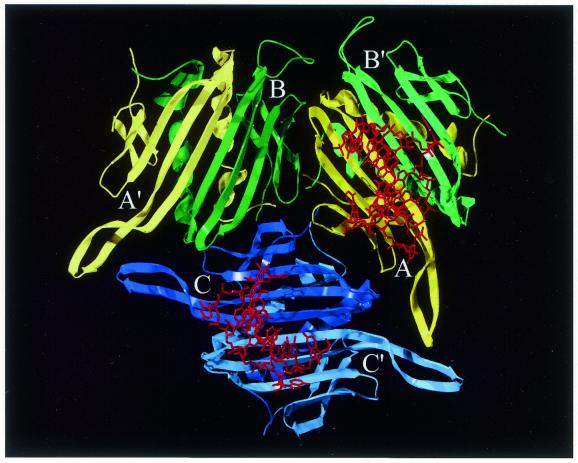
Figure 1.
A schematic drawing of the three coat protein conformers A, B and C in the viral asymmetric unit in yellow, green and blue, respectively. Also shown are the dimer-partners B′, A′ and C′ in paler colours and the wild-type RNA TR hairpin (red) binding to one of the A/B dimers and to the C/C dimer. The view is from inside the virus capsid. This figure and Figures 3–6 were prepared using SwissPdb Viewer (29) (www.expasy.ch/spdbv/) and Pov-Ray™ for Windows (www.povray.org).
The wild-type TR consists of a 7 bp stem, interrupted by one unpaired residue between base pairs five and six, and is closed by a 4-nt loop (Fig. 2A). The crystal structure of the wild-type TR complex (5,6) shows that the RNA binds to the surface of a large β-sheet in the dimer that faces the interior of the capsid. The interaction involves specific contacts with bases as well as contacts with the RNA backbone. The specificity of the interaction has been thoroughly probed by studying the binding of short (8–24 nt) synthetic RNA hairpins to coat protein dimers in solution (7–13). The bulged nucleotide and three of the loop nucleotides have been found to be important for specific, high affinity binding. The –10 base (the bulge) and the –4 base (in the loop) bind similar pockets in the two halves of a dimer, making extensive contacts through hydrogen bonding and hydrophobic interactions (5). Only an adenosine can be accommodated at the –4 position without a significant reduction of binding (7). The wild-type TR sequence has an adenosine at position –10, but binding studies have shown that guanosine gives a similar binding constant to adenosine, if the sequence in the stem is changed so that alternative conformations are avoided (13). Pyrimidines at the –10 position reduce binding at least 1000-fold (13) while modified purines give reduced or similar binding (9,11,13). The –7 base is stacked on top of the RNA helix and does not make any interactions with the protein (5). The wild-type TR has an adenosine at position –7. Hairpins with various purines at this position bind the protein with binding constants similar to the wild-type TR (9,11). Pyrimidines at the –7 position reduce binding at least 100-fold (10). Why purines are needed in this position is not clear, since the base has no direct contact with the protein. The –5 base is stacked between the –7 base and the aromatic ring of TyrA85 of the coat protein. Only pyrimidines at the –5 position result in strong binding (7,12). We have previously determined a number of structures with modifications at the –5 position (6,14,15), including purines. With a single exception (14), all naturally occurring bases and several modified bases bind to the coat protein in a similar manner.
Figure 2.
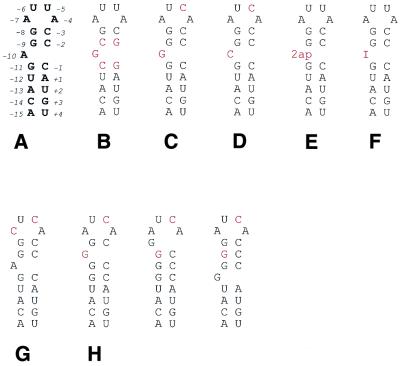
The secondary structures and sequences of: (A) the wild-type TR, numbered relative to the start of the replicase gene; (B) the G-10_2 hairpin with two GC pairs changed to CG; (C) the G-10_1 hairpin without changes in the stem; (D) the C-10 hairpin; (E) the 2AP-10 hairpin; (F) the I-10 hairpin; (G) the C-7 hairpin; and (H) the three possible alternative forms of the hairpin in (C). Bases different from the wild-type TR have been highlighted. To increase the possibility of binding, some hairpins have an additional U to C substitution on position –5. Bottom, adenine, guanine, 2-aminopurine and inosine.
