Abstract
The dimer initiation site (DIS) hairpin of the HIV-2 untranslated leader RNA mediates in vitro dimerization through ‘loop–loop kissing’ of a loop-exposed palindrome sequence. Premature RNA dimerization must be prevented during the retroviral life cycle. A regulatory mechanism has been proposed for the HIV-1 leader RNA that can adopt an alternative conformation in which the DIS motif is effectively masked by long-distance base pairing with upstream leader sequences. We now report that HIV-2 RNA dimerization is also regulated. Sequestering of the DIS motif by base pairing interactions with downstream leader sequences mediates a switch to a dimerization-impaired conformation. The existence of two alternative conformations of the HIV-2 leader RNA is supported by UV melting experiments. Furthermore, the equilibrium between the two conformations can be shifted by annealing of antisense oligonucleotides or by deletion of certain leader regions. These measures have a profound impact on the dimerization properties of the transcript, demonstrating a mutual exclusivity between the alternative conformation and dimerization, similar to what has been described for the HIV-1 leader. The overall resemblance in regulation of HIV-1 and HIV-2 RNA dimerization suggests that a similar mechanism may be operating in other lentiviruses and perhaps all retroviridae.
INTRODUCTION
Retroviral genomes are present in virus particles as two identical RNA molecules that are non-covalently linked near their 5′ ends (1,2). The presence of dimeric viral RNA has been suggested to affect virus replication at different stages of the viral life cycle. First, it has been reported that RNA dimers are preferentially packaged over monomeric RNA genomes (3,4). Second, the dimeric state facilitates template switching during reverse transcription, thereby circumventing the effects of physical damage to the RNA genome. Third, template switching may cause recombination, thereby increasing the genetic diversity and adaptability of retroviruses (reviewed in 5).
Retroviral RNA readily forms dimers in vitro when incubated in cation-containing buffers (6–13). Mutational analysis indicated that a small segment of the HIV-1 untranslated leader, the so-called dimer initiation site (DIS), is an essential motif for in vitro dimerization (6,8,9,14). The HIV-1 DIS consists of a hairpin structure with a loop-exposed 6mer palindromic sequence that is indispenable for dimerization. According to the proposed model for dimer formation, contact between two DIS hairpins is initiated by base pairing of the palindromes, termed loop–loop kissing (6,8,9). Heat treatment or incubation with the viral nucleic acid binding nucleocapsid (NC) protein, which acts as a chaperone, melts the intra-strand base pairs of the DIS stem and triggers the formation of a dimer with extended inter-strand base pairing. This transition is usually referred to as the conversion of the loose dimer (kissing–loop complex) into the tight dimer (extended duplex) (15,16). Although alternative mechanisms have been proposed for dimerization of HIV-2 RNA (17,18), we have recently reported that HIV-2 RNA dimerization is quite similar to that of HIV-1. The HIV-2 DIS hairpin is essential and sufficient for RNA dimerization, which depends on the 6mer palindrome within the hairpin loop (19).
Retroviral RNA dimerization in vivo is likely to be a regulated rather than a constitutive process. Interestingly, it has recently been reported that the HIV-1 leader RNA can adopt two alternative conformations that differ in their ability to form dimers, thus providing an RNA switch mechanism for regulated dimerization (20). The thermodynamically most stable conformation is characterized by a long-distance base pairing interaction (LDI) between DIS nucleotides and upstream leader sequences, which masks the DIS motif. The alternative, metastable conformation is a branched structure with multiple hairpins (BMH), including the DIS hairpin with an exposed palindrome that facilitates dimerization. It has been suggested that this mechanism may serve to prevent premature dimerization, e.g. when the newly synthesized viral RNA is needed for translation of viral proteins (21). Furthermore, it has been demonstrated that the viral nucleic acid binding NC protein causes a rearrangement from the LDI to the BMH conformation, thus promoting dimer formation during virion assembly when NC is released from the Gag precursor protein (21).
In the present study, we characterized HIV-2 DIS-mediated RNA dimerization in further detail. We report inhibition of HIV-2 RNA dimer formation by sequences that are located 3′ of the DIS hairpin. Upstream leader sequences are also required for this inhibition, suggesting that HIV-2 RNA can adopt an alternative conformation that is dimerization incompetent. UV-melting experiments and an antisense oligonucleotide probing analysis support the existence of alternative leader RNA conformations. Our studies point to a mechanism of regulated RNA dimerization that is controlled by the overall leader structure and that is reminiscent of the recently proposed model for regulated HIV-1 RNA dimerization.
MATERIALS AND METHODS
In vitro transcription
The DNA templates for transcription were made by PCR amplification of the plasmid HIV-2 Ψ large, which contains a T7 promoter directly upstream of the +1 position of the wild-type HIV-2 ROD transcript (22). Templates for transcripts starting at the +1 position were generated using the upstream T7 +1 primer and a downstream primer that ends at the position corresponding to the transcript name. Templates for transcription of RNA starting at +126, +185 and +397 were generated using upstream T7-primers starting at these positions, and downstream primers as described above. PCR fragments were excised from agarose gels, purified with the QIAEX II DNA isolation system, and used in transcription with the Ambion Megashortscript T7 Transcription Kit according to the accompanying protocol. In the case of radioactively labeled transcripts, 1 µl of [α−32P]UTP (0.33 MBq/µl) was added to the transcription mixture. Transcripts were purified on 4% denaturing polyacrylamide gels and visualized either by UV shadowing or by autoradiography. Upon overnight elution in TE buffer, the RNA was ethanol precipitated, redissolved in water and quantified by UV-absorbance measurements or scintillation counting. Transcript stock solutions were renatured by heating to 85°C and slow cooling to room temperature, aliquots were stored at –20°C.
