Abstract
Bacterial soft rot causes major crop losses annually and can be caused by several species from multiple genera. These bacteria have a broad host range and often infect produce through contact with soil. The main genera causing bacterial soft rot are Pectobacterium and Dickeya, both of which have widespread geographical distribution. Because of many recent renaming and reclassifications of bacteria causing soft rot, identification and characterization of the causative agents can be challenging. In this work, we surveyed commercially available produce exhibiting typical soft rot symptoms, isolating pectinolytic bacteria and characterizing them genetically and phenotypically. We found that in our sampling, many samples were from the genus Pectobacterium; however, other genera were also capable of eliciting symptoms in potatoes, including an isolate from the genus Chryseobacterium. Genomic analyses revealed that many of the Pectobacterium isolates collected share prophages not found in other soft rot species, suggesting a potential role for these prophages in the evolution or fitness of these isolates. Our Chryseobacterium isolate was most similar to C. scophthalmum, a fish pathogen, suggesting that this isolate may be a crossover pathogen.
Keywords: soft rot, pectinolytic, Pectobacterium, Pseudomonas, Chryseobacterium
1. Introduction
Bacterial soft rot is a disease complex that causes severe crop loss worldwide. Multiple bacterial genera cause this disease, with the genera Pectobacterium and Dickeya being among the most common [1]. These pathogens leave necrotic spots on the soft tissues of many plants, such as stems, leaves, tubers, and flesh, of a wide variety of cultivated vegetables. Soft rot is usually observed as the gradual presence of water-soaked lesions where the tissue becomes depressed, soft, mushy, slimy, and discolored [2]. This results in significant yield losses, affecting both the pre-harvest period and the post-harvest period due to latent infection [3]. Soft rot can occur over a wide temperature range, with most pathogens infecting optimally in the range of 21–27 °C, and is most severe when oxygen is limited [4]. Crops in storage, such as potato tubers, may have restricted access to oxygen when stacked on top of each other with limited airflow [5]. Pectobacterium and Dickeya survive in the soil and on the surface of crops, infecting plants through natural openings and wound sites [6]. Currently, there are no effective treatments once soft rot has infected plant tissues.
Bacteria of the Pectobacteriaceae family release a variety of pectinases as exoenzymes and endoenzymes for pectin degradation [7]. These enzymes break down pectin, a polysaccharide present in the middle lamella of plant cell walls, allowing access to a nutrient-rich environment [8]. Pectinases are classified into hydrolases, esterases, and lyases based on their active sites. Hydrolases, also known as polygalacturonases (PGs), break down pectic substances by catalyzing the hydrolysis of α-1,4-glycosidic bonds between galacturonic monomers that form chains in the pectin molecule. Esterases, also known as pectin methylesterases (PMEs), remove the acetyl and methoxyl groups from pectin. Lyases catalyze the cleavage of glycosidic bonds through β elimination mechanisms [9]. These enzymes have been isolated and used for industrial applications [10]. When Pectobacteriaceae bacteria start to infect the plant and grow quickly in numbers, an overwhelming amount of pectinase enzymes are produced, which results in soft rot. Pectin binds plant cells together, thus causing plant structures to fall apart into mushy, discolored, and sunken pits when infected. Members of the Pectobacterium and Dickeya genera are rod-shaped, facultative anaerobes and are motile by means of peritrichous flagella [11].
Although Pectobacterium and Dickeya are perhaps the most established genera associated with soft rot [12], a variety of other species have been implicated, including Pseudomonas, Bacillus, and Clostridium. Pseudomonas, a large and diverse group of bacteria [13], has been identified through 16S rDNA sequencing as a causative agent of soft rot in potatoes [14]. The versatile, spore-forming genus Bacillus has also exhibited pathogenicity on apples, pears, and other produce [15]. However, it has also been proposed as a potential biological control method against the soft rot pathogen Pectobacterium carotovorum [6]. Clostridium, a spore-forming, primarily anaerobic genus, has been isolated from symptomatic sweet potatoes and other crops [16].
As classification techniques have become more sophisticated and accurate, methods of genome sequencing, such as whole-genome sequencing, have developed, leading to the necessity of the reclassification of previously identified species, including those causing soft rot [17]. For example, the once broad genera Erwinia became divided into subcategories Pectobacterium and Dickeya through 16s rRNA comparative analysis [17,18]. This is a critical differentiation in the field of soft rot and plant disease because Pectobacterium and Dickeya are distinct in their host range and pathogenicity, making proper classification crucial for studying and understanding the virulence mechanisms of pathogenic bacteria. Sophisticated classification efforts will almost certainly yield the identification of novel and previously unknown pathogenic bacteria contributing to soft rot [19]. However, as more novel bacteria are identified, the demand for increasingly advanced molecular techniques will rise to ensure accurate and rapid identification. Technology that is more expensive and less widely available will potentially be needed to make precise classifications between isolates of high similarity, complicating soft rot research endeavors in lower-resource areas and certain laboratories.
Additionally, there may be regulatory concerns implicated with advancements in diagnostic technology, especially in the field of agriculture. In August 2019, the U.S. Department of Agriculture ceased requiring permits for the interstate transfer of Pectobacterium carotovorum strains originating from within the continental United States (Federal Register #2019-13246). However, two months later, a novel species, P. versatile, was formed from isolates formerly considered P. carotovorum [20]. Although the ongoing refinement and revision of pathogen taxonomy can create improved classification and understanding of pathogen behavior, it can also foster confusion around regulatory requirements for newly formed groups. Rapidly changing taxonomy also creates differences in the reporting of diagnostic results from different laboratories, which limits the understanding of which pathogens are present in specific geographic areas.
In our study, we surveyed pectinolytic bacteria in commercial produce and characterized the isolates obtained via both genotypic and phenotypic methods. As expected, we identified Pectobacterium species; however, we also found isolates from several other genera, including isolates pathogenic to potatoes that represent potentially novel species. We further found an isolate of Chryseobacterium scophthalmum, closely related to fish pathogens [21,22], which may represent a crossover pathogen. While the Pectobacterium isolates were highly similar, phenotypic differences and distinct prophage profiles revealed differences between strains. Our survey advances our understanding of the pathogens currently causing soft rot in consumer-available produce and provides a foundation for further work into the mechanisms that make these pathogens successful.
2. Materials and Methods
2.1. Culture and Isolation Conditions
Unless otherwise specified, bacterial isolates were routinely cultured at 30 °C in LB medium (10 g/L tryptone, 5 g/L NaCl, 5 g/L yeast extract; for solid media: 15 g/L agar). Produce samples used in isolations were selected from commercially acquired produce and exhibited symptoms commonly associated with bacterial soft rot, primarily water-soaked tissues and sunken lesions. Samples used for isolation were all collected in 2024 and are specified in Supplemental Table S1. Because the produce was commercially obtained from consumer grocery stores, data regarding the location of cultivation or the specific varieties of plants are unknown.
