Abstract
The feasibility of large-scale genome-wide association studies of complex human disorders depends on the availability of accurate and efficient genotyping methods for single nucleotide polymorphisms (SNPs). We describe a new platform of the invader assay, a biplex assay, where both alleles are interrogated in a single reaction tube. The assay was evaluated on over 50 different SNPs, with over 20 SNPs genotyped in study cohorts of over 1500 individuals. We assessed the usefulness of the new platform in high-throughput genotyping and compared its accuracy to genotyping results obtained by the traditional monoplex invader assay, TaqMan genotyping and sequencing data. We present representative data for two SNPs in different genes (CD36 and protein tyrosine phosphatase 1β) from a study cohort comprising over 1500 individuals with high or low-normal blood pressure. In this high-throughput application, the biplex invader assay is very accurate, with an error rate of <0.3% and a failure rate of 1.64%. The set-up of the assay is highly automated, facilitating the processing of large numbers of samples simultaneously. We present new analysis tools for the assignment of genotypes that further improve genotyping success. The biplex invader assay with its automated set-up and analysis offers a new efficient high-throughput genotyping platform that is suitable for association studies in large study cohorts.
INTRODUCTION
In recent years, single nucleotide polymorphisms (SNPs) have gained widespread interest as markers for association studies in humans. Large numbers of SNPs have been discovered over the past 2 years, and over 1 million sequence variants are publicly available at this point (1). Since SNPs are abundant in the human genome, these polymorphisms offer the opportunity to perform large-scale association studies to identify genes affecting complex traits (2). Major efforts are currently underway to identify genes causing hypertension, diabetes, asthma and other common disorders. In addition, the recent increased interest in haplotype analyses of the human genome will require genome-wide studies of SNPs in cases and controls to define the genetic determinants of complex human disorders (3).
Given the number of SNPs that would have to be genotyped for a large-scale or even genome-wide association or haplotype analysis, an efficient genotyping method is needed that can be easily automated to allow rapid and accurate genotyping of large numbers of samples. To date, several different methods have been developed for this purpose (4). Essentially all current methods are non-gel-based genotyping approaches, and can be grouped according to the basic principle used: allele-specific oligonucleotide ligation; allele-specific primer extension, analyzed in solution (e.g. minisequencing), on tag arrays or by mass spectrometry; allele-specific hybridization, either on solid surfaces (chip-based methods) or in solution (e.g. molecular beacons or 5′-exonuclease assay); and allele-specific cleavage reactions. For a detailed description of the different approaches, see a recent review by Kwok (4).
The Stanford, Asia, Pacific Program for Hypertension and Insulin Resistance (SAPPHIRe) was initiated as part of the Family Blood Pressure Program (FBPP) of the National Heart, Lung and Blood Institute, to identify genetic determinants of essential hypertension in populations of Chinese and Japanese origin. We have begun to systematically analyze candidate genes that are believed to be involved in blood pressure regulation in humans. By identifying SNPs in and around these genes and testing them for association with blood pressure differences in our study population, we hope to identify genetic variants contributing to essential hypertension. The FBPP combines four major networks, all of which study genetic aspects of hypertension in different ethnic groups (Asians, Caucasians and African Americans). This structure facilitates validation of initial associations found by one of the networks in other study cohorts, and requires simple and robust SNP genotyping methods that can be used to genotype over 6000 samples as part of the FBPP.
We have used the TaqMan assay (5) for genotyping SNPs in our study cohort of over 1500 individuals. Several reports have been published using the TaqMan genotyping data (6,7) and we have presented data on the efficiency and accuracy of this method for SNP genotyping (8). Genotyping accuracy was assessed for two SNPs typed on more than 1600 individuals and the error rate was estimated to be <0.05%.
Recently, we have begun to explore the invader technology (Third Wave Technologies, Madison, WI) as an alternative to the TaqMan procedure. A schematic overview of the approach is shown in Figure 1. Briefly, a flap endonuclease (cleavase) recognizes and cleaves a three-dimensional invader structure formed by hybridization of two overlapping oligonucleotides to the target sequence (9). The cleavage of one of the oligonucleotides releases a flap that initiates a secondary cleavage reaction with a fluorescence resonance energy transfer (FRET) label. Both alleles are interrogated using different FRET labels. The fluorescent signals generated are detected at an arbitrary end time point with a traditional fluorescence plate reader. The method can be used with PCR products (10) or genomic DNA as template for the reaction (9,11).