In vitro dimerization assays
Unless indicated otherwise, ∼2 pmol of radiolabeled RNA was incubated in 10 µl dimerization buffer (5 mM MgCl2, 10 mM Tris–HCl pH 7.5, 40 mM NaCl) for 10 min at 65°C, followed by slow cooling to room temperature. An aliquot of 5 µl non-denaturing gel loading buffer (30% glycerol with BFB dye) was added, and samples were analyzed on 4% non-denaturing polyacrylamide gels containing 0.25× TBE (22.5 mM Tris, 22.5 mM boric acid, 0.625 mM EDTA). Approximately 3 pmol of cold transcripts were used and traced with 0.05 pmol of radioactively labeled transcript in dimerization assays with 5′ truncated transcripts. Some samples were also analyzed on a 0.25× TBM gel (22.5 mM Tris, 22.5 mM boric acid, 0.1 mM MgCl2) to measure formation of the loose dimer (kissing–loop complex). Gels were run at 150 V at room temperature, dried and exposed by autoradiography or phosphor imaging. MgCl2 titration experiments with transcripts 397/444 and 1/444 (equal amounts of radioactive label) were performed in dimerization buffer with 0, 0.1, 1, 2.5 or 5 mM MgCl2 as described above. In the temperature variation experiments, transcripts 397/444 and 1/444 were incubated for 10 min in dimerization buffer with 1 or 5 mM MgCl2 at 23, 37, 50 or 65°C, and then slowly cooled to room temperature before gel electrophoresis.
Measurement of the melting temperature of 397/444 and 1/444 dimers
RNA dimers were allowed to form as described above in dimerization buffer with 5 mM MgCl2. Following cooling to room temperature, the RNA was diluted 10-fold in H2O and 10 µl aliquots were incubated for 15 min at 23, 37, 53, 65, 70, 75 and 82°C. Samples were loaded immediately on a non-denaturing 0.25× TBE gel, and dimerization efficiencies were calculated upon phosphor imager quantification of the gel.
The effect of antisense oligonucleotides and NC protein
Approximately 250 ng (2 pmol) of transcript 1/544 in 10 µl dimerization buffer was incubated for 10 min at 65°C in the presence of a 50-fold molar excess of 20mer antisense oligonucleotide (100 pmol). Samples were cooled to room temperature and analyzed on 0.25× TBE gels as described for dimerization assays. The effect of NC protein on RNA conformation and dimerization was investigated by incubating ∼250 ng (2 pmol) of transcript 1/544 for 10 min at 65°C in dimerization buffer either alone or in the presence of a 50-fold molar excess of the antisense oligonucleotide 418/399. Samples were then slowly cooled to room temperature followed by addition of 1.2 µg synthetic HIV-1 NCp7 protein (23) and incubation for 2 h at 37°C. An aliquot of 1.5 µl 10% SDS was added and samples where placed at room temperature for 5 min followed by a phenol extraction and analysis on 0.25× TBE gels.
RNA absorbance melting curves
RNA thermal denaturation was monitored by measuring the absorbance of UV light at 260 nm in a quartz cuvette with a standard 1 cm path length (24,25). Absorbance was measured over a continuous temperature range from 10 to 95°C on a temperature controlled Perkin Elmer Lambda spectrophotometer. The temperature was increased at 0.5°C/min with measurement of the absorbance at 0.1°C intervals. Duplicate measurements with different T7-preparations of the same transcript differed by <0.5°C. Transcript stock solutions (3.0 µg RNA) were incubated in 140 µl 50 mM Na–cacodylate buffer (pH 7.2) for 10 min at 65°C, slowly cooled to 37°C and incubated for another 15 min prior to the measurement. Non-denaturing gel electrophoresis of RNA before the analysis confirmed its monomeric state. Analysis of samples after the absorbance measurement on sequencing gels showed no significant degradation of the RNA.
Mfold RNA secondary structure predictions
The sequence of the HIV-2 ROD isolate was downloaded from the HIV database (http://hiv-web.lanl.gov/). Secondary structure prediction was performed with the Mfold version 3.0 algorithm (26) on the MBCMR Mfold server (http://mfold.edu.burnet.au/) and analyzed with standard settings.
RESULTS
Dimer formation of the isolated HIV-2 DIS hairpin and the 1/444 leader RNA
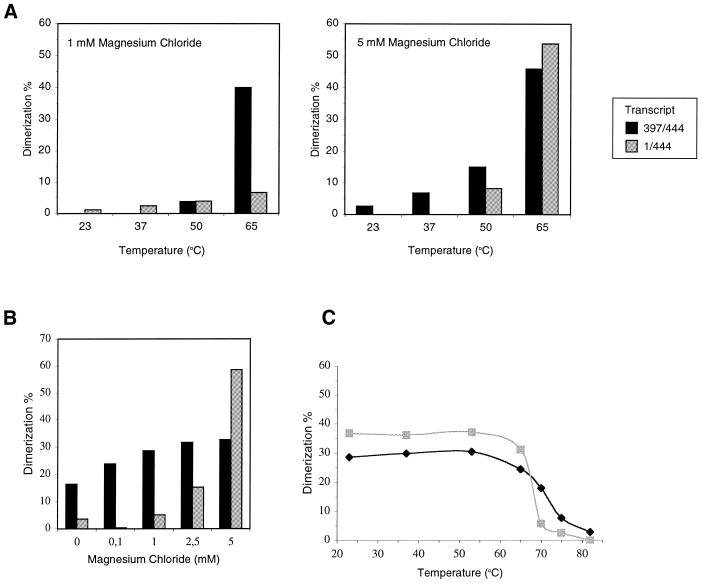
We performed comparative dimerization experiments with the minimal DIS transcript 397/444 and transcript 1/444 that encompasses most of the HIV-2 leader RNA (Fig. 1). We first compared the temperature requirement for dimer formation of the two transcripts at 1 and 5 mM MgCl2 (Fig. 2A). Throughout this study we will show representative experiments, but similar results were obtained in at least three independent assays. RNA was incubated in dimerization buffer for 10 min at 23, 37, 50 or 65°C, followed by slow cooling to room temperature and gel electrophoresis on a non-denaturing 0.25× TBE gel. Monomer and dimer bands were quantified by phophor imager analysis to calculate the percentage of dimers. Dimer formation occured most efficiently at 65°C for both transcripts. Dimerization of the isolated DIS hairpin is almost identical at the two Mg2+ concentrations, which contrasts with the behavior of the longer transcript that dimerizes most efficiently in the presence of 5 mM MgCl2. This difference in Mg2+ requirement of the short versus the long transcript is further illustrated by titration of Mg2+ in the 0–5 mM range (Fig. 2B). Transcript 397/444 dimerizes quite efficiently at all assay conditions, even without MgCl2. These results indicate that Mg2+ is not critical per se for DIS-mediated dimerization. Transcript 1/444 requires at least 2.5 mM MgCl2 for low levels of dimer formation. The strict Mg2+ requirement for dimerization of this transcript suggests that Mg2+ ions influence the overall conformation of the leader RNA and thereby the availability of the DIS hairpin motif for dimerization.