Original isolations of bacteria from symptomatic plant tissues were conducted using crystal violet pectate medium (CVPM) [23]. Briefly, an inoculation loop was used to scrape water-soaked tissue from the plant sample and streaked for isolation on CVPM plates. Colonies forming sunken pits on CVPM were re-isolated using CVPM until pure cultures were obtained.
Genetic identification of the bacteria was conducted via Sanger sequencing of the 16S rDNA gene. Briefly, a fragment of the 16S rDNA gene was PCR-amplified using primers 27F and 1492R [24]. PCR fragments were purified using ethanol precipitation and digestion with exonuclease I and alkaline phosphatase (New England Biolabs, Ipswich, MA, USA). Sanger sequencing reactions and capillary electrophoresis of purified DNA fragments were conducted by the BYU DNA Sequencing Center (Provo, UT, USA) with either the 27F or 1492R primers. A BLAST [25] of the obtained sequences against the NCBI non-redundant database was used to determine the genus and, when possible, the predicted species of the isolate, taking the top BLAST hit as the putative identity of the isolate.
2.2. Phenotypic Characterizations
All pectinolytic isolates were tested using the following phenotypic tests: pathogenicity to potato and Chinese (Napa) cabbage, swimming motility, swarming motility, production of acyl homoserine lactones (AHLs), lactose fermentation, and protease production.
2.2.1. Pathogenicity Assays
For potato pathogenicity, 1 cm slices of russet potatoes were prepared and placed in sterile petri dishes. Potato inoculum was prepared by growing each isolate overnight in LB and normalizing the culture density to OD600 = 0.4. A 2 µL droplet (~2 × 106 CFU) was inoculated on the center of each potato slice. For Chinese (Napa) cabbage pathogenicity, cabbage leaves were placed in plastic totes cleaned with 70% ethanol. For cabbage inoculations, bacterial isolates were grown for 24 h on LB agar, and single colonies were picked with a pipette tip and inoculated by stabbing the tip 2 mm into the surface of the cabbage leaf. Inoculated produce was incubated at 30 °C and evaluated for symptom development every 24 h for 3 days. Symptoms included water soaking, tissue maceration, and discoloration. Potato slices were photographed and analyzed using ImageJ [26] to determine the surface area covered by disease 48 h post-inoculation.
2.2.2. Swimming and Swarming Motility
Swimming and swarming motility assays were conducted as described previously [27]. In short, soft agar plates containing 0.25% agar (swim) or 0.3% agar (swarm) were prepared. Swim plates were inoculated by stab inoculation, and swarm plates were surface inoculated with 3 µL of overnight culture. Plates were photographed 24 h post-inoculation, and the area covered by motile bacteria was quantified using ImageJ [26].
2.2.3. AHL Production
Acyl homoserine lactone production was assessed as described [28,29] using the Agrobacterium fabrum reporter strain NTL4 pZLR4. Briefly, soft agar plates containing the A. fabrum reporter as well as X-gal (5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside) were prepared. A 3 µL droplet of overnight culture of each isolate was dripped onto a plate containing the reporter strain and incubated overnight. A positive reaction was evidenced by the production of a blue color in the soft agar surrounding the test strain.
2.2.4. Lactose Fermentation
LB agar medium was prepared to contain 1% (w/v) lactose and phenol red dye as a pH indicator. At neutral pH, the media is a bright red color. When isolates were grown on this media, a color change from red to yellow was interpreted as a lowering of the pH in the media around colonies fermenting lactose.
2.2.5. Extracellular Protease Activity
LB agar containing 1% (w/v) skim milk powder was prepared, and isolates were cultured in this medium. A halo of clearing around colonies was taken as evidence of extracellular protease activity.
2.2.6. Biofilm Formation
Biofilm formation by each isolate was assayed using 96-well plates as described previously [30]. Briefly, isolates were grown overnight in LB broth. A 5 µL volume of overnight culture, standardized to an absorbance of 0.4, was added to 100 µL of LB in the well of a 96-well plate, and the plate was incubated at 30 °C for 48 h. Following incubation, media and planktonic cells were removed and plates were allowed to dry. Adherent cells were stained with 100 µL of 1% crystal violet, and cells were washed three times with deionized water. The dye was solubilized using 70% ethanol, and the absorbance of the solubilized dye was measured at 595 nm.
2.3. Genome Sequencing
For genome sequencing, select bacterial isolates were cultured in LB medium overnight at 30 °C. Genomic DNA was isolated from 5 mL of culture using the Quick DNA Miniprep Plus Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions. Purified DNA sample concentration and quality were verified spectrophotometrically and by agarose gel electrophoresis. Samples were submitted to Plasmidsaurus (Eugene, OR, USA) for sequencing using long-read sequencing and assembly approach. All genome sequences generated are available on NCBI under BioProject number PRJNA1183984.
2.4. Computational Analyses
Reference genomes of all type strains from the following genera were obtained from the NCBI GenBank database: Pectobacterium, Dickeya, Pseudomonas, and Chryseobacterium. Where type strains were used, all GenBank genomes represent taxonomically named species. The average nucleotide identity between these type strains and our isolates with sequenced genomes was calculated using fastANI [31]. Further comparisons were made using additional genomes from the species Pectobacterium carotovorum and Pectobacterium versatile. Whole genome alignments were generated and visualized using the progressiveMauve aligner [32] with default parameters.
Lysogenic bacteriophage within the sequenced Pectobacterium genomes were identified using Phastest [33]. Extracted sequences of identified lysogenic bacteriophages were further analyzed using fastANI [31]. The Newick tree was generated with ETEToolkit [34] for visualization.
3. Results
3.1. Isolation and Identification
We collected commercial produce samples exhibiting typical soft rot symptoms and plated them onto CVPM to isolate pectinolytic bacteria associated with these samples (Figure S1). From 47 produce samples, we collected 28 isolates of pectinolytic bacteria (Table S1). When apparently distinct isolates were obtained from a single sample, these isolates were designated ‘a’ or ‘b’ isolates. We used 16S rDNA sequencing to identify the genus and species of the pectinolytic isolates, when possible, and found isolates from nine different genera: Achromobacter, Chryseobacterium, Escherichia, Lelliottia, Pantoea, Pectobacterium, Pseudomonas, Sphingobacterium, and Stenotrophomonas (Table 1 and Table S2).
Table 1.