Figure 1.
Schematic of the invader assay. During the primary phase, an Invader® oligo and a primary probe are annealed to target DNA, overlapping at the SNP position (indicated in upper case letters). The black arrow indicates the site of cleavage by the cleavase enzyme. The released 5′ flap anneals to the FRET cassette during the secondary phase and initiates a second cleavage reaction that releases the fluorescent dye. The signal is only released when the invasive structure is formed on the target DNA (‘Perfect Match’, left reaction). If the primary probe does not match the nucleotide at the SNP position, cleavase will not act (reaction on right).
Mein et al. (10) described the use of a monoplex invader assay for SNP genotyping with PCR products as target. In this assay, a target sequence is amplified by PCR from small amounts of genomic DNA and the two alleles of the SNP are interrogated in two separate invader reactions. The authors compared results obtained with invader assays for 36 SNPs with genotyping results for the same SNPs using alternative approaches (e.g. PCR–RFLP). They evaluated the design, the robustness and the success rate of several SNP assays. In their studies, they determined an average failure rate of 2.3%, largely due to PCR failure, and a 99.2% accuracy of invader genotypes when compared to genotypes obtained with other established approaches. However, Mein et al. (10) optimized each assay individually rather than ran them under standard conditions, an approach not feasible for high-throughput applications. Furthermore, the assays were only evaluated on a small set of samples, which did not permit the evaluation of its usefulness for larger studies.
It was the purpose of our study to investigate a new improved invader method for genotyping large numbers of samples and to evaluate accuracy as well as the possibility for automation. To this extent, we describe the use of automation tools for both set-up and analysis of the new biplex invader assays that facilitate high-throughput genotyping and compare representative genotyping results with those obtained from our TaqMan genotyping, traditional monoplex invader assays and sequencing results. Our data verify that the invader assay in its single tube biplex format represents an efficient new platform for high-throughput genotyping of SNPs that is highly accurate and can be run efficiently in a semi-automated set-up.
MATERIALS AND METHODS
SNP discovery
Sequence-tagged sites (STSs) were designed as previously described (12) to cover exons and flanking intronic sequence of the genes CD36 and PTPN1. STSs were 300–600 bp in size. PCR was initially tested under standard conditions using human genomic DNA. Amplification products were separated by agarose gel electrophoresis. Primer pairs that resulted in a single amplification product of the expected size were used for subsequent amplification reactions. The amplification products from 24 individual SAPPHIRe DNA samples were sequenced using Big Dye Terminator chemistry (Applied Biosystems, Foster City, CA) and forward or reverse primer in separate reactions. The resulting sequencing traces were analyzed using phredPhrap (13) and Polyphred (14) to identify SNPs. SNPs with a minor allele frequency of >10% in these 24 samples were used for subsequent genotyping.
Genotyping assays
Invader assays were designed using the InvaderCreator software (Third Wave Technologies). Probe designs are listed in Table 1A. All assays were designed to be run at the same incubation temperature (65°C for biplex assays, 63°C for monoplex assays). Similarly, TaqMan probes were designed as described previously (7). TaqMan probe designs are listed in Table 1B.
Table 1. Invader assay and TaqMan designs used in the study.
