Abstract
We have imaged a non-damaged and UV-damaged DNA fragment and its complexes with human replication protein A (RPA) using tapping mode atomic force microscopy (AFM). For imaging, molecules were immobilized under nearly physiological conditions on mica surfaces. Quantitative sizing of the 538 bp DNA before and after UV light treatment shows a reduction in the contour and persistence lengths and mean square end-to-end distance as a consequence of UV irradiation. Complexes of the UV-damaged DNA with RPA, an essential component of the initial steps of nucleotide excision repair, can be detected at high resolution with AFM and reveal conformational changes of the DNA related to complex formation. By phase image analysis we are able to discriminate between protein and DNA in the complexes. The DNA molecules are found to ‘wrap’ around the RPA, which in turn results in a considerable reduction in its apparent contour length.
INTRODUCTION
The human replication protein A (RPA) is a heterotrimeric protein containing subunits of 70, 32 and 14 kDa and is involved in replication and recombination processes and participates in the regulation of transcription (1,2). An essential role of RPA has been demonstrated for nucleotide excision repair (NER), a pathway that removes a variety of major DNA lesions, including photoproducts, adducts of carcinogens and cisplatin (3–6).
By its DNA-binding properties, RPA can be classified as a single-stranded (ss)DNA-binding protein. It binds with high affinity and low sequence specificity to ssDNA (7,8). In addition, RPA has been shown to bind with high affinity to DNA lesions that cause a distortion of the DNA. The affinity for damaged sites on double-stranded (ds)DNA is by more than one order of magnitude higher than for undamaged dsDNA (9). Since the chemical nature of the DNA lesions recognized by RPA is diverse, it is believed that RPA binds to unpaired regions created at the sites of DNA damage. The stronger binding of RPA to damaged versus undamaged DNA suggests that RPA participates in the damage recognition step in NER.
DNA-binding domains have been identified on the 70 and 32 kDa subunits of RPA, although most of the DNA contacts appear to be mediated by the 70 kDa subunit. Crosslinking experiments have identified a binding site for damaged DNA on this subunit (10). Furthermore, X-ray crystallography of a complex between a fragment of the 70 kDa subunit and (dC)8 has revealed details of the binding site for ssDNA. The DNA is bound in a shallow groove containing several aromatic residues and a surplus of positive charges (11).
Immobilization of DNA on freshly cleaved mica surfaces (e.g. via magnesia or nickel ions) gives the unique possibility to image and size the individual biomolecules by atomic force microscopy (AFM) in their native hydrated state. Thereby artifacts caused by washing and drying of the sample are avoided.
Investigation of the RPA and UV light-treated DNA complexes by AFM may help to characterize their geometry and structure.
MATERIALS AND METHODS
Proteins
RPA was expressed in Escherichia coli BL21 (DE3) using the expression vector pET11d-tRPA (a generous gift from M. S. Wold) and purified following the protocol of Henricksen et al. (12) through Affigel-Blue (Biorad), hydroxyapatite (Biorad) and anion exchange chromatography on EMD-TMAE (Merck, Darmstadt, Germany). RPA eluted from EMD-TMAE at 200 mM KCl at >95% purity as judged by SDS–PAGE and staining with Coomassie Blue. Aliquots containing 10% glycerol were shock-frozen in liquid nitrogen and stored at –78°C until use. At least 70% of the RPA molecules in this preparation were active in ssDNA binding as checked by titrations under stoichiometric conditions followed by fluorescence anisotropy (9).
DNA substrates
A 538 bp DNA fragment was obtained by PCR with the forward primer 5′-CGCCACTTGGCGAGAAATTTGCTCAAAG-3′ and the reverse primer 5′-GGTTGAGCTCGAGTCACAGGAGTTCGTCACGGC-3′ using the plasmid pRN1 from Sulfolobus islandicus (13) as a template. The PCR reaction was purified by spin chromatography on QiaQuick spin columns (Qiagen) and the concentration of the 538 bp fragment was determined by UV spectroscopy. The purity of the DNA preparation was checked by PAGE with subsequent ethidium bromide staining.
