Abstract
Background
Esophageal squamous cell carcinoma (ESCC) is a global cancer related to the sixth largest cause of death. Circular RNAs (circRNAs) have affected the progress of ESCC during recent years, but the mechanism is not completely clear. So here we probed the effects of hsa_circ_0096710 (circ_0096710) in ESCC.
Methods
Relative levels of circ_0096710, miR‐1294, and ADAM10 were quantified by the quantitative real‐time reverse transcription‐polymerase chain reaction in ESCC tissues. Western blot assessed ADAM10, PCNA, MMP2, VEGFA, and OCT4 protein levels. Cell proliferative capacity was assessed by cell counting and cell colony‐forming assays. Transwell assays assessed cell migration and invasion. Angiogenesis was detected by tube formation assays. Stemness of cancer cells was estimated by sphere formation assays. Dual‐luciferin reporter and RNA immunoprecipitation assays determined the targeting relationship between miR‐1294 and circ_0096710 or ADAM10.
Results
Relative levels of circ_0096710 and ADAM10 mRNA were upregulated in ESCC cells, yet miR‐1294 was downregulated. Circ_0096710 silencing repressed ESCC cell proliferation, migration, invasion, angiogenesis, and stem‐like properties. Moreover, circ_0096710 was an upstream target of miR‐1294, and miR‐1294 inhibition reversed the role of circ_0096710 downregulation in ESCC cells. Furthermore, ADAM10 was a downstream target of miR‐1294, and miR‐1294 overexpression suppressed ESCC cell proliferation, migration, invasion, angiogenesis, and stem‐like properties by targeting ADAM10. Meanwhile, circ_0096710 upgraded ADAM10 expression through sponging miR‐1294. Also, circ_0096710 downregulation restrained tumor growth in mouse models.
Conclusion
Circ_0096710 upregulates ADAM10 via mediating miR‐1294 expression so as to accelerate the occurrence of ESCC, suggesting that circ_0096710 may be a potential therapeutic target for ESCC.
Keywords: ADAM10, Circ_0096710, ESCC, miR‐1294
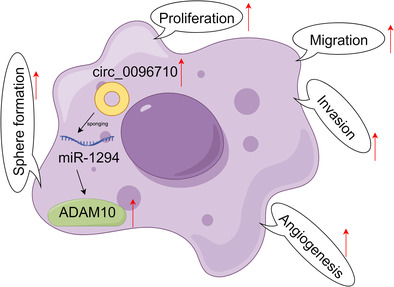
Schematic illustration of the mechanism by which circ_0096710 upregulates ADAM10 expression by interacting with miR‐1294, resulting in promoting esophageal squamous cell carcinoma (ESCC) cell proliferation, migration, invasion, angiogenesis, and stem‐like properties.
INTRODUCTION
Esophageal cancer (EC) was the seventh highest malignant tumor in morbidity and mortality, the highest incidence of EC in our country, and its main pathological subtype is esophageal squamous cell carcinomas (ESCC). 1 , 2 Factors such as consumption of foods containing nitrosamines, poor dietary habits, smoking, alcohol consumption, obesity, poor oral hygiene, human papillomavirus (HPV) infection, nutrient deficiencies, and a family history of EC are risk factors for EC development. 3 Currently, chemotherapy and surgery have no significant effect, with a 5‐year survival rate of <25% in patients with ESCC. 4 Hence, more efficient treatment decisions and new biomarkers are urgently needed to further cure ESCC. 5
Circular RNAs (circRNAs) are closed‐circular RNAs formed by the 3′ and 5′ ends linked by exon or intron cyclization. CircRNAs with a unique ring structure are less susceptible to degradation by nucleic acid exonucleases, thereby having better stability. 6 Studies have shown that circRNAs could play a role as a biomarker in a variety of diseases. 7 , 8 For example, circRNA hydroxysteroid dehydrogenase‐like 2 mediates the hippo pathway to facilitate breast cancer progression. 9 Furthermore, circRNA sorbin and SH3 domain‐containing protein 1 exerts an anti‐tumor role in lung cancer by elevating RUFY3 expression. 10 Similarly, circRNAs are promising therapeutic targets for ESCC. Cao et al. uncovered that circRNA homeodomain‐interacting protein kinase 3 elevates FASN expression and boosts fatty acid metabolism, resulting in promoting tumor growth in ESCC. 11 Also, circRNA FIRRE intergenic repeating RNA element elevates GLI2 protein expression, causing the upregulation of CCNE2, CCNE1, and MYC, thus facilitating the growth of ESCC. 12 Furthermore, circ‐ZDHHC5 was used as a new potential circulating biomarker, and it could promote the occurrence of ESCC. 13 Glucose uptake and cell proliferation for ESCC cells promote by upregulation of circ_0058063. 14 Two circRNAs formed by the NADPH oxidase 4 (NOX4) gene, hsa_circ_0023988 and hsa_circ_0023990, play promoting roles in lung cancer and colorectal cancer, respectively. 15 , 16 Hsa_circ_0096710 (circ_0096710) is generated from 6 to 15 exons of the NOX4 gene, but the exact role of circ_0096710 in ESCC remains uncertain and requires further study.
MicroRNAs (miRs) were single‐stranded noncoding RNA polymers consisting of 20–22 nucleotides. 17 A common mechanism for cytoplasmic circRNAs is to compete with mRNAs so as to bind miRs, thereby regulating downstream genes. 18 For instance, circ‐ZNF609 promotes ESCC progression via repressing miR‐150‐5p expression. 19 Mounting evidence suggests that resistance to cisplatin and TMZ and poorer prognosis in patients with ESCC, gastric cancer, epithelial ovarian cancer, pancreatic ductal adenocarcinoma, or lung cancer are associated with downregulation of miR‐1294. 20 However, the mechanism by which miR‐1294 is downregulated in ESCC has not been well elucidated.
A disintegrin and metalloproteinase 10 (ADAM10) was composed of domains of metalloproteinases, cysteine‐rich, disintegrin, and epidermal growth factor (EGF)‐like, which played a key role in the immune system. 21 , 22 , 23 Numerous studies have confirmed that ADAM10 plays a role in a variety of cancers. 24 , 25 , 26 For example, ADAM10 is involved in the oncogenic process and chemotherapy resistance in breast cancer by regulating the Notch1 signaling pathway, CD44, and PrPc. 27 In addition, ADAM10 is associated with the cleavage of Trop‐2, an activation switch for cancer growth and metastasis, 28 and this cleavage is detected in a variety of tumors including skin, ovarian, colon, and breast cancers. 29 In clinical studies of ESCC, ADAM10 mediated cell invasion and metastasis by regulating E‐cadherin activity. 22 Current explanations for the role of ADAM10 in ESCC are still limited. ADAM10 is predicted as a miR‐1294 target, but their relationship is unclear.
Here, we investigated the mechanism of circ_0096710 in ESCC cells. Our data suggested that circ_0096710, as an oncogene, may promote tumor development via the miR‐1294/ADAM10 axis. Our study may provide new insights into future treatment strategies for ESCC.
MATERIALS AND METHODS
Clinical samples
Specimens of ESCC (n = 60) and matched paracancerous tissues (n = 60) used in the present study were taken from patients with ESCC who underwent surgery at Shanxi Province Cancer Hospital. For all procedures, they were approved and supervised by the Ethics Committee of Shanxi Province Cancer Hospital. The storage of all samples was at a temperature of −80°C.
Cell incubation and transfection
Four ESCC cell lines KYSE450 (#ml‐CC1355, Mlbio, Shanghai, China), KYSE30 (#ml‐CC1377, Mlbio), KYSE150 (#SNL‐344, Sunncell, Wuhan, China), and EC109 (#SNL‐511, Sunncell) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (BIOSUN, Shanghai, China) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and 1% streptomycin (Invitrogen). The natural esophageal epithelial cell line Het‐1A (#ml096307, Mlbio) was cultured in its specialized medium. All cell lines were incubated in a biological incubator containing 5% CO2 at 37°C.