| SNP | Assay type | Oligonucleotide type | Sequence |
|---|---|---|---|
| A | |||
| CD36 | Monoplex | Invader | CCAATGATTAGACGAATTGATTCTTTCTGTGACTCATCAGTTCT |
| Primary probe 1 | CGCGCCGAGGATTTCCTGTAAAATTCATGTCTTG | ||
| Primary probe 2 | CGCGCCGAGGCTTTCCTGTAAAATTCATGTCTT | ||
| Biplex | Invader | CCAATGATTAGACGAATTGATTCTTTCTGTGACTCATCAGTTCT | |
| Primary probe 1 | ATGACGTGGCAGACATTTCCTGTAAAATTCATGTCTTGC | ||
| Primary probe 2 | CGCGCCGAGGCTTTCCTGTAAAATTCATGTCTTG | ||
| PTP03 | Monoplex | Invader | CGAGGACCTGGAGCCCCCACCA |
| Primary probe 1 | CGCGCCGAGGCGAGCATATCCCCCCA | ||
| Primary probe 2 | CGCGCCGAGGTGAGCATATCCCCCCAC | ||
| Biplex | Invader | ACGAGGACCTGGAGCCCCCACCA | |
| Primary probe 1 | ATGACGTGGCAGACCGAGCATATCCCCCCA | ||
| Primary probe 2 | CGCGCCGAGGTGAGCATATCCCCCC | ||
| B | |||
| CD36 | Forward primer | CAGATAGCTTTCCAATGATTAGACGAA | |
| Reverse primer | CCTTATTCACAAATCAACAGCAAGAC | ||
| Probe 1 | 6FAM-TCATCAGTTCATTTCCTGTAAAATT-TAMRA | ||
| Probe 2 | VIC-TCATCAGTTCCTTTCCTGTAAAAT-TAMRA | ||
| PTP03 | Forward primer | GAGCTTTCCCACGAGGACCT | |
| Reverse primer | AAGAACTCCCTGCATTTCCCA | ||
| Probe 1 | 6FAM-AGCCCCCACCCGAGCATATCCC-TAMRA | ||
| Probe 2 | VIC-AGCCCCCACCTGAGCATATCCCC-TAMRA |
Biplex invader assay. PCR amplicons were diluted 1:20 with H2O. Invader reactions (6 µl) were set up with the following final concentrations: 4 pmol each primary probe, 0.4 pmol invader probe and 7.5 mM MgCl2. These reagents were added to a drydown plate containing cleavase VIII enzyme, FRET probes, 4% PEG, 5 mM MOPS, pH 7.5, 2% glycerol, 0.01 mM EDTA, 0.03% NP-40, 0.03% Tween-20, 36 ng BSA and 150 ng tRNA. Reaction plates were sealed, denatured for 5 min at 95°C and then incubated for 20 min at 65°C using a PE9700 thermocycler (PE Biosystems, Foster City, CA). Plates were read using a Cytofluor 4000 fluorescent plate reader (PE Biosystems), first at 485 nm excitation and 530 nm emission and a second time at 560 nm excitation and 620 nm emission.
Monoplex invader assay. Again, PCR amplicons were diluted 1:20 with H2O. Invader reactions (10 µl) included 8 pmol primary probe, 0.8 pmol invader probe, 5 mM MgCl2 and 4.4 mM MOPS pH 7.5. Plates were incubated for 20 min at 63°C and subsequently read at 485 nm excitation and 530 nm emission.
Cluster algorithms
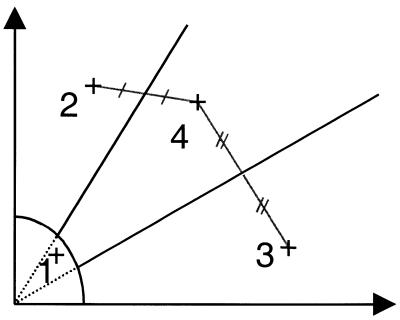
The approach for the CA clustering algorithm is described in detail in the Supplementary Material. In short, the algorithm defines initial centers of gravity for data points falling into four areas (corresponding to the approximate locations of the four clusters; see Fig. 2) and then iteratively determines cluster centroids and probabilities for each data point cluster membership, based on standard deviations of the data point distribution around each centroid.
Figure 2.
Sector definition for CA clustering algorithm. The initial four cluster areas defined by the heuristic part of the clustering algorithm. The crosses denote the centers of gravity of the clusters (corresponding to areas of high density). Lines connecting the centers indicate how the midpoints are determined. The midpoints connect to the origin to define the three initial sectors.
For our k-means clustering, we used the algorithm provided in the S-plus v.3.5 software package (Insightful Corporation, Seattle, WA).
For both algorithms, we used the compiled data from the raw data files obtained directly from the fluorescence plate reader without any prior data modification or normalization.
RESULTS
In this study, we evaluated the usefulness and accuracy of a new invader method, the biplex assay. In this new assay format, the two alleles of a SNP are interrogated in a single reaction tube using two different fluorescent dyes to identify the cleavage products for each allele. Genotyping results were compared with data obtained using the established monoplex PCR invader assay. In contrast to the biplex assay format, this approach investigates the two alleles of a SNP in two separate reactions (10). Here, we present data for SNP genotyping performed as part of several collaborative studies aimed at identifying underlying genetic factors influencing human hypertension, lipid metabolism or obesity. Genotyping data obtained using different invader methods are analyzed in detail for two representative SNPs identified by our group as part of our hypertension study. Results from these invader assays were compared with results that we obtained previously using the same samples and the same SNPs in TaqMan assays.