UV-damaged DNA samples were obtained by irradiation of the purified fragment with germicidal lamps (G8T5, 15 W) at 254 nm. Aliquots of 20 µl reactions in 8 mM HEPES were exposed for the indicated time in Eppendorf cups with the lid open at 15 cm distance from the UV source.
Electrophoretic mobility shift assays (EMSA)
The purified 538 bp fragment was labeled at its 5′-end using T4 polynucleotide kinase (Fermentas) and [γ-32P]ATP (5000 Ci/mmol; Hartmann Analytics, Braunschweig, Germany). Aliquots of this radiolabeled DNA were subjected to UV irradiation as described above. Indicated amounts of RPA and 1 ng labeled DNA were preincubated in 10 µl RPA buffer (25 mM HEPES–KOH, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.01% v/v Nonidet P-40, 10% v/v glycerol, pH 7.8) for 10 min at room temperature. Samples were loaded on native polyacrylamide gels (6%, acrylamide:bisacrylamide 29:1) after addition of loading buffer either following fixation of the complexes by a 5 min incubation with 0.1% (w/v) glutaraldehyde or without any further treatment of the sample. Electrophoresis was carried out in 0.5× TBE (44 mM Tris base, 44 mM boric acid, 1 mM EDTA, pH 8.3) as running buffer for 3 h at 10 V/cm and 4°C. The gels were analyzed by electronic autoradiography using an Instant Imager (Canberra Packard).
AFM imaging and analysis
AFM measurements were performed on a MultiMode AFM (Digital Instruments, Santa Barbara, CA) operated in tapping mode. Silicon nitride oxide sharpened tips were used. The drive frequencies ranged between 3.4 and 34 kHz. All measurements were performed in a buffer containing 8 mM HEPES pH 8, 2 mM NaCl and 2 mM MgCl2. Sample preparation for AFM measurements was improved by further purification of all solutions with a 3 kDa Microfilter (Amicon Centriprep; Millipore Corp.). Immobilization of the biomolecules on mica was achieved via 2.5–5.0 mM NiCl2 without further rinsing of the samples and omitting any drying. Image processing analysis was performed using the Nanoscope III 4.43r8 software package and the in-house software ‘Look’ for automated flattening of large numbers of AFM images. For DNA contour length determination we used ImageJ v.1.20s and DnaCalc6. For quantitative analysis, only molecules that were entirely imaged in the chosen scan area were used. Molecules overlapping with others were also disregarded.
We note that for tapping mode phase imaging, it is important to determine the resonance frequency of the cantilever and the phase zero point with the tip located close to the surface. Therefore, we first measured the tip resonance frequency by Fourier transforming the thermal noise spectrum of the tip ∼1 µm above the surface. Subsequently, the drive frequency was set to the resonance frequency and the phase zero was adjusted ∼20 nm above the surface.
RESULTS
Contour length and persistence length of DNA
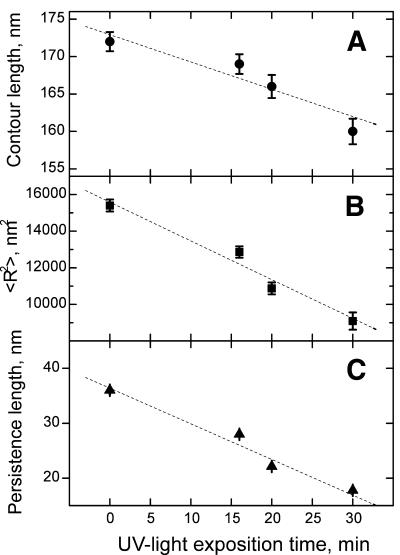
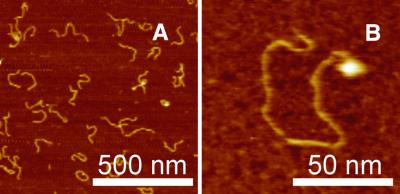
In a first set of AFM experiments we investigated single DNA molecules on mica surfaces and the influence of UV irradiation on the shape of the attached DNA. A 538 bp DNA probe at 1–3 nM in deposition buffer was placed on freshly cleaved mica. After 1 min, needed for DNA equilibration on the mica surface, Ni2+ was added to the sample at a final concentration of 2.5 mM. It proved important to keep the buffer around pH 8 to achieve efficient DNA attachment to the mica surface. Figure 1A shows the image of the undamaged 538 bp DNA. From AFM images of this type the contour length and end-to-end distance of the undamaged and UV-irradiated DNA molecules were determined. From the contour length and mean square end-to-end distance one can calculate the persistence length P, i.e. the distance over which orientational correlations along the DNA molecule persist. P was calculated as described earlier (14) using both the mean square end-to-end distance <R2>2D and the contour length L determined by our experiments (Fig. 2).