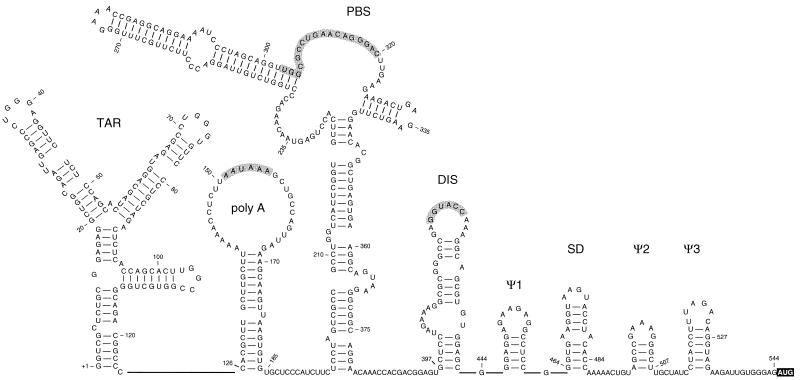
Figure 1.
HIV-2 leader RNA secondary structure model. The model of the 5′ untranslated leader RNA of the HIV-2 isolate ROD is adapted from Berkhout (31). The hairpin structures are named according to their putative function in HIV-1 replication, and several critical sequence elements are highlighted in gray (the polyadenylation signal, the PBS and the DIS palindrome). The Gag start codon at position 545 is boxed. Positions used in this study as 5′/3′ transcript ends are marked.
Figure 2.
Characterization of dimerization requirements for transcripts 397/444 and 1/444. (A) Temperature dependency was assayed by incubation of the RNA samples for 10 min at 23, 37, 50 and 65°C in buffer with 1 mM (left) or 5 mM (right) MgCl2. The RNA was slowly cooled to room temperature and analyzed on a non-denaturing TBE gel. The dimerization efficiency was calculated by phosphor imager quantification and is illustrated in bar graphs. (B) Mg2+ requirement was measured in a 10 min incubation in buffer with 0, 0.1, 1, 2.5 and 5 mM MgCl2, and samples were analyzed as described for (A). (C) Dimer melting curves. RNA dimers were formed at optimal dimerization conditions (10 min at 65°C in 5 mM MgCl2 and slow cooling). After dimer formation, the sample was diluted 10-fold in water to avoid reassociation of melted RNA, and incubated for 15 min at 23, 37, 53, 65, 70, 75 and 82°C. The samples were analyzed on a TBE gel, phosphor imager analysis was performed and the percentage of remaining dimer was calculated. Black and gray coloring represents transcript 397/444 and 1/444, respectively.
We also investigated the stability of the dimers formed by the 397/444 and 1/444 transcripts by determining the dimer melting temperatures (Tm). Transcripts were incubated under optimal dimerization conditions (5 mM MgCl2, 65°C) and extended dimers were allowed to form during slow cooling. Samples were subsequently diluted 10-fold and incubated for 15 min in the 23–82°C temperature range. The dilution prevents reassociation of melted dimers. Samples were analyzed on a non-denaturing TBE gel, and the percentage of dimer was determined (Fig. 2C). The melting curves for the short and long transcripts are very similar with a Tm of ∼70°C. This result indicates that the DIS interaction is solely responsible for the stability of HIV-2 RNA dimers. We also note that melting of the 397/444 dimers is not complete, which may result from (re)dimerization of this RNA at low MgCl2 concentrations (Fig. 2B).
Inhibition of HIV-2 RNA dimerization by sequences downstream of the DIS hairpin
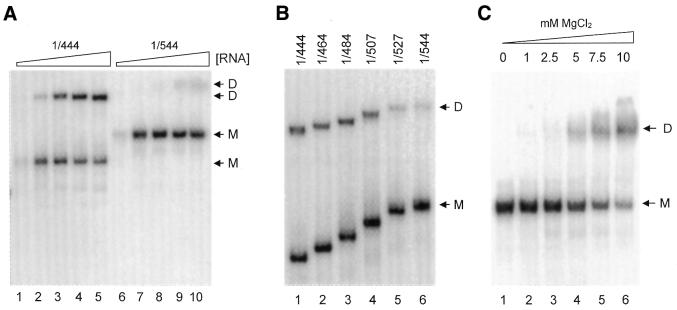
Dimerization of the HIV-1 leader RNA can be repressed by a base pairing interaction between nucleotides of the DIS element and the upstream poly(A) region (21). We previously performed an antisense DNA oligonucleotide scan in the context of transcript 1/444 to locate sequences that inhibit dimerization in the HIV-2 leader RNA (19). Unlike in the HIV-1 system, we identified no oligonucleotides that could stimulate HIV-2 RNA dimerization. We therefore decided to search for repressive sequences in the downstream leader region, and synthesized transcript 1/544 that runs up to the Gag start codon (Fig. 1). Transcripts 1/444 and 1/544 were incubated at optimal dimerization conditions (5 mM MgCl2, 65°C) at increasing RNA concentrations in the 10–1000 ng range (Fig. 3A). Whereas the 1/444 transcript dimerizes efficiently, transcript 1/544 is almost completely inactive. A low level of dimer formation is observed at high RNA concentrations of transcript 1/544 (Fig. 3A, lanes 9 and 10), indicating that the ability to dimerize is not completely abolished. To map the repressive sequences in more detail, we constructed a nested set of truncated transcripts with 3′ termini at positions 464, 484, 507 and 527 (Fig. 3B). Although there is a gradual decrease in dimerization efficiency with increasing transcript length, the most severe decline is observed by including sequences between position 507 and 527 (the dimerization efficiency drops from 40 to 25% as quantified by phosphor imager analysis).
