Abstract
HhaI DNA methyltransferase belongs to the C5-cytosine methyltransferase family, which is characterized by the presence of a set of highly conserved amino acids and motifs present in an invariant order. HhaI DNA methyltransferase has been subjected to a lot of biochemical and crystallographic studies. A number of issues, especially the role of the conserved amino acids in the methyltransferase activity, have not been addressed. Using sequence comparison and structural data, a structure-guided mutagenesis approach was undertaken, to assess the role of conserved amino acids in catalysis. Site-directed mutagenesis was performed on amino acids involved in cofactor S-adenosyl-l-methionine (AdoMet) binding (Phe18, Trp41, Asp60 and Leu100). Characterization of these mutants, by in vitro /in vivo restriction assays and DNA/AdoMet binding studies, indicated that most of the residues present in the AdoMet-binding pocket were not absolutely essential. This study implies plasticity in the recognition of cofactor by HhaI DNA methyltransferase.
INTRODUCTION
HhaI DNA methyltransferase (EC 2.1.1.37) is a part of the restriction–modification system of Haemophilus haemolyticus (1). The gene for the HhaI methylase was cloned, sequenced and found to code for a 327 amino acid protein (2). The protein has been overexpressed and purified in Escherichia coli (3–5). HhaI DNA methyltransferase (M.HhaI) is a C5-cytosine methyltransferase that recognizes the sequence 5′-GCGC-3′ in a double-stranded context and methylates the internal cytosine at the C5 position. Sequence alignment with the members of the C5-methyltransferase family revealed the presence of 10 conserved motifs, each containing about 5–10 amino acids, present in an invariant order in nearly all methyltransferases (6–8) including HhaI methyltransferase.
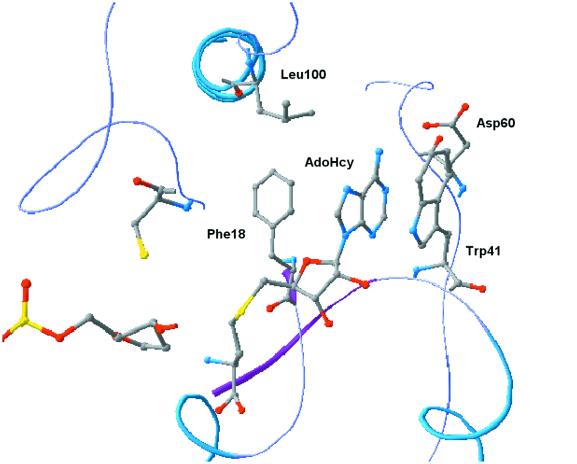
Among the 10 conserved motifs present in M.HhaI, two have been assigned a functional role: motif I, the FxGxG motif, found in all S-adenosyl-l-methionine (AdoMet)-dependent methyltransferases and implicated in cofactor AdoMet binding (9); and motif IV, the PC motif involved in catalysis, where the sulfhydryl group of the conserved cysteine forms the covalent link with the target cytosine (10–12). Apart from motif I, the AdoMet-binding pocket is composed of a number of other amino acids from motifs II–V. The key interactions between the cofactor and the protein are mediated through a set of conserved amino acid residues, as seen from the ternary complex (12,13). The adenine ring of the AdoMet molecule is surrounded by hydrophobic amino acids, Phe18 (motif I) and Leu100 (motif V) on one side and Trp41 on the other. The FxGxG motif forms a tight loop in the first turn of a β1–αa–β2 structural unit, with the first glycine being important in allowing a tight loop to form. Phe18 in this motif interacts with the adenosyl moiety of AdoMet, with its aromatic ring perpendicular to the plane of the purine ring. The ribose hydroxyl groups of AdoMet are hydrogen bonded by the acidic side chain of an invariant Glu40 from motif II. Asp60 (motif III) forms hydrogen bonds with the N6-amino group of adenine (8,14,15). The adenine ring of the cofactor is in hydrophobic interaction with Trp41, which lies parallel to the cofactor ring (π–π interaction) on one side and Leu100 and Phe18 on the other (8,12). The entire region surrounding the cofactor (motifs I, II and III) has resemblance to the Rossmann fold of the dinucleotide-binding motif (Fig. 1) (8,14). The DNA-binding region of M.HhaI is present within the variable region between motifs VIII and IX, and lacks any of the classical DNA-binding motifs.
Figure 1.
AdoMet binding pocket of HhaI DNA methyltransferase. The amino acid residues labeled have been subjected to mutagenesis.
Various crystal structures of M.HhaI, in binary complex with AdoMet or the reaction end product, S-adenosyl-l-homocysteine (AdoHcy) or in a ternary complex with cofactor and different DNA substrates, have been determined (12–16). Based on these structures and those available for other methyltransferases, it was seen that the methyltransferases are organized into a two-domain structure. The larger domain comprises nine of the 10 conserved motifs (in the case of M.HhaI) and has the cofactor-binding domain and the catalytic motif. The variable region, between motifs IX and X, serves as the target recognition domain and forms the bulk of the smaller domain (17). Analysis of the crystal structures also revealed that the cofactor could be found in two different orientations. Based on kinetic analysis it was proposed that the two orientations could be related to the catalytically inactive unprimed and catalytically active primed orientations, as seen in the binary M.HhaI–AdoMet complex and ternary M.HhaI–DNA–AdoHcy complex or the binary M.HhaI–AdoMet complex in the presence of non-specific DNA, respectively (12,14,18).
For the M.HhaI reaction mechanism, which involves the formation of covalent enzyme–DNA complex and a dihydrocytosine intermediate, to be operational, a major distortion in the DNA would have to be evoked. It was indeed seen from the ternary complex of HhaI–DNA–AdoHcy (12) that the target cytosine was completely flipped out of the DNA double helix and placed in the enzyme active site.
HhaI DNA methyltransferase has been one of the most extensively studied methyltransferases in terms of the numerous crystal structures available (12–16,19). However, adequate mutational data to biochemically validate the crystallographic data is lacking. To date, only a few residues have been subjected to mutagenesis, namely Cys81, which forms a part of the conserved PC motif (20), Glu237, which fills the void created in the DNA as a result of base flipping (21), and Thr250, which is involved in DNA binding (22). Therefore, a structure-guided mutational analysis study of some of the conserved residues in M.HhaI was initiated. Site-directed mutagenesis was performed on residues present in the AdoMet-binding region. These mutants were analyzed in terms of their methylation activity, kinetic parameters and stability, using filter binding assays, fluorescence quenching and temperature dependence studies.
