Abstract
Background
The incidence of glioblastoma in the elderly population is increasing as the worldwide population ages. The differential and poorer survival in the elderly population compared to younger patients is partially explained. The present study aimed to investigate the clinical impact of epidermal growth factor receptor EGFR-altered glioblastoma in a real-life elderly glioblastoma population.
Patients and Methods
A bicentric and retrospective study was conducted. Patients were 70 years or older and suffering from histomolecularly confirmed glioblastoma. Single nucleotide variants (SNV), amplification, or chromosome 7 polysomy were sought. The primary endpoint was the comparison of overall survival (OS) in patients with or without EGFR alteration. Secondary objectives were to determine other clinical parameters correlated with EGFR alteration status.
Results
Seventy-three patients were analyzed: 41.1% had at least one EGFR alteration. The presence of EGFR alteration did not impact overall survival: HR 0.97 [0.6–1.57], p = 0.9; the median overall survival was 6.5 months [5.3–9.3] in the EGFR-altered group versus 7 months [4.5–10] in the EGFR wild-type group, p = 0.75. In multivariate analysis, tumor resection was associated with a significant overall survival improvement: the median OS in the resected group (n = 20) was 11 months [95% CI 7.8–22] versus a median OS of 5.5 months [4.6–7.8] in the unresected group (n = 53), without correlation to EGFR alteration status.
Conclusion
In the modern era of molecular characterization and improved treatment modalities, the presence of at least one EGFR alteration did not influence survival outcomes in an elderly population of glioblastoma patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11060-024-04879-w.
Keywords: Glioblastoma, Elderly, EGFR, Survival, Gene amplification
Introduction
Glioblastoma is the most common and aggressive primary central nervous system malignancy in adults [1]. The incidence of glioblastoma in the elderly population is rising steadily as the world population ages, and it is already an increasingly frequent clinical situation [2]. The recognized prognostic factors for glioblastoma are age, extent of initial resection, and Karnofsky index at diagnosis [1, 3, 4]. The median overall survival for elderly patients in general population is lower compared to younger patients: 6 months versus 15–18 months [5, 6]. This survival difference is partly explained by non-tumoral factors (performance status at diagnosis, comorbidities, polymedication, treatment toxicity), but could also be due to different and specific glioblastoma molecular profiles in elderly patients. Batchelor et al. studied the survival impact of tumor protein 53 (TP53) and epidermal growth factor receptor (EGFR) genes alterations in glioblastoma patients according to age groups [7]. EGFR amplification was associated with survival in patients over 46 years old and was more unfavorable in younger patients. Another study in 2005, focusing specifically on elderly patients (over 75 years old), also demonstrated longer survival in 20 patients with EGFR amplification [8]. In the past two decades, the WHO classification of glioblastoma [9–11] and molecular characterization have evolved [12], as have the therapeutic approaches for elderly patients [5]. Current guidelines recommend that patients older than 70 years with good performance status undergo tumor resection followed by concomitant hypofractionated radiotherapy (40 Gy in 15 fractions) with continuous and maintenance temozolomide for patients with MGMT promoter-methylated glioblastoma [13]. In cases of frail general condition, three options can be discussed: hypofractionated radiotherapy alone, chemotherapy alone, or best supportive care, taking into account the MGMTp status [14]. This renders the survival results for the current elderly population obsolete. More recently, Johnson et al. investigated the potential differences in the molecular profile of the tumor and tumor microenvironment between glioblastoma patients aged 65 years or more versus those less than 65 [15]. A proper survival difference was observed according to age, but no association with gene expression according to bulk RNA sequencing data. To date, no study has been published dedicated to the evaluation of the clinical impact of EGFR alterations in an elderly population of glioblastoma patients.
Patient and methods
Study population and objectives
A retrospective, bicentric study was conducted at the Comprehensive Cancer Centre Henri Becquerel and the University Hospital of Rouen. Patients were selected from the neuropathology department database of the University Hospital of Rouen. The selected patients were aged 70 or over at the time of diagnosis and had histomolecularly-confirmed glioblastoma. The diagnostic neurosurgical procedure (biopsy, partial or total tumor resection) was performed between January 1, 2013, and December 31, 2022. All tumors were reviewed and classified according to the CNS5 WHO classification 2021 [11]; patients with gliosarcoma, H3K27M-altered diffuse midline glioma, or grade 4 IDH-mutant astrocytoma were excluded. Clinical and biological data collected included: demographic characteristics, presence or absence of comprehensive geriatric assessment at diagnosis, comorbidities according to the Cumulative Illness Rating Scale—Geriatric (CIRS-G) score [16], number of comedications, type of surgery (resection versus biopsy alone), type of treatment received, and date of death from any cause, as well as hospitalization during or after oncologic treatment. CIRS-G was calculated at the time of the study based on medical records. The primary objective was to study the impact on overall survival (OS) of EGFR alterations, taking into account the type of surgery (resection versus biopsy), age at diagnosis (70–79 versus > = 80), MGMT promoter methylation status (methylated versus unmethylated), radiotherapy (radiotherapy versus no radiotherapy), and chemotherapy (chemotherapy versus no chemotherapy). Secondary objectives were to investigate correlation between clinical features and EGFR alterations status in the studied population.