In this study, we have determined the structure of an RNA hairpin with a guanosine at position –10 (Fig. 2B), in order to explore how this base is accommodated in the binding pocket. We have also determined the structures of two RNA hairpins with cytidine at positions –10 and –7, respectively (Fig. 2D and G) to probe the purine specificity at these positions. Data were collected for an additional complex with an RNA containing a guanosine at position –10, but with multiple potential alternative conformations of the stem–loop (Fig. 2C and H). This hairpin has low affinity for the coat protein according to binding studies (13) and we wished to establish whether binding to the coat protein would induce a conformation similar to the wild-type. We also collected data on two complexes with modified bases at the –10 position, 2′-deoxy-2-aminopurine (Fig. 2E) or inosine (Fig. 2F), to study the importance of the various hydrogen bonds involved in the –10 interaction.
MATERIALS AND METHODS
RNA hairpins of the wild-type TR sequence, but with the A-10 replaced by a guanosine (G-10_1), a 2′-deoxy-2-aminopurine (2AP-10) or an inosine (I-10) (Fig. 2C, E and F), were produced by solid phase synthesis and purified to homogeneity by HPLC as described (16). It has been shown that a variant hairpin with cytidine at position –5 instead of the wild-type uracil binds up to 50 times stronger to the MS2 coat protein (11,17), due to an internal hydrogen bond within the RNA (6). In order to maximise the binding, an additional U to C substitution was introduced at position –5 in the G-10_1 hairpin (Fig. 2C). An RNA hairpin with two of the wild-type GC base pairs in the stem replaced by CG base pairs and the wild-type A-10 replaced by a guanosine (G-10_2) (Fig. 2B) was also produced. The purity and expected molecular weights of the synthesised RNAs were confirmed by electrospray mass spectrometry, using negative ionisation at a concentration of 10 pmol/µl in aqueous methanol with 1% (v/v) triethylamine. Two RNA hairpins, with the wild-type A-7 replaced by a cytidine (C-7) (Fig. 2G) and the wild-type A-10 replaced by a cytidine (C-10) (Fig. 2D), were purchased from DNA Technology A/S, Denmark. Both these hairpins also contained a U to C substitution at position –5. All RNA hairpins were 19 nt in length.
The wild-type coat protein was over-expressed in E.coli (4) and the capsids purified and crystallised as described (18). Crystals grew in space group R32 with cell dimensions a = b = 288.0 Å and c = 653.0 Å in the hexagonal setting. The crystals used were 0.7–1.5 mm in the longest direction.
The crystals were soaked with RNA at a final concentration of 3 mg/ml at 30°C for 4–6 days before data collection. The crystals were mounted in glass capillaries and data collected at 5–8°C at beamlines ID14-3 and ID14-1 at ESRF, Grenoble, France, beamline 9.6 at SERC Daresbury Synchrotron Radiation Source, UK and beamline 7-11 at MAX-lab, Lund, Sweden. Data were collected at several positions on each crystal to maximum resolutions of 2.2 Å (G-10_1), 2.56 Å (G-10_2), 2.8 Å (2AP-10), 2.5 Å (I-10) 2.7 Å (C-10) and 3.3 Å (C-7). The resolution obtained was mainly dependent on the size and quality of the crystals available on each occasion. Generally, more than one crystal was used for each complex. The data were processed using the HKL package (19). Statistics for the data collection and scaling are given in Table 1. The structures were solved by molecular replacement using the empty MS2 particle as the initial model for phasing. The phases were refined by five cycles of density averaging with the program RAVE (20) using the 10-fold non-crystallographic symmetry. As a result of the averaging, the maps are of very good quality, in spite of the incompleteness of the data. The three models, G-10_2, C-10 and C-7, were built using the program O (21). Positional refinement and refinement of the restrained individual temperature factors were carried out using the program CNS (22). Statistics from the model building and refinement are shown in Table 2. The RNA occupancies were chosen so that atoms in the RNA molecule that have direct interactions with atoms in the protein have similar, but not lower, temperature factors than the protein atoms with which they interact. The coordinates have been deposited at the Protein Data Bank with ID codes 1GKW (G-10_2), 1KUO (C-10) and 1GKV (C-7).
Table 1. Statistics from data collection and scaling.
| G-10_1 | G-10_2 | 2AP-10 | I-10 | C-10 | C-7 | |
|---|---|---|---|---|---|---|
| Resolution (Å) |
30.0–2.20 |
30.0–2.56 |
28.0–2.80 |
35.0–2.50 |
40.0–2.70 |
30.0–3.30 |
| High resolution bin (Å) |
2.30–2.20 |
2.65–2.56 |
2.85–2.80 |
2.54–2.50 |
2.80–2.70 |
3.36–3.30 |
| No. of reflections |
246 981 |
254 630 |
108 577 |
191 842 |
134 487 |
123 591 |
| No. of crystals |
3 |
3 |
3 |
5 |
3 |
1 |
| Scaling R-factor |
0.196 |
0.183 |
0.153 |
0.138 |
0.210 |
0.143 |
| (high resolution bin) |
0.325 |
0.367 |
0.209 |
0.188 |
0.207 |
0.183 |
| Completeness (%) |
36 |
77 |
43 |
54 |
48 |
79 |
| (high resolution bin) |
9.4 |
8.2 |
8.4 |
8.5 |
9.4 |
30 |
| Synchrotron |
SERC, Daresbury |
ESRF, Grenoble |
MaxII, Lund |
MaxII, Lund |
MaxII, Lund |
ESRF, Grenoble |
| Wavelength (Å) |
0.877 |
0.933 |
0.894 |
0.894/1.02 |
1.085 |
0.934 |
| Oscillation angle (°) | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
Table 2. Statistics from model building and refinement.
