Abstract
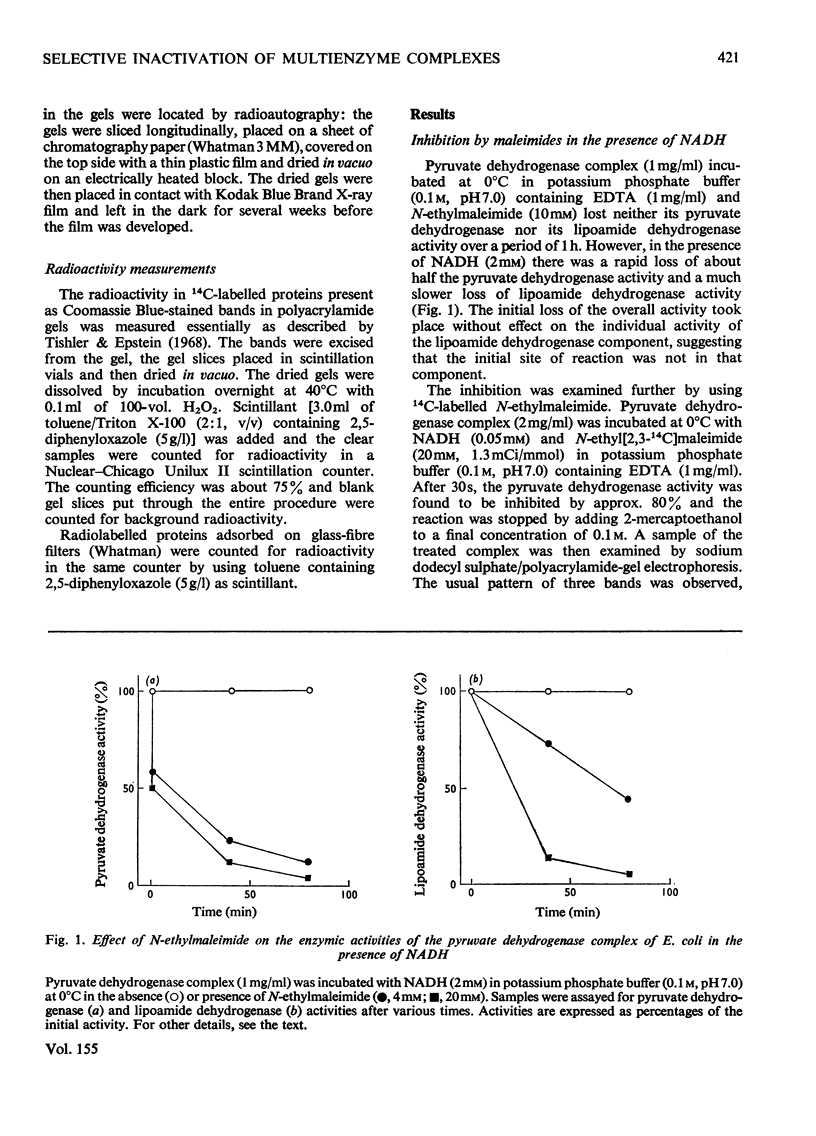
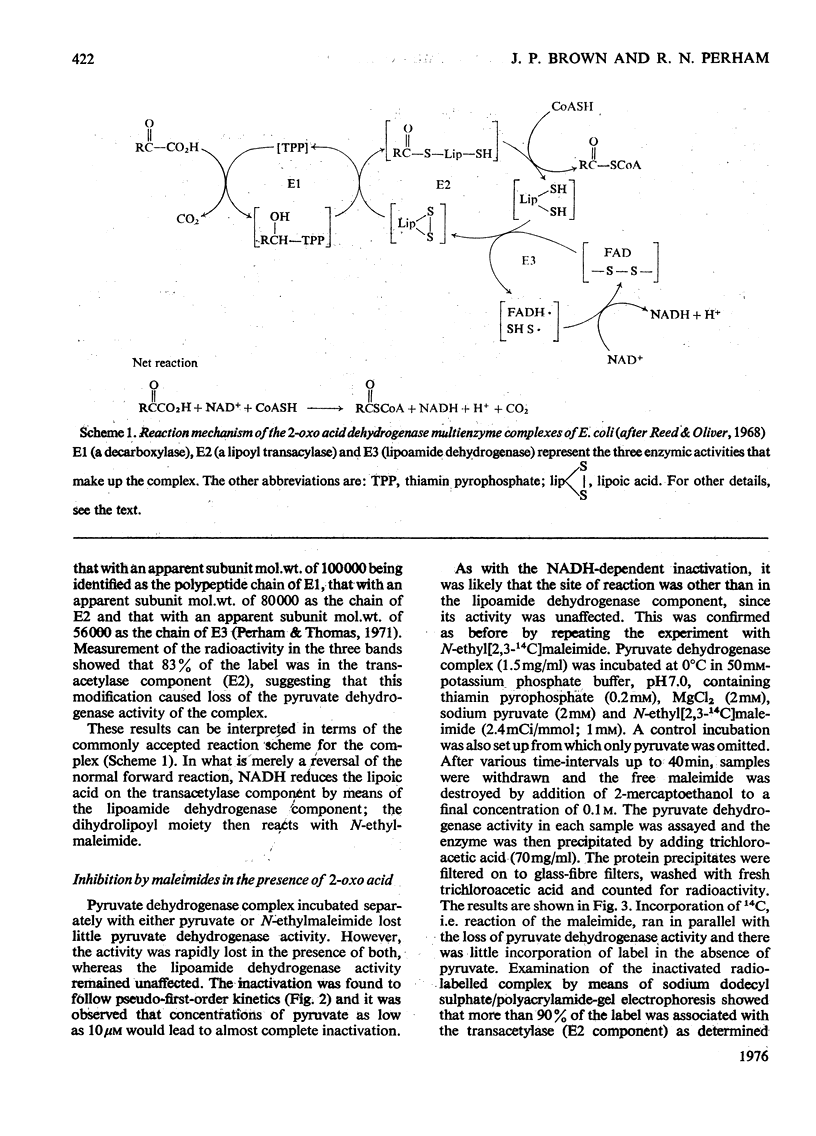
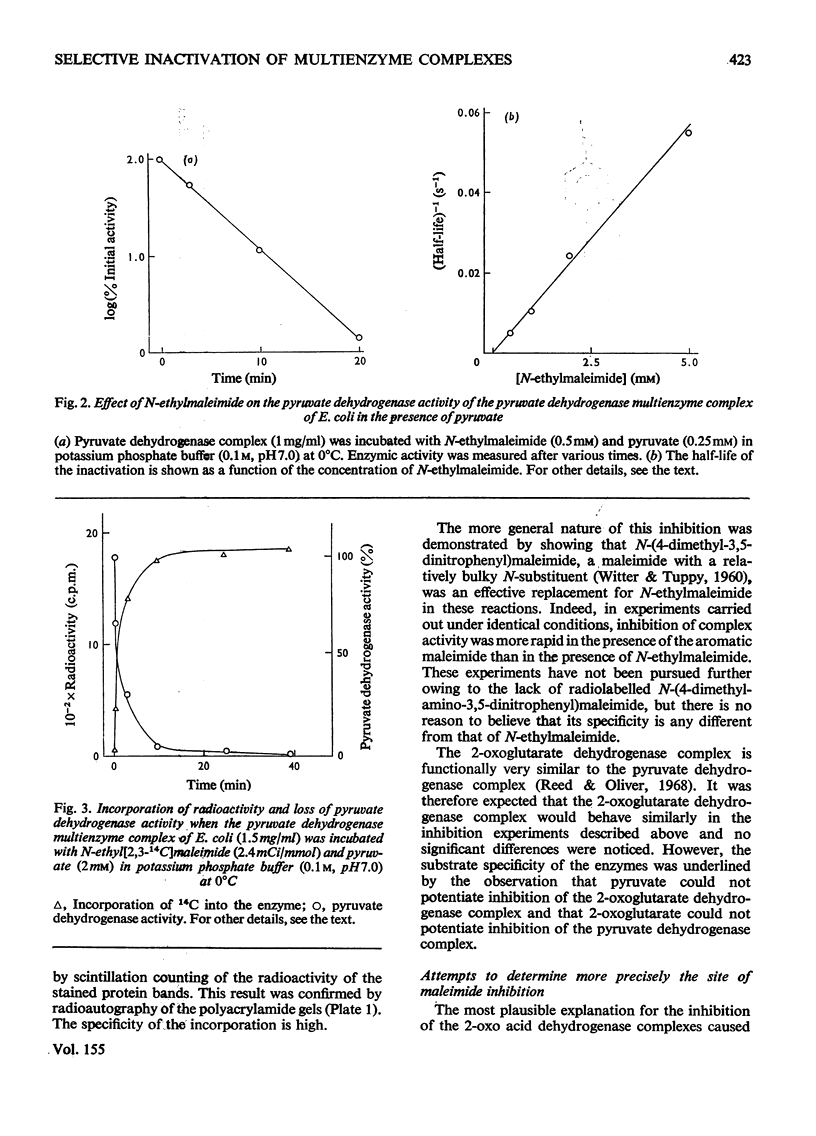
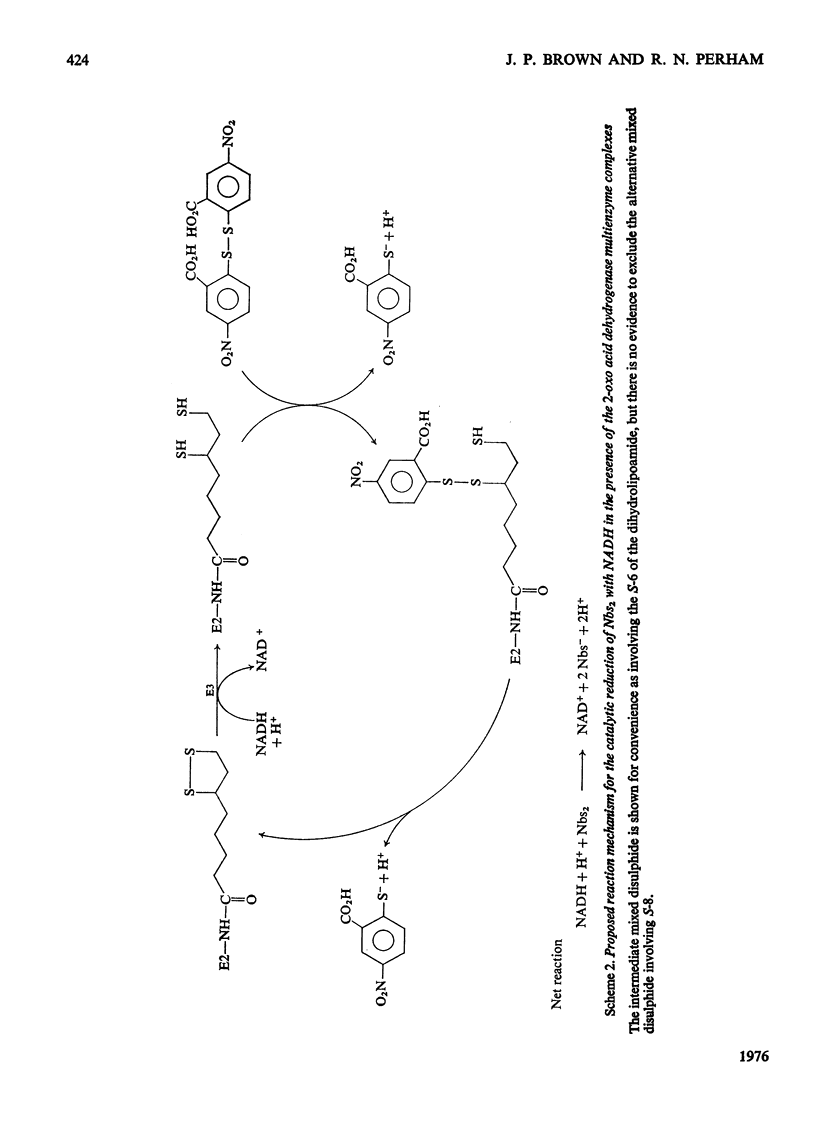
1. The reaction of the pyruvate dehydrogenase multienzyme complex of Escherichia coli with maleimides was examined. In the absence of substrates, the complex showed little or no reaction with N-ethylmaleimide. However, in the presence of pyruvate and N-ethylmaleimide, inhibition of the pyruvate dehydrogenase complex was rapid. Modification of the enzyme was restricted to the transacetylase component and the inactivation was proportional to the extent of modification. The lipoamide dehydrogenase activity of the complex was unaffected by the treatment. The simplest explanation is that the lipoyl groups on the transacetylase are reductively acetylated by following the initial stages of the normal catalytic cycle, but are thereby made susceptible to modification. Attempts to characterize the reaction product strongly support this conclusion. 2. Similarly, in the presence of N-ethylmaleimide and NADH, much of the pyruvate dehydrogenase activity was lost within seconds, whereas the lipoamide dehydrogenase activity of the complex disappeared more slowly: the initial site of the reaction with the complex was found to be in the lipoyl transacetylase component. The simplest