Molecular experiments
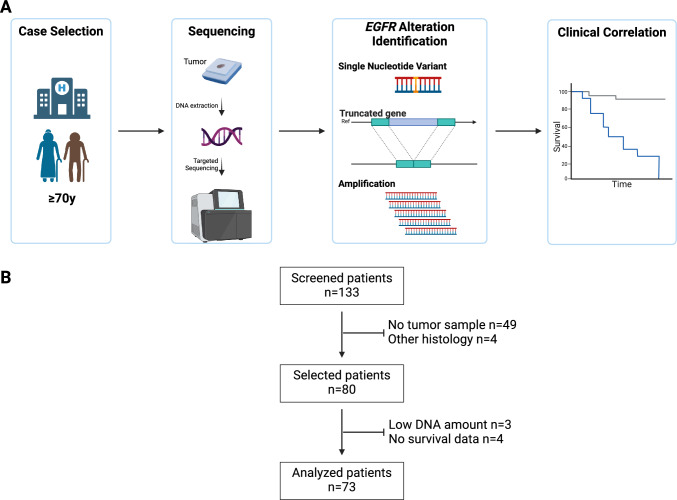
EGFR alterations were investigated using next-generation sequencing (Illumina NextSeq® platform). The types of EGFR alterations examined were the following: single nucleotide variations (SNV), copy number alterations, and splicing variants (Fig. 1A). DNA extraction, library preparation, targeted sequencing, pre-processing workflow, and variant calling were previously described [17]. In brief, targeted sequencing was performed using a custom panel GLIOPANEL-v2 [18], and sequencing reads were aligned to the GRCh37 genome. GLIOPANEL-v2 allows the detection of genomic alterations within recurrent altered regions of 20 genes, including all exons of the EGFR gene. SNVs were considered somatic mutations if they had a pathogenic or likely pathogenic status according to the COSMIC database. Only variants with a variant allele fraction (VAF) higher than 2% and a deduped depth equal to or greater than × 1000 were considered in the final analysis. Copy number variations were detected using mCNA [19] (https://gitlab.com/pierrejulien.viailly/mcna/). EGFR amplification (EGFRamp) was defined by a number of copies gain equal to or higher than 5. Chromosome 7 polysomy was considered using the PI3KCG gene (chromosome 7q22.3) copy number variation; if the CNV was higher than 2.5 copies, chromosome 7 was considered polysomic. Truncated mutated EGFR were identified by detecting differential CNV between exons for a unique sample: deletion of exon 2 to exon 7 (EGFRvIII) or exon 26 to exon 27 were particularly studied [20]. EGFR was considered altered (EGFRalt) if at least one mutation and/or amplification and/or truncated gene was identified; otherwise, the tumor was considered as EGFR wild-type (EGFRwt). Finally, MGMT promoter methylation status was determined using the pyrosequencing method (Therascreen MGMT Pyro®, Qiagen®) after DNA bisulfite reaction. A mean of methylated CpG islands greater than 12% was considered as methylated MGMTp.
Fig. 1.
Description of the study. Schematic representation of the workflow of the study (A) and flow chart (B) of the population based on the database of the University Hospital of Rouen. The screened patients were aged 70 or over at the time of diagnosis (between January 1, 2013, and December 31, 2022)
TCGA database
Data from The Cancer Genome Atlas Program (TCGA) were utilized to enhance the cohort for survival analysis. The data were downloaded from cBioPortal using the ‘Glioblastoma Multiforme (TCGA, PanCancer Atlas)’ dataset [21]. The filtering criteria were as follows: patients had to be 70 years of age or older at diagnosis, and genomic data (including mutations and structural variants) along with copy number alteration information (both detection and log2 values of CNA) had to be available.
Statistical analyses
Continuous variables are expressed as mean and standard deviation (sd) or median and interquartile range according to parametric or non-parametric distribution respectively. Comparison between two groups were done using Student test or Mann–Whitney test. Comparison for qualitative variables were performed by using Chi-Square test. Overall survival (OS) curves were obtained using Kaplan Meier method. Survival curves were compared with log-rank test. Impact on survival of the studied variable were estimated using the Cox regression model (hazard ration – HR). Each variable was explored in univariate analysis and then all variables with p-value equal or lower than 0.1 were considered for multivariate analysis. OS was the time from the day of diagnosis until death from any cause. All tests were two-sided and a p value of 0.05 or less was considered as statistically significant, apart for variable selection for the multivariate analysis of the Cox model. Figures and analyses were performed in R version 4.3.1 using several packages (trackViewer_1.36.2 [22], survival_3.7–0, survminer_0.4.9, tableone_0.13.2, dplyr_1.1.4 and corrplot_0.92) with the default settings.
Results
Characteristics of the study cohort
From 2013 to 2022, 73 patients were included in the final analysis out of the 133 patients identified in the database (Fig. 1)(Fig. 1B). The median age was 75 years (range 70–87), and the sex ratio was 1.9 (Table 1). A vast majority of the patients underwent biopsy-alone as the diagnostic procedure (n = 53/73, 72.6%). No significant differences were observed between the resected and unresected groups in terms of age (mean 74.8 versus 76.1, p = 0.215), CIRS-G score (mean score 4.7 versus 4.5, p = 0.78), or Karnofsky index score for those available (n = 31/70, 42.5%, mean 69% versus 77%, p = 0.138). Only 5 patients (6.8%) of the entire population had an oncogeriatric evaluation before treatment. Regarding treatment after the neurosurgical procedure, data were available for 74% of the population (n = 54/73). Among these patients, 79.6% (n = 43/54) had radiotherapy-based first-line treatment, and 46.3% (n = 25/54) received temozolomide, either concurrently with radiotherapy or as maintenance therapy. No patient had chemotherapy alone.