| G-10_2 | C-10 | C-7 | |
|---|---|---|---|
|
R-factora (%) |
19.8 |
24.6 |
18.3 |
| RMS deviation of |
|
|
|
| Bond lengths (Å) |
0.008 |
0.007 |
0.010 |
| Bond angles (°) |
1.4 |
1.4 |
1.4 |
| Dihedral angles (°) |
24.3 |
24.6 |
24.0 |
| Improper angles (°) |
1.01 |
1.08 |
1.06 |
| Average B-factor of |
|
|
|
| Protein (Å2) |
40.1 |
40.1 |
23.1 |
| RNA (Å2) |
64.9 |
58.3 |
49.4 |
| H2O (Å2) |
46.1 |
49.8 |
– |
| Ramachandran outliersb (%) |
3.1 |
2.0 |
4.5 |
| Occupancy of |
|
|
|
| A/B RNA |
0.5 |
0.5 |
0.6 |
| C/C RNA |
0.4 |
0.5 |
0.4 |
| No. of nucleotides in |
|
|
|
| A/B RNA |
15 |
13 |
16 |
| C/C RNA |
11 |
13 |
13 |
| No. of H2O molecules | 136 | 147 | – |
aR-factor = Σ││Fobs│ – k│Fcalc││/Σ│Fobs│, summation over all hkl for all reflections in the working set, no cut-off was used.
bRamachandran outliers defined according to Kleywegt and Jones (30).
RESULTS
Maps and models
Electron density maps were calculated to the resolution indicated in Table 1 for the G-10_2, C-10 and C-7 complexes. The resulting maps clearly showed density for both the RNA (Fig. 3A and B) and the protein. All 129 amino acids of the three protein chains A, B and C were included in the models of these complexes.
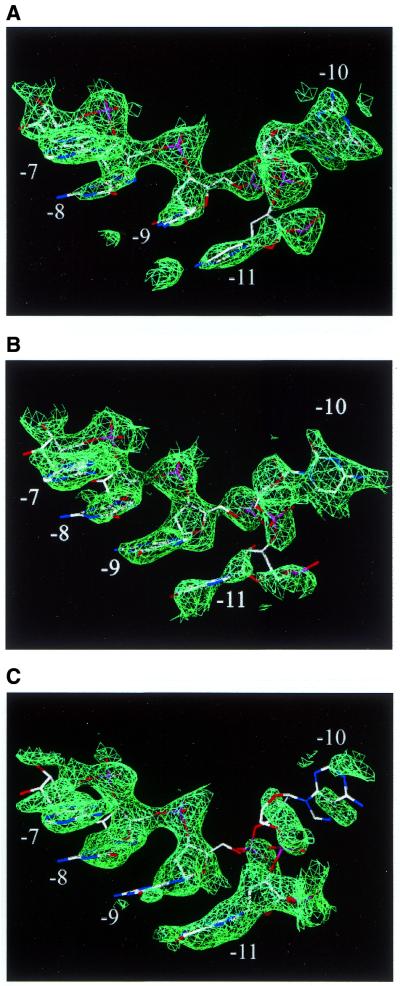
Figure 3.
Typical examples of density versus no density in the –10 binding pocket. All the examples have RNA hairpins bound to the coat protein, but the 2AP-10 does not have a base in the binding pocket. The densities are contoured at 1.6σ around parts of the (A) G-10_2 hairpin, (B) C-10 hairpin, (C) 2AP-10 hairpin (model shown is wild-type). For clarity, protein is not shown.
Since the capsid is composed of two slightly different types of coat protein dimers, there is a possibility that the RNA conformation is different at the A/B and C/C dimers. The C/C dimer is symmetric, so single RNA molecules bound to this dimer can have two different orientations, leading to a 2-fold disorder of the electron density. This disorder makes the density for the C/C RNA hairpin harder to interpret than for the RNA at the A/B dimer, which binds asymmetrically (5). Both RNA conformers were modelled for all three complexes. The models discussed below are of the RNA molecules at the A/B dimers.