SNP discovery
The two SNPs described in detail in this paper as well as others were discovered as part of SAPPHIRe. SNPs were identified through direct sequencing of PCR products covering exons and the flanking intronic sequence of candidate genes from 16 unrelated individuals with hypertension and eight individuals with low-normal blood pressure from our SAPPHIRe study cohort. The first SNP, CD36, is located in exon 14 past the stop codon in the gene of the same name and the second SNP, PTP03, is a synonymous change in exon 8 of the gene for protein tyrosine phosphatase 1β (PTPN1).
All SNPs were genotyped for 1593 samples of the SAPPHIRe study cohort using both invader assay formats and TaqMan genotyping. An additional 23 SNPs were genotyped on the same cohort or cohorts of similar size using the biplex invader assay platform only. Furthermore, an additional 34 SNPs were genotyped on a small set of 90 DNA samples using the biplex platform, six of these SNPs with each sample in duplicate. The results for the additional assays confirm the representative results shown here for two assays.
Biplex invader SNP genotyping
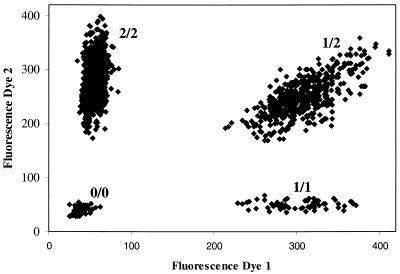
Genotyping for all invader assays was performed in 384-well plates. Assays were set up using a Biomek2000 robotics system (Beckman, Palo Alto, CA). After incubation, assay plates were analyzed using a fluorescence plate reader and fluorescence data were automatically transferred and analyzed using two different clustering algorithms once the entire sample set was genotyped. Samples genotyped in duplicate were analyzed individually to assess PCR failure and consistency for each assay. A scatter plot of representative data for SNP PTP03 is shown in Figure 3.
Figure 3.
Scatter plot of fluorescence data from biplex invader assay for PTP03. Raw fluorescence data are plotted for each sample. The x-axis depicts the fluorescence intensity for dye 1, corresponding to SNP allele 1, while the y-axis indicates the fluorescence intensity for the second dye, corresponding to the alternative SNP allele. Four clusters can be identified, one consisting primarily of no-DNA control samples and samples that failed to generate signal (0/0) and the remaining three clusters indicating the three possible genotypes (homozygous for allele 1, 1/1; homozygous for allele 2, 2/2; heterozygous, 1/2).
Evaluation of biplex invader assay performance
In total, 57 SNPs were analyzed for a panel of 90 independent DNA samples. For all assays, the average failure rate was 1.64%. Six additional SNPs were typed in duplicate for the same 90 DNA samples. Here, six samples failed for both duplicates. For an additional nine DNA samples, one of the duplicate calls failed while the other assay was successful. Of the remaining 525 samples (1050 genotypes), no discrepant calls were observed and the duplicate genotype calls agreed.
To assess the failure rate of the biplex invader assay in using samples from study cohorts, 23 different SNPs were genotyped in study cohorts similar to the SAPPHIRe cohort. A total of 23 940 genotypes were performed. Of these, 838 reactions failed (3.50%). In comparison, 2016 genotypes performed in duplicate for four SNPs using the monoplex invader assay resulted in a failure rate of 4.37%.
Comparison of biplex invader assay with monoplex invader format
A total of 1630 genotype calls were compared for two different invader assays. Only 13 discrepancies (0.80%) were found between the biplex genotype calls and the monoplex assay results (Table 2A). All samples resulting in discrepant results were subsequently sequenced and the genotype for each sample was determined using the automated software tools phredPhrap (13) and Polyphred (14). For three samples, sequencing confirmed the biplex results, for eight other samples the monoplex assay gave the correct genotype. Two samples could not be resequenced. For six of the 13 discrepancies, one of the two assays correctly identified the sample as heterozygous while the other method failed to do so.