MATERIALS AND METHODS
Bacterial strains and plasmids
HhaI DNA methyltransferase was expressed in E.coli K12 strain ER1727 [(mcrBC-hsdRMS-mrr)2::Tn10, mcrA1272::Tn10, F′lac proAB lacIq (lacZ)-M15]. Escherichia coli strain ER1546 (McrBC+) was used to check for the phenotype of the M.HhaI mutants. The M.HhaI gene was cloned into an IPTG inducible vector, pUHE25, (4). For carrying out site-directed mutagenesis the M.HhaI gene was cloned into pGEM-3Zf(+) (as a SphI–HindIII fragment from pUHE25) which contains the f1 origin of replication. Single-stranded DNA for site-directed mutagenesis was prepared from E.coli strain ER2380 (dut– ung– strain).
Protein purification
ER1727 cells containing the plasmid pUHE25 were grown to an absorbance (600 nm) of 0.8–1.0 at 37°C and subsequently induced with 0.15 mM IPTG. The cells were allowed to grow for 2 h after induction. The cells were harvested and either stored at –20°C or used immediately.
The protocol followed for the purification of M.HhaI was a slight modification of the earlier protocol (5). Cells were sonicated and the M.HhaI protein extracted from the cell pellet using buffer containing high salt (400 mM NaCl). The protein was then dialyzed with buffer containing 100 mM NaCl. The dialyzed protein was purified on Q-Sepharose and SP-Sepharose ion exchange columns. The purity of the protein was checked by SDS–PAGE with Coomassie brilliant blue staining (23). Protein was estimated using Coomassie G-250 and bovine serum albumin as standard (24).
Methylation activity
Filter binding. Methyltransferase activity of the M.HhaI was analyzed in vitro by measuring the incorporation of 3H-labeled methyl groups from [3H]AdoMet into substrate DNA (25). The reactions were carried out in 20 µl of methylase buffer, 50 mM Tris–HCl pH 7.5, 10 mM EDTA and 5 mM β-mercaptoethanol. Typically the reactions were performed with 1 µg λ DNA, 1–10 nM purified wild-type or mutant M.HhaI and 10–400 nM [3H]AdoMet at 37°C for 30 min. Reactions were stopped by snap chilling in liquid nitrogen and then spotted onto DE-81 (Whatmann) anion exchange filters. These filters were then washed three times with cold 0.2 M ammonium bicarbonate and once with 100% ethanol. The filters were dried and analyzed by liquid scintillation counting.
In vivo Mcr restriction assay. Methyltransferase activity was also determined by the in vivo restriction assay (21,26). Plasmids harboring the wild-type or mutant M.HhaI gene were transformed into a McrBC+ (ER1564) or McrBC– (ER1727) strain of E.coli. The number of transformants obtained on a McrBC+ background was compared with those obtained on a McrBC– background.
DNA methylated by wild-type M.HhaI would be a substrate for the McrBC gene product and would therefore get restricted. Hence, no colonies would be obtained on a McrBC+ background upon transformation with wild-type or an active mutant methyltransferase.
In vitro restriction assay. The sensitivity of the plasmid DNA containing the wild-type or mutant M.HhaI gene to restriction digestion with its cognate endonuclease HhaI restriction endonuclease (R.HhaI) was also used to qualitatively assay for methylase activity. Plasmid DNA was isolated using the alkaline lysis method and subjected to digestion with R.HhaI at 37°C for 2 h. Samples were electrophoresed on a 1% agarose gel in TAE buffer and the DNA visualized by ethidium bromide staining and UV illumination. Plasmids expressing wild-type or active mutant methyltransferases would be resistant to digestion with R.HhaI due to methylation of the recognition sites.
Site-directed mutagenesis
The following oligonucleotides (Bangalore Genei, India) were used for site-directed mutagenesis: W41I, 5′-TTCTAATGAAATCGATAAATATG-3′; W41A, 5′-TTCTAATGAATACGATAAATATG-3′; W41Y, 5′-TTCTAATGAAGCGGATAAATATG-3′; W41R, 5′-TTCTAATGAAAGGGATAAATATG-3′; D60P, 5′-CCTGAGGGCCCCATTACCCA-3′; D60N, 5′-CCTGAGGGCAACATTACCCA-3′; L100S, 5′-AGAGGTACAAGCTTTTTTGAT-3′; F18L, 5′-TTTATTGACCTTTTAGCAGGATTAGGT-3′; F18H, 5′-TTTATTGACCTTCATGCAGGATTAGGT-3′; F18X (X = Cys, Tyr or Ser), 5′-TTTATTGACCTTT(AGC)TGCAGGATTAGGT-3′. The underlined sequences represent the site of mutation and the sequence in bold is the restriction site created for that particular mutation.
Mutagenesis was performed according to Kunkel’s protocol for mutagenesis (27). Single-stranded DNA having uracil incorporated into it was made from E.coli strain ER2380 (dut– ung–). Briefly, ER2380 was transformed with the pGEM3Z-f(+) clone of M.HhaI. Transformants were grown to an absorbance (600 nm) of 0.5 and infected with M13 helper phage, M13K07, with a multiplicity of infection of 10. The infected cultures were allowed to grow for another 8–10 h. M13K07 phage, packaged with single-stranded pGEM DNA, was subjected to PEG precipitation. Single-stranded DNA was isolated by phenol/chloroform treatment, followed by alcohol precipitation.
Phosphorylated oligonucleotides (using T4 polynucleotide kinase) were annealed to the single-stranded DNA by heating in a water bath to 80°C and subsequently cooling the mixture to room temperature. Following annealing of the oligonucleotide, extension and ligation was carried out at 16°C for 12–16 h, using Klenow fragment of DNA polymerase I and T4 DNA ligase. This mix was transformed into an E.coli strain (dut+ ung+). The colonies were screened for the presence of mutations by the creation or loss of the restriction site as a result of mutation and by sequencing the appropriate region.
Immunoprecipitation of M.HhaI
Anti-HhaI antibodies were raised in rabbit using denatured M.HhaI protein. Immunoprecipitation was carried out essentially as described earlier (21). Escherichia coli ER1727 transformed with overexpression clones of wild-type or mutant M.HhaI were grown to an absorbance (600 nm) of 0.8 and induced with 0.15 mM IPTG. After 2 h the cells were harvested and the cell pellet resuspended in high salt buffer and briefly sonicated. The lysed cells were centrifuged for 10 min at 10 000 r.p.m. The supernatant was then collected and incubated with anti-M.HhaI antibodies for 3 h at 4°C with constant mixing. Protein A beads were added to this mix and the incubation continued for another 3 h. The beads were then pelleted and the immunoprecipitated protein was washed three times with low salt buffer. The amount of immunoprecipitated protein was estimated by western blotting using known amounts of purified wild-type protein. Protein A beads with bound HhaI methyltransferase were stored at 4°C and used directly in methylation assays.
Electrophoretic mobility shift assays
The following oligonucleotides (NEB, USA) were synthesized and used as a substrate for the mobility shift assays: 5′-GACTGGTACAGTATCAGGCGCTGACCCACAACATCCG-3′; 5′-TCGGATGTTGTGGGTCAGCGCCTGATACTGTACCAGT-3′. The HhaI recognition sequence is in bold.