Pectinolytic isolates, their genetic identities, and phenotypic characteristics.
| Isolate | Isolation Host | Genus/Species | Pathogenic to Potato | Pathogenic to Chinese (Napa) Cabbage) | Swim | Swarm | AHL Production | Lactose | Protease |
|---|---|---|---|---|---|---|---|---|---|
| M1 | Onion | not determined | − | − | + | + | − | + | − |
| M2 | Lettuce | Pseudomonas sp. | − | − | + | + | − | − | + |
| M3 | Lettuce | Pectobacterium carotovorum | + | + | + | + | + | + | + |
| M4a | Lettuce | Pectobacterium carotovorum | + | − | + | + | + | + | + |
| M4b | Lettuce | not determined | − | − | + | + | − | − | + |
| M5 | Lettuce | Chryseobacterium indoltheticum | + | − | + | + | − | + | + |
| M6 | Lettuce | Pseudomonas cedrina | − | − | + | + | − | − | + |
| M7 | Cilantro | Pectobacterium carotovorum | + | − | + | + | + | + | + |
| M8a | Cilantro | Pectobacterium carotovorum | + | − | + | + | + | + | + |
| M8b | Cilantro | Pectobacterium carotovorum | + | − | + | + | + | + | + |
| M9 | Cilantro | Lelliottia amnigena | − | − | + | + | − | − | + |
| M10 | Cilantro | Pectobacterium carotovorum | + | + | + | + | + | + | − |
| M11 | Spinach | Pseudomonas azotoformans | − | − | + | + | − | − | + |
| M12a | Zucchini | Pseudomonas allopoutida | − | − | + | + | − | − | − |
| M12b | Zucchini | Escherichia fergusonii | − | − | + | + | − | − | − |
| M13 | Onion | Pseudomonas marginalis | + | − | + | + | − | − | + |
| M14 | Onion | Pseudomonas petroselini | − | − | + | + | − | + | + |
| M15 | Potato | Stenotrophomonas maltophila | − | − | + | + | − | − | + |
| M16 | Potato | not determined | − | − | + | + | − | + | + |
| M17 | Spinach | Achromobacter spanius | − | − | + | + | − | − | + |
| M18 | Spinach | Pseudomonas cyclaminis | + | − | + | + | − | − | + |
| M19 | Spinach | Stenotrophomonas rhizophila | − | − | + | + | − | − | + |
| M20 | Spinach | Pseudomonas sp. | + | − | + | + | − | − | + |
| M21 | Celery | Pseudomonas composti | − | − | + | + | + | + | + |
| M22a | Celery | Stenotrophomonas maltophila | − | + | + | + | − | − | + |
| M22b | Celery | Pectobacterium carotovorum | + | − | + | + | + | + | + |
| M23 | Carrot | Pseudomonas grimontii | − | − | + | + | − | − | + |
| M24 | Carrot | Pantoea sp. | + | − | + | + | + | − | + |
+ represents a positive reaction for each phenotype, and − represents a negative reaction for the phenotype.
3.2. Phenotypic Characterization
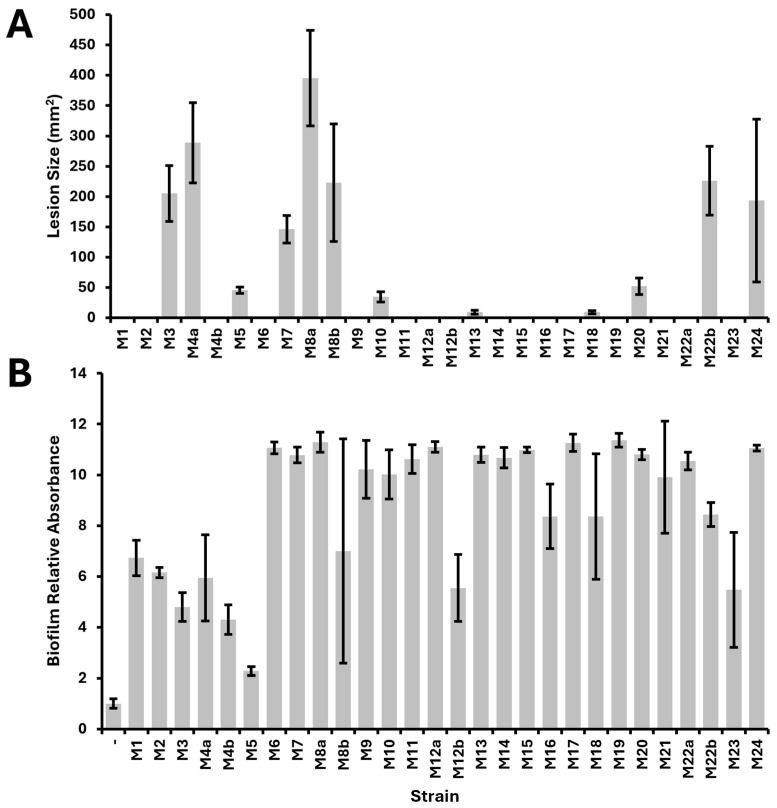
Because each of our isolates was collected from plant tissue exhibiting soft rot symptoms, we tested each of the isolates against potato and Chinese (napa) cabbage to determine whether these isolates were pathogenic to these hosts (Figures S2 and S3). We found that only 12 of the 28 isolates (43%) were pathogenic to potatoes, and only three (11%) were pathogenic to Chinese (napa) cabbage (Table 1). The Pectobacterium isolates, which were mainly isolated from different hosts, were all pathogenic to potatoes as they caused soft rot symptoms as they grew on the potatoes. We observed a high degree of variability in the size of lesions caused by our different isolates when used to inoculate potato slices (Figure 1A).
Figure 1.
Virulence of isolates as measured by area of lesion on inoculated potato slices (A) and biofilm formation in 96-well plates following static growth for 48 h (B).
We assessed whether each of the 28 isolates exhibited various phenotypes associated with virulence, including swimming and swarming motility, biofilm formation, production of acyl-homoserine lactones, fermentation of lactose, and extracellular protease activity (Table 1). All of the isolates exhibited both swimming and swarming motility. All of the isolates appeared to form biofilms, with crystal violet staining higher than the negative control (Figure 1B). Several strains appeared to form strong biofilms with staining 10-fold higher than the negative control; however, strain M5 exhibited the lowest biofilm formation of all the isolates, at only about twice the crystal violet staining of the negative control. Nine of the isolates (32%) produced acyl homoserine lactones detectable with our A. fabrum reporter system [28,29]. Twelve of the isolates (43%) fermented lactose, and 24 of the isolates (86%) exhibited extracellular protease activity.
We used linear regression and computed F-statistics for pairwise correlation between the pathogenicity-to-potato trait and the other virulence-associated phenotypic traits we tested. The additional traits included motility, lactose fermentation, production of acyl homoserine lactones, and extracellular protease activity. We found a significant pairwise correlation (p < 0.001) between the production of acyl homoserine lactones and pathogenicity to potatoes. The correlations for all other virulence traits were not statistically significant.