Short hairpin RNAs (sh‐circ_0096710#1, sh‐circ_0096710#2, sh‐circ_0096710#3) against circ_0096710 and a matched control (sh‐NC), miR‐1294 mimic (miR‐1294) and the matched control (miR‐NC), miR‐1294 inhibitor (inh‐miR‐1294) and a matched control (inh‐NC), as well as ADAM10 overexpression vector (ADAM10) and its control (vector) were gained from RiBoBio (Guangzhou, China). Transfection of KYSE30 and KYSE150 cells with oligonucleotides and plasmids was done using Lipofectamine 3000 (Invitrogen).
Quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR)
Extraction of total RNA from cells and clinical specimens was undertaken using Trizol reagent (Beyotime, Shanghai, China) as per the manufacturer's instructions. Evaluation of RNA quality was carried out using a NanoDrop ND‐1000, and validation of RNA integrity was undertaken by conventional agarose electrophoresis. Degradation of RNase R was determined by treatment with RNase R (Tiangen, Beijing, China) at 37°C for 10 min. Reverse transcription experiments were conducted using a Prime Script RT reagent Kit (TaKaRa, Kusatsu, Japan) or a TaqMan MicroRNA RT Kit (Thermo, Waltham, MA, USA) in accordance with the manufacturer's instructions. The SYBR Green kit (Takara, Tokyo, Japan) was used for qPCR reaction. Twenty microliters of the PCR system consisted of 10 μL of SYBR Green Mixture, 1 μL of forward primer (10 μM), 1 μL of reverse primer (10 μM), 2 μL of cDNA, and 6 μL of ddH2O. The amplification conditions for miRs were performed with the following thermocycling conditions: 95°C for 20 s, then 40 cycles of 95°C for 1 s, and 60°C for 20 s. The amplification conditions for mRNAs were as follows: 95°C for 10 min, 95°C for 15 s, 60°C for 1 min, and 72°C for 30 s, for a total of 40 cycles. Relative expression was calculated by the 2−ΔΔCt method under conditions where GAPDH or U6 was used as a control. The sequences of primers are demonstrated in Table 1.
TABLE 1.
Primer sequences used for qPCR.
| Names | Primers for PCR (5′–3′) | |
|---|---|---|
| circ_0096710 | Forward | GCAACATTTGGGGTTCATCT |
| Reverse | GTTCGGCACATGGGTAAAAG | |
| NOX4 | Forward | GCCAGAGTATCACTACCTCCAC |
| Reverse | CTCGGAGGTAAGCCAAGAGTGT | |
| miR‐1294 | Forward | GCCGAGTGTGAGGTTGGCAT |
| Reverse | CAGTGCGTGTCGTGGAGT | |
| ADAM10 | Forward | GAGGAGTGTACGTGTGCCAGTT |
| Reverse | GACCACTGAAGTGCCTACTCCA | |
| GAPDH | Forward | CTGACTTCAACAGCGACACC |
| Reverse | GTGGTCCAGGGGTCTTACTC | |
| U6 | Forward | CTCGCTTCGGCAGCACATATACT |
| Reverse | ACGCTTCACGAATTTGCGTGTC |
Abbreviation: qPCR, quantitative polymerase chain reaction.
Cell Counting Kit‐8 (CCK‐8) assay
For proliferation analysis, transfected ESCC cells (5 × 103) were inoculated into 96‐well plates and incubated for 48 h. Ten microliters of CCK‐8 solution (Beyotime) was added at different time points and cultured for 3 h. Absorbance at 450 nm was determined using a microplate reader (Thermo).
Cell colony‐forming assay
Transfected ESCC cells (5 × 103) were placed in 6‐well plates and cultured for 10–12 days until visible clones appeared in the culture dish, then fixed by 4% paraformaldehyde (Beyotime), and stained by 0.05% crystal violet (Beyotime). These colonies were photographed and counted.
Transwell assay
Matrigel (Yubo Biotech Co., Ltd., Shanghai, China) melted at 4°C was diluted with 200 μL of serum‐free medium at a 1:1 ratio. Fifty microliters of the diluted Matrigel was added to the upper chamber of each Transwell plate, followed by incubation in an incubator for 2.5 h to solidify. Transwell assay (Costar, Corning, NY, USA) was used to determine cell migration and invasion. ESCC cells and serum‐free RPMI 1640 were inoculated into the upper chamber precoated with (for detecting cell invasion) or without (for detecting cell migration) Matrigel (BD Biosciences, Franklin, NJ, USA). RPMI 1640 (600 μL) containing 10% serum was put into the lower chamber and incubated in 5% CO2 incubator at 37°C for 48 h. The cells on the lower surface of the membrane were fixed and stained; finally, the number of cells was counted.
Tube formation assay
The supernatant was collected as a conditioned medium when transfected cells (KYSE30 and KYSE150) reached 80% confluence. Twenty‐four‐well plates were coated with Matrigel (BD Biosciences) in each well and incubated for 0.5 h at 37°C for polymerization. Next, human umbilical vein endothelial cells (HUVECs; Procell, Wuhan, China) were inoculated at a density of 1 × 105 cells per well. After incubation at 37°C for 6 h, tube images were taken using a microscope (Nikon, Tokyo, Japan).
Sphere formation assay
Cultivation of KYSE30 and KYSE150 cells in RPMI 1640 medium containing insulin (4 ng/mL; Sigma), basic fibroblast growth factor (10 ng/mL; Sigma), EGF (100 ng/mL; Sigma), and B‐27 (2%; Invitrogen) was undertaken in an ultra‐low attachment 6‐well plate (Costar). Renewal of the medium was conducted every 2–3 days. Ten days later, spheroids were visualized using a microscope (Nikon).
Western blot
Lysis of tissue specimens or cells in radioimmunoprecipitation assay lysis buffer (Beyotime) was undertaken for the production of total proteins. The extracted protein samples were separated using sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE; Beyotime), followed by transferring to polyvinylidene fluoride membranes (Invitrogen). Following sealing with 5% milk (Beyotime), these membranes were probed with unique primary antibodies against proliferating cell nuclear antigen (PCNA; ab29, 1:1000, Abcam), matrix metalloproteinase 2 (MMP2; ab97779, 1:1000, Abcam), vascular endothelial growth factor A (VEGFA; ab46154, 1:1000, Abcam), organic cation/carnitine transporter 4 (OCT4; ab200834, 1:1000, Abcam), and ADAM10 (ab227172, 1:1000, Abcam) overnight at 4°C, with anti‐β‐actin antibody (ab227387, 1:3000, Abcam) as a control. Imaging observation was displayed with RapidStep ECL Reagent (Millipore, Billerica, MA, USA) following incubation with a secondary antibody (1:4000, ab205718, Abcam).
CircRNA subcellular localization assay
Cytoplasm and nucleus fractions were isolated using a PARIS™ Kit (Invitrogen), and qRT‐PCR was applied to detect the expression level of RNA in the nuclear and cytoplasm to further determine the location of circ_0096710.
Dual‐luciferase reporter assay
The candidate target miRNAs of circ_0096710 were predicted using CircInteractome (https://circinteractome.nia.nih.gov/). The candidate downstream targets of miR‐1294 were predicted using Starbase (http://starbase.sysu.edu.cn/). The wild‐type (WT) or mutant (MUT) sequences of circ_0096710 and ADAM10 within the putative binding sequence of miR‐1294 were inserted into the pmirGLO vector (YouBia, Changsha, China) to establish luciferase reporter vectors. KYSE30 and KYSE150 cells were cotransfected with a corresponding reporter vector and miR‐1294 or miR‐NC. Finally, the Dual‐Luciferase Reporter Gene Assay Kit (Yeasen, Shanghai, China) was employed to measure luciferase activities.
RNA immunoprecipitation assay (RIP assay)
An EZ‐Magna RIP™ RNA‐Binding Protein Immunoprecipitation Kit (Millipore) was applied. The lysed ESCC cells were incubated with magnetic beads conjugated to anti‐Ago2 or anti‐IgG at 4°C for 12 h. The immunoprecipitated RNA was extracted following treatment with protease K for another 30 min. Expression of RNA was assessed by qRT‐PCR.