interpretation of these experiments is that NADH reduces the covalently bound lipoyl groups on the transacetylase by means of the associated lipoamide dehydrogenase component, thereby rendering them susceptible to modification. However, the dependence of the rate and extent of inactivation on NADH concentration was complex and it proved impossible to inhibit the pyruvate dehydrogenase activity completely without unacceptable modification of the other component enzymes. 3. The catalytic reduction of 5,5'-dithiobis-(2-nitrobenzoic acid) by NADH in the presence of the pyruvate dehydrogenase complex was demonstrated. A new mechanism for this reaction is proposed in which NADH causes reduction of the enzyme-bound lipoic acid by means of the associated lipoamide dehydrogenase component and the dihydrolipoamide is then oxidized back to the disulphide form by reaction with 5,5'-dithiobis-(2-nitrobenzoic acid). 4. A maleimide with a relatively bulky N-substituent, N-(4-diemthylamino-3,5-dinitrophenyl)maleimide, was an effective replacement for N-ethylmaleimide in these reactions with the pyruvate dehydrogenase complex. 5. The 2-oxoglutarate dehydrogenase complex of E. coli behaved very similarly to the pyruvate dehydrogenase complex, in accord with the generally accepted mechanisms of the two enzymes. 6. The treatment of the 2-oxo acid dehydrogenase complexes with maleimides in the presence of the appropriate 2-oxo acid substrate provides a simple method for selectively inhibiting the transacylase components and for introducing reporter groups on to the lipoyl groups covalently bound to those components.
Full text
PDF

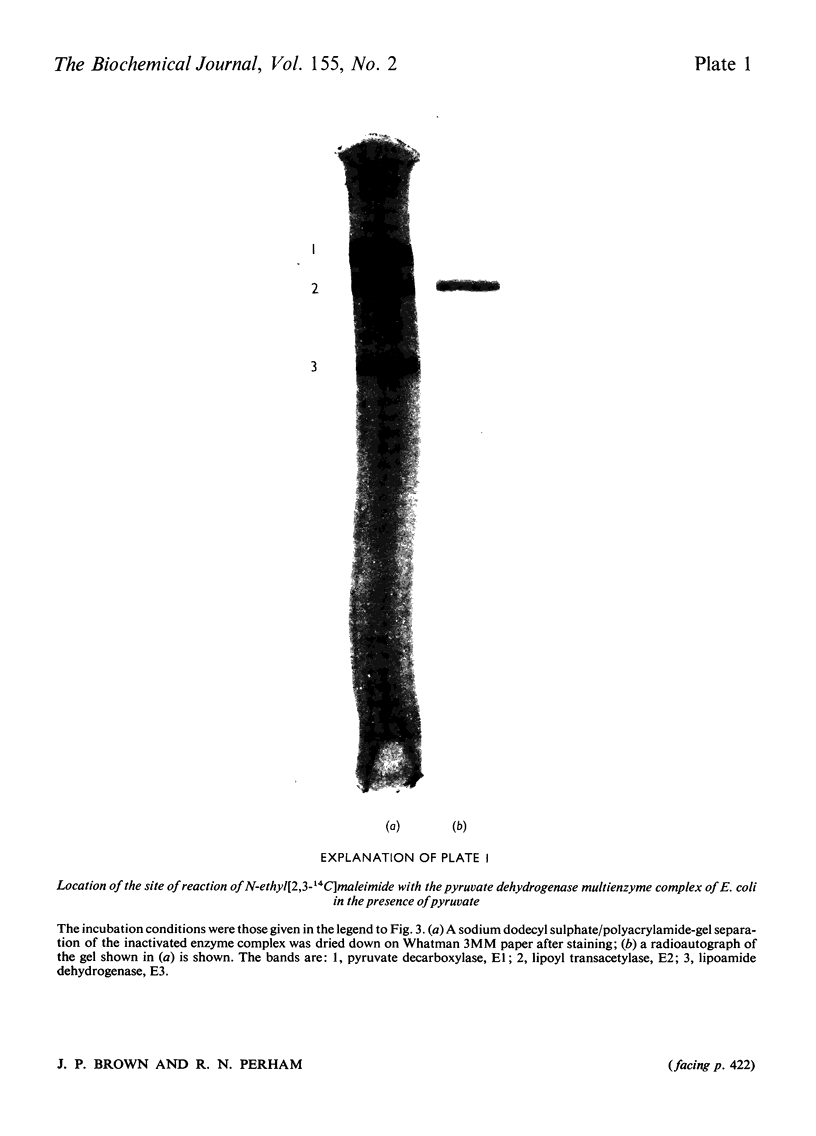



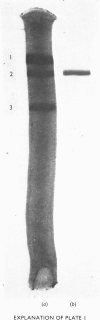
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrose M. C., Perham R. N. Spin-label study of the mobility of enzyme-bound lipoic acid in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Biochem J. 1976 May 1;155(2):429–432. doi: 10.1042/bj1550429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. L., Harrison R. A., Perham R. N. The stoichiometry of polypeptide chains in the pyruvate dehydrogenase multienzyme complex of E. coli determined by a simple novel method. FEBS Lett. 1975 Dec 15;60(2):427–430. doi: 10.1016/0014-5793(75)80764-2. [DOI] [PubMed] [Google Scholar]
- Brown J. P., Perham R. N. A highly sensitive method for amino-acid analysis by a double-isotope-labelling technique using dansyl chloride. Eur J Biochem. 1973 Nov 1;39(1):69–73. doi: 10.1111/j.1432-1033.1973.tb03104.x. [DOI] [PubMed] [Google Scholar]