Table 2. Discrepancies in genotype calls between the different types of genotyping assays used.
| Discrepant call | Monoplex correct | TaqMan correct | Biplex correct | Inconclusive | |
|---|---|---|---|---|---|
| A Comparison of monoplex and biplex invader assays | |||||
| CD36 | 9 | 7 | 1 | 1 | |
| PTP03 | 4 | 1 | 2 | 1 | |
| B Comparison of biplex invader assay and TaqMan | |||||
| CD36 | 6 | 6 | 0 | 0 | |
| PTP03 | 4 | 2 | 2 | 0 | |
| C Comparison of TaqMan and monoplex invader assay | |||||
| CD36 | 2 | 1 | 1 | 0 | |
| PTP03 | 5 | 1 | 3 | 1 |
Comparison of cluster analysis methods
All genotype data were analyzed independently using two different cluster algorithms. The k-means clustering algorithm is based on nearest-centroid sorting where data are assigned to a predetermined number of clusters or groups by iteratively determining the cluster centroid nearest to a given data point. The CA algorithm similarly assigns individual data points to a predetermined number of clusters, but initially defines the cluster centroids through a heuristic approach and then reassigns cluster memberships using these initial centroid coordinates, based on standard deviations of the distribution of data points around each centroid.
In our dataset, duplicates of each sample were analyzed as separate genotypes. The results are summarized in Table 3. For the biplex invader assay data, the CA algorithm was able to assign more samples to one of the three genotype clusters, leaving a smaller number of samples in the NTC cluster (NTC = no target control/no reaction cluster). Here, only 31 of 1800 samples (1.72%) could not be assigned to a cluster (both duplicates were not assigned) and only one of the duplicates was assigned to a cluster for another 17 samples (0.94%), while the other duplicate fell into the NTC cluster. In contrast, the k-means algorithm (KM) failed to assign 51 of the 1800 samples (2.83%) to a cluster (both duplicates failed). Furthermore, the KM gave discrepant cluster assignments for one sample, while the duplicate calls agreed for all assignments made by CA. The results for the monoplex invader assays were similar (Table 3). Only 33 of 3186 samples (1.04%) could not be assigned to a cluster (both duplicates were not assigned) and only one of the duplicates was assigned to a cluster for another two samples (0.06%), while the other duplicate fell into the NTC cluster. Similarly, the KM failed to assign 39 of the 3186 samples (1.22%) to a cluster (both duplicates failed), but again one sample resulted in discrepant cluster assignments.
Table 3. Comparison of k-means and CA cluster analysis results.
| Number of samples | CA cluster analysis | k-means | ||||||
|---|---|---|---|---|---|---|---|---|
| NTC | NTC/call | Discrepant | NTC | NTC/call | Discrepant | |||
| PTP03 | Monoplex | 1593 | 10 | 1 | 0 | 11 | 0 | 0 |
| Biplex | 900 | 20 | 3 | 0 | 25 | 3 | 1 | |
| CD36 | Monoplex | 1593 | 23 | 1 | 0 | 28 | 0 | 1 |
| Biplex | 900 | 11 | 14 | 0 | 26 | 0 | 0 | |
| Total | Monoplex | 3186 | 33 | 2 | 0 | 39 | 0 | 1 |
| Biplex | 1800 | 31 | 17 | 0 | 51 | 3 | 1 | |
Overall, there is a significant difference between the number of failed genotype calls and discrepant calls made by the two algorithms (paired Student’s t-test, P < 0.05). This difference is even more pronounced for data from SNP assays that do not result in perfectly separated clusters: here, 31.6% of the genotypes assigned to the failed cluster by the KM algorithm were successfully assigned to one of the other clusters by CA (data not shown). Furthermore, while k-means clustering resulted in 2.6% of discrepant calls between duplicate genotypes, CA did not result in any discrepant calls (data not shown).
Finally, we compared the actual cluster assignments between the two different algorithms. Of all samples successfully assigned to one of the three genotypes after initial comparison of the duplicate calls, only one genotype disagreed for the biplex data (0.04%) and only five genotypes disagreed for the monoplex assay (0.1%).