RNA pull‐down assay
MiR‐1294 mimic‐biotin (Bio‐miR‐1294) and its corresponding control (Bio‐miR‐NC) were designed by RiboBio. The lysate samples of ESCC cells were incubated with Bio‐miR‐1294 or Bio‐miR‐NC. Then, the probe‐bead compound was incubated with streptavidin magnetic beads. After elution, circ_0096710 and ADAM10 mRNA levels were measured by qRT‐PCR.
Immunohistochemistry (IHC) analysis
Post‐fixation in formaldehyde (10%) and embedding in paraffin, slices of the tissues were made into 5 μm thickness, followed by incubation with antibodies against Ki‐67 (ab16667; 1:300, Abcam) or ADAM10 (ab227172; 1:200, Abcam). Next, slices were probed with the corresponding secondary antibody (ab205718; 1:2000, Abcam). Thereafter, staining of these sections was undertaken using hematoxylin–eosin (Maxim, Fuzhou, China). Eventually, a microscope (Nikon) was used to observe these sections.
In vivo experiment
This study followed the instructions of the Ethics Committee of Shanxi Province Cancer Hospital. All nude mice (6 weeks old, female; n = 10) were supplied by (Vital River, Beijing, China) and randomly divided into two groups (n = 5/group). KYSE30 cells transfected with sh‐circ_0096710 or sh‐NC were subcutaneously injected into mice. Volume for these tumors was checked every 7 days using calipers, with the formula: 0.5 × length × width2. Thirty‐five days later, tumor specimens were collected and weighed for further study after euthanasia of the mice.
Statistical analyses
At least three independent replicates were performed for all data. SPSS (version 17.0; SPSS Inc., Chicago, IL, USA) and GraphPad Prism version 9.0 (GraphPad Software, La Jolla, CA, USA) were used for statistical analyses. The normality of the observed data distribution was assessed using the Shapiro–Wilk test. Student's t‐test was utilized to analyze the significance of differences between two groups. One‐way and two‐way analyses of variance followed by Tukey's post hoc test were used for analyzing significant differences among more than two groups. p < 0.05 was considered to be statistically significant.
RESULTS
Circ_0096710 was overexpressed in ESCC
To evaluate the action of circ_0096710, circ_0096710 expression in ESCC samples (n = 60) was evaluated. Figure 1a,b presents the upregulation of circ_0096710 in tumor samples as compared to corresponding paraneoplastic tissues (n = 60). The diagnostic potential of circ_0096710 in ESCC was determined. Receiver operating characteristic curve showed that circ_0096710 could effectively distinguish ESCC from normal tissues, improving the efficiency and specificity of ESCC diagnosis (Figure 1c). Also, circ_0096710 was expressed at high levels in four ESCC cell lines (KYSE450, KYSE30, KYSE150, and EC109) with respect to Het‐1A (Figure 1d). Importantly, circ_0096710 expression was higher in KYSE30 and KYSE150 cells. And circ_0096710 expression was associated with tumor size, TNM stage, and lymph node metastasis (Table 2). Moreover, patients with high circ_0096710 expression showed poor survival compared to the low expression group (Figure 1e). Circ_0096710 is generated from 6 to 15 exons of the NOX4 gene and located at chr11:89106599‐89185063, possessing a mature sequence that is 982 bp in length (Figure 1f). RNase R degradation assays showed that circ_0096710 was resistant to RNase R treatment, while NOX4 was significantly degraded (Figure 1g). Collectively, circ_0096710 might be associated with ESCC progression.
FIGURE 1.
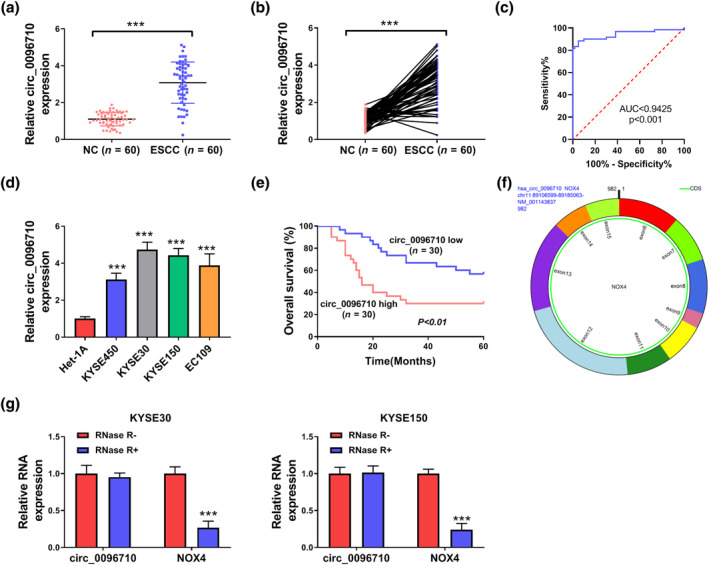
Circ_0096710 was highly expressed in ESCC tissues and cells. (a, b) The expression levels of circ_0096710 in ESCC tissue (n = 60) and normal tissue (n = 60) were detected by qRT‐PCR. (c) ROC curve was used to analyze the diagnostic potential of circ_0096710. (d) The expression levels of circ_0096710 in four ESCC cell lines (KYSE450, KYSE30, KYSE150, and EC109) and normal cells (Het‐1A). (e) Relationship between circ_0096710 expression level and survival rate of patients. (f, g) The expression levels of circ_0096710 and NOX4 were determined after treatment of RNase R by qRT‐PCR in KYSE30 and KYSE150. ***p < 0.001. ESCC, esophageal squamous cell carcinoma; qRT‐PCR, quantitative real‐time reverse transcription‐polymerase chain reaction; ROC, receiver operating characteristic.
TABLE 2.
Association between clinicopathologic features and circ_0096710 expression in 60 patients with ESCC.
| Features | circ_0096710 expression | p‐value | |
|---|---|---|---|
| High (n = 30) | Low (n = 30) | ||
| Age (years) | |||
| ≥50 | 22 | 19 | 0.405 |
| <50 | 8 | 11 | ‐ |
| Gender | |||
| Female | 17 | 20 | 0.426 |
| Male | 13 | 10 | ‐ |
| Tumor size (cm) | |||
| ≥5 | 20 | 9 | 0.004 |
| <5 | 10 | 21 | ‐ |
| TNM stage | |||
| I–II | 15 | 27 | 0.001 |
| III | 15 | 3 | ‐ |
| Differentiation | |||
| Well‐moderate | 12 | 18 | 0.121 |
| Poor | 18 | 12 | ‐ |
| Lymph node metastasis | |||
| Yes | 19 | 6 | 0.001 |
| No | 11 | 24 | ‐ |
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Circ_0096710 downregulation repressed the malignant characteristics of ESCC cells
Interference with circ_0096710 was executed to interrogate the function of circ_0096710. We observed that circ_0096710 expression was sharply decreased by sh‐circ_0096710, with sh‐circ‐_0096710#1 working better for knocking down circ_0096710 (Figure 2a). The activity and colony formation ability of these two ESCC cells were significantly suppressed by sh‐circ_0096710#1 (Figure 2b,c). Also, the migration and invasion abilities of KYSE30 and KYSE150 cells were obviously impeded after transfection with sh‐circ_0096710#1 (Figure 2d,e). Moreover, circ_0096710 interference decreased tube formation in KYSE30 and KYSE150 cells, indicating inhibition of angiogenesis (Figure 2f). Furthermore, cell sphere formation assay suggested that downregulation of circ_0096710 reduced sphere formation efficiency in KYSE30 and KYSE150 cells (Figure 2g). Finally, western blot demonstrated that sh‐circ_0096710#1 critically constrained the expression of PCNA (a proliferation marker), MMP2 (a metastasis marker), VEGFA (a pro‐angiogenic factor), and OCT4 (a cancer stem cells marker) in KYSE30 and KYSE150 cells (Figure 2h). Therefore, circ_0096710 might act as a tumor promoter in ESCC.