Figure 3.
Mapping of sequences 3′ of the HIV-2 DIS that inhibit dimerization. (A) Transcripts 1/444 and 1/544 were analyzed for their ability to dimerize at varying RNA concentrations. We incubated 10 ng (lanes 1 and 6), 50 ng (lanes 2 and 7), 250 ng (lanes 3 and 8), 500 ng (lanes 4 and 9) and 1000 ng (lanes 5 and 10) transcript at optimal dimerization conditions (65°C, 5 mM MgCl2), followed by gel electrophoresis on a non-denaturing 4% TBE gel and autoradiography. The monomer (M) and dimer (D) bands of both transcripts are marked. (B) Approximately 2 pmol of transcripts 1/444, 1/464, 1/484, 1/507, 1/527 and 1/544 was incubated at dimerization conditions as described in (A). Monomer and dimer bands are marked M and D. The dimerization efficiencies for the transcripts measured by phophorimager quantification are 43.6, 41.8, 44.8, 40.2, 25.0 and 17.9%, respectively. (C) Approximately 2 pmol of radiolabeled transcript 1/544 was incubated at optimal dimerization conditions (65°C, 5 mM MgCl2) in buffer containing 0, 1, 2.5, 5, 7.5 and 10 mM MgCl2.
We tried to rescue dimerization of the 1/544 transcript by increasing the MgCl2 concentration. Indeed, recovery of the dimerization efficiency was measured at 7.5 and 10 mM MgCl2 (Fig. 3C). These combined results are consistent with the notion that alternative foldings exist for the HIV-2 leader RNA. The equilibrium between the different RNA conformations is apparently influenced by variation of the transcript 3′ end and the Mg2+ concentration, thereby affecting DIS-mediated dimerization.
Both upstream and downstream leader sequences are required for repression of RNA dimerization
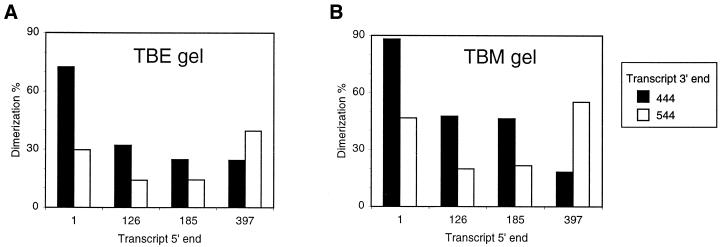
Having identified the downstream region required for inhibition of HIV-2 RNA dimerization, we next examined the involvement of upstream leader sequences. This was done in reference to the HIV-1 system in which dimerization is repressed by a long-distance base pairing interaction between upstream and downstream leader sequences. We synthesized a nested set of 5′ truncated HIV-2 transcripts starting at positions 1, 126, 185 and 397 and ending at either position 444 or 544. To assess both the kissing–loop and extended dimer complexes, the samples were loaded on TBM and TBE gels, and the dimerization efficiency was calculated (Fig. 4). Inhibition by the 444–544 region is apparent for transcripts starting at positions 1, 126 and 185, but not for the transcript starting at 397. Thus, inhibition of dimerization by the 444–544 domain is indeed dependent on the presence of upstream leader sequences. This suggests that a dimerization incompetent conformer of the HIV-2 leader RNA is formed by a long distance RNA–RNA interaction, which is analogous to HIV-1.
Figure 4.
Both 5′and 3′ leader sequences are required for inhibition of dimerization. Approximately 3 pmol of transcripts starting at position 1, 126, 185 and 397 and ending at position 444 or 544 were incubated at optimal dimerization conditions as described in the legend to Figure 2. Samples were loaded on a non-denaturing TBE gel (A) and a non-denaturing TBM gel (B). After gel drying, phosphor imager analysis was performed to calculate the dimerization efficiencies that are plotted in bar graphs. Black and white bars represent dimerization of transcripts terminating at position 444 and 544.
In addition, a significant drop in dimerization efficiency is observed upon deletion of the TAR element, which is located at nucleotide postions 1–126. A previous study demonstrated that the TAR element does not confer potent dimerization capacity (19), ruling out the possibility that a secondary dimerization site is lost upon deletion of TAR. In fact, this result is again reminiscent of HIV-1 dimerization, in which TAR strongly influences the equilibrium between alternative conformations of the leader RNA (21,27).