Table 1.
Clinical, molecular characteristics at diagnosis and treatment received according to EGFR alteration
| Entire cohort n = 73 |
EGFRwt group n = 43 |
EGFRalt group n = 30 |
p-value | |
|---|---|---|---|---|
| Age – mean (SD) | 75.8y (3.9) | 75.72 (3.50) | 75.87 (4.52) | 0.877 |
| Male sex – no. (%) | 39 (53.4) | 25 (58.1) | 14 (46.7) | 0.466 |
| Brain Location1 – no. (%) | ||||
| Internal capsule | 2 (2.7) | 1 (2.3) | 1 (3.3) | 0.664 |
| Frontal | 13 (17.8) | 7 (16.3) | 6 (20.0) | |
| Insula | 1 (1.4) | 0 (0.0) | 1 (3.3) | |
| Occipital | 2 (2.7) | 1 (2.3) | 1 (3.3) | |
| Parietal | 13 (17.8) | 7 (16.3) | 6 (20.0) | |
| Temporal | 25 (34.2) | 14 (32.6) | 11 (36.7) | |
| More than 2 | 17 (23.4) | 13 (30.2) | 4 (13.3) | |
| Laterality – no. (%) | ||||
| Left | 45 (61.6) | 27 (62.8) | 18 (60.0) | 0.429 |
| Right | 26 (35.6) | 14 (32.6) | 12 (40.0) | |
| Both | 2 (2.8) | 2 (4.7) | 0 (0.0) | |
| CIRS-G score median [IQR] | 4 (3, 6) | 5.00 [2.50, 5.50] | 4.00 [3.00, 6.75] | 0.946 |
| Biopsy-alone – no. (%) | 53 (72.6) | 32 (74.4) | 21 (70.0) | 0.881 |
| Radiotherapy2 – no. (%) | 43 (79.6) | 22 (51.2) | 21 (70.0) | 0.171 |
| Temozolomide2 – no. (%) | 25 (46.3) | 13 (44.8) | 12 (48.0) | 1 |
| MGMT promoter unmethylated3 – no. (%) | 36 (51.4) | 19 (46.3) | 17 (58.6) | 0.441 |
1Brain location was considered as the main lobe involved by the tumor, in case of multiple lobes involvement without evidence of one the tumor was considered as more than 2
2Data available for 54 patients
3Data available for 70 patients
Impact of EGFR alterations on survival
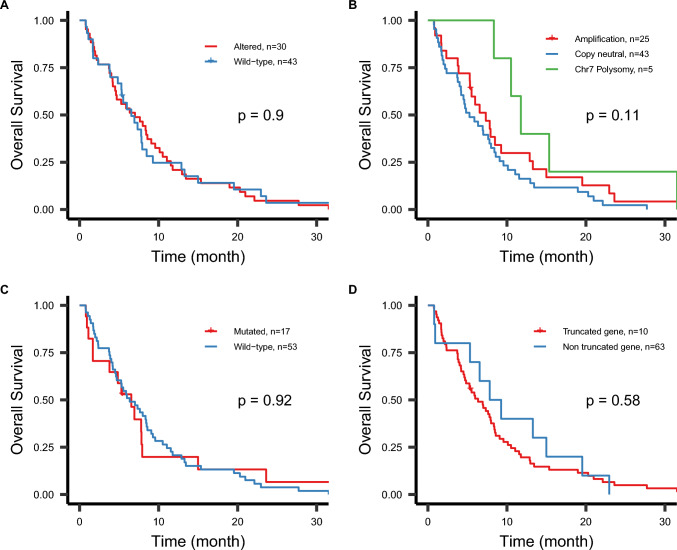
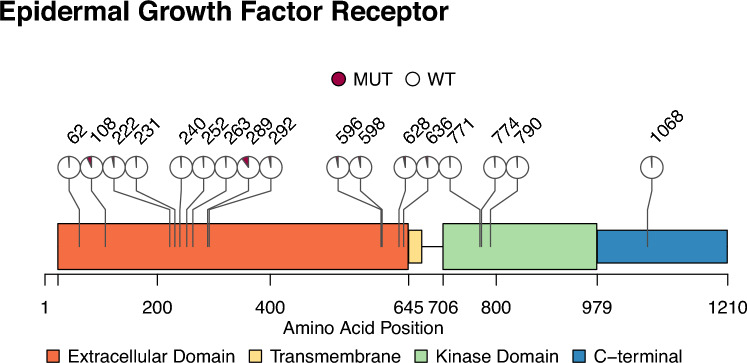
In the entire population, 30 patients (41.1%) had at least one EGFR alteration, namely a mutation (n = 17, 23.3%), an amplification (n = 25, 34.2%) and/or a truncated gene (n = 10, 13.7%) (Table 1). No clinical differences at baseline or treatment schedule between the EGFRalt group and the EGFRwt group were observed (Table 1). The median overall survival in the entire population was 6.9 months [95% CI 5.3–8.4]. The overall survival rate at one year was 22% [95% CI 15–35%] and at two years was 4.2% [95% CI 1.4–13%]. The presence of EGFR alterations did not impact overall survival in the studied population: HR 0.97 [95% CI 0.6–1.57], p = 0.9. The median overall survival was 6.5 months [95% CI 5.3–9.3] in the EGFRalt group versus 7 months [95% CI 4.5–10] in the EGFRwt group, p = 0.9 (Fig. 2A). In detail, neither copy number variation (Fig. 2B), mutation (Fig. 2C), nor truncated gene (Fig. 2D) had an impact on OS. For the CNV analysis, HR was equal to 1.42 [95% CI 0.85–2.37], p = 0.2, for the copy neutral and 0.59 [95% CI 0.22–1.55], p = 0.3, for chromosome 7 polysomy compared to EGFRamp group. When pooling patients that carried EGFRamp glioblastoma (n = 25) and those with chromosome 7 polysomy (n = 5), a trend for a favorable outcome was observed compared to EGFR copy neutral in univariate analysis, but this was not confirmed when adjusting for other prognostic factors such as resection status (Table 2). Tumor resection was associated with a significant overall survival improvement: the median OS in the resected group (n = 20) was 11 months [95% CI 7.8–22] versus a median OS of 5.5 months [95% CI 4.6–7.8] in the unresected group (n = 53), log rank p < 0.0001. EGFR alteration status was not associated with survival outcome in the biopsy-alone group: HR 1.3 [0.72–2.32], p = 0.4. Eighty-eight percent of the EGFR mutations occurred in the extracellular domain of EGFR protein (Fig. 3) as previously described in the TCGA cohort [20]. The small number of mutations in our cohort did not allow to explore potential different prognostic impact of EGFR mutation’s location.