These complementary oligonucleotides were annealed to form duplexes by heating to 80°C and gradual cooling to room temperature. Either one of the oligonucleotides was end-labeled using T4 polynucleotide kinase and [γ-32P]ATP, prior to annealing. Gel mobility shift assays were carried out in binding buffer (50 mM Tris–HCl pH 8.0, 10 mM EDTA, 13% glycerol, 7 mM β-mercaptoethanol and 100 µM AdoHcy). The assay conditions were essentially similar to those described previously (28). In a 10 µl reaction mix, M.HhaI was incubated with 32P-labeled oligonucleotide in binding buffer for 10–15 min at 20°C. The protein–DNA complexes were analyzed on a 6% native polyacrylamide gel, which was pre-run for 30 min. Electrophoresis was performed in 90 mM Tris–borate pH 8.3, 2 mM EDTA, at room temperature for 1.5–2 h. The gels were dried on Whatmann 3 paper and autoradiographed on X-ray film or quantitated using a PhosphorImager.
Photoaffinity labeling of M.HhaI with [methyl-3H]AdoMet
Crosslinking of AdoMet to M.HhaI was done as described earlier (29). Wild-type or mutant M.HhaI (5 µM) was incubated with [methyl-3H]AdoMet (7.3 µM) in 10 µl of buffer (10 mM potassium phosphate, 5 mM EDTA, 10% glycerol and 0.1% 2-mercaptoethanol) for 10 min in a microtiter plate. A hand-held UV lamp was directly placed on the microtiter plate and the samples irradiated with UV for 60 min at 4°C. The samples were boiled in SDS loading buffer. The protein–AdoMet complex was analyzed by 12% SDS–PAGE, followed by fixing in methanol:acetic acid:water (2.5:1:6.5) for 1 h. The fixed gel was subjected to fluorography (Amplify) for 30 min, dried and exposed to Kodak Hyperfilm at –70°C for 5 days.
Fluorescence titrations
The fluorescence emission spectra and the fluorescence intensities from titrations of M.HhaI were measured at 25°C on a FluoroMax3 spectrofluorimeter. The emission spectra were recorded over a wavelength of 300–400 nm with an excitation wavelength of 290 nm. Slit widths of 10 nm for excitation and 10 nm for emission were used. Titration of M.HhaI with AdoMet or AdoHcy was performed in low salt buffer. M.HhaI was allowed to equilibrate (5 min) in buffer before the beginning of the titration. Small aliquots of cofactor (final concentration 0.1–90 µM) were added to HhaI methyltransferase (1 µM) and incubated for 2 min before the spectra were recorded. The binding of AdoMet or AdoHcy to M.HhaI resulted in quenching of the Trp41 fluorescence, a unique amino acid present only at the AdoMet-binding site. Each spectrum recorded was an average of three scans. The fluorescence intensities were plotted against the total cofactor concentration and the data analyzed according to the Stern–Volmer and modified Stern–Volmer equations (30).
The Stern–Volmer relationship is represented by
Fo /F = 1 + Ksv[Q]
where Fo and F are the fluorescent intensities in the absence and presence of cofactor, respectively, Ksv is the collision Stern–Volmer constant and Q is the quencher (cofactor) concentration. In the case where there is a heterogeneous population of fluorophores, the modified Stern–Volmer relationship is used,
Fo /(Fo – F) = 1/{[Q] faKQ} + 1/fa
where fa is the fractional number of fluorophores accessible to the quencher and KQ is the quenching constant. The dissociation constants were calculated graphically using the modified Stern–Volmer plot (a plot of Fo/(Fo – F) versus 1[Q]) where KQ = 1/Kd (31,32).
Isothermal calorimetry (ITC) measurements
Titration calorimetric measurements were performed with a Microcal Omega titration calorimeter as described earlier (33). Briefly, small aliquots of AdoMet solutions at 15–40 times the M.HhaI concentration were added to a solution of M.HhaI (0.02–0.04 mM). The titrations were done under conditions that allowed for the precise estimate of the stoichiometry (n), binding constant (Kb) and enthalpy (ΔH°b) simultaneously in a single ITC experiment. This was achieved by carrying out the titration under optimal ‘c’ value (the c value is an arbitrary number defined as c = Kb[HhaI] and for an optimal titration 100 < c < 1000). Similar experiments were done with both the wild-type and mutant M.HhaI (W41I and W41Y).
The thermodynamic parameters were calculated using the following equations:
Qt = n[M.HhaI]t ΔH°bV{1 + [L]t /n[M.HhaI]t + 1/nKb [M.HhaI]t – [(1 + [L]t /n[M.HhaI]t + 1/nKb[M.HhaI]t )2 – 4[L]t /n [M.HhaI]t]1/2}/2 1
ΔG°b = ΔH°b – TΔS°b 2
ΔG°b = –nRT lnKb 3
where Qt is the heat content of a solution, n is the stoichiometry, Kb is an intrinsic binding constant, ΔH°b is an intrinsic heat of binding, [M.HhaI]t is the total M.HhaI concentration and V is the cell volume.
Specificity of methylation
The specificity of the mutant methyltransferase was determined by measuring the incorporation of [3H] label from AdoMet into premethylated DNA (by wild-type M.HhaI). DNA (λ) was methylated using excess cold AdoMet and wild-type enzyme. The methylation status of the DNA was examined by performing a restriction digestion of the DNA with R.HhaI. Methylation reactions were carried out as described above using the mutant methyltransferases (100 nM) and premethylated DNA or unmethylated DNA. The incorporation of radioactivity into premethylated DNA would suggest a change in specificity.
Stability of mutant methyltransferases
Stability of the wild-type and mutant methyltransferases was monitored by incubating the enzymes at 40°C in low salt buffer. Incubation was carried out at a protein concentration of 1 µM. At various time points, aliquots (2 µl) were withdrawn and assayed for methyltransferase activity at 37°C, using the DE81 filter binding assay as described earlier. The percent activity remaining was calculated and plotted against the incubation time. The stability of the mutant methyltransferases was also monitored by their rate of inactivation in the presence of urea. Wild-type or mutant M.HhaI (100 nM) was incubated with various concentrations of urea (0.1–4 M) in low salt buffer and methylation assays carried out at various concentrations of urea. The DE81 filter binding assay, described earlier, was used to measure the activity of these enzymes. The experiments were performed three times in duplicate and the errors were within 5%.
RESULTS AND DISCUSSION
All C5-methyltransferases share a similar structural organization and contain motifs that are absolutely conserved throughout the family. It was, therefore, of interest to know whether the conserved residues are essential for methylase activity and, if so, to assign a possible role for these amino acids in catalysis. Mutations were made in the AdoMet-binding pocket of M.HhaI. The AdoMet-binding pocket comprises residues from the highly conserved motif I (FxGxG) and from motifs II–V (8). Residues involved in cofactor binding, Phe18, Trp41, Asp60 and Leu100, were subjected to mutagenesis (Fig. 1). Phe18 was changed to Ser, Cys, Tyr, Trp and His, Trp41 changed to Ala, Arg, Ile and Tyr, Asp60 changed to Pro and Asn, and Leu100 changed to Ser. The DNA-binding region of M.HhaI is present within the variable region between motifs VIII and IX, and lacks any of the classical DNA-binding motifs.