FIGURE 2.
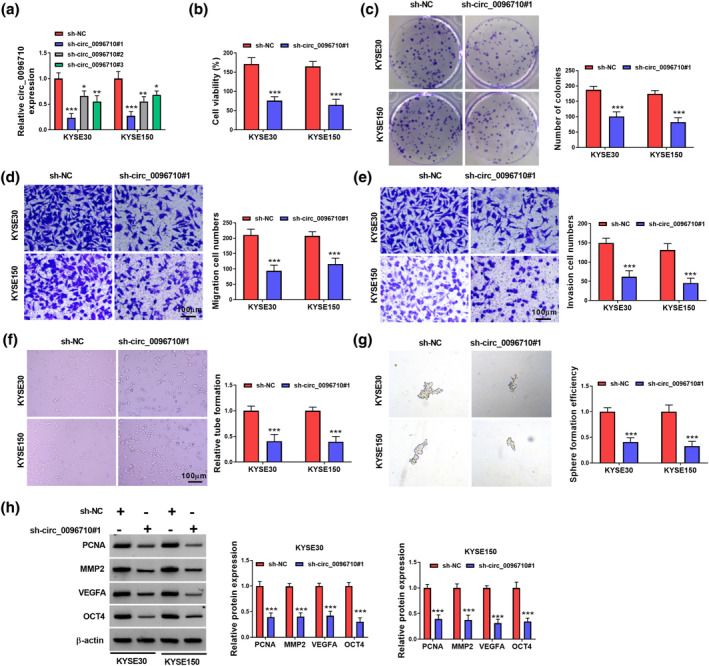
The effect of circ_0096710 knockdown on viability, migration, invasion, angiogenesis, and stem‐like properties of ESCC cells. (a) The expression efficiency of circ_0096710 knockdown. (b) The cell viabilities of KYSE30 and KYSE150 cells by transfection of sh‐NC and sh‐circ_0096710 were detected with CCK‐8 assay. (c) Cloning ability of KYSE30 and KYSE150 cells transfected with sh‐NC and sh‐circ_0096710. (d, e) Detection of migration and invasion of ESCC cells transfected with sh‐circ_0096710. (f, g) Detection of tubule‐forming ability and sphere formation efficiency of ESCC cells transfected with sh‐NC or sh‐circ_0096710. (h) The protein levels of PCNA, MMP2, VEGFA, and OCT4 were detected by western blot. ***p < 0.001. CCK‐8, Cell Counting Kit‐8; ESCC, esophageal squamous cell carcinoma.
Circ_0096710 interacted with miR‐1294
Identification of the location of circ_0096710 in KYSE30 and KYSE150 cells was made. Figure 3a displays that the majority of circ_0096‐710 is present in the cytoplasm. A total of 32 miRs with complementary binding sites to circ_0096710 were predicted using the CircInteractome database. Among them, six miRs (miR‐1294, miR‐140‐3p, miR‐153, miR‐197, miR‐409‐3p, and miR‐873) have been reported to be associated with EC, and the knockdown of circ_0096710 had the most obvious effect on miR‐1294 expression in ESCC cells (Figure S1a,b). Therefore, miR‐1294 drew our attention. The binding sites between circ_0096710 and miR‐1294 are exhibited in Figure 3b. Furthermore, miR‐1294 mimic‐transfected KYSE30 cells and KYSE150 cells showed overexpression of miR‐1294 (Figure 3c). Dual‐luciferase reporter assays showed that transfection of ESCC cells with circ_0096710‐WT and miR‐1294 co‐transfected ESCC cells resulted in significantly lower luciferase activity than that of the miR‐NC group, whereas no difference was detected in the circ_0096‐710‐MUT group (Figure 3d). A direct interaction between circ_0096710 and miR‐1294 was further verified by RIP analysis (Figure 3e). RNA pull‐down showed an enhancement of circ_0096710 enrichment following transfection of Bio‐miR‐1294 in ESCC cells (Figure 3f). Moreover, ESCC tissues and cells exhibited lower levels of miR‐1294 (Figure 3g–i). Collectively, miR‐1294 interacted with circ_0096710.
FIGURE 3.
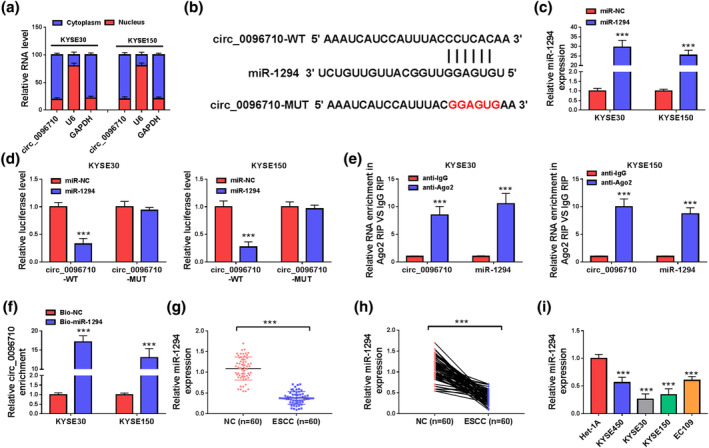
Circ_0096710 was related to the targeting of miR‐1294 in ESCC cells. (a) Circ_0096710 distribution in ESCC cells. (b) Prediction of binding sites between circ_0096710 and miR‐1294. (c) The expression level of miR‐1294 in ESCC cells transfected with miR‐1294. (d) The interaction between circ_0096710 and miR‐1294 was identified by dual‐luciferase reporter assay. (e) RNA enrichment levels of circ_0096710 and miR‐1294 were determined by RIP. (f) RNA pull‐down assays validated the interaction between circ_0096710 and miR‐1294. (g–i) qRT‐PCR was used to detect the expression level of miR‐1294 in ESCC tissues and cells. Each experiment was independently repeated at least three times. ***p < 0.001. ESCC, esophageal squamous cell carcinoma; qRT‐PCR, quantitative real‐time reverse transcription‐polymerase chain reaction; RIP, RNA immunoprecipitation assay.
Circ_0096710 exerted its role by interacting with miR‐1294
We measured miR‐1294 expression in KYSE30 and KYSE150 cells transfected with inh‐NC or inh‐miR‐1294, and miR‐1294 expression was sharply inhibited following transfection compared with inh‐NC (Figure 4a). The introduction of sh‐circ_0096710#1 + inh‐NC inhibited the activity and colony formation ability of these two ESCC cells; inh‐miR‐1294 transfection alleviated the effect of sh‐circ_0096710#1 on ESCC cells (Figure 4b,c). And the migration and invasion abilities of KYSE30 and KYSE150 cells were obviously suppressed after transfection with sh‐circ_0096710#1 + inh‐NC, while the introduction of inh‐miR‐1294 rescued the situation (Figure 4d,e). In addition, the inhibitory effects of circ_0096710 knockdown on tube formation ability and sphere formation efficiency were reversed by inhibiting miR‐1294 in KYSE30 and KYSE150 cells (Figure 4f,g). Subsequently, western blot demonstrated that sh‐circ_0096710#1 + inh‐NC remarkably suppressed the expressions of PCNA, MMP2, VEGFA, and OCT4 in ESCC cells; however, inh‐miR‐1294 co‐transfection completely reversed these changes in protein expressions (Figure 4h). Together, these results indicated that circ_0096710 facilitated ESCC cell malignant characteristics by sponging miR‐1294.
FIGURE 4.