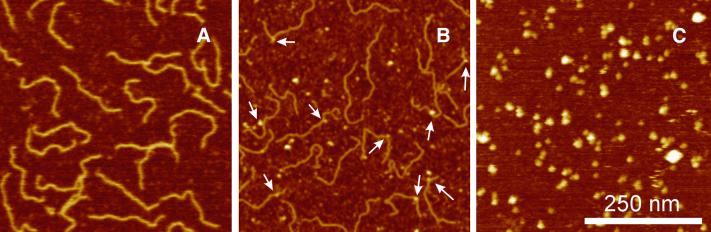
Figure 1.
AFM images of 538 bp DNA fragment deposited on mica. DNA molecules in 2 mM NaCl, 2 mM MgCl2, 2.5 mM NiCl2, 8 mM HEPES, pH 8, prior to (A) and after 20 (B) and 40 min (C) UV light exposure. Knot-like structures on the DNA molecules appear as a consequence of UV damage (marked with arrows). In the images of the DNA molecules exposed for 40 min to UV light (C) rod shaped molecules are not seen (z range is 10 nm).
Figure 2.
Dependence of the DNA contour length, mean square end-to-end distance and persistence length on the UV light exposure time. Circles correspond to the DNA contour length (A), squares to the mean square end-to-end distance (B) and triangles to the persistence length (C). In all cases deposition was carried out in the same way (in 2 mM NaCl, 2 mM MgCl2, 8 mM HEPES, 2.5 mM NiCl2 without glutaraldehyde addition). The lines are linear fits that are used as a guide to the eye.
<R2>2D = 4PL[1 – 2P/L(1 – e–L /2P)].
Similar experiments were performed after UV irradiation and subsequent immobilization. The AFM images of the UV light-damaged DNA molecules (Fig. 1B) show numerous knot-like structures and sharp kinks in the DNA molecules (see white arrows in Fig. 1B), which do not occur prior to UV irradiation. The influence of the irradiation time on the end-to-end distance, the contour length and the persistence length is summarized in Figure 2. The contour length is found to decrease with irradiation time up to times of some 30 min. After longer irradiation times contour length determination becomes increasingly difficult, because the attached DNA molecules were no longer linear but rather spherical (Fig. 1C).
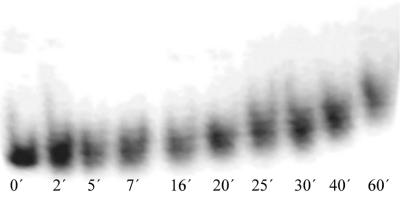
The same DNA probes were examined by gel electrophoresis (Fig. 3). We find a broadening of the bands and a lower mobility of the DNA molecules as a consequence of UV irradiation. The decreased mobility and the line broadening indicates considerable changes in the shape of the DNA molecules with increasing UV damage. This finding is in agreement with the appearance of kinks and knots in the AFM images of UV-damaged DNA.
Figure 3.
Effect of the UV light treatment on the 538 bp DNA molecules. In comparison to the intact DNA (without UV light treatment) electrophoretic mobility of the DNA molecules decreases proportionally to the UV light exposure time.
The number of damaged sites produced by UV irradiation is difficult to estimate, since there is not only an influence of UV dose but also of the sequence context. Based on data published by Yeung et al. (15) we expect the presence of about 10 photoproducts/DNA molecule for 30 min UV exposure time.