Activity of mutant methyltransferases
All the mutants were screened either by the loss or creation of a restriction site as a result of mutation. The mutations were confirmed by sequencing the appropriate regions of the M.HhaI gene.
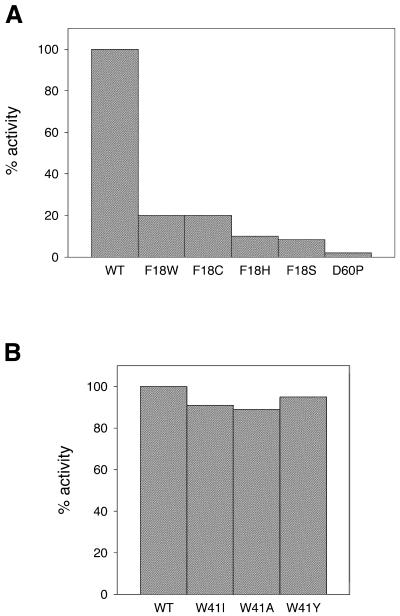
The activity of the mutant methyltransferases was determined by both in vivo and in vitro restriction assays. Plasmid DNA encoding the wild-type or mutant methylase was simultaneously transformed into strains ER1727 (mcrBC–) and ER1564 (mcrBC+) and the number of colonies obtained on each strain compared. The E.coli McrBC system encodes a methylation-sensitive restriction endonuclease, whose recognition sequence overlaps with the HhaI recognition sequence. Hence, plasmids carrying an active methylase would be restricted in E.coli strain ER1564 and not in strain ER1727. As can be seen from Table 1, all mutations at the Trp41 residue were active, and therefore no colonies were obtained on the McrBC+ strain. Mutations at the Phe18 residue also did not affect the activity of the methyltransferase, except for the Phe18→Cys mutant, which showed an inactive phenotype. Leu100→Ser and Asp60→Asn also did not have any affect on activity. However, mutation Asp60→Pro resulted in an inactive phenotype.
Table 1. In vivo wild-type and mutant HhaI DNA methyltransferase activities.
| M.HhaI | Colonies (mcr –/mcr+) |
|---|---|
| Wild-type | 102/0 |
| F18S | 58/0 |
| F18C | 72/88 |
| F18Y | 66/0 |
| F18W | 81/0 |
| F18H | 110/0 |
| L100S | 84/0 |
| D60P | 109/89 |
| D60N | 96/0 |
| W41A | 75/0 |
| W41I | 79/0 |
| W41Y | 72/0 |
Plasmids containing wild-type and mutant M.HhaI genes were transformed into an E.coli strain that expresses the mcrBC restriction system or a strain mutant for the mcrBC system, (ER1564 and ER1727, respectively). The number of colonies obtained on both the strains was compared. The numbers denoted are the average of three different experiments.
These results were further confirmed by the in vitro restriction assay. Plasmid DNA, from cells containing wild-type or mutant methylase, was isolated and its sensitivity to digestion by R.HhaI monitored. Plasmids harboring an active methylase would result in methylation of all HhaI sites in vivo and hence would be resistant to subsequent in vitro cleavage by R.HhaI. The results of the in vitro assay (Table 2) corroborated the in vivo McrBC restriction assay, except for the Phe18→Cys mutant, which was completely protected from endonuclease digestion. All other Phe18 mutants seemed to be partially protected. This suggested that all Phe18 mutants except Phe18→Cys were partially active. Plasmid DNA from cells expressing the Trp41 mutants, Asp60→Asn and Leu100→Ser mutants were protected from R.HhaI digestion and therefore are active methyltransferases, whereas the Asp60→Pro mutant was completely digested by R.HhaI, suggesting it to be an inactive methylase (Table 2).
Table 2. In vivo methylation of HhaI recognition sequence by wild-type and mutant HhaI DNA methyltransferases.
| Enzyme | Sensitivity to R.HhaIa |
|---|---|
| Wild-type | – |
| W41A | – |
| W41R | – |
| W41I | – |
| W41Y | – |
| F18C | – |
| F18H | +/– |
| F18L | +/– |
| F18S | +/– |
| F18W | +/– |
| F18Y | +/– |
| L100S | – |
| D60P | + |
| D60N | – |
Plasmid DNA containing the wild-type or mutant M.HhaI methyltransferase was subjected to digestion by R.HhaI. Plasmids containing an active methyltransferase would be resistant to digestion by R.HhaI whereas plasmids harboring an inactive methyltransferase would be cleaved by R.HhaI.
a –, resistance to cleavage by R.HhaI; +, complete digestion by R.HhaI; +/–, partial digestion by R.HhaI.
Biochemical analysis
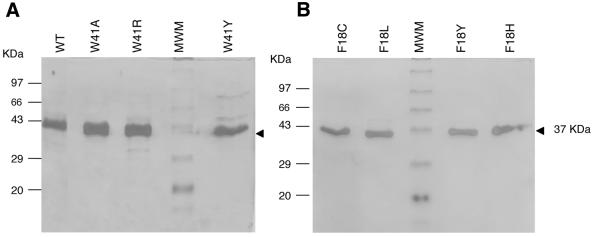
In order to assess the methylation activity of the mutants in a more quantitative manner, filter binding assays were performed to monitor incorporation of the [3H]methyl group into DNA. Either the purified or immunoprecipitated M.HhaI protein was used for the assays. Wild-type, Trp41 and Phe18 mutants were purified to homogeneity and subjected to biochemical analysis. The proteins were purified using a protocol similar to that used for the wild-type protein (5) (Fig. 2). The purified proteins were analyzed for methylation activity and AdoMet- and DNA-binding functions. The Asp60 and Leu100 mutants could not be purified due to either poor induction or insolubility of these mutants during the high salt extraction step of purification.
Figure 2.
SDS–PAGE gel of purified wild-type and mutant HhaI DNA methyltransferases. Silver staining of 8–10 µg purified proteins run on 12% SDS–PAGE. (A) Trp41 mutants, (B) Phe18 mutants. MWM stands for standard molecular weight markers.