Fig. 2.
Kaplan–Meier curves according to the different EGFR alterations in the studied cohort (n = 73). The proportion of alive patients over time is presented according to the presence of at least one EGFR alteration in the tumor (A), the status of copy number variation (B), the presence of at least one mutation (C), and the presence of a truncated gene (D). Chromosome 7 polysomy is defined as a copy number gain of chromosome 7 higher than 2.5, based on the PI3KCG gene (7q22.3). Samples having both chromosome 7 polysomy and EGFR amplification were considered as EGFR-amplified glioblastoma. p-value is from the log-rank test
Table 2.
Cox model for overall survival in the studied population
| n | Univariate Analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | CI95% inf | CI95% sup | p.value | HR | CI95% inf | CI95% sup | p.value | ||
| Tumor resection vs biopsy | 73 | 0.37 | 0.21 | 0.67 | 0.001 | 0.41 | 0.22 | 0.76 | 0.005 |
| Age >= 80y vs 70-79y | 73 | 1.28 | 0.68 | 2.40 | 0.5 | ||||
| MGMTp unmethylated vs methylated | 70 | 1.44 | 0.89 | 2.34 | 0.14 | ||||
| CIRS-G score <4 vs >= 4 | 73 | 0.66 | 0.4 | 1.08 | 0.1 | 0.69 | 0.42 | 1.13 | 0.14 |
| EGFR wild-type vs EGFR alteration | 73 | 0.97 | 0.6 | 1.57 | > 0.9 | ||||
| EGFR copy neutral vs EGFR amplification or chr7 polysomy | 73 | 1.59 | 0.98 | 2.59 | 0.062 | 1.38 | 0.83 | 2.28 | 0.2 |
Fig. 3.
Schematic Representation of the EGFR Protein and the Location of the Altered Amino Acids Identified in the Studied Population (n = 73). Overall, 35 mutations were identified in 17 samples (23.3%) of the cohort. Of these mutations, 91.4% (n = 31/35) were missense mutations, one was a nonsense mutation, one was an insertion, and one was a splice variant. The mutations affecting the amino acids of the EGFR protein are represented. The pie plot by location represents the proportion of mutated samples within this amino acid among the entire cohort
Integrated survival outcomes from TCGA data
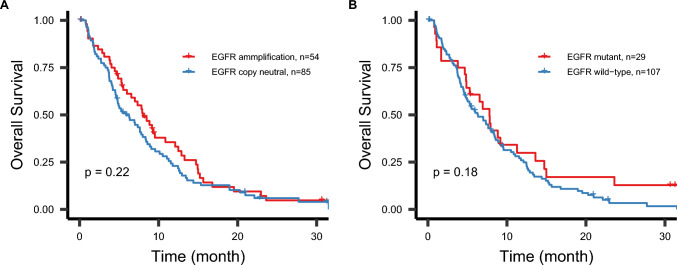
Sixty-six patients were selected from the TCGA dataset, with a median age of 76 years (range 70–89) and a sex ratio of 1.2. Twenty-nine patients had glioblastoma with EGFR amplification (n = 29/66, 43.9%), while 12 patients had glioblastoma with EGFR mutation (n = 12/66, 18.2%). The median overall survival was 7.4 months [95% CI 4.9–11]. When combining the two cohorts, the one-year survival rate was 28% [95% CI 21–37%] and the two-year survival rate was 5.3% [95% CI 2.4–11.2%], Supplementary Table 1. Neither EGFR amplification nor EGFR mutation status influenced overall survival: the median survival was 8 months [95% CI 6.0–12] for glioblastoma with EGFR amplification compared to 5.9 months [95% CI 4.6–8.4] for EGFR copy-neutral glioblastoma, with a log-rank p-value of 0.22; and 7.8 months [95% CI 4.9–14] for glioblastoma with EGFR mutation versus 6.3 months [95% CI 4.9–8.4] for glioblastoma with wild-type EGFR, with a log-rank p-value of 0.18 (Fig. 4). These results were corroborated by univariate Cox regression analysis: hazard ratio (HR) of 1.26 [95% CI 0.87–1.82] for EGFR amplification and HR of 1.37 [95% CI 0.87–2.17] for EGFR mutation.