EMSA investigations of RPA–DNA complexes
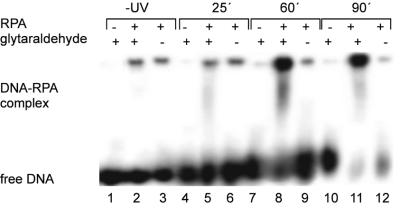
The binding of RPA to UV-damaged DNA was studied by both EMSA and AFM. The EMSA experiments served as a control to verify complex formation. In the EMSA experiments, RPA and radioactively labeled DNA (after UV irradiation for different time intervals) were incubated at a ratio of 1:10. After incubation for 15 min, glutaraldehyde (final concentration 0.1% v/v) was added and 10 min later samples were loaded onto the gels. Figure 4 shows the binding of RPA to the 538 bp DNA for different irradiation times. Clearly, RPA forms complexes with UV-damaged DNA under these conditions. The addition of glutaraldehyde was necessary to stabilize the complexes during electrophoresis. In the absence of glutaraldehyde no binding of RPA to undamaged DNA or to the UV-damaged DNA could be observed even for long irradiation times (see lanes 6, 9 and 12 in Fig. 4).
Figure 4.
Binding of RPA to intact and UV-damaged 538 bp DNA. RPA was incubated with intact DNA and the DNA (protein to DNA ratio 10:1) exposed to UV light for different times with and without glutaraldehyde addition.
AFM of the DNA–RPA complexes
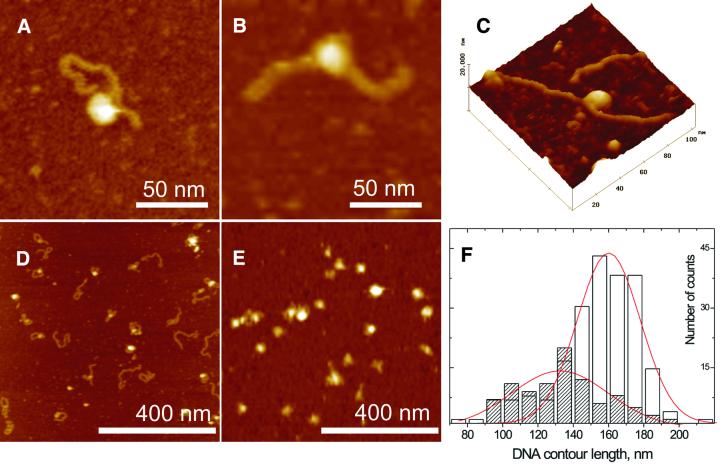
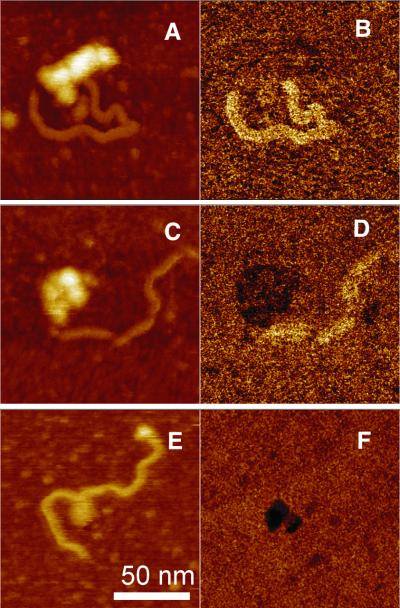
For AFM measurements, DNA and RPA were preincubated at a concentration ratio of 1:1 or 1:3. We find that reasonable data interpretation of the AFM images becomes difficult at a larger excess of protein, because the surface of mica is fully covered with the protein molecules. The preformed complexes were placed on the freshly cleaved mica and allowed to equilibrate on the surface for a couple of minutes. Nickel chloride was added to immobilize the molecules on the surface only after this equilibration step in order not to affect the RPA–DNA complex structure. The addition of RPA to undamaged DNA led to almost no change in the morphology of the DNA. Only rare cases of RPA binding to DNA termini could be observed (Fig. 5A and B). The situation is quite different, though, for UV-damaged DNA. When RPA was added to damaged DNA, globular objects sitting on the rod shaped DNA strands were regularly observed in the AFM images (Fig. 6A–C). The globular objects were already visible at a concentration ratio of 1:1 (Fig. 6D) and appeared to be distributed over the whole length of the DNA, with a slight preference for the termini of the DNA. The DNA molecules formed loop-like structures around globular objects in the majority of cases. When RPA was present in 3-fold excess over DNA, the number of irregular globular particles increased significantly (Fig. 6E) and the number of free DNA molecules reduced to 1:10. A significant decrease in contour length was observed for complexes where both ends of the DNA probe could be identified in the image (Fig. 6A–C). After complexation, the DNA contour length appeared to be 27.7 ± 7.6 nm shorter in comparison to DNA molecules damaged under identical conditions (Fig. 6F). The AFM images are in agreement with the notion of DNA wrapping around the protein (Fig. 6A–C). To allow a more rigorous scanning of the complexes by AFM it proved useful to stabilize the complexes with glutaraldehyde. The presence of glutaraldehyde (at the concentrations used for ESMA) did not increase RPA binding to the undamaged DNA, but merely improved high resolution imaging with AFM. We note that one (Fig. 6A–C) or more (Fig. 7A and C) globular objects were observed to bind to damaged DNA molecules. While high resolution AFM images show a certain sub-structure of the UV-damaged DNA–RPA complexes, quantitative results on the stoichiometry of the complex could not be achieved, as it is difficult to distinguish between RPA subunits and separate protein molecules.