3.3. Genome Sequencing and Assemblies
We selected a subset of 11 isolates obtained in our survey for whole genome sequencing, with a focus on isolates pathogenic to potatoes. The isolates sequenced and sequencing results are summarized in Table 2. The average sequencing depth was 76× across all samples. All of the Pectobacterium and the Chryseobacterium genomes were assembled to a single contig with no plasmids. The Pectobacterium isolates appeared to have the smallest genome size, ranging between 4.57 Mb and 5.14 Mb, while the Pseudomonas isolates appeared to have larger genome sizes, ranging between 6.27 Mb and 6.93 Mb. All of the Pseudomonas genomes sequenced had at least one plasmid present, suggesting that their genomes may be more complex.
Table 2.
Sequencing data for isolates selected for genome sequencing.
| Isolate | Species | Genome Size | Sequencing Coverage | Contigs | Putative Plasmids | Accession |
|---|---|---|---|---|---|---|
| M3 | Pectobacterium carotovorum | 4.80 Mb | 103× | 1 | 0 | GCA_045038015.1 |
| M4a | Pectobacterium carotovorum | 4.79 Mb | 46× | 1 | 0 | GCA_045038025.1 |
| M5 | Chryseobacterium scophthalmum | 4.57 Mb | 42× | 1 | 0 | GCA_045037995.1 |
| M8a | Pectobacterium carotovorum | 4.80 Mb | 96× | 1 | 0 | GCA_045038005.1 |
| M8b | Pectobacterium carotovorum | 4.80 Mb | 102× | 1 | 0 | GCA_045037985.1 |
| M10 | Pectobacterium versatile | 4.88 Mb | 101× | 1 | 0 | GCA_045037945.1 |
| M12a | Pseudomonas alloputida | 6.27 Mb | 46× | 4 | 3 | GCA_045037975.1 |
| M13 | Pseudomonas marginalis | 6.70 Mb | 85× | 4 | 3 | GCA_045037965.1 |
| M14 | Pseudomonas petroselini | 6.93 Mb | 57× | 2 | 1 | GCA_045037875.1 |
| M20 | Pseudomonas spp. within P. koreensis group | 6.76 Mb | 62× | 2 | 1 | GCA_045037865.1 |
| M22b | Pectobacterium versatile | 5.14 Mb | 100× | 1 | 0 | GCA_045037855.1 |
3.4. Genome Alignments
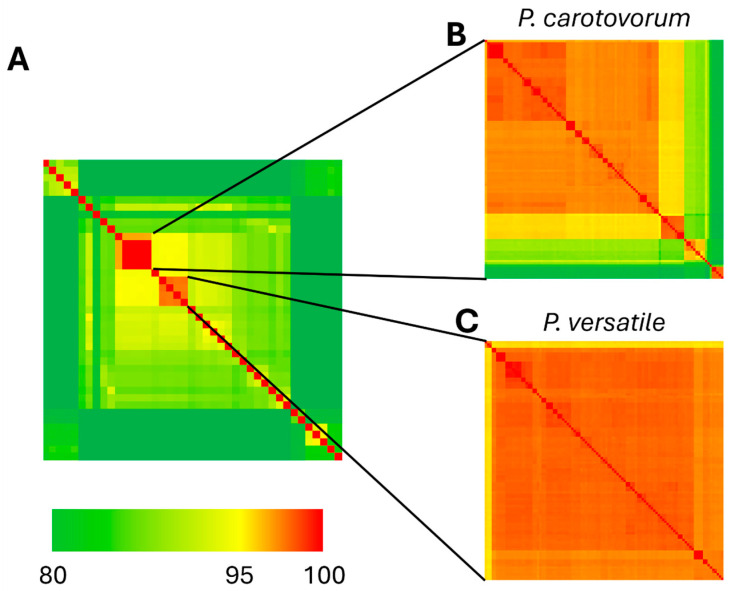
We compared the genomes sequenced in our study with genomes available in the RefSeq database by calculating average nucleotide identity (ANI). We first compared our Pectobacterium genomes to the 35 type strain genomes available from the genera Pectobacterium and Dickeya, which confirmed that isolates M3, M4a, M8a, and M8b are P. carotovorum and isolates M10 and M22b are P. versatile (Figure 2A, Table S3). We then calculated ANI across M3, M4a, M8a, M8b, and all 99 available P. carotovorum genomes (Figure 2B, Table S4), which indicated that our isolates formed their own highly related cluster, more similar to each other than any other P. carotovorum genome. We similarly calculated ANI for M10, M22b, and all 103 available P. versatile genomes (Figure 2C, Table S5) and found M10 and M22b to be closely related to P. versatile strains isolated from cabbage, potato, and surface water, and from geographically diverse areas across the United States and Europe.
Figure 2.
Average nucleotide identity of Pectobacterium isolates represented as a heatmap. (A) ANI across all type strains of Pectobacterium and Dickeya species in RefSeq database (n = 41 genomes). (B) ANI across all genomes entered in RefSeq database as P. carotovorum (n = 103 genomes). (C) ANI across all genomes entered in RefSeq database as P. versatile (n = 105 genomes).
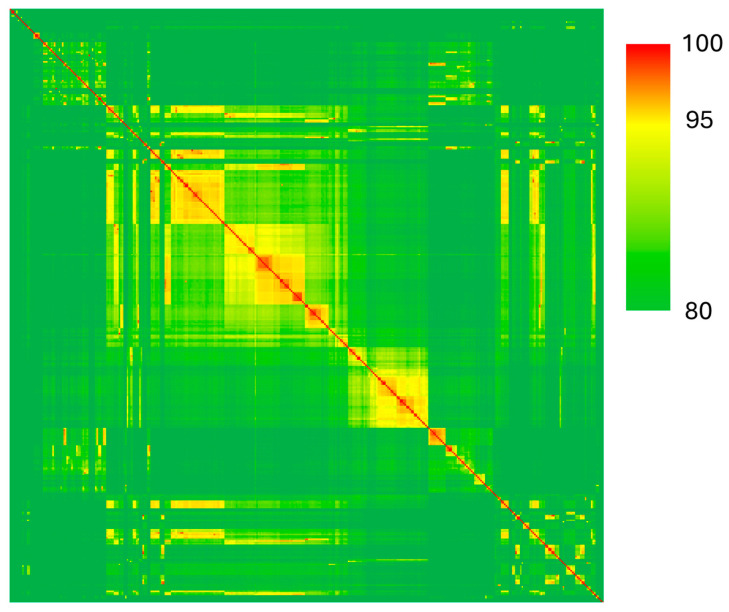
For our four genomes from the genera Pseudomonas, we calculated ANI to compare these isolates to the 369 type strain genomes available (Figure 3, Table S6). This analysis confirmed that strain M12a is Pseudomonas alloputida (ANI = 96.71%), M13 is Pseudomonas marginalis (ANI = 99.22%), and M14 is Pseudomonas petroselini (ANI = 98.81%). The highest ANI found for strain M20 was for Pseudomonas tensinigenes (ANI = 93.27%), a member of the P. koreensis group.