In the G-10_2 complex, nucleotides –13 to +2 were built at the A/B dimer and nucleotides –11 to –1 were built at the C/C dimer. The density clearly shows a guanine in the –10 position (Fig. 3A). The number of water molecules included in the model is 137. In the C-10 complex, nucleotides –12 to +1 were built at both the A/B dimer and the C/C dimer. The density at position –10 is consistent with a cytosine (Fig. 3B). The number of water molecules included in the model is 147. In the C-7 complex, nucleotides –13 to +3 were built at the A/B dimer and nucleotides –12 to +1 were built at the C/C dimer. The density at position –7 is consistent with a cytosine at this position and in addition there is clear density for the uracil the –6 position, which is poorly defined in the wild-type TR complex. Due to the low resolution of the map, no water molecules were added. The R-factors of the refined models are 19.8% for G-10_2, 24.6% for C-10 and 18.3% for C-7. The quality of the models is summarised in Table 2.
Electron density maps were also calculated for the G-10_1, 2AP-10 and I-10 complexes. For all these complexes, the protein part of the maps was clear and essentially identical to the empty virus capsid. In the G-10_1 map, only very weak density for the RNA could be seen. The bases and phosphates in the loop were visible, but there was no density for the riboses. There was density for a few phosphates in the RNA stem, but no significant density in the –10 binding pocket. The 2AP-10 map has good density for the upper part of the hairpin, especially nucleotides –9 to –4. There is well defined density in the positions corresponding to the G-9 and G-11 bases in the wild-type complex, but no density to indicate binding of the 2AP-10 in the binding pocket (Fig. 3C). There is density close (∼1.5 Å) to where phosphates –9 and –10 are found in the wild-type complex, possibly indicating a different mode of binding or flexibility of the –10 2AP. The I-10 map showed no evidence of binding of the RNA hairpin to the protein. There is isolated density at the positions of the phosphates of the loop bases, a base stacking on TyrA85 and a base bound in the –4 binding pocket, but there is no indication of binding in the –10 pocket. The density is similar to the density found in capsids that have not been soaked with RNA (K.Valegård, N.J.Stonehouse, P.G.Stockley and L.Liljas, unpublished results) and appears to be caused by cellular E.coli RNA that binds weakly to the coat protein when the capsids are formed (23).
Effect of the A to G substitution at position –10
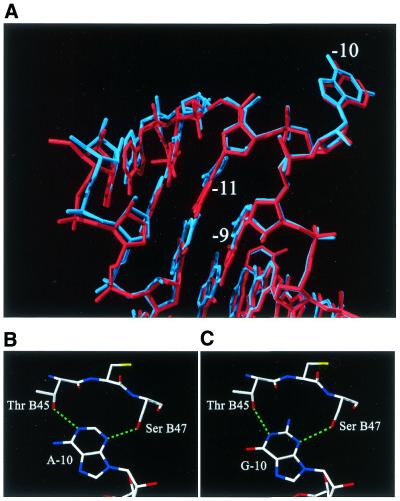
In the G-10_2 complex, the conformation of the protein as well as the RNA molecule is the same in the complex with the wild-type TR. The exchange of the two GC pairs on each side of the bulge to CG pairs is clearly visible in the map (Fig. 3A). The lower part of the stem–loop is slightly rotated towards the protein compared with the wild-type position (Fig. 4A) and the atoms of nucleotides –12, –13, +1 and +2 have moved up to 1 Å. In the wild-type complex, the adenine at position –10 has two hydrogen bonds to the protein, between N3 and Oγ of SerB47 and between N1 and Oγ1 of ThrB45. The amino group at position 6 is also close to Oγ1 of ThrB45, but the distance (3.2 Å) and angle are not optimal for a strong hydrogen bond (Fig. 4B). The two hydrogen bonds are retained in the G-10_2 complex. The base is rotated in the plane of the base to move it slightly out of the pocket (Fig. 4A). The carbonyl oxygen is displaced by 0.6–0.9 Å from the protein compared with N6 of the adenine in the wild-type or other complexes. The 2-amino group is at the bottom of the binding pocket and is close to both the carbonyl oxygen and Oγ1 of ThrB45 (Fig. 4C). The position and orientation of these groups are not favourable for the formation of a strong hydrogen bond.
Figure 4.
(A) A superposition of the A-10 (wild-type) hairpin and the G-10_2 hairpin. G-10 (blue) is somewhat rotated in comparison with A-10 (red). For clarity, protein is not shown. (B) The detailed interactions of A-10 with the coat protein. Hydrogen bonds are in green. (C) The detailed interactions of G-10 with the coat protein.