Fig. 4.
Kaplan–Meier curves stratified by EGFR amplification or mutation status in the studied cohort (n = 73) and the TCGA cohort (n = 66). Overall survival is displayed based on the type of alteration. The p-value is derived from the log-rank test
Contributing elements to clinical outcomes
In the studied population, the in-patient rate was obtained for 60.3% (n = 44) of the entire cohort. Among this subgroup, almost half of the patients (n = 21/44, 47%) were hospitalized twice or more during active or palliative management. The three most frequent causes for hospitalization were: deterioration in general condition (35%), neurological impairment including deficit, intracranial hypertension, or seizures (26%), and treatment-related toxicity (19%). The in-patient rate was not associated with the EGFR alteration status of the tumor: 90% in the EGFRalt group (n = 18/20) versus 95.8% in the EGFRwt group (n = 23/24), p = 0.870.
When considering EGFR CNV as a continuous variable, the mean copy number gain was 37 copies (min. 2.82 – max. 111.55) in the EGFR gained glioblastoma population (EGFR amplification + chromosome 7 polysomy, n = 30). In the EGFRamp group (n = 25), all tumors harbored a copy gain higher than 10 copies. No correlation between EGFR CNV and clinical features was identified (Fig. 5).
Fig. 5.
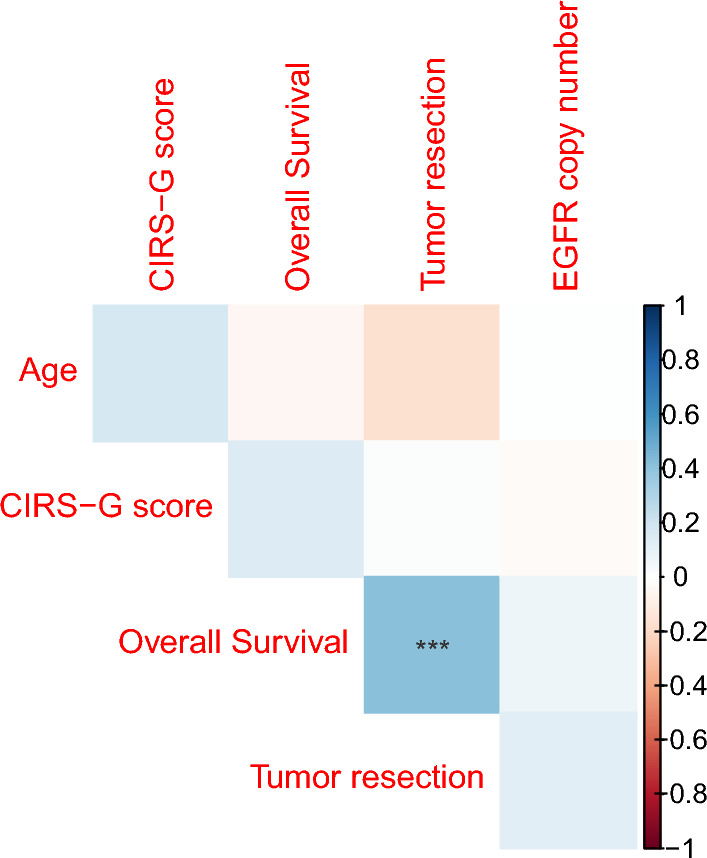
Correlation plot between clinical, therapeutic features and EGFR copy number in the studied cohort (n = 73). EGFR copy number is set at 2 copies for non-amplified and non-polysomic samples. Tumor resection variable was transformed as binary variables. No correlation between those variables and EGFR copy number was identified. The only significant correlation was identified between overall survival (months) and tumor resection status (p < 0.01)
In the subgroup of patients with adjuvant treatment data available (n = 54), radiotherapy, regardless of the delivered dose, or temozolomide administration were identified as favorable prognostic factors: HR 0.55 [0.34–0.89], p = 0.015 and HR 0.15 [0.07–0.31], p < 0.001, respectively. The rate of patients receiving radiotherapy and/or temozolomide was equivalent between the EGFRalt subgroup and the EGFRwt subgroup: 70% versus 51.2% for radiotherapy, p = 0.171, and 48% versus 44.8% for temozolomide, p = 1.