Figure 3.
Average nucleotide identity of Pseudomonas isolates from our survey and type strains of all species in the RefSeq database (n = 375 genomes).
We also calculated ANI for our Chryseobacterium isolate, M5, and the 117 type strain genomes available for this genus (Figure 4, Table S7). We found that isolate M5 is similar to Chryseobacterium scophthalmum (ANI = 95.31%), which is a fish pathogen.
Figure 4.
Average nucleotide identity of the Chryseobacterium isolate and all type strains from Chryseobacterium species in the RefSeq database (n = 118 genomes).
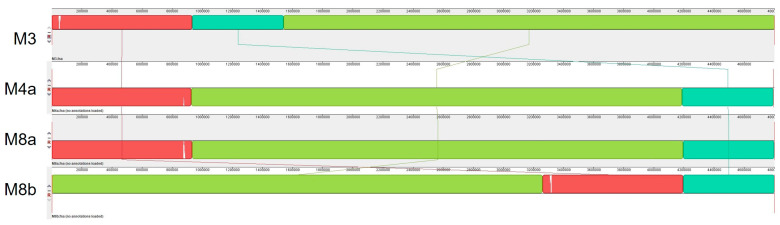
Because we found that average nucleotide identity had little power to resolve differences between our closely related Pectobacterium carotovorum isolates, we sought to better understand the relationships between these genomes and isolates. We first aligned the full-length genomes of these isolates and found that strains M3 and M8b had large rearrangements, but strains M4a and M8a shared the same structure (Figure 5).
Figure 5.
Whole genome alignments of P. carotovorum isolates M3, M4a, M8a, and M8b. Alignment and visualization generated by Mauve.
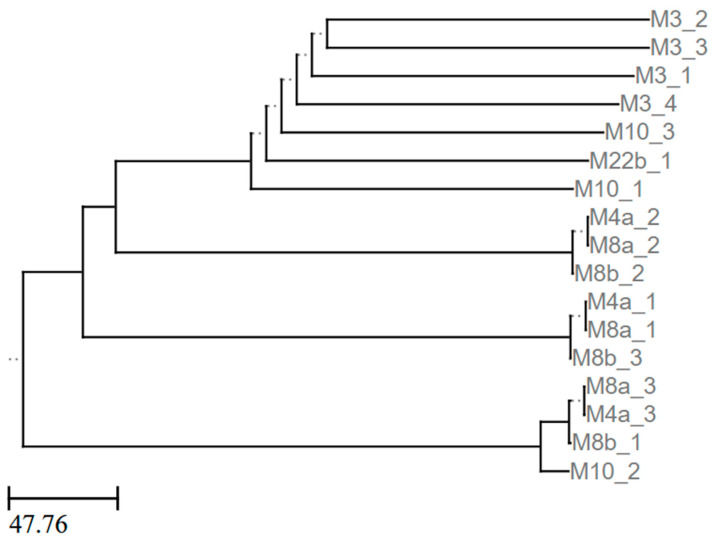
To test whether the Pectobacterium strains also shared horizontally acquired genetic elements or if unique DNA fragments could serve to separate these strains, we computationally predicted prophage elements in each of the Pectobacterium genomes we sequenced (Table S8) and then compared the sequences of the predicted prophages (Figure 6). We found that all of the genomes contained at least one predicted prophage. We further found that although some isolates, such as M4a and M8a, shared their predicted prophages, other isolates, such as M3, carried almost entirely distinct prophages from their otherwise genomically close relatives. Top BLAST hits for each of the predicted prophages show that in addition to P. carotovorum and P. versatile, similar prophages are found in P. brasiliense, P. parvum, P. odoriferum, and P. wasabiae (Table S9).
Figure 6.
Prophage dendrogram. Newick tree generated by ANIclustermap using sequences of prophages predicted by Phastest in newly sequenced Pectobacterium genomes. Representation rendered by ETEToolkit.
4. Discussion
In this study, we isolated 28 pectinolytic bacteria from various produce samples exhibiting typical bacterial soft rot symptoms. These isolates came from nine different genera, but only isolates from three of these genera, Chryseobacterium, Pectobacterium, and Pseudomonas, were pathogenic to potatoes. Only isolates from Pectobacterium elicited disease symptoms on Chinese (Napa) cabbage. Although several species from the genera Pectobacterium and Dickeya are typically associated with bacterial soft rot, in our survey, we only obtained isolates from P. carotovorum and P. versatile. P. versatile represents a group of strains that was recently elevated to its own species and was formerly classified with P. carotovorum [20]. Our genomic observations that P. versatile and P. carotovorum have average nucleotide identity below 96% align with their division into two species [20]. Furthermore, our two P. versatile isolates, M10 and M22b, both infect Chinese (Napa) cabbage, but strain M3 was the only one of four P. carotovorum isolates to infect this host. Interestingly, strains M3, M10, and M22b each carry prophages in their genome distinct from the other Pectobacterium isolates and their predicted prophages. BLAST searches showed that nine of the predicted prophages are most similar to prophages found in P. carotovorum and P. versatile, while the remaining prophages are more similar to prophages from P. brasiliense, P. odoriferum, P. parvum, and P. wasabiae. Interestingly, one cluster of phages (M4a_2, M8a_2, and M8b_2) only had matches along 28% of the length of the prophage sequence. Another five phages had matches less than 80% of the length of the sequence. Future work is needed to address the underlying mechanisms of host specificity and the potential roles of horizontally acquired genetic elements, such as phage, in contributing to host range.
Phenotypic characterization of our isolates showed that many of them exhibited traits often associated with virulence in pathogens. Although the traits assessed are considered virulence factors for several pathogens, only acyl homoserine lactone production was significantly associated with pathogenicity in potatoes. All isolates exhibited swimming and swarming motility and most expressed extracellular protease activity. Because our isolates came from diverse host plants and represent various taxa, it is unclear whether these isolates use these phenotypic traits for virulence in other host plants but do not include potatoes as hosts. Thus, we hypothesize that the significant correlation between acyl homoserine lactone production and potato pathogenicity is because most of the potato pathogenic isolates are Pectobacterium isolates and all of the Pectobacterium isolates produce acyl homoserine lactones.