Figure 5.
Interaction of RPA with undamaged DNA. AFM images do not show distinct complex formation of RPA with intact DNA (A) (5 mM NiCl2, without glutaraldehyde addition); only random cases of terminal binding were noticed (B) (z range 20 nm, 2.5 mM NiCl2, 0.1% glutaraldehyde).
Figure 6.
Interaction of RPA with DNA after UV treatment. High resolution AFM shows that UV-damaged DNA shows shortening of the DNA contour length (A and B) (2.5 mM NiCl2, 0.1% glutaraldehyde, z range 20 nm) and makes distinct turns around the protein molecule (C) (three-dimensional representation of UV-damaged DNA and RPA complex deposited on mica, 2.5 mM NiCl2, 0.1% glutaraldehyde, z range 20 nm). UV-damaged (30 min exposure) DNA was incubated with RPA in the ratios 1:1 (D) (5 mM NiCl2, without glutaraldehyde, z range 20 nm) and 1:3 (E) (5 mM NiCl2, without glutaraldehyde, z range 20 nm). (F) Contour length distributions evaluated from the AFM images. Empty bars correspond to free DNA molecules exposed to UV light for 30 min and filled bars to the same DNA measured in DNA–RPA complexes. The red lines represent the Gaussian fit of the distribution.
Figure 7.
AFM images of UV-damaged DNA and RPA complexes immobilized on mica, scanned in a buffer environment. RPA molecules sitting on the split ends of the DNA molecule (A) (height image, 2.5 mM NiCl2, 0.1% glutaraldehyde, z range 15 nm); (B) corresponding to the (A) phase image (z range 60°). RPA molecules sitting near the end of the DNA molecule: (C) height image (2.5 mM NiCl2, 0.1% glutaraldehyde, z range 15 nm); (D) corresponding to (C) phase image (z range 60°). The AFM image (E) shows two objects of increased height sitting on one DNA molecule (z range 30 nm). Only one of them is seen in the phase image while the other is not (F) (z range 30°).
AFM phase imaging
It has been shown in earlier studies that the phase signal in tapping mode AFM can be used for a faster and less invasive imaging of biological samples in liquids (16). The phase signal is known to be quite sensitive to attractive and repulsive forces acting on the tip. Therefore, in addition to the topography signal, it can be useful to differentiate between different materials. We have recorded the phase signal along with the height image to differentiate between DNA and RPA. With the procedure outlined in Materials and Methods, we observe a well-defined phase contrast between DNA and RPA molecules. As an example, Figure 7 shows different height and phase images of single DNA–RPA complexes. DNA exhibits a positive phase shift (i.e. it appears brighter in the phase image) and proteins show a negative phase shift (i.e. they appear darker in the phase image). Scanning with AFM at the resonance frequency of the tip enables visualization of either DNA (Fig. 7B) or RPA (Fig. 7F) or both molecules in the phase image (Fig. 7D). Figure 7E shows two objects of increased height along the DNA molecule. While the feature at the upper end of the molecule is not visible in the corresponding phase image (Fig. 7F) and possibly relates to a knot in the molecule, the feature in the center of the DNA most likely represents a protein. Moreover, the phase image (Fig. 7F) indicates a small bright line on the surface of the protein, in agreement with the notion that the DNA molecule coils itself around the RPA molecule.