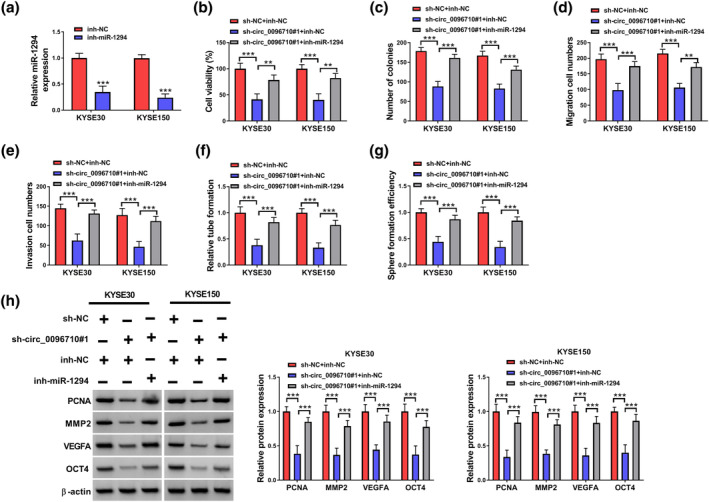
Inhibition of miR‐1294 partially reversed the impact of circ_0096710 knockdown on ESCC. (a) The expression level of miR‐1294 in ESCC cells transfected with inh‐miR‐1294 was detected. (b) CCK8 was used to detect the viability of ESCC cells transfected with sh‐NC + inh‐NC, sh‐circ_0096710#1 + inh‐NC, or sh‐circ_0096710#1 + inh‐miR‐1294. (c) Cell colony‐forming assays detected colony formation ability of ESCC cells transfected with sh‐NC + inh‐NC, sh‐circ_0096710#1 + inh‐NC, or sh‐circ_0096710#1 + inh‐miR‐1294. (d, e) Transwell assays were used to detect the cell migration and invasion ability of different transfected ESCC cells. (f, g) The detection of tubule‐forming ability and sphere formation efficiency of ESCC cells transfected with different treatments. (h) Western blot was used to detect the protein expressions of PCNA, MMP2, VEGFA, and OCT4 in ESCC cells. **p < 0.01, *** p < 0.001. ESCC, esophageal squamous cell carcinoma.
MiR‐1294 targeted ADAM10 in ESCC cells
The binding sites of ADAM10‐3′UTR and miR‐1294 were predicted by Starbase software (Figure 5a). Also, miR‐1294 could block the luciferase activity of ADAM10‐3′UTR‐WT reporter vector in ESCC cells without affecting the luciferase activity of ADAM10‐3′UTR‐MUT reporter vector (Figure 5b). Also, ADAM10 could be pulled down by Bio‐miR‐1294 but not Bio‐NC (Figure 5c). In KYSE30 and KYSE150 cells, miR‐1294 overexpression repressed the protein expression level of ADAM10, and inh‐miR‐1294 enhanced ADAM10 expression (Figure 5d). We further found that circ_0096710 knockdown remarkably suppressed ADAM10 expression but inhibition of miR‐1294 reverted the protein expression level of ADAM10 (Figure 5e). Then, we discovered that ADAM10 protein levels were greatly elevated in ESCC tissues (Figure 5f,g). IHC analysis also exhibited the upregulation of ADAM10 in ESCC tissues (Figure 5h). Furthermore, ADAM10 protein levels in four ESCC cell lines were substantially elevated as compared to Het‐1A (Figure 5i). All in all, ADAM10 was a target of miR‐1294.
FIGURE 5.
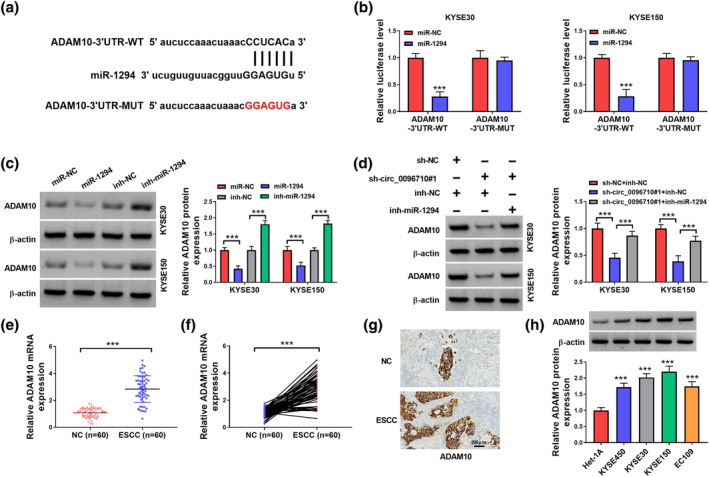
MiR‐1294 targeted ADAM10 in ESCC cells. (a) The binding sites of miR‐1294 and ADAM10 were predicted by Starbase. (b) The luciferase activities of ADAM10‐3′UTR‐WT and ADAM10‐3′UTR‐MUT were detected by dual‐luciferase reporter assay. (c) RNA pull‐down assays analyzed the enrichment of ADAM10 mRNA in the Bio‐miR‐1294 and Bio‐NC groups. (d) Effects of knockdown or overexpression of miR‐1294 on ADAM10 protein expression. (e) Protein levels of ADAM10 in ESCC cells transfected with sh‐NC + inh‐NC, sh‐circ_0096710#1 + inh‐NC, or sh‐circ_0096710#1 + inh‐miR‐1294. (f, g) mRNA expression level of ADAM10 in ESCC cells. (h) Expression level of ADAM10 in ESCC cells was detected by IHC. (i) Western blot was used to detect the protein expression level of ADAM10 in ESCC cells. Each experiment was repeated at least three times independently, ***p < 0.001. ESCC, esophageal squamous cell carcinoma.
MiR‐1294 exerted its role via repressing ADAM10 expression
As illustrated in Figure 6a, ADAM10 protein levels were higher in ADAM10‐transfected ESCC cells than in the empty vector. In KYSE30 and KYSE150 cells, miR‐1294 overexpression repressed the cell activity and cell colony formation ability, while ADAM10 introduction recovered miR‐1294‐mediated suppressive influence on ESCC cells (Figure 6b,c). Similarly, the migration and invasion abilities of KYSE30 and KYSE150 cells were obviously repressed after transfection with miR‐1294 + vector, while co‐transfection of ADAM10 recovered these situations (Figure 6d,e). In addition, miR‐1294 overexpression reduced tube formation ability and sphere formation efficiency, which could be reversed by upregulating ADAM10 in KYSE30 and KYSE150 cells (Figure 6f,g). Finally, western blot data demonstrated that miR‐1294 overexpression remarkably blocked the expressions of PCNA, MMP2, VEGFA, and OCT4 in ESCC cells, while ADAM10 co‐transfection could neutralize these effects (Figure 6h). Together, miR‐1294 exerted its anti‐cancer role in ESCC cells via targeting ADAM10.
FIGURE 6.
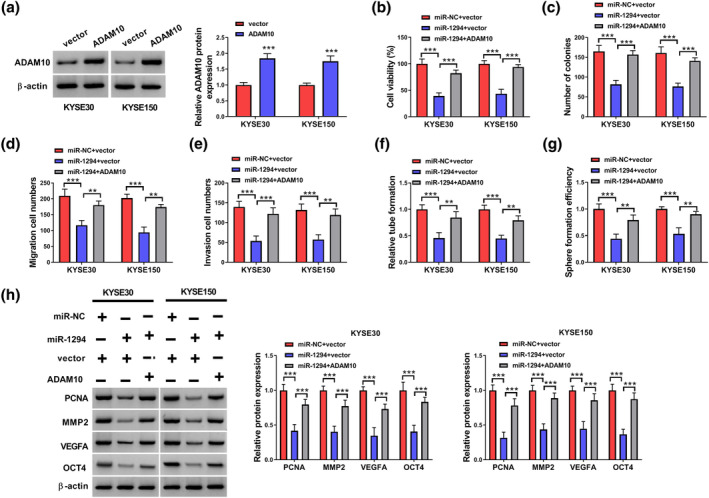
Overexpression of ADAM10 partially reversed the inhibitory effect of miR‐1294 on ESCC. (a) Expression level of ADAM10 in ESCC cells. (b) CCK8 was used to detect the viability of ESCC cells transfected with miR‐NC + vector, miR‐1294 + vector, or miR‐1294 + ADAM10. (c) Cell colony‐forming assay was used to detect colony formation ability of ESCC cells transfected with miR‐NC + vector, miR‐1294 + vector, or miR‐1294 + ADAM10. (d, e) Transwell assays were used to detect the cell migration and invasion ability of different transfected ESCC cells. (f, g) The detection of tubule‐forming ability and sphere formation efficiency of ESCC cells transfected with different treatments. (h) The protein expression levels of PCNA, MMP2, VEGFA, and OCT4 in ESCC cells were detected by western blot. **p < 0.01; ***p < 0.001. ESCC, esophageal squamous cell carcinoma.