Antisense oligonucleotide probing of the HIV-2 leader RNA
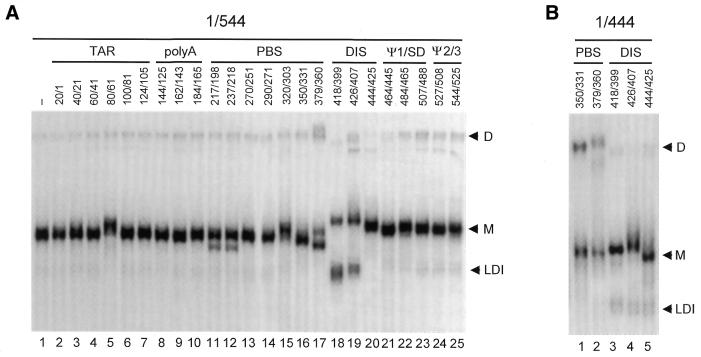
As an alternative way to probe for sequence motifs that are critical for regulation of RNA dimerization, we incubated transcript 1/544 in dimerization buffer with a set of antisense oligonucleotides. The transcript and a 50-fold molar excess of DNA oligonucleotide were incubated at 65°C and DNA/RNA hybrids were allowed to form during slow cooling. The annealing positions of the oligonucleotides on the HIV-2 transcript are indicated in Figure 5A. Annealing of most of the antisense oligonucleotides causes a slight change in the migration of the monomeric RNAs, and this band-shift is more pronounced with extended electrophoresis times (results not shown). Striking RNA migration effects are observed with oligonucleotides that target the DIS domain. Antisense oligonucleotides 418/399 and 444/425 that target the DIS hairpin inhibit dimerization, which is consistent with previous results (19). Furthermore, a fast-migrating band, marked LDI in Figure 5A, is strongly induced by the anti-DIS oligonucleotides 418/399 and 426/407 (Fig. 5A, lanes 18 and 19). For oligonucleotide 418/399, this effect coincides with reduced RNA dimerization. Interestingly, inhibition of HIV-1 dimerization also correlates with formation of a fast-migrating RNA species (21). Oligonucleotides 217/198, 237/218 and 379/360 that target the lower stem of the primer binding site (PBS) domain have a similar effect on the monomeric RNA, causing part of the population to migrate with slightly increased mobility in the gel (Fig. 5A, lanes 11, 12 and 17). Oligonucleotide 379/360 stimulates RNA dimerization slightly (Fig. 5A, lane 17), and this result was confirmed for other transcripts terminating at positions 464–527 (results not shown). A slight increase in dimerization levels is also apparent for oligonucleotides that target the transcript at positions 488–527, which corresponds to the domain that mediates inhibition of dimerization (Fig. 5A, lanes 23 and 24).
Figure 5.
Antisense oligonucleotide scanning of the HIV-2 leader RNA. (A) Approximately 2 pmol of radiolabeled transcript 1/544 was incubated with a 50-fold molar excess of antisense DNA oligonucleotide at optimal dimerization conditions (see Fig. 2 legend) and subjected to non-denaturing gel electrophoresis. The annealing position of the antisense oligonucleotides and the domains of the HIV-2 transcript are indicated on top of the autoradiogram. (B) Excerpt of an antisense oligonucleotide scan of transcript 1/444 shown in Dirac et al. (19), which was performed as described in panel (A). The positions of dimer (D), monomer (M) and fast-migrating monomer (LDI) conformations are indicated.
We also tested the effect of the viral NC protein on the conformation and the dimerization properties of HIV-2 leader RNA. Transcript 1/544 showed increased dimerization in the presence of this RNA chaperone protein (Fig. 6, compare lanes 1 and 2). For HIV-1 RNA, which adopts the dimerization-incompetent LDI conformation, we previously demonstrated that NC protein facilitates the LDI-to-BMH switch and subsequent RNA dimerization (21). To mimic this scenario for HIV-2 RNA, we annealed the anti-DIS oligonucleotide 418/399 that forces the transcript into the fast-migrating LDI form and subsequently added NC protein. Indeed, NC triggers the LDI-to-BMH conformational switch and subsequent dimerization (Fig. 6, lanes 3 and 4, which are shown at two intensities). A corresponding transcript with a mutated DIS palindrome did not dimerize under these conditions (results not shown), indicating that NC-mediated RNA dimerization is dependent on an intact DIS element.
Figure 6.
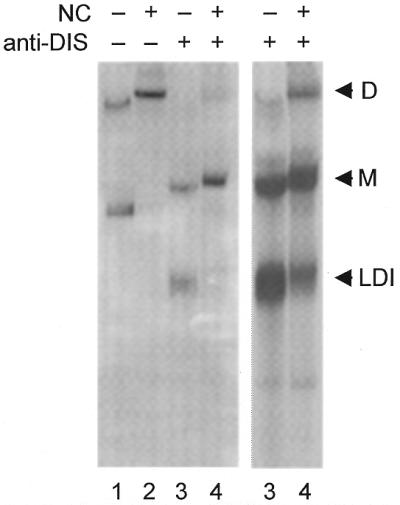
The effect of NC protein on the HIV-2 leader RNA conformation. Approximately 2 pmol of radiolabeled transcript 1/544 was incubated either alone (lanes 1–2) or with a 50-fold molar excess of the antisense DNA oligonucleotide 418/399 (lanes 3–4) at optimal dimerization conditions (see Fig. 2 legend). After slow cooling to room temperature, 1.2 µg synthetic HIV-1 NCp7 protein was added (lanes 2 and 4) and all samples were incubated for 2 h at 37°C, followed by treatment with SDS and a phenol extraction. The positions of dimer (D), monomer (M) and fast-migrating monomer (LDI) conformations are indicated. Lanes 3 and 4 are shown at two intensities.
Different HIV-2 leader RNA conformations observed by absorbance melting
The experiments described above indicate that inhibition of HIV-2 RNA dimerization occurs through a LDI between sequence elements that are located upstream and downstream of the DIS hairpin region. In this respect the mechanism of regulation of HIV-2 RNA dimerization is analogous to that of HIV-1 RNA. The HIV-1 leader RNA can adopt an alternative LDI conformation in which the DIS is base paired with upstream sequences. This structure is mutually exclusive with dimer formation and shows a typical fast migration on non-denaturing gels (21). Interestingly, anti-DIS oligonucleotides that inhibit dimerization of HIV-2 RNA also cause the appearance of a fast-migrating RNA band (Fig. 5A), which may in fact correspond to the LDI structure. We decided to perform RNA absorbance melting experiments with HIV-2 RNA because this technique has proven very useful in the study of the alternative HIV-1 RNA conformations. The method takes advantage of the change in absorbance measured at 260 nm of non-denaturing RNA relative to denatured RNA. Monitoring of the UV-absorbance over a temperature range yields the thermal denaturation profile of an RNA, in which increases in absorbance correspond to the melting of structured domains.