- Brown J. P., Perham R. N. An amino acid sequence in the active site of lipoamide dehydrogenase from pig heart. Biochem J. 1974 Mar;137(3):505–512. doi: 10.1042/bj1370505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. P., Perham R. N. An amino acid sequence in the active site of lipoamide dehydrogenase from the 2-oxoglutarate dehydrogenase complex of E. coli (Crookes strain). FEBS Lett. 1972 Oct 1;26(1):221–224. doi: 10.1016/0014-5793(72)80577-5. [DOI] [PubMed] [Google Scholar]
- Burleigh B. D., Jr, Williams C. H., Jr The isolation and primary structure of a paptide containing the oxidation-reduction active cystine of Escherichia coli lipoamide dehydrogenase. J Biol Chem. 1972 Apr 10;247(7):2077–2082. [PubMed] [Google Scholar]
- Eley M. H., Namihira G., Hamilton L., Munk P., Reed L. J. -Keto acid dehydrogenase complexes. 18. Subunit composition of the Escherichia coli pyruvate dehydrogenase complex. Arch Biochem Biophys. 1972 Oct;152(2):655–669. doi: 10.1016/0003-9861(72)90262-7. [DOI] [PubMed] [Google Scholar]
- Erfle J. D., Sauer F. An NADH dependent cleavage of DTNB by the alpha-ketoglutarate dehydrogenase complex. Biochem Biophys Res Commun. 1968 Aug 13;32(3):562–567. doi: 10.1016/0006-291x(68)90700-6. [DOI] [PubMed] [Google Scholar]
- GUIDOTTI G., KONIGSBERG W. THE CHARACTERIZATION OF MODIFIED HUMAN HEMOGLOBIN. I. REACTION WITH IODOACETAMIDE AND N-ETHYLMALEIMIDE. J Biol Chem. 1964 May;239:1474–1484. [PubMed] [Google Scholar]