Knocking down circ_0096710 inhibited the growth of ESCC cells in vivo
To investigate the effects of circ_0096710 on ESCC cells in vivo, xenograft assays were performed. The tumor volume and weight were hindered in the sh‐circ_0096710#1 group relative to the control (Figure 7a,b). In addition, IHC showed that Ki67 and ADAM10 protein expressions in nude mouse tumor tissues in the sh‐circ_0096710#1 group were lower (Figure 7c). Tumor samples from the sh‐circ_0096710#1 group showed lower levels of circ_0096710 and ADAM10, accompanied by a high expression of miR‐1294 (Figure 7d). Levels of related proteins (ADAM10, PCNA, MMP2, VEGFA, and OCT4) in the sh‐circ_0096710#1 group were reduced (Figure 7e). Together, circ_0096710 knockdown inhibited the growth of xenograft tumors.
FIGURE 7.
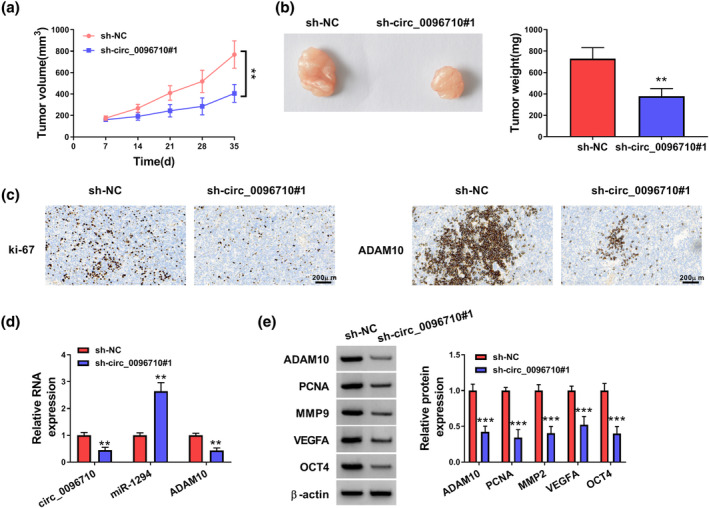
The effects of knockdown circ_0096710 on tumor growth in vivo. (a) Changes in tumor volume after subcutaneous injection of KYSE30 cells containing sh‐circ_0096710#1 or sh‐NC in immunodeficient mice. (b) Changes in tumor weight after subcutaneous injection of KYSE30 cells containing sh‐circ_0096710#1 or sh‐NC in immunodeficient mice. (c) The determination of Ki67 and ADAM10 in sh‐circ_0096710#1 group was carried out by IHC assay. (D) RNA expression levels of circ_0096710, miR‐1294, and ADAM10 in tumor tissue of nude mice in the sh‐circ_0096710#1 or the sh‐NC group. (e) Protein expression levels of ADAM10, PCNA, MMP2, VEGFA, and OCT4 in tumor tissue of nude mice in the sh‐circ_0096710#1 or the sh‐NC group. **p < 0.01; ***p < 0.001. IHC, immunohistochemistry.
DISCUSSION
EC was a malignant tumor with a very low 5‐year survival rate. Although many breakthroughs have been made in recent years, the improvement in ESCC was limited. 30 , 31 In this research, we studied the role of circ_0096710 in ESCC.
CircRNAs are characterized by stable structure, tissue‐specific and conservative evolution. 32 Previous studies have found that there were many pivotal circRNAs in ESCC. 33 , 34 , 35 For example, circ‐101 491 upregulation elevates cancer cell radioresistance in ESCC. 36 Lei et al. revealed that repression of PI3K/AKT signaling by the circPDE5A‐encoded protein PDE5A‐500aa fosters USP14‐mediated deubiquitination of PIK3IP1, therefore restraining ESCC progression. 37 Moreover, hsa_circ_0000700 knockdown inhibited the migratory capacities of ESCC cells. 38 Also, circ‐0000592 mediates FZD5 expression through interaction with miR‐155‐5p, contributing to ESCC progression. 39 In the research, we identified the effects of circ_0096710 on ESCC. Our results showed that circ_0096710 was overexpressed in ESCC samples and cell lines, and circ_0096710 expression was associated with tumor size, TNM stage, and lymph node metastasis. Unfortunately, the small clinical sample size is a limitation of this study, which may make the results more susceptible to chance. Accordingly, a larger sample size is needed to further validate the results to address the effects of selection bias. Also, ESCC patients with high levels of circ_0096710 showed a worse survival. Furthermore, circ_0096710 knockdown inhibited the proliferation, migration, invasion, angiogenesis, and stem‐like properties of ESCC cells. In addition, circ_0096710 silencing repressed tumor growth in mouse xenograft models, manifesting that circ_0096710 exerts a tumor‐promoting role in ESCC.
Given that circRNAs in the cytoplasm can regulate the expression of corresponding target genes by competing for the same miR response elements (MREs) as mRNA. 40 Here, the location of circ_0096710 in the cytoplasm was validated, implying that circ_0096710 may serve as a miR sponge. Our research identified the interaction between miR‐1294 and circ_0096710 in ESCC. Available evidence suggests an anti‐tumor effect of miR‐1294, including gastric cancer, 41 acute myeloid leukemia, 42 papillary thyroid cancer, 43 hepatocellular cancer, 44 and glioblastoma. 45 Wang et al. disclosed that downregulation of miR‐1294 mediated by circ‐STRBP increases E2F2 expression, thus contributing to gastric cancer growth. 41 In acute myeloid leukemia, circ‐FN1 binds to miR‐1294 to upregulate ARHGEF10L, thus accelerating cell invasion and proliferation. 42 Chen et al. unmasked that circ‐LDLR elevates HMGB3 expression via repressing miR‐1294, causing cancer cell invasion in papillary thyroid cancer. 43 Similarly, miR‐1294 possesses a tumor‐repressive role in ESCC. 46 , 47 , 48 Liu et al. exposed that c‐MYC upregulation mediated by miR‐1294 silencing facilitates ESCC growth. 47 Also, circ_0023984 enhances the invasive and migrated capacities of ESCC cells by repressing miR‐1294 expression. 46 In addition, circ‐ATIC represses the miR‐1294/PBX3 pathway, thus promoting cell invasion in ESCC. 48 Here, circ_0096710 interacted with miR‐1294 in ESCC cells, and miR‐1294 knockdown impaired circ_0096710 knockdown‐mediated repressive effects on ESCC cell proliferation, migration, invasion, angiogenesis, and stem‐like properties, suggesting the regulatory role of the circ_0096710/miR‐1294 axis in ESCC.
As is known, miRs are able to participate in multiple biological and pathological processes through directly interacting with target mRNAs. 49 Herein, ADAM10 was demonstrated as a target of miR‐1294. Studies have shown that ADAMs may be closely related to many human tumors. 50 , 51 ADAMs are strongly associated with gastroesophageal reflux disease and esophageal adenocarcinoma. 52 Patients with cervical cancer with human papillomavirus E6‐induced elevated ADAM10 expression possess a worse prognosis. 53 Moreover, ADAM10 was overexpressed in ESCC, and ADAM10 silencing reduced ESCC cell invasion, migration, and proliferation. 22 In our study, ADAM10 was also significantly upregulated in ESCC, and ADAM10 overexpression could impair the inhibitory effects of miR‐1294 on ESCC cell proliferation, migration, invasion, angiogenesis, and stem‐like properties. All results revealed that circ_0096710 adsorbs miR‐1294 by acting as a sponge molecule, causing the deregulation of the inhibitory effect of miR‐1294 on ADAM10 and upregulation of ADAM10, thus facilitating the proliferation, migration, invasion, angiogenesis, and stem‐like properties of ESCC cells (Figure 8). Orme et al. unearthed that ADAM10 mediates cell resistance to PD‐L1 inhibitors through cleavage of PD‐L1 on the surface of malignant cells and generation of sPD‐L1, leading to the induction of apoptosis and impairment of anti‐tumor immunity for CD8+ T cells. 54 ADAM10 is associated with the cleavage of Trop‐2, which is an activation switch for cancer growth and metastasis. 28 Therefore, we speculated that circ_0096710 may regulate ESCC immunosuppression and progression by modulating ADAM10‐mediated PD‐L1 and/or Trop‐2 cleavage. In the future, we would explore whether circ_0096710 modulates ESCC immunosuppression by regulating ADAM10‐mediated PD‐L1 and/or Trop‐2 cleavage.