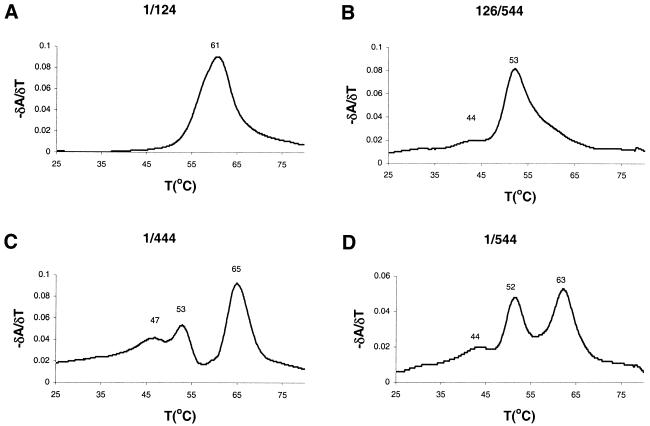
Figure 7 shows the UV-melting data for transcripts 1/124, 126/544, 1/444 and 1/544 as δA/δT plots that highlight the melting transitions. The melting profile of transcript 1/124, corresponding to the TAR element, shows a single transition at Tm = 61°C. A similar transition is observed for transcripts 1/444 (Tm = 65°C) and 1/544 (Tm = 63°C), but this peak is absent for transcript 126/544 that lacks TAR. The variation in the Tm of TAR in the different transcripts may be caused by interactions of TAR with downstream leader sequences.
Figure 7.
Thermal melting profiles of HIV-2 leader RNAs. Melting was monitored by UV absorption at 260 nm. The curves show the first order derivative (δA/δT) to highlight the melting transitions. We analyzed transcripts 1/124 (A), 126/544 (B), 1/444 (C) and 1/544 (D). The melting temperature Tm is indicated at the top of the peaks.
The transcripts containing downstream leader sequences have two melting transitions below 60°C in common, but there is some variability in their relative intensities. A clear transition at ∼53°C is apparent in transcripts 1/444, 126/544 and 1/544. In addition, all three transcripts share a broad transition with a Tm = ∼44°C. Interestingly, the relative intensities of the 44 and 53°C transitions are very different for the three transcripts. Both transitions are of comparable intensity for transcript 1/444, whereas the transition at Tm = 53°C is much more prominent for transcript 1/544. The transition at Tm = 44°C is nearly absent for transcript 126/544. We think that these transitions correspond to alternative conformations of the HIV-2 leader RNA. The drastic increase in intensity of the transition at Tm = 53°C in the melting profile of transcript 1/544 coincides with the observed decrease in dimer formation efficiency of this transcript. Interestingly, the two-peak melting profile of the 1/544 transcript closely resembles the spectrum of HIV-1 RNA in its LDI conformation. It is our interpretation that the transition in the 44°C range is a metastable conformation of the HIV-2 leader RNA. This transition is more pronounced in transcript 1/444 which dimerizes efficiently. The transition at 53°C is more pronounced in transcripts 1/544 and 126/544 which dimerize poorly (Fig. 4), suggesting that this transition corresponds to an RNA conformation that down-regulates dimerization.
DISCUSSION
We set out to study regulation of HIV-2 RNA dimerization because there is recent evidence that dimerization of HIV-1 RNA is strictly controlled by alternative RNA conformations (21,27). We first performed a comparative analysis of HIV-2 RNA dimerization of the minimal DIS transcript 397/444 and the extended transcript 1/444. Both transcripts require pre-incubation at 65°C for efficient dimerization, indicating that a certain level of energy is needed to open the base paired DIS stems and allow formation of the extended dimer complex. In agreement with this idea, more energy is required for mutant HIV-2 transcripts with a stabilized DIS stem (results not shown). HIV-1 extended RNA dimers are formed efficiently at 50–55°C (28). This may indicate that the HIV-2 DIS stem is more stable than its HIV-1 counterpart, which is fully consistent with the recently proposed extension of the HIV-2 DIS hairpin by 5 additional bp as shown in Figure 1 (19). These additional base pairs in the DIS hairpin will form additional intermolecular base pairs in the extended dimer complex. Indeed, the HIV-2 RNA dimer melts at 68–72°C and is more stable than the HIV-1 dimer that melts at 53–65°C, depending on the concentration of mono- and divalent cations (14,28). Secondary structure modeling of the extended dimers with the Mfold algorithm supports this notion (results not shown). The HIV-1 and HIV-2 complexes have a predicted thermodynamic stability of –37.3 and – 47.2 kcal/mol, respectively.
The first indication that alternative HIV-2 RNA conformations may exist came from dimerization studies with varying Mg2+ concentrations. Numerous studies have reported the importance of Mg2+ for HIV-1 RNA dimerization (6–13). Similarly, we measured efficient dimerization of the extended HIV-2 transcript only at 5 mM MgCl2. However, the minimal HIV-2 DIS transcript readily formed dimers in buffers lacking MgCl2. This result suggests that it is not RNA dimerization itself that requires Mg2+ ions. The divalent cations may instead influence dimerization indirectly by affecting the conformation of the leader RNA. For HIV-1, it has been reported that certain DIS palindromic sequences are more dependent on Mg2+ than others (29). We therefore tested several palindromes in the minimal HIV-2 context, including sequences from HIV-1 and SIV isolates (GCGCGC, GUGCAC and GUCGAC). All palindromes support dimerization in the absence of Mg2+ (results not shown). We therefore think that dimer formation of the extended HIV-2 leader requires Mg2+ to mediate a structural reorganization that exposes the DIS hairpin.