All of the Pectobacterium isolates, both P. carotovorum (M3, M4a, M8a, M8b) and P. versatile (M10, M22b), exhibited all measured phenotypic characteristics, with the exception of M10, which did not display protease activity. While strain M10 was pathogenic to potatoes, the lesion size caused by M10 was less than 50 mm2, whereas the lesions caused by the other Pectobacterium isolates were closer to 200 mm2. Additional work will reveal whether the reduced lesion size is directly or indirectly linked to the lack of extracellular protease activity in strain M10, as extracellular proteases are known to be important for full virulence in some plant-pathogenic bacteria but dispensable for others [35,36,37]. More broadly, there was no significant correlation between protease activity and pathogenicity to potatoes among all of our isolates.
Most of the Pseudomonas isolates we obtained were not pathogenic to potatoes, except for isolates M13, M18, and M20. Based on our average nucleotide identity analyses, strain M13 is a new isolate of Pseudomonas marginalis, a species that has previously been associated with soft rot disease [14,38]. For strain M20, the highest average nucleotide identity observed with another genome was with Pseudomonas tensinigenes, a member of the P. koreensis group [39]. However, the average nucleotide identity between these strains was only 93.27%, well below the typical species-level thresholds of 95–96%. We hypothesize that M20 represents a novel species that is capable of causing soft rot disease in potatoes and potentially other diverse hosts, as it was isolated from spinach. On potatoes, the lesion size caused by M20 was comparable to that of the Pectobacterium isolate M10. It is recognized that various species of Pseudomonas can cause soft rot, and the species most commonly associated therewith are P. glycinae, P. cichorii, P. marginalis, and P. viridiflava [38,40,41,42]. Of these species, we recovered one isolate of P. marginalis (M13) and a putatively novel species associated with soft rot (M20).
The genus Chryseobacterium is diverse, and its members occupy several environmental niches [43]. Many Chryseobacteria are not pathogenic to humans and animals, and of those that are, many tend to be weakly pathogenic, such as C. indologenes, formerly Flavobacterium indologenes [44]. C. indologenes has also been reported as a plant pathogen, causing root rot in Panax ginger [45]. On the other hand, Chryseobacterium isolates have been reported to exhibit plant-beneficial effects [46,47]. Some Chryseobacterium isolates also exhibit the ability to digest complex organic molecules, such as herbicides [48]. Our Chryseobacterium strain M5 contains predicted pectate lyase (ACI513_RS09935) and pectin esterase (ACI513_RS09930) genes located adjacent to each other in the genome, supporting a role for this strain in host cell wall degradation. Other strains of Chryseobacterium have previously been reported to express pectinolytic activity [10,49]. By our calculations, Chryseobacterium strain M5 was most similar to Chryseobacterium scophthalmum based on average nucleotide identity. C. scophthalmum is pathogenic to fish, being originally isolated from turbots in the Atlantic Ocean [21,22]. Because a fish pathogen is the closest relation to M5, this isolate may represent a crossover pathogen, evolving from an animal to a plant pathogen. It will be interesting to determine in future work whether this particular isolate also exhibits any animal pathogenic activity. However, because the average nucleotide identity between M5 and C. scophthalmum is 95.3%, we hypothesize that M5 may represent a novel species of Chryseobacterium. Further work to understand the evolution, host range, and virulence of this isolate and its relatives is warranted.
Among the other isolates obtained, both Stenotrophomonas isolates (M15 and M19), which were obtained from different hosts, displayed the same phenotypic characteristics and were not pathogenic to potatoes. Neither of the isolates that came from symptomatic potatoes (M15 and M16) were pathogenic to potatoes under our experimental conditions and time frame. Our isolation approach used crystal violet pectate medium to isolate from tissues. Because of the crystal violet component of the medium, it was anticipated that Gram-positive pectinolytic bacteria would not be isolated in this approach. Indeed, our isolations only yielded bacteria from Gram-negative taxonomic groups. We successfully isolated pectinolytic bacteria from more than half of our plant samples and may have found additional isolates had our methods also targeted Gram-positive pectinolytic bacteria; thus, some of our pectinolytic isolates may have been non-pathogenic because they are members of disease complexes in which multiple diverse bacteria work together to cause soft rot.
Altogether, our work demonstrates that there are abundant and diverse bacteria associated with and causing post-harvest soft rot disease in commercial produce, including a potential crossover pathogen. It is important to be aware of this diversity in future work to develop and improve disease control methods. Our survey work and the genomic resources we have developed lay an initial foundation for work on these pathogens to better understand their virulence mechanisms and host specificities. Furthermore, a lack of redundancy in the isolates we collected from our survey indicates that there is value in ongoing work to monitor the bacteria associated with soft rot disease.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens13121096/s1, Table S1: Produce samples collected and tested, Table S2: Top BLAST hits for 16S sequences, Table S3: Average nucleotide identity of Pectobacterium and Dickeya species, Table S4: Average nucleotide identity of Pectobacterium carotovorum, Table S5: Average nucleotide identity of Pectobacterium versatile, Table S6: Average nucleotide identity of Pseudomonas species, Table S7: Average nucleotide identity of Chryseobacterium species, Table S8: Sequences of predicted prophage elements in Pectobacterium genomes, Table S9: Top BLAST hits of prophages using all Pectobacterium genomes in NCBI; Figure S1: Representative CVP plate from isolation of pectinolytic bacteria, Figure S2: Potato slices inoculated with pectinolytic isolates, Figure S3: Chinese (napa) cabbage leaves inoculated with pectinolytic isolates.