Effect of the A to C substitution at position –10
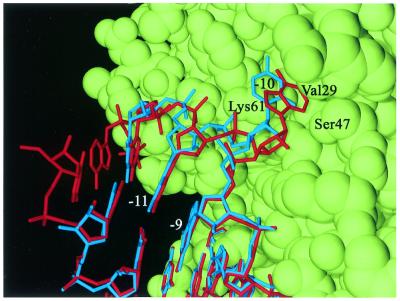
In the C-10 complex, the substitution of adenosine to cytidine at the –10 position has had no effect on conformation of the protein, except for a conformational difference of LysA61 (see Discussion). The electron density for nucleotide –10, as well as for the preceding nucleotides –11 and –12, is weaker than the density for the A-10 variant, but similar to the G-10 variant. The overall conformation of the RNA molecule is similar to the wild-type TR, but the riboses and phosphates of nucleotides –7 to –9 have moved 0.5 Å closer to the protein. At the –10 base, the base and the sugar have moved up to 1.5 Å away from the protein, while the phosphate is slightly (0.5 Å) closer (Fig. 5). This causes the cytidine base at position –10 to only partly occupy the binding pocket, and it does not come close enough to the protein to form any hydrogen bonds. The riboses and phosphates of nucleotides –11 and –12 have moved 0.6 Å away from the –10 base, parallel to the protein surface, while the bases are essentially in the same place as in the wild-type TR. The largest difference as compared with the wild-type TR model is the position of the uracil (U) at position –6, which has poorly defined density in the wild-type structure. The base, ribose and phosphate have moved up to 1.5 Å in a direction parallel to the protein surface. There is relatively good density for U-6 in the structures of two other complexes with TRs modified at the –5 position (15), and the position of U-6 in the C-10 TR is similar to the one found in these complexes. In addition, it is identical to the position of U-6 in the C-7 TR described below.
Figure 5.
A superposition of the A-10 (wild-type) hairpin in red and the C-10 hairpin in blue. The RNA hairpins are shown bound to an AB dimer of coat protein in green (VdW surface). The A-10 is inserted in a pocket formed mainly by LysB61 and ValB29. C-10 is partly inserted into the pocket, but not as deep as A-10. It does not come close enough to form hydrogen bonds to the protein.
Effect of the A to C substitution at position –7
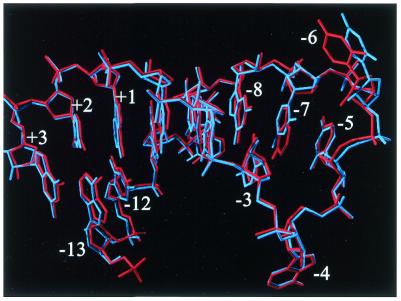
In the C-7 complex, apart from a conformational difference of LysA61, the substitution of adenosine to cytidine at the –7 position does not affect the conformation of the protein. The RNA molecule is also very similar to the wild-type TR structure. The cytosine at –7 occupies the same position as the wild-type TR adenine (Fig. 5). The rings of G-8, which C-7 stacks onto, and C-3, which base pairs with G-8, are slightly tilted compared with the wild-type model. Again, the largest difference as compared with the wild-type TR model is the position of the uracil (U) at position –6, which in this complex is identical to the position in the C-10 TR. This difference is probably not caused by –7 substitution.
DISCUSSION
In both the C-7 and the C-10 complexes, LysA61 has a conformation different from that in the wild-type complex. The electron density for the lysine side chain is weak in the wild-type complex and shows evidence of alternative conformations, where the amino group interacts with ribose –5, as previously seen for other complexes (15,24). In all these complexes, the RNA backbone is stabilised by this interaction as judged by comparatively low temperature factors for large portions of the backbone, especially for nucleotides –6 and –5. The electron density for the –6 base is also much clearer in these complexes, and slightly different from the wild-type structure. It is very difficult to correlate this conformational difference to any features of the RNA, and it might be caused by unknown factors during RNA soaking.
For some of the complexes in this study, the binding affinity observed in solution is very low. In spite of that, the RNA molecules bind to the MS2 capsids in our experiment. The affinity measurements, however, are performed with protein in vast excess but at very low concentrations. In the crystallographic experiment, RNA is added in excess. It is therefore possible also for RNA molecules with apparent low affinity to bind in sufficient amounts to be observed in the electron density maps.
Experiments with base substitutions of the bulged adenosine at position –10 in the translational TR have shown that if adenosine is substituted with guanosine and the rest of the sequence is unchanged, the binding is reduced by more than a 1000-fold (13,25). However, the binding constants for adenosine and guanosine variants were similar if the sequence in the stem was changed to avoid possible alternative conformations of the RNA hairpin (13). The attempt to produce a structure of a hairpin with the A-10 changed to guanosine and the rest of the sequence identical to the wild-type TR (hairpin G-10_1) gave poor electron density for the RNA part of the complex. There are no less than four potential alternative conformations (Fig. 2H) of the RNA stem–loop in this case. The weak density may be caused by only a small fraction of the RNA molecules having a conformation that allows binding, which results in only a fraction of the binding sites being occupied. Alternatively, all, or most of, the conformers could to some extent be able to bind to the coat protein. The electron density of the different conformers would be averaged out in the crystal and by the non-crystallographic averaging, resulting in a weak density.