We next focused on possible mechanisms for regulated HIV-2 RNA dimerization. We will compare HIV-2 with the HIV-1 leader RNA that adopts alternative conformations (21). The thermodynamically most stable HIV-1 RNA structure masks the DIS in a LDI with upstream leader sequences of the poly(A) region (Fig. 8). Dimer formation requires that HIV-1 RNA shifts into the BMH conformation that exposes the DIS hairpin. Thus, LDI folding and RNA dimerization are mutually exclusive processes. For HIV-2, we previously reported that the upstream poly(A) region does not modulate RNA dimerization (19), which prompted us to examine the downstream leader sequences in this study. Inclusion of the downstream 444–544 domain caused a profound loss of dimerization, and further deletion analysis indicated that the 507–527 segment is largely responsible for this repression. We performed detailed RNA structure probing experiments and we analyzed the secondary structure by the Mfold algorithm (26), but we were not able to resolve the actual base pairing interactions of the alternative HIV-2 RNA folding. Nevertheless, we present a model in Figure 8 that incorporates several of the experimental findings and Mfold predictions. This working model should form the basis for additional experimentation. An alternative conformation in which the DIS nucleotides are base paired with downstream sequences is predicted by Mfold for the 1/527 transcript that has lost the ability to dimerize efficiently, but not for shorter transcripts that dimerize efficiently. This DIS masking is visualized in the alternative HIV-2 folding (Fig. 8). In analogy with the HIV-1 system, we use the LDI and BMH nomenclature for the leader RNA structures that differ in folding of the DIS region. Dimerization of the minimal DIS transcript is not inhibited by the downstream 444–544 domain, suggesting that upstream leader sequences are also involved in this repression. This result is extrapolated in the long-distance interaction in the LDI fold (purple segments in Fig. 8).
Figure 8.
An RNA switch regulates dimerization in HIV-1 and HIV-2 RNA. The different structural elements within the leaders are color coded: TAR bulges and loops (green), polyadenylation signals (orange), PBS (blue) and DIS palindrome (pink). The linear presentation is rearranged to show the equivalent BMH structure. A structural BMH-to-LDI rearrangement is proposed that results in disappearance of the DIS hairpin. In HIV-2, the DIS region is proposed to base pair with 3′ sequences, and the stem of the PBS domain is extended by base pairing of sequences that are located far upstream and downstream of the DIS (sequence elements are marked in purple). Details of the DIS hairpin in the BMH conformation are shown in Figure 1. The HIV-2 LDI model is based on several experimental findings, but we were unable to unequivocally assign the new base pairing scheme (see Discussion). Many more details are available for the HIV-1 RNA, in which both the DIS and the upstream poly(A) hairpin are opened to facilitate the extension of the PBS stem to form the LDI. In both viral systems, the RNA switch is proposed to provide a mechanism for the regulated dimerization of the viral genomes.
There is additional evidence for the existence of alternative conformations of the HIV-2 leader RNA. Antisense oligonucleotides are sensitive tools to shift the equilibrium between alternative leader RNA conformations. The antisense oligonucleotide 379/360, which targets the lower right side of the extended PBS stem, stimulates HIV-2 RNA dimerization. Interestingly, an oligonucleotide targeting the corresponding region in the HIV-1 leader also stimulates RNA dimerization (30). Inspection of the structure models (Fig. 8) provides a possible explanation. Targeting of the lower PBS stem, which is the extended form of the LDI interaction in HIV-1, affects the stability of the LDI conformation in both HIV-1 and HIV-2 RNA, thus triggering the alternative BMH folding that allows subsequent dimerization.
The LDI conformation of HIV-1 RNA is characterized by fast migration on non-denaturing gels (20). We did not observe this for HIV-2 RNA, but annealing of anti-DIS oligonucleotides causes a profound shift of the monomeric 1/544 RNA to a fast-migrating form (Fig. 5A, lanes 18 and 19). In addition to promoting a conformational change, oligonucleotide 418/399 also inhibits dimerization. We propose that targeting of the left side of the DIS stem liberates the right side for base pairing in the alternative LDI conformation (Fig. 8). Thus, inhibition of HIV-2 RNA dimerization correlates with the appearance of the LDI RNA conformer with increased electrophoretic mobility, which is reminiscent of the HIV-1 LDI structure (21).
The HIV-2 leader RNA conformation was investigated further by UV-melting experiments. The melting profile of the complete leader (1–544) reveals two major transitions, one of which corresponds to the TAR hairpin. This two-peak melting profile is reminiscent of the HIV-1 leader RNA in the LDI conformation, suggesting that the second transition represents an extended PBS stem segment in the HIV-2 LDI conformation (Fig. 8). However, the HIV-2 LDI conformation appears not as dominant as the equivalent HIV-1 conformation, both in the UV-melting experiments and on non-denaturing gels. As discussed previously, the HIV-2 DIS hairpin is much more stable than its HIV-1 counterpart, and this may shift the equilibrium in the direction of the BMH conformation. Deletion of the 5′ terminal TAR hairpin favors the HIV-2 LDI conformation as observed by UV melting (Fig. 7) and causes a concomitant dimerization defect (Fig. 4). In contrast, deletion of TAR in HIV-1 RNA has an opposite effect on the BMH–LDI equilibrium, with a concomitant increase in dimerization (27). These opposing effects indicate that one should be extremely careful when interpreting the results of a mutational analysis of the regulatory domain of a retroviral genome.
In conclusion, the wild-type HIV-2 leader RNA is in equilibrium between two alternative conformations and this delicate balance is likely to be exploited in the viral replication cycle. The LDI structure prevents premature RNA dimerization, which may be important for the execution of other early replication steps. The BMH structure allows efficient RNA dimerization and perhaps also exposes the downstream leader signals that control packaging of the RNA into virions. Thus, the existence of alternative conformations of HIV-1 RNA appears to correlate with the dual role of retroviral transcripts as messenger and genomic RNA. DIS-mediated dimerization is repressed in HIV-1 and HIV-2 by an alternative conformation that refolds the DIS motif. The NC protein was shown to mediate the LDI-to-BMH transition of the HIV-1 RNA (21) and HIV-2 RNA (this study). Regulation of RNA genome dimerization by means of alternative RNA structures is therefore likely to be a more general property of the entire HIV–SIV family, and possibly of all retroviruses.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jean-Luc Darlix for the kind donation of synthetic HIV-1 NCp7 protein and Wim van Est for the artwork. This work was funded by the European Molecular Biology Organization, the Netherlands Organization for Scientific Research (NWO-CW), the Norfa Foundation, Plasmid Fondet, Aarhus University and SSVF (Danish National Foundation of Health).