DNA binding and kinetic properties
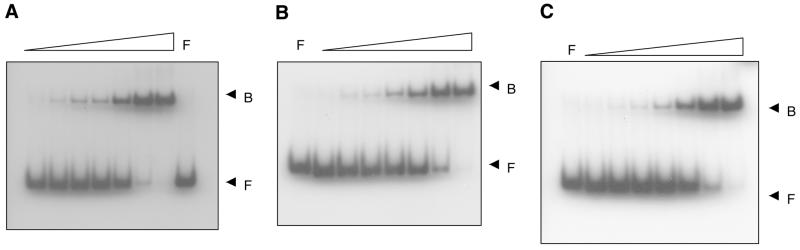
To determine whether the Trp41 mutations had any effect on the DNA-binding properties, gel shift assays were performed using a 37mer oligonucleotide containing a single M.HhaI recognition site. End-labeled oligonucleotide was titrated with increasing concentrations of HhaI methyltransferase. The complex formed was resolved on native polyacrylamide gels and quantitated using a PhosphorImager. It was seen that the Trp41 mutants did not have any altered DNA-binding capabilities (Fig. 3).
Figure 3.
Binding of wild-type and mutant HhaI DNA methyltransferases to DNA. Electrophoretic mobility shift assays were performed using 5′-end-labeled duplex, containing a single recognition site for M.HhaI. Increasing concentrations of protein (1–250 nM) were added to a fixed amount of oligonucleotide (10 nM). The bound and the free oligonucleotides were separated by 6% native PAGE, in 0.5× TBE. The analysis was done for wild-type and Trp41 mutants. (A) Wild-type M.HhaI; (B) W41Y M.HhaI; (C) W41I M.HhaI. The arrows indicate protein–DNA complex (B) and free DNA (F).
As structural data implied an important role for Trp41 in cofactor binding, we investigated the effect of mutation at Trp41 on the kinetic properties of M.HhaI. Steady-state kinetic experiments were performed to obtain Km(AdoMet) and kcat values using λ DNA and [methyl-3H]AdoMet as substrates (Table 3). Methyltransferase activity of Trp41 mutants (Ile, Tyr and Arg) did not vary appreciably from that of the wild-type protein. These mutations did not have a drastic effect on the Km(AdoMet) value (a maximum of 2-fold difference was seen with Trp41Arg). The Km values reflect the ability of AdoMet to interact with M.HhaI, whereas kcat values are a reflection of the catalysis step. Substitution of Trp41 with Tyr did not change the enzyme properties significantly, as it is a conserved change, which can still maintain the π–π interaction with the adenine ring of AdoMet. Substitution with Ile however had an effect only on kcat and not on Km, i.e. only the catalysis step was affected by this mutation and there was no effect on cofactor binding. Ile could be maintaining the hydrophobic interactions necessary for cofactor binding, thus having no effect on Km. The Arg mutation resulted in lowered Km and kcat values (Table 3), suggesting that the charged residue could be interfering with both AdoMet binding and catalysis.
Table 3. Kinetic parameters of wild-type and Trp41 mutants of HhaI DNA methyltransferasea.
| M.HhaI | Km(AdoMet) (nM) | kcat (min–1) | kcat/Km (× 10–3) (nM–1 min–1) |
|---|---|---|---|
| Wild-type | 68.3 | 0.56 | 8.2 |
| W41I | 60.4 | 0.15 | 2.5 |
| W41Y | 70.9 | 0.57 | 8.0 |
| W41R | 25.0 | 0.24 | 9.5 |
aThe average value of two independent experiments is reported.
Both kinetic and gel shift assays indicate that the Trp residue at position 41 is dispensable as far as AdoMet binding and catalysis are concerned. This is not surprising as this Trp residue is not an absolutely conserved amino acid among the C5-methyltransferase family and other residues have been found to occupy a similar position (Met, Ile, Lys, Tyr, Asn, Phe, Leu and Gly) (12,15). The fact that this residue in not conserved undermines the importance of the hydrophobic interactions between Trp41 and the adenosyl moiety of AdoMet.
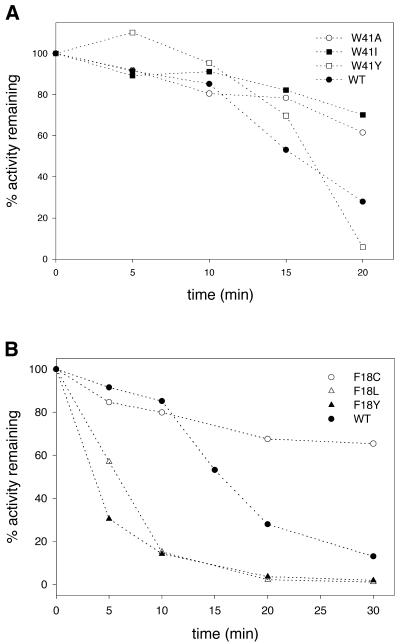
Methyltransferase activity was also measured for the Phe18 and Asp60 mutants. Since purification of some of these mutants proved to be difficult, immunoprecipitated protein was used for activity measurements (21). The activity of the immunoprecipitated protein (wild-type) was comparable with that of the normal soluble protein, indicating that the antibodies did not have an inhibitory effect on the methyltransferase activity. A comparison of the activities is shown in Figure 4. As can be seen, Phe18 mutants do retain some methyltransferase activity (5–20%). While the Asp60→Pro mutation led to a complete loss of activity (Fig. 4A), all Trp41 mutants retained wild-type-like activity (Fig. 4B). Subsequently, we were able to purify some of the Phe18 mutants (Cys, Leu, His and Tyr). The purified proteins were used in later experiments.
Figure 4.
Activity of immunoprecipitated HhaI methyltransferases. Wild-type or mutant methyltransferases were precipitated from crude extract using anti-HhaI antibodies and protein A–Sepharose beads. The immunoprecipitated proteins were assayed for methyltransferase activity using the DE81 filter binding assay, as described in Materials and Methods. The figure shows activity for the mutant methyltransferases: (A) F18W, F18C, F18H, F18S and D60P; (B) W41I, W41A and W41Y. The activity shown is percentages normalized against wild-type M.HhaI. WT stands for wild-type.
AdoMet binding
Site-directed mutagenesis was carried out on residues involved in AdoMet binding. Most of the mutants do not result in the loss of methyltransferase activity (except Asp60→Pro), as seen by in vivo and in vitro restriction assay (Tables 1 and 2). As these mutations were in the AdoMet-binding pocket, they would be expected to have an effect, at least in their capacity to bind AdoMet and AdoHcy. The ability of these mutants to bind AdoMet was assessed by fluorescence quenching and isothermal titration calorimetry.