Author Contributions
Conceptualization, K.R., B.R., M.S. and J.K.S.; Investigation: K.R., B.R., M.S., J.H. and J.K.S.; Writing—original draft preparation, K.R., B.R., M.S., J.H. and J.K.S.; Writing—review and editing, K.R., B.R., M.S., J.H. and J.K.S.; project administration and supervision: J.K.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All genome sequences generated as part of this study are available through NCBI under BioProject number PRJNA1183984.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Charkowski A., Sharma K., Parker M.L., Secor G.A., Elphinstone J. Bacterial diseases of potato. In: Campos H., Ortiz O., editors. The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind. Springer; Cham, Switzerland: 2020. pp. 351–388. [Google Scholar]
- 2.Kamau J.W., Ngaira J., Kinyua J., Gachamba S., Ngundo G., Janse J., Macharia I. Occurence of pectinolytic bacteria causing blackleg and soft rot of potato in Kenya. J. Plant Pathol. 2019;101:689–694. doi: 10.1007/s42161-018-00219-w. [DOI] [Google Scholar]
- 3.Czajkowski R., Perombelon M., Jafra S., Lojkowska E., Potrykus M., van der Wolf J., Sledz W. Detection, identification and differentiation of Pectobacterium and Dickeya species causing potato blackleg and tuber soft rot: A review. Ann. Appl. Biol. 2015;166:18–38. doi: 10.1111/aab.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toth I.K., Bell K.S., Holeva M.C., Birch P.R. Soft rot erwiniae: From genes to genomes. Mol. Plant Pathol. 2003;4:17–30. doi: 10.1046/j.1364-3703.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 5.Pinhero R.G., Coffin R., Yada R.Y. Advances in Potato Chemistry and Technology. Elsevier; Amsterdam, The Netherlands: 2009. Post-harvest storage of potatoes; pp. 339–370. [Google Scholar]
- 6.Maung C.E.H., Choub V., Cho J.Y., Kim K.Y. Control of the bacterial soft rot pathogen, by CE 100 in cucumber. Microb. Pathog. 2022;173:105807. doi: 10.1016/j.micpath.2022.105807. [DOI] [PubMed] [Google Scholar]
- 7.Murata H., Chatterjee A., Liu Y., Chatterjee A.K. Regulation of the production of extracellular pectinase, cellulase, and protease in the soft rot bacterium Erwinia carotovora subsp. carotovora: Evidence that aepH of E. carotovora subsp. carotovora 71 activates gene expression in E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and Escherichia coli. Appl. Environ. Microbiol. 1994;60:3150–3159. doi: 10.1128/aem.60.9.3150-3159.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patidar M.K., Nighojkar S., Kumar A., Nighojkar A. Pectinolytic enzymes-solid state fermentation, assay methods and applications in fruit juice industries: A review. 3 Biotech. 2018;8:199. doi: 10.1007/s13205-018-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu P., Yang S.H., Zhan Z.C., Zhang G.M. Origins and features of pectate lyases and their applications in industry. Appl. Microbiol. Biot. 2020;104:7247–7260. doi: 10.1007/s00253-020-10769-8. [DOI] [PubMed] [Google Scholar]
- 10.Haile S., Ayele A. Pectinase from Microorganisms and Its Industrial Applications. Sci. World J. 2022;2022:1881305. doi: 10.1155/2022/1881305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toth I.K., Barny M.-a., Czajkowski R., Elphinstone J.G., Li X., Pédron J., Pirhonen M., Van Gijsegem F. Pectobacterium and Dickeya: Taxonomy and evolution. In: Van Gijsegem F., van der Wolf J., Toth I.K., editors. Plant Diseases Caused by Dickeya and Pectobacterium Species. Springer Nature; Cham, Switzerland: 2021. pp. 13–37. [Google Scholar]
- 12.Charkowski A.O. The Changing Face of Bacterial Soft-Rot Diseases. Annu. Rev. Phytopathol. 2018;56:269–288. doi: 10.1146/annurev-phyto-080417-045906. [DOI] [PubMed] [Google Scholar]
- 13.Sawada H., Fujikawa T., Tsuji M., Satou M. Pseudomonas allii sp. nov., a pathogen causing soft rot of onion in Japan. Int. J. Syst. Evol. Micr. 2021;71:004582. doi: 10.1099/ijsem.0.004582. [DOI] [PubMed] [Google Scholar]
- 14.Li J., Chai Z., Yang H., Li G., Wang D. First report of Pseudomonas marginalis pv. marginalis as a cause of soft rot of potato in China. Australas. Plant Dis. Notes. 2007;2:71–73. doi: 10.1071/DN07029. [DOI] [Google Scholar]
- 15.Elbanna K., Elnaggar S., Bakeer A. Characterization of Bacillus altitudinis as a New Causative Agent of Bacterial Soft Rot. J. Phytopathol. 2014;162:712–722. doi: 10.1111/jph.12250. [DOI] [Google Scholar]
- 16.da Silva W.L., Yang K.T., Pettis G.S., Soares N.R., Giorno R., Clarks C.A. Flooding-Associated Soft Rot of Sweetpotato Storage Roots Caused by Distinct Isolates. Plant Dis. 2019;103:3050–3056. doi: 10.1094/PDIS-03-19-0548-RE. [DOI] [PubMed] [Google Scholar]
- 17.Hauben L., Moore E.R., Vauterin L., Steenackers M., Mergaert J., Verdonck L., Swings J. Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst. Appl. Microbiol. 1998;21:384–397. doi: 10.1016/S0723-2020(98)80048-9. [DOI] [PubMed] [Google Scholar]
- 18.Samson R., Legendre J.B., Christen R., Saux M.F., Achouak W., Gardan L. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int. J. Syst. Evol. Microbiol. 2005;55:1415–1427. doi: 10.1099/ijs.0.02791-0. [DOI] [PubMed] [Google Scholar]
- 19.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Portier P., Pedron J., Taghouti G., Fischer-Le Saux M., Caullireau E., Bertrand C., Laurent A., Chawki K., Oulgazi S., Moumni M., et al. Elevation of Pectobacterium carotovorum subsp. odoriferum to species level as Pectobacterium odoriferum sp. nov., proposal of Pectobacterium brasiliense sp. nov. and Pectobacterium actinidiae sp. nov., emended description of Pectobacterium carotovorum and description of Pectobacterium versatile sp. nov., isolated from streams and symptoms on diverse plants. Int. J. Syst. Evol. Microbiol. 2019;69:3207–3216. doi: 10.1099/ijsem.0.003611. [DOI] [PubMed] [Google Scholar]
- 21.Mudarris M., Austin B., Segers P., Vancanneyt M., Hoste B., Bernardet J.F. Flavobacterium-scophthalmum sp. nov., a Pathogen of Turbot (Scophthalmus-maximus L.) Int. J. Syst. Bacteriol. 1994;44:447–453. doi: 10.1099/00207713-44-3-447. [DOI] [PubMed] [Google Scholar]
- 22.Shahi N., Sharma P., Pandey J., Bisht I., Mallik S.K. Characterization and pathogenicity study of Chryseobacterium scophthalmum recovered from gill lesions of diseased golden mahseer, Tor putitora (Hamilton, 1822) in India. Aquaculture. 2018;485:81–92. doi: 10.1016/j.aquaculture.2017.11.018. [DOI] [Google Scholar]