DISCUSSION
Undamaged DNA
Direct sizing of more than 1000 intact DNA molecules yields an average contour length of 172 ± 10 nm, which is in agreement with the theoretically calculated value of 182 nm for 538 bp long fragments of B-DNA using 0.338 nm/bp. This level of precision of AFM sizing seems reasonable given the potential error introduced by the finite size of the AFM tip. This error is particularly relevant when short molecules are investigated.
It was shown recently that the polymer worm-like chain (WLC) theory is applicable to the DNA molecules (14,17). The value of the DNA persistence length indicates the state of equilibration of the molecule on the mica surface, i.e. the immobilization conditions. With AFM it is inherently impossible to distinguish between the intrinsic conformation of the DNA molecule in solution and conformational changes induced by surface immobilization. From our AFM measurements we find a persistence length of the intact DNA molecules of 36 ± 1 nm. This value can be compared with the results of Rivetti et al., who determined the persistence length of undamaged DNA equilibrated on a surface as 52.3 ± 0.3 nm (14). Given their result, one may anticipate that the DNA molecules in our measurements are in an intermediate state of adsorption between trapping and full two-dimensional equilibrium. One has to realize, though, that both the particular route of sample preparation as well as the particular method used for persistence length evaluation can also explain the difference in persistence length.
With regard to the former, Bustamante and co-workers (14) performed measurements on dried samples, which renders direct comparison to our measurements in a physiological environment questionable. Hansma et al. (18) measured the persistence length of DNA fragments of approximately the same length (500 bp) in aqueous buffer on a Ni2+-treated mica surface and found a value of ∼30 nm, in closer agreement with our results. However, as stated by the authors of this work, the DNA molecules were only loosely attached to the underlying surface, rendering AFM imaging difficult. In the present work, we managed to improve immobilization of the DNA molecules on the mica surface to the extent that they could reproducibly be scanned with AFM. It proved essential to keep the deposition buffer at pH ≈8 to realize efficient attachment of the DNA molecules.
In addition to the matter of sample preparation, different methods for persistence length determination vary considerably as well. This fact makes is difficult to compare absolute values of the persistence length. Here, however, we are mainly interested in following changes in the persistence length induced by UV treatment. Such changes are reliably monitored when a single numerical method is used for persistence length evaluation throughout the work.
UV treatment
Systematic comparison of the values of the contour length and mean square end-to-end distance of the DNA molecules prior to and after UV irradiation showed that both values decrease with increasing exposure time, i.e. increasing UV dose (Fig. 2). UV irradiation leads to the formation of photoproducts, of which cyclobutane pyrimidine dimers and pyrimidine(6–4)pyrimidone adducts are most prominent (19,20). Both adducts cause slight distortions of the B-helix and lead to local destabilization of the double-strand, although structural studies on these photoproducts in solution (21) did not reveal major bends in the DNA due to the damage sites. Detailed structural information on long DNA molecules containing multiple photoproducts is, however, not available. As judged from denaturing gel electrophoresis, there was no significant degradation of the DNA probe under the irradiation conditions applied. Systematic shortening of the DNA molecules may be partially explained by UV-induced destabilization and melting at the termini of the DNA with the production of single-stranded regions. This destabilization may lead to compaction and length reduction. Additional effects due to a collapse of DNA molecules carrying a critical number of UV photoproducts or an influence of the adsorption process cannot be excluded. Reduction of the mean square end-to-end distance with UV exposure indicates a reduced elasticity of the molecules. This interpretation is in line with the reduction of the migration speed observed in the gel electrophoretic experiments (Fig. 3). AFM measurements show an increase in the number of kinks on the DNA chain, which increased roughly proportional to UV light exposure time. At very high UV dose (UV exposure >30 min) severe damage seems to be produced in the DNA strands, which in turn makes it impossible to determine the characteristic lengths of the DNA molecules by AFM.