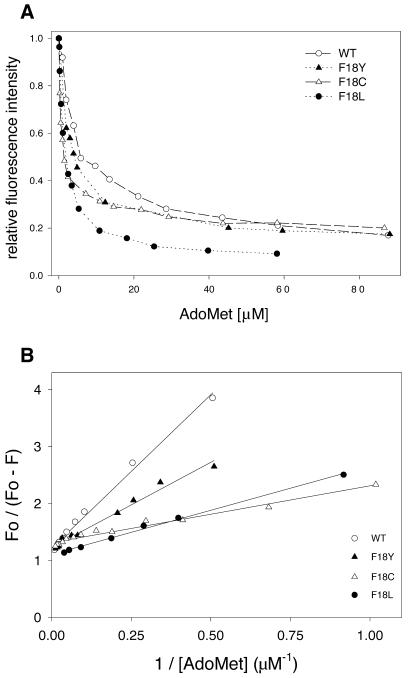
Fluorescence quenching. The presence of a Trp residue in the AdoMet-binding pocket of HhaI has been used as a handle to study cofactor binding. As can be seen, there was significant quenching of the fluorescence intensity of M.HhaI upon addition of AdoMet (Fig. 5A). This allowed for the quantitative measurement of the interaction of cofactor with HhaI methyltransferase. Wild-type or mutant M.HhaI (1 µM) was titrated with increasing concentrations of AdoMet (0.1–90 µM) and the fluorescence emission spectra were recorded between 300 and 400 nm. The fluorescence intensity at 354 nm was monitored and plotted against AdoMet concentration (Fig. 5A). The data were analyzed using the modified Stern–Volmer plot (see Materials and Methods) (Fig. 5B). The dissociation constant (Kd) estimated for the wild-type HhaI was found to agree with the previously determined value using calorimetric (33) and fluorescence quenching studies (34). Similarly, the dissociation constants were estimated for the Phe18 mutants (Table 4). The affinity of these mutants for AdoMet increased 2- to 6-fold, the maximum being for the Phe18→Cys mutant.
Figure 5.
Tryptophan fluorescence analysis of the interaction of HhaI DNA methyltransferase with AdoMet. (A) Titration of 1 µM M.HhaI with AdoMet (0.1–90 µM). The relative fluorescence intensity (at 354 nm) (F/Fo ) is plotted against AdoMet concentration for various mutant methyltransferases. (B) Fluorescence quenching data were analysed using the modified Stern–Volmer equation (see Materials and Methods) to obtain dissociation constants (Kd) for AdoMet.
Table 4. Dissociation constants for wild-type and mutant HhaI DNA methyltransferases as determined by fluorescence quenching and ITC studies.
| Enzyme | Dissociation constant, Kd (µM) | |
|---|---|---|
| Fluorescencea | ITC | |
| Wild-type | 4.5 | 6.34 |
| F18C | 0.76 | ND |
| F18L | 1.28 | ND |
| F18Y | 2.45 | ND |
| W41I | ND | 12.8 |
| W41Y | ND | 12.1 |
aFluorescence measurements were not performed with the Trp41 mutants as M.HhaI has a single Trp residue, the loss of which results in decreased fluorescence.
ND, not determined.
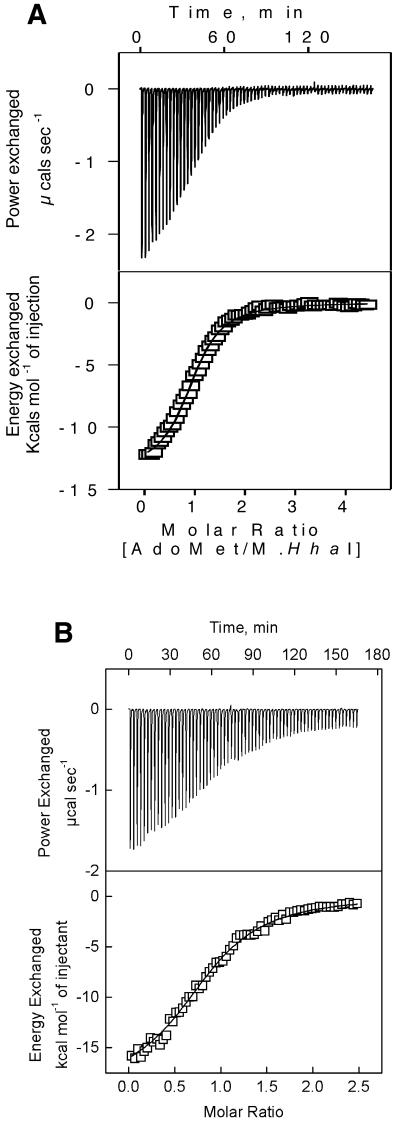
ITC measurements. ITC measurements were carried out to characterize the thermodynamics of the interaction of cofactor (AdoMet or AdoHcy) with M.HhaI protein. ITC analysis done with wild-type M.HhaI indicated a dual mode of interaction with the cofactors (33). The primary high affinity mode could easily be saturated, while the secondary low affinity mode was not saturated within the concentration range of AdoMet used for titration. Hence, the thermodynamic parameters were estimated by analyzing only the primary mode (33). The dissociation constant obtained from such an analysis was similar to that obtained by fluorescence quenching studies (34). Two Trp41 mutants, Trp41→Ile and Trp41→Tyr, were also subjected to calorimetric analysis. Both these mutants displayed a dual mode interaction, as seen for the wild-type enzyme. A representative binding isotherm, for the primary mode, for the wild-type and Trp41→Ile mutant is shown in Figure 6. A similar binding isotherm was obtained for Trp41→Tyr. The binding affinities estimated from the isothermal titration calorimetry experiments for both these mutants were 2-fold lower compared with the wild-type protein (Table 4).
Figure 6.
Isothermal titration calorimetric analysis of wild-type and Trp41 mutant of HhaI DNA methyltransferase. Titrations were performed as mentioned in Materials and Methods. The primary endothermic binding mode depicting the heat absorbed as a function of increasing AdoMet/M.HhaI ratio for (A) wild-type M.HhaI and (B) Trp41Ile mutant. The top panel represents the raw data obtained for each addition of AdoMet to M.HhaI. The bottom panel is the non-linear least squares fit of the incremental heat per mole of added ligand (AdoMet). Using these heats, the enthalpy, free energy and binding constants were derived.
It can, therefore, be concluded from the AdoMet binding studies that mutations at the Trp41 residue do not significantly affect the M.HhaI–cofactor interaction. However, mutations at Phe18 resulted in an enhanced binding of AdoMet, as seen from the fluorescence quenching data (Table 4).
Methyltransferase specificity and stability
In order to check whether the mutant methyltransferases have any altered specificities, methylation reactions were carried out using premethylated DNA. Incorporation of radioactivity into DNA premethylated by wild-type HhaI would indicate a change in specificity. Typically, 100 nM enzyme was incubated with premethylated λ DNA in the presence of 200 nM [3H]AdoMet. The methylation reaction with unmethylated DNA was used as a control. As can be seen (Table 5), the wild-type M.HhaI was not capable of methylating premethylated DNA. Similarly, among all the mutant methylases analyzed, those which were partially active were incapable of further methylating the premethylated DNA, thus confirming that there was no change in specificity of the mutant methyltransferases.