- KOIKE M., REED L. J., CARROLL W. R. alpha-Keto acid dehydrogenation complexes. IV. Resolution and reconstitution of the Escherichia coli pyruvate dehydrogenation complex. J Biol Chem. 1963 Jan;238:30–39. [PubMed] [Google Scholar]
- Moe O. A., Jr, Lerner D. A., Hammes G. G. Fluorescence energy transfer between the thiamine diphosphate and flavine adenine dinucleotide binding sites on the pyruvate dehydrogenase multienzyme complex. Biochemistry. 1974 Jun 4;13(12):2552–2557. doi: 10.1021/bi00709a012. [DOI] [PubMed] [Google Scholar]
- Perham R. N. Self-assembly of biological macromolecules. Philos Trans R Soc Lond B Biol Sci. 1975 Nov 6;272(915):123–136. doi: 10.1098/rstb.1975.0075. [DOI] [PubMed] [Google Scholar]
- Perham R. N., Thomas J. O. The subunit molecular weights of the alpha-ketoacid dehydrogenase multienzyme complexes from E. coli. FEBS Lett. 1971 Jun 2;15(1):8–12. doi: 10.1016/0014-5793(71)80066-2. [DOI] [PubMed] [Google Scholar]
- Pettit F. H., Reed L. J. Alpha-keto acid dehydrogenase complexes. 8. Comparison of dihydrolipoyl dehydrogenases from pyruvate and alpha-ketoglutarate dehydrogenase complexes of Escherichia coli. Proc Natl Acad Sci U S A. 1967 Sep;58(3):1126–1130. doi: 10.1073/pnas.58.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. J., Oliver R. M. The multienzyme alpha-keto acid dehydrogenase complexes. Brookhaven Symp Biol. 1968 Jun;21(2):397–412. [PubMed] [Google Scholar]