DNA–protein complex
Both the electrophoretic mobility experiments and the AFM measurements of the undamaged DNA molecules did not show complex formation with RPA no matter whether or not glutaraldehyde was added. At higher RPA concentrations some rare cases of terminal binding of the RPA to undamaged DNA were observed. Since RPA is known to have a high affinity for ssDNA, this finding may be explained by RPA binding to unpaired nucleotides at the termini of the DNA molecules (22). The situation is very different if RPA is added to UV-damaged DNA. In this case, globular complexes are observed, the shape of which render the determination of contour length and end-to-end distances difficult if not impossible. The observation of globular complexes can be attributed to RPA binding to UV-damaged sites. The stoichoimetry of the complex is uncertain. Most studies on the binding of RPA to damaged DNA have shown binding of a single RPA heterotrimer to a damaged site (9) and we therefore assume that these globular objects represent single RPA molecules bound to a photodamaged region. Furthermore, the AFM experiments have been performed at a RPA:DNA ratio of 1:1, with each DNA molecule carrying about 10 UV photoproducts. The formation of higher RPA oligomers is unlikely under these conditions. In many of the complexes, loop-like structures are observed, indicating that RPA contacts at least two regions of a damaged DNA molecule. DNA-binding sites have been identified both on RPA70 and RPA32 (23) and the loop-like structures may be due to DNA binding to both of these subunits via the damaged site and via the termini.
In cases where contour length measurements were yet possible after complex formation, we observe a contour length reduction of 27.7 ± 7.6 nM (Fig. 6F). From AFM measurements we find that a single RPA molecule has a diameter DRPA of ∼6–7 nm while the diameter DDNA of a DNA chain is ∼2 nm. Therefore a 27.7 nm loss of apparent DNA contour length is in good agreement with the notion that the DNA molecules coil around the RPA molecule, which would require Lcoil = π × (DRPA + DDNA) ≈ 27 nm.
CONCLUSION
In the present work, we were able to follow complex formation between undamaged and UV-damaged DNA molecules and the human replication protein RPA both by electrophoretic and AFM measurements. AFM measurements enabled us to access the contour length, the mean square end-to-end distance and the persistence length of the DNA molecules prior to and after UV damage. All lengths decreased on UV exposure. Efficient complex formation between DNA and RPA was possible only after UV exposure of the DNA, indicating that the latter efficiently creates binding sites on the DNA. AFM measurements of the complexes showed that the DNA tends to wrap around the protein. The use of AFM therefore enabled us to reveal additional information on the complex structure. It is indispensable, though, to combine the microscopy approach with a well-established analytical technique such as electrophoresis, in order to cross-check the microscopy results and reveal supporting information for AFM image interpretation.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful for financial support through the Deutsche Forschungsgemeinschaft (Kr1369/11).
REFERENCES
- 1.Lao Y., Lee,C.G. and Wold,M.S. (1999) Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry, 38, 3974–3984. [DOI] [PubMed] [Google Scholar]
- 2.Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- 3.Burns L., Guzder,S., Sung,P., Prakash,S. and Prakash,L. (1996) An affinity of human replication protein A for ultraviolet-damaged DNA. J. Biol. Chem., 271, 11607–11610. [DOI] [PubMed] [Google Scholar]
- 4.Wood R.D. (1999) DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie, 81, 39–44. [DOI] [PubMed] [Google Scholar]
- 5.Patrick S.M. and Turchi,J.J. (1999) Replication protein A (RPA) binding to duplex cisplatin-damaged DNA is mediated through the generation of single-stranded DNA. J. Biol. Chem., 274, 14972–14978. [DOI] [PubMed] [Google Scholar]