Table 5. Specificity of methyltransferases.
| Enzyme | c.p.m. (DNAme) | c.p.m. (DNA) |
|---|---|---|
| Wild-type | 105 | 14 048 |
| W41A | 182 | 1482 |
| W41I | 115 | 8131 |
| W41Y | 76 | 13 752 |
| F18C | 60 | 6055 |
| F18H | 61 | 169 |
| F18L | 113 | 2024 |
| F18Y | 110 | 1322 |
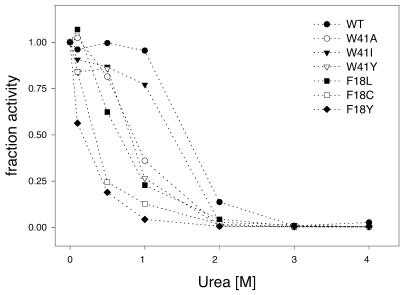
Having seen that there was no altered specificity of these mutant methyltransferases, we next went ahead to see if the thermal stability of the mutants was affected. The stability of the mutant methyltransferases was assayed by incubating the enzymes at 40°C and assaying for methyltransferase activity at 37°C at the indicated time points. The fraction inactivation was plotted against the incubation time (Fig. 7). Results indicate that the Trp41 mutant (Ile, Ala and Tyr) methyltransferases had slightly increased stability as compared with the wild-type enzyme (Fig. 7A). In contrast, the Phe18 mutants were found to be unstable (except Phe18→Cys) compared with the wild-type protein (Fig. 7B). The Phe18→Tyr and Phe18→His mutants were very unstable and by 20 min lost activity completely. Stability of the mutant methyltransferases was also determined by their resistance to urea denaturation. Methylation activity of the mutants was measured in the presence of various concentrations of urea. As can be seen (Fig. 8), all the mutants (Trp41 and Phe18) were less stable compared with the wild-type protein. The effect was more prominent in the case of the Phe18 mutants. The Phe18→Cys and Phe18→Tyr mutants were the least stable and, at a concentration of 0.5 M urea, lost 75% of their activity.
Figure 7.
Kinetics of inactivation of wild-type and mutant HhaI DNA methyltransferases. Wild-type or mutant M.HhaI (1 µM) were incubated at 40°C in low salt buffer (see Materials and Methods). Aliquots (2 µl) were removed at various times and assayed for methylase activity, as described in Materials and Methods. A plot of activity versus incubation time is shown: (A) Trp41 mutant methyltransferases; (B) Phe18 mutant methyltransferases. The figure shows activity as percentages normalized against the wild-type M.HhaI. WT stands for wild-type.
Figure 8.
Denaturation of wild-type and mutant HhaI DNA methyltransferases in the presence of urea. M.HhaI (100 nM) was incubated with urea (0.1–4 M) for 5 min at 37°C and the activity of the protein measured in the presence of denaturant. Fractional activity plotted against urea concentration for Trp41 mutants (Ala, Ile and Tyr) and Phe18 mutants (Leu, Cys and Tyr) is shown.
Phe18 is a part of motif I, FxGxG, and, except in one or two cases, is absolutely conserved among all AdoMet-dependent methyltransferases. It is, therefore, proposed to play a role in cofactor binding (8,9). In the case of M.HhaI, the Phe18 ring lies perpendicular to the adenine ring of AdoMet, thus interacting with the cofactor through hydrophobic interactions. Results obtained in this study show that changes at Phe18 result in partial loss of activity. No apparent correlation could be drawn between the nature of the side chains and enzymatic activity. Substitutions with either a bulky or a smaller side chain could result in disruption of the interaction with AdoMet, thus affecting the methyltransferase activity. On the other hand, substitutions at Phe18 also seem to have an effect on the stability of the mutant methylase (Fig. 8). These results, therefore, suggest that the loss of stability could also be responsible for the reduced activity seen. However, it is not clear why the affinity for the cofactor AdoMet is increased. One possible reason for the reduced activity of the mutants is the inhibitory effect due to the increased affinity for cofactors. Although Kd values for AdoHcy are not available, they would be expected to follow a trend similar to the wild-type enzyme, i.e. the values would be higher for the wild-type compared with the mutants. This increased affinity for the reaction end product, AdoHcy, could affect the product release step during catalysis. Among the Phe18 mutants, Phe18→Cys showed anomalous behavior, as seen by the in vitro and in vivo restriction assays, and was found to be more stable to heat denaturation. At present we are unable to explain these observations.
In summary, this mutational study indicates that amino acid residues involved in the binding of the cofactor AdoMet are not absolutely essential and suggests flexibility in the AdoMet-binding pocket (Table 6). The hydrophobic pocket formed by Phe18, Leu100 and Trp41 may not be as important as expected from structural analysis and this could be a peculiarity of M.HhaI. It is possible that the size of the side chain plays a more important role than the specific nature of interactions with the cofactor. On the other hand, the stability of some of the Phe18 mutants was affected, suggesting an additional structural role for this amino acid apart from a functional one. The absence of absolute conservation at these (expected) important residues suggests to us that the enzyme may be using an alternative mode to interact with the cofactor. Our mutational analysis and those done earlier (21,22) suggest that M.HhaI is more tolerant to changes at the conserved residues or residues thought to have an important functional role.
Table 6. Properties of wild-type and mutant M.HhaI.
| Enzyme | In vitro assaya | In vivo assaya | Methylase activityb | AdoMet bindingc | Stability to heat/uread | |
|---|---|---|---|---|---|---|
| Crosslinking | Kdb | |||||
| Wild–type | + | + | 1 | + | 1 | ++/++ |
| Trp41 | ||||||
| W41A | + | + | 0.74 | +/– | ND | ++/+ |
| W41I | + | + | 0.58 | +/– | 2 | ++/+ |
| W41R | + | + | 0.52 | +/– | ND | |
| W41Y | + | + | 0.97 | +/– | 1.9 | ++/+ |
| Phe18 | ||||||
| F18C | + | – | 0.43 | +/– | 0.17 | ++/+ |
| F18L | +/– | + | 0.14 | + | 0.28 | +/+ |
| F18Y | +/– | + | 0.73 | +/– | 0.54 | +/+ |
| F18H |
+/– |
+ |
0.01 |
+/– |
ND |
ND |
| F18S | +/– | + | ||||
| F18W | +/– | + | ||||
| Asp60 | ||||||
| D60P | – | – | These mutant methyltransferases could not be purified due to the insoluble nature of these proteins | |||
| D60N | – | – | ||||
| Leu100 | ||||||
| L100S | + | + | ||||
a+, –, +/–, active, inactive and partially active, respectively.
bValues relative to wild-type M.HhaI.
cAdoMet binding studied qualitatively by crosslinking (+/–, weak; +, strong) or quantitatively by ITC (Trp41 mutants) or fluorescence quenching (Phe18 mutants).
d++, stable; + unstable; ND, not determined.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Richard Roberts (New England Biolabs) for kindly providing us with the M.HhaI-expressing clone. We thank Dr E. Raleigh for E.coli strains ER1727, ER1564 and ER2380 and Prof. A. Surolia for the ITC experiments. We thank S. Arathi for technical assistance and Prof. K. Muniyappa for allowing us to use the FluoroMax3 spectrofluorimeter. One of us (D.N.R.) gratefully acknowledges the Rockefeller Foundation for a Biotechnology Career Award fellowship. This work was supported by a grant from the Department of Biotechnology, Government of India.