REFERENCES
- 1.Coffin J.M. (1990) Retroviridae and their replication. In Fields,B.N. and Knipe,D.M. (eds), Virology. Raven Press Ltd, NY. [Google Scholar]
- 2.Greatorex J. and Lever,A. (1998) Retroviral RNA dimer linkage. J. Gen. Virol., 79, 2877–2882. [DOI] [PubMed] [Google Scholar]
- 3.Fu W. and Rein,A. (1993) Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol., 67, 5443–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darlix J.L., Gabus,C., Nugeyre,M.T., Clavel,F. and Barre-Sinoussi,F. (1990) Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol., 216, 689–699. [DOI] [PubMed] [Google Scholar]
- 5.Gotte M., Li,X. and Wainberg,M.A. (1999) HIV-1 reverse transcription: a brief overview focused on structure-function relationships among molecules involved in initiation of the reaction. Arch. Biochem. Biophys., 365, 199–210. [DOI] [PubMed] [Google Scholar]
- 6.Laughrea M. and Jette,L. (1994) A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry, 33, 13464–13474. [DOI] [PubMed] [Google Scholar]
- 7.Marquet R., Baudin,F., Gabus,C., Darlix,J.L., Mougel,M., Ehresmann,C. and Ehresmann,B. (1991) Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res., 19, 2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paillart J.C., Marquet,R., Skripkin,E., Ehresmann,B. and Ehresmann,C. (1994) Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem., 269, 27486–27493. [PubMed] [Google Scholar]
- 9.Skripkin E., Paillart,J.C., Marquet,R., Ehresmann,B. and Ehresmann,C. (1994) Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl Acad. Sci. USA, 91, 4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sundquist W.I. and Heaphy,S. (1993) Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc. Natl Acad. Sci. USA, 90, 3393–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieth E., Gabus,C. and Darlix,J.L. (1990) A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res., 18, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clever J., Sassetti,C. and Parslow,T.G. (1995) RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol., 69, 2101–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haddrick M., Lear,A.L., Cann,A.J. and Heaphy,S. (1996) Evidence that a kissing loop structure facilitates genomic RNA dimerisation in HIV-1. J. Mol. Biol., 259, 58–68. [DOI] [PubMed] [Google Scholar]
- 14.Muriaux D., Girard,P.M., Bonnet-Mathonière,B. and Paoletti,J. (1995) Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J. Biol. Chem., 270, 8209–8216. [DOI] [PubMed] [Google Scholar]
- 15.Laughrea M. and Jette,L. (1996) Kissing–loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensable for dimer formation. Biochemistry, 35, 1589–1598. [DOI] [PubMed] [Google Scholar]
- 16.Muriaux D., Fosse,P. and Paoletti,J. (1996) A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry, 35, 5075–5082. [DOI] [PubMed] [Google Scholar]
- 17.Berkhout B., Essink,B.B. and Schoneveld,I. (1993) In vitro dimerization of HIV-2 leader RNA in the absence of PuGGAPuA motifs. FASEB J., 7, 181–187. [DOI] [PubMed] [Google Scholar]
- 18.Jossinet F., Lodmell,J.S., Ehresmann,C., Ehresmann,B. and Marquet,R. (2001) Identification of the in vitro HIV-2/SIV RNA dimerization site reveals striking differences with HIV-1. J. Biol. Chem., 276, 5598–5604. [DOI] [PubMed] [Google Scholar]
- 19.Dirac A.M., Huthoff,H., Kjems,J. and Berkhout,B. (2001) The dimer initiation site hairpin mediates dimerization of the human immunodeficiency virus, type 2 RNA genome. J. Biol. Chem., 276, 32345–32352. [DOI] [PubMed] [Google Scholar]
- 20.Berkhout B. and van Wamel,J.L. (2000) The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA, 6, 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huthoff H. and Berkhout,B. (2001) Two alternating structures of the HIV-1 leader RNA. RNA, 7, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oude Essink B.B., Das,A.T. and Berkhout,B. (1996) HIV-1 reverse transcriptase discriminates against non-self tRNA primers. J. Mol. Biol., 264, 243–254. [DOI] [PubMed] [Google Scholar]
- 23.De Rocquigny H., Gabus,C., Vincent,A., Fournie-Zaluski,M.C., Roques,B. and Darlix,J.L. (1992) Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl Acad. Sci. USA, 89, 6472–6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brion P. and Westhof,E. (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct., 26, 113–137. [DOI] [PubMed] [Google Scholar]
- 25.Puglisi J.D. and Tinoco,I.,Jr (1989) Absorbance melting curves of RNA. Methods Enzymol., 180, 304–325. [DOI] [PubMed] [Google Scholar]
- 26.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 27.Huthoff H. and Berkhout,B. (2001) Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res., 29, 2594–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laughrea M. and Jette,L. (1996) HIV-1 genome dimerization: formation kinetics and thermal stability of dimeric HIV-1Lai RNAs are not improved by the 1–232 and 296–790 regions flanking the kissing–loop domain. Biochemistry, 35, 9366–9374. [DOI] [PubMed] [Google Scholar]
- 29.Jossinet F., Paillart,J.C., Westhof,E., Hermann,T., Skripkin,E., Lodmell,J.S., Ehresmann,C., Ehresmann,B. and Marquet,R. (1999) Dimerization of HIV-1 genomic RNA of subtypes A and B: RNA loop structure and magnesium binding. RNA, 5, 1222–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berkhout B., Ooms,M., Beerens,N., Huthoff,H., Southern,E. and Verhoef,K. (2002) In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem., 277, 19967–19975. [DOI] [PubMed] [Google Scholar]
- 31.Berkhout B. (1996) Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol., 54, 1–34. [DOI] [PubMed] [Google Scholar]