When two GC base pairs in the stem are changed to CG base pairs (hairpin G-10_2), only a single conformation at the bulge is possible (Fig. 2B). The electron density clearly showed binding of this RNA hairpin (Fig. 3A). The changes of two base pairs have only minor effects on the RNA conformation and the guanine at –10 is easily accommodated in the binding pocket. The base is slightly rotated compared with the wild-type adenine to avoid a close contact between Oγ1of ThrB45 and O6 of the base and between the 2-amino group and the protein backbone at the bottom of the pocket (Fig. 4A–C). It is important to note, however, that a guanosine in the –10 position, or indeed the changes made in the stem could never be tolerated in a live phage since it would destroy the GAGG (–11 to –8) Shine–Dalgarno sequence of the replicase gene.
It is possible for both inosine and 2-aminopurine to form the same two hydrogen bonds as G-10, but the lack of density in the binding pocket suggests that that has not been the case. The similarity in binding mode and affinity of G-10 and A-10 indicates that the functional groups at C2 and C6 are not very important for the binding. A 2-aminopurine or an inosine at –10 in the context of the wild-type stem sequence could to some extent lead to alternative base pairing. In the study of Wu and Uhlenbeck (13), the affinity of RNA hairpins with these bases at position –10 and various combinations of GC and CG base pairs in the upper part of the stem follows the same pattern as the guanine variants, indicating poor binding due to multiple alternative conformations. This is consistent with the absence of RNA density for the I-10 complex. In the 2AP-10 case, however, the RNA molecule is bound to the coat protein, although the –10 base is not bound in the protein binding pocket. The weak and non-continuous density around –10 suggests that the bound molecule has conformational flexibility at this nucleotide. It has been shown that an RNA aptamer with a 2AP substitution at –10 binds with the highest affinity of any RNA in this system to date (9), but the structure of the 2AP –10 complex does not suggest any explanation of this result. It is possible that the 2AP and I modifications with the alternative stem–loop used in the G-10_2 hairpin would lead to improved binding and interpretable electron density maps.
In the structure of the C-10 complex, the –10 base is bulged out and partly inserted into the protein-binding pocket, even though no hydrogen bonds are formed between the base and ThrB45 or SerB47. Nucleotides –12 to –11 are partly disordered as an effect of the poor binding of the –10 base. The lack of interactions between the protein and the –10 base probably causes the low binding affinity of this complex. A bulged base at –10 is not necessary for binding, however, as shown by the structure of an RNA aptamer without unpaired bases in the stem (26). In the wild-type RNA in solution, the –10 base appears to be mainly intercalated in the stem (9,27). The correct positioning of the –10 base in the binding pocket in the protein stabilises the observed RNA conformation in the case of A-10 (or G-10), resulting in a high affinity. Possibly, intercalation of any base in the stem leads to a conformation of the lower part of the stem that is not compatible with binding. The C-10 RNA is therefore bound with a bulged base even if the base does not make any favourable interactions with the protein.
In the structure of the C-7 complex, the cytidine occupies the same position as the wild-type adenine. Neither this structure nor the structure of the wild-type TR complex gives an obvious explanation to why a purine is preferred at the –7 site. The base does not form any interactions with the coat protein, but is stacked onto the top of the stem. In the case of a uracil at –7, pairing with A-4 would add a new base pair to the stem. This kind of base pair is commonly seen in RNA tetraloops and stabilises the loop. Since A-4 is involved in a crucial interaction with the coat protein in the complex, such a base pair would favour a conformation unable to bind to the protein and result in lower affinity. This is supported by binding experiments where the affinity of an U-7 variant hairpin was at least 1000-fold less than wild-type, and at least 10-fold less than a C-7 variant (10). Although a cytosine at –7 could not lead to a base pair with A-4, it might lead to a loop conformation where A-4 is unavailable for binding to the coat protein. A purine, however, might, by its larger size, force A-4 to be exposed (Fig. 6). The NMR structure of an RNA hairpin similar to the MS2 TR indicates that the base –4 might be in an extended conformation in solution (28), as do fluorescence measurements of a 2AP-4 hairpin (9). An alternative explanation for the purine specificity at position –7 is that it is important for the relative orientation of the loop and stem of the bound form of the RNA, since its larger size allows good stacking both to the –5 and the –8 base. The –5 base, which is stacking to TyrA85, is important for the binding and stacks on top of the –7 base. Consistent with this explanation, there is a visibly smaller interaction surface between the cytosine at –7 and the –5 base in the C-7 complex than for the adenine in the wild-type complex, but the positions of the –5 base are identical in the two structures.