REFERENCES
- 1.Roberts R.J., Myers,P.A., Morrison,A. and Murray,K. (1976) A specific endonuclease from Haemophilus haemolyticus. J. Mol. Biol., 103, 199–208. [DOI] [PubMed] [Google Scholar]
- 2.Caserta M., Zacharias,W., Nwankwo,D., Wilson,G.G. and Wells,R.D. (1987) Cloning, sequencing, in vivo promoter mapping and expression in Escherichia coli of the gene for the HhaI methyltransferase. J. Biol. Chem., 262, 4770–4777. [PubMed] [Google Scholar]
- 3.Wu J.C. and Santi,D.V. (1988) High level expression and purification of HhaI methyltransferase. Nucleic Acids Res., 16, 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klimasauskas S., Nelson,J.L. and Roberts,R.J. (1991) The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res., 19, 6183–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar S., Cheng,X., Pflugrath,J.W. and Roberts,R.J. (1992) Purification, crystallization and preliminary X-ray diffraction analysis of an M.HhaI–AdoMet complex. Biochemistry, 31, 8648–8653. [DOI] [PubMed] [Google Scholar]
- 6.Posfai J., Bhagwat,A.S., Posfai,G. and Roberts,R.J. (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res., 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng X., Kumar,S., Klimasauskas,S. and Roberts,R.J. (1993) Crystal structure of the HhaI DNA methyltransferase. Cold Spring Harbor Symp. Quant. Biol., 58, 331–338. [DOI] [PubMed] [Google Scholar]
- 8.Kumar S., Cheng,X., Klimasauskas,S., Mi,S., Posfai,J., Roberts,R.J. and Wilson,G.G. (1994) The DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 22, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingrosso D., Fowler,A.V., Bleibaum,J. and Clarke,S. (1989) Sequence of the D-aspartyl/L-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA and small molecule S-adenosylmethionine-dependent methyltransferases. J. Biol. Chem., 264, 20131–20139. [PubMed] [Google Scholar]
- 10.Wu J.C. and Santi,D.V. (1987) Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem., 262, 4778–4786. [PubMed] [Google Scholar]
- 11.Chen L., MacMillan,A.M., Chang,W., Ezaz-Nikpay,K., Lane,W.S. and Verdine,G.L. (1991) Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry, 30, 11018–11025. [DOI] [PubMed] [Google Scholar]
- 12.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 13.O’Gara M., Klimasauskas,S., Roberts,R.J. and Cheng,X. (1996) Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaI methyltransferase–DNA–AdoHcy complexes. J. Mol. Biol., 261, 634–645. [DOI] [PubMed] [Google Scholar]
- 14.O’Gara M., Zhang,X., Roberts,R.J. and Cheng,X. (1999) Structure of a binary complex of HhaI methyltransferase with S-adenosyl-L-methionine formed in the presence of a short non-specific DNA oligonucleotide. J. Mol. Biol., 287, 201–209. [DOI] [PubMed] [Google Scholar]
- 15.Cheng X., Kumar,S., Posfai,J., Pflugrath,J.W. and Roberts,R.J. (1993) Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell, 74, 299–307. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S., Horton,J.R., Jones,G.D., Walker,R.T., Roberts,R.J. and Cheng,X. (1997) DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res., 25, 2773–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng X. (1995) Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct., 24, 293–318. [DOI] [PubMed] [Google Scholar]
- 18.Lindstrom W.M. Jr, Flynn,J. and Reich,N.O. (2000) Reconciling structure and function in HhaI DNA cytosine-C-5 methyltransferase. J. Biol. Chem., 275, 4912–4919. [DOI] [PubMed] [Google Scholar]
- 19.O’Gara M., Horton,J.R., Roberts,R.J. and Cheng,X. (1998) Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nature Struct. Biol., 5, 872–877. [DOI] [PubMed] [Google Scholar]
- 20.Mi S. and Roberts,R.J. (1993) The DNA binding affinity of HhaI methylase is increased by a single amino acid substitution in the catalytic center. Nucleic Acids Res., 21, 2459–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mi S., Alonso,D. and Roberts,R.J. (1995) Functional analysis of Gln-237 mutants of HhaI methyltransferase. Nucleic Acids Res., 23, 620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vilkaitis G., Dong,A., Weinhold,E., Cheng,X. and Klimasauskas,S. (2000) Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem., 275, 38722–38730. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 24.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 25.Mi S. and Roberts,R.J. (1992) How M.MspI and M.HpaII decide which base to methylate. Nucleic Acids Res., 20, 4811–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raleigh E.A., Trimarchi,R. and Revel,H. (1989) Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics, 122, 279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel T.A. (1985) The mutational specificity of DNA polymerases-alpha and -gamma during in vitro DNA synthesis. J. Biol. Chem., 260, 12866–12874. [PubMed] [Google Scholar]
- 28.Dubey A.K. and Roberts,R.J. (1992) Sequence-specific DNA binding by the MspI DNA methyltransferase. Nucleic Acids Res., 20, 3167–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad I. and Rao,D.N. (1994) Photolabeling of the EcoP15 DNA methyltransferase with S-adenosyl-l-methionine. Gene, 142, 67–71. [DOI] [PubMed] [Google Scholar]
- 30.Lehrer S.S. (1971) Solute perturbation of protein fluorescence. The quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry, 10, 3254–3263. [DOI] [PubMed] [Google Scholar]
- 31.Samworth C.M., Degli Esposti,M. and Lenaz,G. (1988) Quenching of the intrinsic tryptophan fluorescence of mitochondrial ubiquinol–cytochrome-c reductase by the binding of ubiquinone. Eur. J. Biochem., 171, 81–86. [DOI] [PubMed] [Google Scholar]
- 32.Lackowicz J.R. (1983) Principles of Fluorescence Spectroscopy. Plenum Press, NY.
- 33.Swaminathan C.P., Sankpal,U.T., Rao,D.N. and Surolia,A. (2002) Water-assisted dual mode cofactor recognition by HhaI DNA methyltransferase. J. Biol. Chem., 277, 4042–4049. [DOI] [PubMed] [Google Scholar]
- 34.Vilkaitis G., Merkiene,E., Serva,S., Weinhold,E. and Klimasauskas,S. (2001) The mechanism of DNA cytosine-5 methylation. Kinetic and mutational dissection of HhaI methyltransferase. J. Biol. Chem., 276, 20924–20934. [DOI] [PubMed] [Google Scholar]