- Shapiro A. L., Maizel J. V., Jr Molecular weight estimation of polypeptides by SDS-polyacrylamide gel electrophoresis: further data concerning resolving power and general considerations. Anal Biochem. 1969 Jun;29(3):505–514. doi: 10.1016/0003-2697(69)90335-2. [DOI] [PubMed] [Google Scholar]
- Tishler P. V., Epstein C. J. A convenient method of preparing polyacrylamide gels for liquid scintillation spectrometry. Anal Biochem. 1968 Jan;22(1):89–98. doi: 10.1016/0003-2697(68)90262-5. [DOI] [PubMed] [Google Scholar]
- Vogel O., Hoehn B., Henning U. Molecular structure of the pyruvate dehydrogenase complex from Escherichia coli K-12. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1615–1619. doi: 10.1073/pnas.69.6.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WITTER A., TUPPY H. N-(4-Dimethylamino-3,5-dinitrophenyl)maleimide: a coloured sulfhydryl reagent. Isolation and investigation of cysteine-containing peptides from human and bovine serum albumin. Biochim Biophys Acta. 1960 Dec 18;45:429–442. doi: 10.1016/0006-3002(60)91480-3. [DOI] [PubMed] [Google Scholar]
- Wassarman P. M., Major J. P. The reactivity of the sulfhydryl groups of lobster muscle glyceraldehyde 3-phosphate dehydrogenase. Biochemistry. 1969 Mar;8(3):1076–1082. doi: 10.1021/bi00831a039. [DOI] [PubMed] [Google Scholar]