Discussion
Seventy-three elderly patients with glioblastoma were retrospectively analyzed in our study. The observed median OS was 6.9 months, and the 1-year survival rate was 22%. This OS is in accordance with those observed in previous studies [14] or the CBTRUS database in the USA [1]: 14.1% for the population of glioblastoma patients older than 75. EGFR alterations (EGFRalt) status in the tumor did not influence survival in our cohort: HR 0.97 [0.6–1.57], p = 0.9. In detail, EGFR amplification occurred in one-third of the patients (34.2%) and was the most frequent EGFR alteration. The EGFR amplification (EGFRamp) group had similar survival compared to the copy neutral group, HR 1.42, p = 0.2. This result is in accordance with a meta-analysis conducted by Chen et al. by pooling the results of three studies, HR 1.101 [0.845–1.434], p = 0.475 [23]. The population of the studies included all ages and not solely elderly patients. Studies focusing on EGFR amplification as an independent prognostic factor in the elderly in the modern era of tumor molecular characterization [11] and with up-to-date post-surgical treatment such as hypofractionated radiotherapy [5] are still lacking. The negative impact of EGFR amplification on survival in young patients (less than 45 years old) is controversial [24–26]. One limitation is the homogeneity of the method to consider a glioblastoma as “EGFR amplified”. EGFR amplification could be accurately determined by fluorescence in situ hybridization (FISH) or RNA sequencing (RNAseq). Tumor DNA sequencing of the EGFR locus is now considered a valuable surrogate marker of EGFR amplification and is highly correlated with FISH [27]. Moreover, the threshold to consider EGFR as amplified was set at a gain of at least 5 copies [28], especially for clinical trials. This more stringent threshold that we integrated into our study could partially explain the lower frequency of EGFR-amplified tumors compared to the frequency of amplified chr7p11.2 where EGFR is located in the TCGA database [20] (n = 276/543, 50.8%). In fact, all EGFRamp glioblastomas in our cohort harbored a copy gain higher than 10 copies. A more recent study also identified an overall frequency of EGFRamp glioblastoma of 34.5% without a difference between patients younger or older than 65 years of age [15]. Three different CNV situations could be distinguished: EGFR gene amplification, a polysomy of chromosome 7 when the gain of chromosome 7 is higher than 2 copies, and the copy neutral situation, where neither of the other situations occurred [29]. The situation of chromosome7/EGFR polysomy without amplification should be interesting to explore as a prognostic factor because it seems to be an independent entity [30]. The low number of patients carrying this polysomy in our cohort (n = 5) prevented us from identifying any prognostic signal.
When incorporating the TCGA dataset (n = 66) for patients over 69 years old, no survival differences were observed based on EGFR amplification status or mutation status. This finding aligns with earlier results from the TCGA glioblastoma cohort (n = 380), which indicated equivalent median overall survival for both the EGFR-amplified and non-amplified groups, as well as for the EGFR-mutated and wild-type groups: 15 months versus 13 months, with a log-rank p-value of 0.36, and 15 months versus 15 months, with a p-value of 1 [21]. The lack of impact from EGFR alterations, particularly amplification, has recently been reaffirmed in a large cohort of IDH-wildtype grade 4 gliomas [31]. However, a negative impact of EGFR amplification was noted in IDH-mutant gliomas. IDH-mutant gliomas were not included in our cohort as we aimed to focus solely on glioblastoma, in accordance with the CNS5 WHO classification. The incidence of IDH-mutant gliomas in the elderly population is very rare, and a dedicated study investigating the prognostic impact of EGFR alterations in this specific group could be valuable.
No study has addressed the age threshold to define what constitutes an elderly population of glioblastoma. The threshold varies from >60 to >75 [2]. The threshold of equal to or higher than 70 was chosen. The exploratory analysis performed on the subgroup of patients being “older old”, i.e., higher than 80 years of age, did not reveal any influence of EGFR alterations on survival. A dedicated study investigating the impact of EGFR alterations on survival in this “older old” population should be interesting and complementary to recently published clinical cases cohort [32].
A significant improvement in OS was observed in the tumor-resected group compared to the biopsy-alone group. The survival impact of tumor resection in patients over 70 is still lacking robust prospective clinical trials. Most studies are retrospective or prospective with a low number of participants or are not randomized. Nevertheless, resection seems to improve OS for fit patients since a phase III trial published in 2003 by Vuorinen et al. has shown a median survival of 171 days for the resected group versus 85 days for the non-resected group [33]. In 2021, the French neuro-oncology society (ANOCEF) conducted a multicenter randomized controlled trial comparing resection surgery versus biopsy [34]. The study showed that surgery did not significantly increase overall survival but did increase progression-free survival (PFS) and quality of life in patients. It should be noted that in this study, the median survival in each group was 8 and 9 months, which is slightly higher than the usual survival in this population, and all patients had at least radiotherapy after diagnosis. In this context, our results should be interpreted with caution as potential confounding factors such as performance status at diagnosis were missing and could possibly influence the OS in the resected group. The slightly lower proportion of resections in our cohort (27.4%) compared to the SEER database (35% for patients aged 65 to 79, and 28% for patients aged 80 and older) [35] may also be attributed to other clinical factors, such as performance status in this frail and unselected population. Regarding MGMT promoter methylation, a non-significant difference in OS was observed. MGMTp methylation is known in the literature to be a favorable prognostic factor in glioblastoma and a predictive biomarker of response to alkylating agents, especially temozolomide [36]. The lack of significance for MGMTp methylation is probably due to the small number of patients exposed to temozolomide (n = 24, 31%). As in phase III studies in the elderly, we have shown that patients who received radiotherapy or radiochemotherapy had an improvement in overall survival [5, 6].