- 6.Lao Y., Gomes,X.V., Ren,Y., Taylor,J.S. and Wold,M.S. (2000) Replication protein A interactions with DNA. III. Molecular basis of recognition of damaged DNA. Biochemistry, 39, 850–859. [DOI] [PubMed] [Google Scholar]
- 7.Mitsis P.G., Kowalczykowski,S.C. and Lehman,I.R. (1993) A single-stranded DNA binding protein from Drosophila melanogaster: characterization of the heterotrimeric protein and its interaction with single-stranded DNA. Biochemistry, 18, 5257–5266. [DOI] [PubMed] [Google Scholar]
- 8.Kim C., Paulus,B.F. and Wold,M.S. (1994) Interactions of human replication protein A with oligonucleotides. Biochemistry, 33, 14197–14206. [DOI] [PubMed] [Google Scholar]
- 9.Hey T., Lipps,G. and Krauss,G. (2001) Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy. Biochemistry, 40, 2901–2910. [DOI] [PubMed] [Google Scholar]
- 10.Schweizer U., Hey,T., Lipps,G. and Krauss,G. (1999) Photocrosslinking locates a binding site for the large subunit of human replication protein A to the damaged strand of cisplatin-modified DNA. Nucleic Acids Res., 27, 3183–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochkarev A., Pfuetzner,R.A., Edwards,A.M. and Frappier,L. (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature, 385, 176–181. [DOI] [PubMed] [Google Scholar]
- 12.Henricksen L.A., Umbricht,C.B. and Wold,M.S. (1994) Recombinant replication protein A: expression, complex formation and functional characterization. J. Biol. Chem., 269, 11121–11132. [PubMed] [Google Scholar]
- 13.Keeling P.J., Klenk,H.P., Singh,R.K., Feeley,O., Schleper,C., Zillig,W., Doolittle,W.F. and Sensen,C.W. (1996) Complete nucleotide sequence of the Sulfolobus islandicus multicopy plasmid pRN1. Plasmid, 35, 141–144. [DOI] [PubMed]
- 14.Rivetti C., Guthold,M. and Bustamante,C. (1996) Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol., 264, 919–932. [DOI] [PubMed] [Google Scholar]
- 15.Yeung A.T., Mattes,W.B. and Grossman,L. (1986) Protein complexes formed during the incision reaction catalyzed by the Escherichia coli UvrABC endonuclease. Nucleic Acids Res., 14, 2567–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argaman M., Golan,R., Thomson,N.H. and Hansma,H.G. (1997) Phase imaging of moving DNA molecules and DNA molecules replicated in the atomic force microscope. Nucleic Acids Res., 25, 4379–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivetti C., Walker,C. and Bustamante,C. (1998) Polymer chain statistics and conformational analysis of DNA molecules with bends or sections of different flexibility. J. Mol. Biol., 280, 41–59. [DOI] [PubMed] [Google Scholar]
- 18.Hansma H.G., Kim,K.J., Laney,D.E., Garcia,R.A., Argaman,M., Allen,M.J. and Parsons,S.M. (1997) Properties of biomolecules measured from atomic force microscope images: a review. J. Struct. Biol., 119, 99–108. [DOI] [PubMed] [Google Scholar]
- 19.Spivak G., Leadon,S.A., Vos,J.M., Meade,S., Hanawalt,P.C. and Ganesan,A.K. (1988) Enhanced transforming activity of pSV2 plasmids in human cells depends upon the type of damage introduced into the plasmid. Mutat. Res., 193, 97–108. [DOI] [PubMed] [Google Scholar]
- 20.Bourre F., Renault,G. and Sarasin,A. (1987) Sequence effect on alkali-sensitive sites in UV-irradiated SV40 DNA. Nucleic Acids Res., 15, 8861–8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J.K., Soni,S.D., Arakali,A.V., Wallace,J.C. and Alderfer,J.L. (1995) Solution structure of a nucleic acid photoproduct of deoxyfluorouridylyl-(3′-5′)-thymidine monophosphate (d-FpT) determined by NMR and restrained molecular dynamics: structural comparison of two sequence isomer photoadducts (d-U5p5T and d-T5p5U). Nucleic Acids Res., 23, 1810–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgaki A. and Hubscher,U. (1993) DNA unwinding by replication protein A is a property of the 70 kDa subunit and is facilitated by phosphorylation of the 32 kDa subunit. Nucleic Acids Res., 21, 3659–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iftode C., Daniely,Y. and Borowiec,J.A. (1999) Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol., 34, 141–180. [DOI] [PubMed] [Google Scholar]