FIGURE 8.
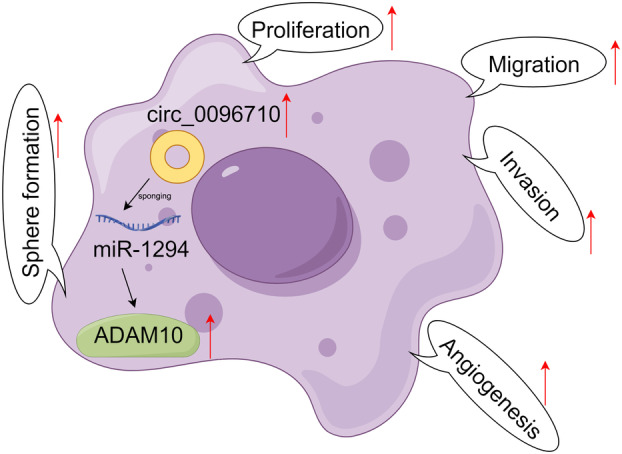
Schematic illustration of the mechanism by which the circ_0096710/miR‐1294/ADAM10 pathway mediates ESCC progression. ESCC, esophageal squamous cell carcinoma.
In brief, the present evidence revealed that circ_0096710 and ADAM10 levels were higher and miR‐1294 expression was lower in ESCC. Meanwhile, circ_0096710 knockdown represses the malignant property of ESCC cells by regulating the miR‐1294/ADAM10 axis. However, the network relationships of circRNAs are complex and other circRNAs may also play important roles in ESCC, so more studies should be performed to gain a fuller understanding of circRNAs in ESCC. We believe that these findings will draw an important stroke to attack ESCC.
AUTHOR CONTRIBUTIONS
Chaoqun Dong designed and performed the research; Zhilong Li analyzed the data; Chaoqun Dong wrote the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
Figure S1. qRT‐PCR detected the expression levels of six miRs (miR‐1294, miR‐140‐3p, miR‐153, miR‐197, miR‐409‐3p, and miR‐873) in circ_0096710‐knockdown ESCC cells.
Dong C, Li Z. Circ_0096710 facilitates tumor growth via controlling ADAM10 expression in esophageal squamous cell carcinoma. Thorac Cancer. 2025;16(1):e15483. 10.1111/1759-7714.15483
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- 1. Then EO, Lopez M, Saleem S, Gayam V, Sunkara T, Culliford A, et al. Esophageal cancer: an updated surveillance epidemiology and end results database analysis. World J Oncol. 2020;11:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020;13:1010–1021. [DOI] [PubMed] [Google Scholar]
- 3. Lander S, Lander E, Gibson MK. Esophageal cancer: overview, risk factors, and reasons for the rise. Curr Gastroenterol Rep. 2023;25:275–279. [DOI] [PubMed] [Google Scholar]
- 4. Wang Z, Li H, Li F, Su X, Zhang J, Silvestris N. Bioinformatics‐based identification of a circRNA‐miRNA‐mRNA Axis in esophageal squamous cell carcinomas. J Oncol. 2020;2020:8813800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li J, Yu H, Peng L, Li L, Wang X, Hao J, et al. Novel nomogram predicting cancer‐specific survival and overall survival in patients with primary esophageal small‐cell carcinoma: a surveillance, epidemiology, and end results‐based study. J Cancer Res Ther. 2021;17:630–637. [DOI] [PubMed] [Google Scholar]
- 6. Cortes‐Lopez M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89:527–537. [PMC free article] [PubMed] [Google Scholar]
- 7. Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong Y, Zhang L, Chang H. Regulation of exosomes‐mediated circNR4A1 on Chemoresistance and biological effects of oral squamous cell carcinoma cells. Lett Drug Des Discov. 2023;20:921–929. [Google Scholar]
- 9. Wang D, Yang S, Lyu M, Xu L, Zhong S, Yu D. Circular RNA HSDL2 promotes breast cancer progression via miR‐7978 ZNF704 axis and regulating hippo signaling pathway. Breast Cancer Res. 2024;26:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu H, Zheng Y, Wu J, Zhang R, Zhao Q, Chen S, et al. circSORBS1 inhibits lung cancer progression by sponging miR‐6779‐5p and directly binding RUFY3 mRNA. J Transl Med. 2024;22:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao SQ, Xue ST, Li WJ, Hu GS, Wu ZG, Zheng JC, et al. CircHIPK3 regulates fatty acid metabolism through miR‐637/FASN axis to promote esophageal squamous cell carcinoma. Cell Death Discov. 2024;10:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Li Z, Xu S, Li W, Chen M, Jiang M, et al. LncRNA FIRRE functions as a tumor promoter by interaction with PTBP1 to stabilize BECN1 mRNA and facilitate autophagy. Cell Death Dis. 2022;13:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Q, Yang L, Fan Y, Tang W, Sun H, Xu Z, et al. Circ‐ZDHHC5 accelerates esophageal squamous cell carcinoma progression in vitro via miR‐217/ZEB1 Axis. Front Cell Dev Biol. 2020;8:570305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng Y, Chen Y, Jiang H, Zhang H, Wang H, Xu J, et al. Circ_0058063 upregulates GLUT1 expression and promotes glucose‐uptake in esophageal squamous‐cell carcinomas. J Thorac Dis. 2020;12:925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao Y, Jia Y, Wang J, Chen X, Han J, Zhen S, et al. circNOX4 activates an inflammatory fibroblast niche to promote tumor growth and metastasis in NSCLC via FAP/IL‐6 axis. Mol Cancer. 2024;23:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X, Tao G, Huang D, Liang S, Zheng D. Circular RNA NOX4 promotes the development of colorectal cancer via the microRNA‐485‐5p/CKS1B axis. Oncol Rep. 2020;44:2009–2020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Brunetti O, Russo A, Scarpa A, Santini D, Reni M, Bittoni A, et al. MicroRNA in pancreatic adenocarcinoma: predictive/prognostic biomarkers or therapeutic targets? Oncotarget. 2015;6:23323–23341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen L, Wang C, Sun H, Wang J, Liang Y, Wang Y, et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform. 2021;22:1706–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mao Y, Wang J, Wang Y, Fu Z, Dong L, Liu J. Hypoxia induced exosomal Circ‐ZNF609 promotes pre‐metastatic niche formation and cancer progression via miR‐150‐5p/VEGFA and HuR/ZO‐1 axes in esophageal squamous cell carcinoma. Cell Death Discov. 2024;10:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mao Y, Shen J, Fang L, Zhu F, Duan S. The tumor suppressor role and ceRNA network of miR‐1294 in cancer. Oncol Res. 2023;31:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. [DOI] [PubMed] [Google Scholar]
- 22. Ma B, Zhang HY, Bai X, Wang F, Ren X‐H, Zhang L, et al. ADAM10 mediates the cell invasion and metastasis of human esophageal squamous cell carcinoma via regulation of E‐cadherin activity. Oncol Rep. 2016;35:2785–2794. [DOI] [PubMed] [Google Scholar]
- 23. Manzine PR, Ettcheto M, Cano A, Busquets O, Marcello E, Pelucchi S, et al. ADAM10 in Alzheimer's disease: pharmacological modulation by natural compounds and its role as a peripheral marker. Biomed Pharmacother. 2019;113:108661. [DOI] [PubMed] [Google Scholar]
- 24. Erin N, Akdeniz Ö. ADAM10 and Neprilysin level decreases in immune cells of mice bearing metastatic breast carcinoma: possible role in cancer inflammatory response. Int Immunopharmacol. 2024;127:111384. [DOI] [PubMed] [Google Scholar]
- 25. Gao JH, He AD, Liu LM, Zhou YJ, Guo YW, Lu M, et al. Direct interaction of platelet with tumor cell aggravates hepatocellular carcinoma metastasis by activating TLR4/ADAM10/CX3CL1 axis. Cancer Lett. 2024;585:216674. [DOI] [PubMed] [Google Scholar]