Figure 6.
A superposition of the A-7 (wild-type) hairpin in red and the C-7 hairpin in blue. The cytidine at –7 superimposes nicely with the adenosine at the corresponding position. The greatest change is at the U-6. For clarity, protein is not shown.
The results presented in this paper emphasise the complex nature of the binding of RNA to the MS2 coat protein. The structures of G-10_2 and C-10 indicate that if a bulged nucleotide is present at –10 position, it needs to bind in an extended conformation, associated with the protein, to obtain order of the lower part of the stem. From the C-7 complex, it is clear that the interaction of bound RNA and coat protein is not the only determinant of binding affinity. The conformation of the C-7 complex is essentially the same as for the wild-type TR, but the binding to the coat protein is reduced by 100-fold. This is probably due to differences in the preferred conformation and flexibility of the RNA molecules in solution.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the staff at ID14-3 and ID14-1 at the ESRF, Grenoble, at 9.6 at the SERC, Daresbury and at 7-11 at MAX-lab, Lund, for assistance with data collection. Kerstin Fridborg, Martin Hällberg, Sabah Mahdi, EvaLena Andersson and Mats Sandgren are acknowledged for help with the data collection. We also thank Alison Ashcroft and Chris J. Adams for assistance with mass spectrometry and RNA synthesis, respectively. This work was supported by the Swedish Natural Science Research Council, the Foundation for Strategic Research in Sweden, the UK BBSRC and MRC and the Leverhulme Trust, UK.
PDB nos 1GKV, 1GKW, 1KUO
REFERENCES
- 1.Beckett D. and Uhlenbeck,O.C. (1988) Ribonucleoprotein complexes of R17 coat protein and a translation operator analog. J. Mol. Biol., 204, 927–938. [DOI] [PubMed] [Google Scholar]
- 2.Beckett D., Wu,H.-N. and Uhlenbeck,O.C. (1988) Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J. Mol. Biol., 204, 939–947. [DOI] [PubMed] [Google Scholar]
- 3.Ling C.M., Hung,P.P. and Overby,L.R. (1970) Independent assembly of Qβ and MS2 phages in doubly infected Escherichia coli.Virology, 40, 920–929. [DOI] [PubMed] [Google Scholar]
- 4.Mastico R.A., Talbot,S.J. and Stockley,P.G. (1993) Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J. Gen. Virol., 74, 541–548. [DOI] [PubMed] [Google Scholar]
- 5.Valegård K., Murray,J.B., Stockley,P.G., Stonehouse,N.J. and Liljas,L. (1994) Crystal structure of an RNA bacteriophage coat protein–operator complex. Nature, 371, 623–626. [DOI] [PubMed] [Google Scholar]
- 6.Valegård K., Murray,J.B., Stonehouse,N.J., van den Worm,S., Stockley,P.G. and Liljas,L. (1997) The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein–RNA interactions. J. Mol. Biol., 270, 724–738. [DOI] [PubMed] [Google Scholar]
- 7.Carey J., Lowary,P. and Uhlenbeck,O.C. (1983) Interaction of R17 coat protein with synthetic variants of its ribonucleic acid binding site. Biochemistry, 22, 4723–4730. [DOI] [PubMed] [Google Scholar]
- 8.Lago H., Fonseca,S.A., Murray,J.B., Stonehouse,N.J. and Stockley,P.G. (1998) Dissecting the key recognition features of the MS2 bacteriophage translational repression complex. Nucleic Acids Res., 26, 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parrott A.M., Lago,H., Adams,C.J., Ashcroft,A.E., Stonehouse,N.J. and Stockley,P.G. (2000) RNA aptamers for the MS2 bacteriophage coat protein and the wild-type RNA operator have similar solution behaviour. Nucleic Acids Res., 28, 489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romaniuk P.J., Lowary,P.T., Wu,H.N., Stormo,G. and Uhlenbeck,O.C. (1987) RNA binding site of R17 coat protein. Biochemistry, 26, 1563–1568. [DOI] [PubMed] [Google Scholar]
- 11.Stockley P.G., Stonehouse,N.J., Murray,J.B., Goodman,S.T.S., Talbot,S.J., Adams,C.J., Liljas,L. and Valegård,K. (1995) Probing sequence-specific RNA recognition by the bacteriophage MS2 coat protein. Nucleic Acids Res., 23, 2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talbot S.J., Goodman,S., Bates,S.R.E., Fishwick,C.W.G. and Stockley,P.G. (1990) Use of synthetic oligoribonucleotides to probe RNA–protein interactions in the MS2 translational operator complex. Nucleic Acids Res., 18, 3521–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H.N. and Uhlenbeck,O.C. (1987) Role of a bulged A residue in a specific RNA–protein interaction. Biochemistry, 26, 8221–8227. [DOI] [PubMed] [Google Scholar]