Finally, other oncogeriatric evaluations should be explored in the specific population of glioblastoma patients. To our knowledge, no available oncogeriatric scale is validated in glioblastoma. The CIRS-G score partially reflects the frailty of elderly patients and was not found to influence OS in multivariate analysis or to be correlated with EGFR alterations.
Conclusion
When considering the characterization of somatic EGFR alterations using next-generation sequencing in a real-life elderly glioblastoma population, the presence of at least one EGFR alteration did not influence survival outcomes. Aggressive treatment, such as tumor resection, is significantly associated with improved survival outcomes. However, predictive clinical or biological factors prior to tumor-related intervention are still lacking in this specific population.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- TCGA
The Cancer Genome Atlas
- CIRS-G
Cumulative Illness Rating Score – Geriatric
- EGFR
Epidermal growth factor receptor
- HR
Hazard Ratio
- IDH
Isocitrate deshydrogenase
- KI
Karnofsky Index
- MGMT
Methyl Guanine Methyl Transferase
- OS
Overall Survival
- SNV
Single Nucleotide Variant
- WHO
World Health Organization
Author contributions
SP collected the data, conducted the analyses, and wrote the manuscript. LBT and NSV performed the sequencing experiments and contributed to the data analysis. FM selected the cases and reviewed all of them. PJV handled the data sequencing analysis. OL, CA, IT, and FDF contributed to the collection and curation of clinical data. MF and NSV had the lead to conduct the study. MF created the figures, assisted with the data analysis, and wrote the final manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This study was funded by the internal funds of the INSERM U1245 Cancer and Brain Genomics research laboratory, Team 2, IRON group.
Data availability
Survival data are available in Supplementary Table 1. Deidentified sequencing data are available on request.
Declarations
Competing Interests
The authors declare no competing interests.
Ethical approval
All patients provided written informed consent for the use of de-identified tumor material and demographic data for research purposes.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nasrin Sarafan-Vasseur and Maxime Fontanilles contribute equally to the work.
References
- 1.Ostrom QT, Price M, Neff C et al (2023) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2016–2020. Neuro Oncol. 10.1093/neuonc/noad149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazarakis NK, Robinson SD, Sinha P et al (2024) Management of glioblastoma in elderly patients: a review of the literature. Clin Transl Radiat Oncol 46:100761. 10.1016/j.ctro.2024.100761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simpson JR, Horton J, Scott C et al (1993) Influence of location and extent of surgical resection on survival of patients with glioblastoma multiforme: results of three consecutive Radiation Therapy Oncology Group (RTOG) clinical trials. Int J Radiat Oncol Biol Phys 26(2):239–244. 10.1016/0360-3016(93)90203-8 [DOI] [PubMed] [Google Scholar]
- 4.Lacroix M, Abi-Said D, Fourney DR et al (2001) A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg 95(2):190–198. 10.3171/jns.2001.95.2.0190 [DOI] [PubMed] [Google Scholar]
- 5.Perry JR, Laperriere N, O’Callaghan CJ et al (2017) Short-course radiation plus temozolomide in elderly patients with glioblastoma. N Engl J Med 376(11):1027–1037. 10.1056/NEJMoa1611977 [DOI] [PubMed] [Google Scholar]
- 6.Malmström A, Grønberg BH, Marosi C et al (2012) Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol 13(9):916–926. 10.1016/S1470-2045(12)70265-6 [DOI] [PubMed] [Google Scholar]
- 7.Batchelor TT, Betensky RA, Esposito JM et al (2004) Age-dependent prognostic effects of genetic alterations in glioblastoma. Clin Cancer Res 10(1 Pt 1):228–233. 10.1158/1078-0432.ccr-0841-3 [DOI] [PubMed] [Google Scholar]
- 8.Kleinschmidt-DeMasters BK, Lillehei KO, Varella-Garcia M (2005) Glioblastomas in the older old. Arch Pathol Lab Med 129(5):624–631. 10.5858/2005-129-0624-GITOO [DOI] [PubMed] [Google Scholar]
- 9.Louis DN, Ohgaki H, Wiestler OD et al (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114(2):97–109. 10.1007/s00401-007-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 11.Louis DN, Perry A, Wesseling P et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23(8):1231–1251. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brat DJ, Aldape K, Colman H et al (2018) cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV.” Acta Neuropathol 136(5):805–810. 10.1007/s00401-018-1913-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weller M, van den Bent M, Preusser M et al (2021) EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol 18(3):170–186. 10.1038/s41571-020-00447-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ironside SA, Sahgal A, Detsky J, Das S, Perry JR (2021) Update on the management of elderly patients with glioblastoma: a narrative review. Ann Palliat Med 10(1):899–908. 10.21037/apm-20-1206 [DOI] [PubMed] [Google Scholar]