- 26. Hong Y, Wei Z, Wang Y. Proteolytic cleavage of amyloid precursor protein by ADAM10 promotes proliferation and migration via activating MAPKs pathway in tongue squamous cell carcinoma in vitro. Cell Mol Biol (Noisy‐le‐Grand). 2023;69:167–173. [DOI] [PubMed] [Google Scholar]
- 27. Cheng Y, Lin L, Li X, Lu A, Hou C, Wu Q, et al. ADAM10 is involved in the oncogenic process and chemo‐resistance of triple‐negative breast cancer via regulating Notch1 signaling pathway, CD44 and PrPc. Cancer Cell Int. 2021;21:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trerotola M, Guerra E, Ali Z, Aloisi AL, Ceci M, Simeone P, et al. Trop‐2 cleavage by ADAM10 is an activator switch for cancer growth and metastasis. Neoplasia. 2021;23:415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lombardi P, Filetti M, Falcone R, Altamura V, Paroni Sterbini F, Bria E, et al. Overview of trop‐2 in cancer: from pre‐clinical studies to future directions in clinical settings. Cancers (Basel). 2023;15:1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ilson DH, van Hillegersberg R. Management of patients with adenocarcinoma or squamous cancer of the esophagus. Gastroenterology. 2018;154:437–451. [DOI] [PubMed] [Google Scholar]
- 31. Yu J, Wang W, Yao W, Yang Z, Gao P, Liu M, et al. Gambogic acid affects ESCC progression through regulation of PI3K/AKT/mTOR signal pathway. J Cancer. 2020;11:5568–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haddad G, Lorenzen JM. Biogenesis and function of circular RNAs in health and in disease. Front Pharmacol. 2019;10:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu J, Xue N, Guo Y, Niu K, Gao L, Zhang S, et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR‐217/Wnt3 pathway. Aging (Albany NY). 2019;11:12412–12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu X, Wu D, He X, Zhao H, He Z, Lin J, et al. circGSK3β promotes metastasis in esophageal squamous cell carcinoma by augmenting β‐catenin signaling. Mol Cancer. 2019;18:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fan L, Cao Q, Liu J, Zhang J, Li B. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol Cancer. 2019;18:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin C, Huang X, Qian Y, Li J, He Y, Su H. CircRNA_101491 regulated the radiation sensitivity of esophageal squamous cell carcinomas via sponging miR‐125a‐5p. Radiat Oncol. 2024;19:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lei K, Liang R, Liang J, Lu N, Huang J, Xu K, et al. CircPDE5A‐encoded novel regulator of the PI3K/AKT pathway inhibits esophageal squamous cell carcinoma progression by promoting USP14‐mediated de‐ubiquitination of PIK3IP1. J Exp Clin Cancer Res. 2024;43:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fang J, Ji WH, Wang FZ, Xie TM, Wang L, Fu ZF, et al. Circular RNA hsa_circ_0000700 promotes cell proliferation and migration in esophageal squamous cell carcinoma by sponging miR‐1229. J Cancer. 2021;12:2610–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He J, Yu K, Liang G, Shen W, Tian H. circ_0000592 facilitates the progression of esophageal squamous cell carcinoma via miR‐155‐5p/FZD5 axis. J Biochem Mol Toxicol. 2024;38:e23742. [DOI] [PubMed] [Google Scholar]
- 40. Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18:780–788. [DOI] [PubMed] [Google Scholar]
- 41. Wang Y, Zou R, Li D, Gao X, Lu X. Exosomal circSTRBP from cancer cells facilitates gastric cancer progression via regulating miR‐1294/miR‐593‐3p/E2F2 axis. J Cell Mol Med. 2024;28:e18217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang S, Zhang BS, Yang Y, Fu LL. CircFN1 promotes acute myeloid leukemia cell proliferation and invasion but refrains apoptosis via miR‐1294/ARHGEF10L axis. Kaohsiung J Med Sci. 2024;40:221–230. [DOI] [PubMed] [Google Scholar]
- 43. Chen G, Han P, Zhang Q, Li M, Song T, Chen Z, et al. Circ_LDLR promotes the progression of papillary thyroid carcinoma by regulating miR‐1294/HMGB3 axis. J Biochem Mol Toxicol. 2023;37:e23498. [DOI] [PubMed] [Google Scholar]
- 44. Qin X, Wang S. LncASAP1‐IT1 promotes hepatocellular carcinoma progression through the regulation of the miR‐1294/TGFBR1 pathway in vitro and in vivo. J Gastrointest Oncol. 2023;14:1451–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li Y, Zheng X, Wang J, Sun M, Li D, Wang Z, et al. Exosomal circ‐AHCY promotes glioblastoma cell growth via Wnt/β‐catenin signaling pathway. Ann Clin Transl Neurol. 2023;10:865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liang W, Wang C, Wang J, Zhang M. Hsa_circ_0023984 regulates cell proliferation, migration, and invasion in esophageal squamous cancer via regulating miR‐1294/PI3K/Akt/c‐Myc pathway. Appl Biochem Biotechnol. 2022;194:1–16. [DOI] [PubMed] [Google Scholar]
- 47. Liu K, Li L, Rusidanmu A, Wang Y, Lv X. Down‐regulation of MiR‐1294 is related to dismal prognosis of patients with esophageal squamous cell carcinoma through elevating C‐MYC expression. Cell Physiol Biochem. 2015;36:100–110. [DOI] [PubMed] [Google Scholar]
- 48. Zhou Q, Lei C, Cui F, Chen H, Cao X. Circ‐ATIC regulates esophageal squamous cell carcinoma growth and metastasis through miR‐1294/PBX3 pathway. Heliyon. 2023;9:e12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Felekkis K, Touvana E, Stefanou C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236–241. [PMC free article] [PubMed] [Google Scholar]
- 50. Herrlich P, Herrlich A. ADAM metalloprotease‐released cancer biomarkers. Trends Cancer. 2017;3:482–490. [DOI] [PubMed] [Google Scholar]
- 51. Lu X, Lu D, Scully M, Kakkar V. ADAM proteins ‐ therapeutic potential in cancer. Curr Cancer Drug Targets. 2008;8:720–732. [DOI] [PubMed] [Google Scholar]
- 52. Kauttu T, Mustonen H, Vainionpaa S, Krogerus L, Ilonen I, Räsänen J, et al. Disintegrin and metalloproteinases (ADAMs) expression in gastroesophageal reflux disease and in esophageal adenocarcinoma. Clin Transl Oncol. 2017;19:58–66. [DOI] [PubMed] [Google Scholar]
- 53. Guo X, Dou Y, Liu S, du Y, Guo R, Yue Y, et al. Elevated expression of ADAM10 induced by HPV E6 influences the prognosis of cervical cancer. Genet Test Mol Biomarkers. 2023;27:165–171. [DOI] [PubMed] [Google Scholar]
- 54. Orme JJ, Jazieh KA, Xie T, Harrington S, Liu X, Ball M, et al. ADAM10 and ADAM17 cleave PD‐L1 to mediate PD‐(L)1 inhibitor resistance. Onco Targets Ther. 2020;9:1744980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. qRT‐PCR detected the expression levels of six miRs (miR‐1294, miR‐140‐3p, miR‐153, miR‐197, miR‐409‐3p, and miR‐873) in circ_0096710‐knockdown ESCC cells.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.