- 23.Hélias V., Hamon P., Huchet E., Wolf J.V.D., Andrivon D. Two new effective semiselective crystal violet pectate media for isolation of Pectobacterium and Dickeya. Plant Pathol. 2012;61:339–345. doi: 10.1111/j.1365-3059.2011.02508.x. [DOI] [Google Scholar]
- 24.Hongoh Y., Yuzawa H., Ohkuma M., Kudo T. Evaluation of primers and PCR conditions for the analysis of 16S rRNA genes from a natural environment. FEMS Microbiol. Lett. 2003;221:299–304. doi: 10.1016/S0378-1097(03)00218-0. [DOI] [PubMed] [Google Scholar]
- 25.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abràmoff M.D., Magalhães P.J., Ram S.J. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 27.Schachterle J.K., Sundin G.W. The Leucine-Responsive Regulatory Protein Lrp Participates in Virulence Regulation Downstream of Small RNA ArcZ in Erwinia amylovora. mBio. 2019;10:10–1128. doi: 10.1128/mBio.00757-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Z.Q., Su S.C., Farrand S.K. In situ activation of the quorum-sensing transcription factor TraR by cognate and noncognate acyl-homoserine lactone ligands: Kinetics and consequences. J. Bacteriol. 2003;185:5665–5672. doi: 10.1128/JB.185.19.5665-5672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw P.D., Ping G., Daly S.L., Cha C., Cronan J.E., Rinehart K.L., Farrand S.K. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santander R.D., Biosca E.G. Erwinia amylovora psychrotrophic adaptations: Evidence of pathogenic potential and survival at temperate and low environmental temperatures. PeerJ. 2017;5:e3931. doi: 10.7717/peerj.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain C., Rodriguez-R L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darling A.E., Mau B., Perna N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wishart D.S., Han S., Saha S., Oler E., Peters H., Grant J.R., Stothard P., Gautam V. PHASTEST: Faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023;51:W443–W450. doi: 10.1093/nar/gkad382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huerta-Cepas J., Dopazo J., Gabaldon T. ETE: A python Environment for Tree Exploration. BMC Bioinform. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dow J.M., Clarke B.R., Milligan D.E., Tang J.L., Daniels M.J. Extracellular Proteases from Xanthomonas-Campestris Pv Campestris, the Black Rot Pathogen. Appl. Environ. Microb. 1990;56:2994–2998. doi: 10.1128/aem.56.10.2994-2998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gouran H., Gillespie H., Nascimento R., Chakraborty S., Zaini P.A., Jacobson A., Phinney B.S., Dolan D., Durbin-Johnson B.P., Antonova E.S., et al. The Secreted Protease PrtA Controls Cell Growth, Biofilm Formation and Pathogenicity in Xylella fastidiosa. Sci. Rep. 2016;6:31098. doi: 10.1038/srep31098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou H.S., Song X., Zou L.F., Yuan L., Li Y.R., Guo W., Che Y.Z., Zhao W.X., Duan Y.P., Chen G.Y. EcpA, an extracellular protease, is a specific virulence factor required by Xanthomonas oryzae pv. oryzicola but not by X. oryzae pv. oryzae in rice. Microbiology. 2012;158:2372–2383. doi: 10.1099/mic.0.059964-0. [DOI] [PubMed] [Google Scholar]
- 38.Janse J.D., Derks J.H.J., Spit B.E., Vandertuin W.R. Classification of Fluorescent Soft Rot Pseudomonas Bacteria, Including P. marginalis Strains, Using Whole Cell Fatty-Acid Analysis. Syst. Appl. Microbiol. 1992;15:538–553. doi: 10.1016/S0723-2020(11)80114-1. [DOI] [Google Scholar]
- 39.Girard L., Lood C., Höfte M., Vandamme P., Rokni-Zadeh H., van Noort V., Lavigne R., De Mot R. The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group. Microorganisms. 2021;9:1766. doi: 10.3390/microorganisms9081766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godfrey S.A.C., Marshall J.W. Identification of cold-tolerant Pseudomonas viridiflava and P. marginalis causing severe carrot postharvest bacterial soft rot during refrigerated export from New Zealand. Plant Pathol. 2002;51:155–162. doi: 10.1046/j.1365-3059.2002.00679.x. [DOI] [Google Scholar]
- 41.Wright P.J., Grant D.G. Evaluation of Allium germplasm for susceptibility to foliage bacterial soft rot caused by Pseudomonas marginalis and Pseudomonas viridiflava. N. Z. J. Crop Hort. 1998;26:17–21. doi: 10.1080/01140671.1998.9514034. [DOI] [Google Scholar]
- 42.Liu K., Sun W.S., Li X.L., Shen B.Y., Zhang T.J. Isolation, identification, and pathogenicity of Pseudomonas glycinae causing ginseng bacterial soft rot in China. Microb. Pathog. 2024;186:106497. doi: 10.1016/j.micpath.2023.106497. [DOI] [PubMed] [Google Scholar]
- 43.Bernardet J.F., Hugo C.J., Bruun B. Chryseobacterium. In: Whitman W.B., editor. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2015. pp. 1–35. [Google Scholar]
- 44.Mukerji R., Kakarala R., Smith S.J., Kusz H.G. Chryseobacterium indologenes: An emerging infection in the USA. BMJ Case Rep. 2016;2016:bcr2016214486. doi: 10.1136/bcr-2016-214486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou H., Ding Y.F., Shang J.J., Ma C.L., Li J.H., Yang Y., Cui X.M., Zhang J.H., Ji G.H., Wei Y.L. Isolation, characterization, and genomic analysis of a novel bacteriophage MA9V-1 infecting Chryseobacterium indologenes: A pathogen of Panax notoginseng root rot. Front. Microbiol. 2023;14:1251211. doi: 10.3389/fmicb.2023.1251211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jung H., Lee D., Lee S., Kong H.J., Park J., Seo Y.S. Comparative genomic analysis of Chryseobacterium species: Deep insights into plant- growth- promoting and halotolerant capacities. Microb. Genom. 2023;9:001108. doi: 10.1099/mgen.0.001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh A.V., Chandra R., Goel R. Phosphate solubilization by Chryseobacterium sp. and their combined effect with N and P fertilizers on plant growth promotion. Arch. Agron. Soil Sci. 2013;59:641–651. doi: 10.1080/03650340.2012.664767. [DOI] [Google Scholar]
- 48.Zhao H.H., Xu J., Dong F.S., Liu X.G., Wu Y.B., Wu X.H., Zheng Y.Q. Characterization of a novel oxyfluorfen-degrading bacterial strain Chryseobacterium aquifrigidense and its biochemical degradation pathway. Appl. Microbiol. Biot. 2016;100:6837–6845. doi: 10.1007/s00253-016-7504-x. [DOI] [PubMed] [Google Scholar]
- 49.Roy K., Dey S., Uddin M.K., Barua R., Hossain M.T. Extracellular Pectinase from a Novel Bacterium Chryseobacterium indologenes Strain SD and Its Application in Fruit Juice Clarification. Enzym. Res. 2018;2018:3859752. doi: 10.1155/2018/3859752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genome sequences generated as part of this study are available through NCBI under BioProject number PRJNA1183984.