- 14.Grahn E., Stonehouse,N.J., Adams,C.J., Fridborg,K., Beigelman,L., Matulic-Adamic,J., Warriner,S.L., Stockley,P.G. and Liljas,L. (2000) Deletion of a single hydrogen bonding atom from the MS2 RNA operator leads to dramatic rearrangements at the RNA–coat protein interface. Nucleic Acids Res., 28, 4611–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grahn E., Moss,T., Helgstrand,C., Fridborg,K., Sundaram,M., Tars,K., Stonehouse,N.J., Stockley,P.G. and Liljas,L. (2001) Structures of MS2–RNA complexes with variant bases at position –5. RNA, 7, 1616–1627. [PMC free article] [PubMed] [Google Scholar]
- 16.Murray J.B., Collier,A.K. and Arnold,J.R.P. (1994) A general purification procedure for chemically synthesized oligoribonucleotides. Anal. Biochem., 218, 177–184. [DOI] [PubMed] [Google Scholar]
- 17.Lowary P.T. and Uhlenbeck,O.C. (1987) An RNA mutation that increases the affinity of an RNA-protein interaction. Nucleic Acids Res., 15, 10483–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valegård K., Unge,T., Montelius,I., Strandberg,B. and Fiers,W. (1986) Purification, crystallization and preliminary X-ray data of the bacteriophage MS2. J. Mol. Biol., 190, 587–591. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z. and Minor,W. (1996) Processing of X-ray diffraction data collected in oscillation mode. In Carter,C.W.,Jr and Sweet,R.M. (eds), Methods in Enzymology. Academic Press, New York, Vol. 276, pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 20.Kleywegt G.J. and Jones,T.A. (1994) Halloweenºmasks and bones. In Bailey,S., Hubbard,R. and Waller,D. (eds), From First Map to Final Model. Proceedings of the CCP4 Study Weekend. SERC Daresbury Laboratory, Daresbury, pp. 59–66.
- 21.Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr., A47, 110–119. [DOI] [PubMed] [Google Scholar]
- 22.Brünger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.-S., Kuszewski,J., Nilges,M., Pannu,N.S., Read,R.J., Rice,L.M., Simonson,T. and Warren,G.L. (1998) Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr., D54, 905–921. [DOI] [PubMed] [Google Scholar]
- 23.Pickett G.G. and Peabody,D.S. (1993) Encapsidation of heterologous RNAs by bacteriophage MS2 coat protein. Nucleic Acids Res., 21, 4621–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grahn E., Stonehouse,N.J., Murray,J.B., van den Worm,S., Valegård,K., Fridborg,K., Stockley,P.G. and Liljas,L. (1999) Crystallographic studies of RNA hairpins in complexes with recombinant MS2 capsids and implications for binding requirements. RNA, 5, 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlenbeck O.C., Carey,J., Romaniuk,P., Lowary,P.T. and Beckett,D. (1983) Interaction of R17 coat protein with its RNA binding site for translational repression. J. Biomol. Struct. Dyn., 1, 539–552. [DOI] [PubMed] [Google Scholar]
- 26.Rowsell S., Stonehouse,N.J., Convery,M.A., Adams,C.J., Ellington,A.D., Hirao,I., Peabody,D.S., Stockley,P.G. and Phillips,S.E.V. (1998) Crystal structure of a series of RNA aptamers complexed to the same protein target. Nature Struct. Biol., 5, 970–975. [DOI] [PubMed] [Google Scholar]
- 27.Borer P.N., Lin,Y., Wang,S., Roggenbuck,M.W., Gott,J.M., Uhlenbeck,O.C. and Pelczer,I. (1995) Proton NMR and structural features of a 24-nucleotide RNA hairpin. Biochemistry, 34, 6488–6503. [DOI] [PubMed] [Google Scholar]
- 28.Smith J.S. and Niconowicz,E.P. (1998) NMR structure and dynamics of an RNA motif common to the spliceosome branch-point helix and the RNA-binding site for phage GA coat protein. Biochemistry, 37, 13486–13498. [DOI] [PubMed] [Google Scholar]
- 29.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modelling. Electrophoresis, 18, 2718–2723. [DOI] [PubMed] [Google Scholar]
- 30.Kleywegt G.J. and Jones,T.A. (1996) Phi/psi-chology: Ramachandran revisited. Structure, 4, 1395–1400. [DOI] [PubMed] [Google Scholar]