- 15.Johnson M, Bell A, Lauing KL et al (2023) Advanced age in humans and mouse models of glioblastoma show decreased survival from extratumoral influence. Clin Cancer Res 29(23):4973–4989. 10.1158/1078-0432.CCR-23-0834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salvi F, Miller MD, Grilli A et al (2008) A manual of guidelines to score the modified cumulative illness rating scale and its validation in acute hospitalized elderly patients. J Am Geriatr Soc 56(10):1926–1931. 10.1111/j.1532-5415.2008.01935.x [DOI] [PubMed] [Google Scholar]
- 17.Noeuveglise A, Sarafan-Vasseur N, Beaussire L et al (2023) Impact of EGFRA289T/V mutation on relapse pattern in glioblastoma. ESMO Open 8(1):100740. 10.1016/j.esmoop.2022.100740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontanilles M, Deniel A, Marguet F et al (2022) Usefulness of circulating tumor DNA from cerebrospinal fluid in recurrent high-grade glioma. Rev Neurol (Paris). 10.1016/j.neurol.2022.02.462 [DOI] [PubMed] [Google Scholar]
- 19.Viailly PJ, Sater V, Viennot M et al (2021) Improving high-resolution copy number variation analysis from next generation sequencing using unique molecular identifiers. BMC Bioinform 22(1):120. 10.1186/s12859-021-04060-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennan CW, Verhaak RGW, McKenna A et al (2013) The somatic genomic landscape of glioblastoma. Cell 155(2):462–477. 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Lichtenberg T, Hoadley KA et al (2018) An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173(2):400-416.e11. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou J, Zhu LJ (2019) trackViewer: a bioconductor package for interactive and integrative visualization of multi-omics data. Nat Methods 16(6):453–454. 10.1038/s41592-019-0430-y [DOI] [PubMed] [Google Scholar]
- 23.Chen JR, Xu HZ, Yao Y, Qin ZY (2015) Prognostic value of epidermal growth factor receptor amplification and EGFRvIII in glioblastoma: meta-analysis. Acta Neurol Scand 132(5):310–322. 10.1111/ane.12401 [DOI] [PubMed] [Google Scholar]
- 24.Hoffman DI, Abdullah KG, McCoskey M et al (2019) Negative prognostic impact of epidermal growth factor receptor copy number gain in young adults with isocitrate dehydrogenase wild-type glioblastoma. J Neurooncol 145(2):321–328. 10.1007/s11060-019-03298-6 [DOI] [PubMed] [Google Scholar]
- 25.Armocida D, Pesce A, Frati A, Santoro A, Salvati M (2020) EGFR amplification is a real independent prognostic impact factor between young adults and adults over 45yo with wild-type glioblastoma? J Neurooncol 146(2):275–284. 10.1007/s11060-019-03364-z [DOI] [PubMed] [Google Scholar]
- 26.Labussière M, Boisselier B, Mokhtari K et al (2014) Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology 83(13):1200–1206. 10.1212/WNL.0000000000000814 [DOI] [PubMed] [Google Scholar]
- 27.Lassman AB, Roberts-Rapp L, Sokolova I et al (2019) Comparison of biomarker assays for EGFR: implications for precision medicine in patients with glioblastoma. Clin Cancer Res 25(11):3259–3265. 10.1158/1078-0432.CCR-18-3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.French PJ, Eoli M, Sepulveda JM et al (2019) Defining EGFR amplification status for clinical trial inclusion. Neuro Oncol 21(10):1263–1272. 10.1093/neuonc/noz096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bieńkowski M, Piaskowski S, Stoczyńska-Fidelus E et al (2013) Screening for EGFR amplifications with a novel method and their significance for the outcome of glioblastoma patients. PLoS ONE 8(6):e65444. 10.1371/journal.pone.0065444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilcock DM, Goold E, Zuromski LM, Davidson C, Mao Q, Sirohi D (2024) EGFR/CEP7 high polysomy is separate and distinct from EGFR amplification in glioblastoma as determined by fluorescence in situ hybridization. J Neuropathol Exp Neurol 83(5):338–344. 10.1093/jnen/nlae028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh HS, Patel RV, Woodward E et al (2024) Contemporary prognostic signatures and refined risk stratification of gliomas: an analysis of 4,400 tumors. Neuro Oncol. 10.1093/neuonc/noae164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stadler C, Gramatzki D, Le Rhun E et al (2024) Glioblastoma in the oldest old: clinical characteristics, therapy, and outcome in patients aged 80 years and older. Neurooncol Pract 11(2):132–141. 10.1093/nop/npad070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vuorinen V, Hinkka S, Färkkilä M, Jääskeläinen J (2003) Debulking or biopsy of malignant glioma in elderly people - a randomised study. Acta Neurochir (Wien) 145(1):5–10. 10.1007/s00701-002-1030-6 [DOI] [PubMed] [Google Scholar]
- 34.Laigle-Donadey F, Metellus P, Guyotat J et al (2023) Surgery for glioblastomas in the elderly: an Association des Neuro-oncologues d’Expression Française (ANOCEF) trial. J Neurosurg 138(5):1199–1205. 10.3171/2022.8.JNS221068 [DOI] [PubMed] [Google Scholar]
- 35.Horowitz MA, Ghadiyaram A, Mehkri Y et al (2024) Surgical resection of glioblastoma in the very elderly: an analysis of survival outcomes using the surveillance, epidemiology, and end results database. Clin Neurol Neurosurg 245:108469. 10.1016/j.clineuro.2024.108469 [DOI] [PubMed] [Google Scholar]
- 36.Hegi ME, Diserens AC, Gorlia T et al (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352(10):997–1003. 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Survival data are available in Supplementary Table 1. Deidentified sequencing data are available on request.