Comparison with TaqMan assay results
Genotypes from the TaqMan assays were assigned as described previously (7). Genotype calls were compared with the genotypes obtained from the duplicate calls in the invader assays. The comparison of the biplex invader assays with genotypes determined by the TaqMan assay are summarized in Table 2B; the comparison of the TaqMan genotypes with the monoplex results are shown in Table 2C. Again, only very few discrepancies were detected between the genotype calls. TaqMan and biplex invader disagreed for only 12 samples (0.32%) and the monoplex assay results disagreed with the TaqMan genotypes for only nine samples (0.28%).
DISCUSSION
Over the past few years, several new methods have been developed that promise to improve on existing methods of SNP genotyping. Given the large number of SNPs publicly available at this time and the growing interest in using these sequence variants for large-scale genome-wide linkage disequilibrium and haplotype studies, the method needs to be highly accurate, since false genotyping results may seriously impact on the success of an association analysis, and the genotyping procedure needs to be automated to allow for the high throughput needed to type thousands of SNPs on hundreds or thousands of samples. It was the purpose of our study to assess these requirements for invader assays, a recently developed new method for SNP genotyping. Although other studies have previously evaluated invader assays, one of the primary concerns has always been that the invader assay investigated the two alleles of a given SNP in separate invader reactions (3). These concerns have been addressed, and our study presents a new biplex invader platform for SNP genotyping that has been optimized for use with a standard reagent plate under uniform conditions regardless of the SNP that is being assayed. This eliminates the time-consuming optimization for each individual SNP assay, and thus increases throughput.
The set-up and preparation of invader assays have been simplified dramatically. A 384-well drydown reaction plate contains all generic reagents for the assays (enzyme, buffers and FRET probe). Target DNA (PCR product or genomic DNA) and three unlabeled oligonucleotides (invader probe and primary probe for each of the two alleles of the SNP) are added to the plate by a pipetting robot. We used PCR amplicons as targets in our reactions, and a 10 µl PCR reaction volume is usually sufficient for dozens of invader reactions. Since we design our PCR primers to be used under standard reaction conditions (12), both the PCR set-up as well as the invader reaction set-up can be completely automated. Given the short assay time for our invader reactions (20 min), a large number of samples can easily be genotyped in 1 day using a robotics platform for PCR and invader assay set-up. In our laboratory, one technician routinely performs up to 10 000 genotypes per day using only a single pipetting robot and standard laboratory equipment (PCR machines and a fluorometer). Additional robotics tools and individuals can significantly increase this number.
In order to minimize human error in the analysis of assay data and in the genotype assignment, we developed tools to automatically transfer raw data from the fluorescence reader and analyze them using clustering algorithms. Initially, we performed the analysis using traditional k-means clustering software. However, we quickly realized that the KM algorithm often placed two centroids within one group of data that would be assigned manually to a single cluster. This is particularly apparent when one of the homozygote clusters had only very few data points and when there was no sharp distinction between one of the genotype clusters and the no-reaction cluster. For every k-means analysis, the coordinates for the centroids of each cluster had to be examined manually. This problem arises from the fact that the algorithm does not use any method to initially define the approximate location of the centroids of the four clusters. Rather, the algorithm iteratively reduces the number of clusters based exclusively on the distance between the centroids and a given data point until the predetermined number of clusters remains. To improve on this clustering, we developed a new algorithm that initially examines all data points and determines the approximate location of the centroid for each cluster by dividing the space into sections for each expected cluster. Since we always expect four clusters for our genotyping (we only use SNPs with an estimated minor allele frequency of >10% and we type a large number of samples), this heuristic approach to predetermining the approximate area of each cluster eliminates the problems seen with k-means clustering. Nevertheless, the algorithm can be adjusted to fit data of low frequency SNPs or small sample sizes where less than four clusters are present. The CA algorithm is very quick and the analysis of SNP genotyping data of 2000 or more individual samples requires only seconds.
In our analysis of the genotyping data, we found that CA clustering assigns more genotypes successfully to one of three clusters, while k-means assigns these to the no-reaction cluster. This is particularly apparent in datasets where the individual clusters and the no-reaction cluster are not clearly separated, due to technical problems in the assay or large variation in the amount of PCR template generated from different DNA samples. Over 30% of the genotypes assigned to the no-reaction cluster by k-means clustering can be assigned to one of the three genotypes using the CA algorithm. Thus, the CA algorithm is able to assign significantly more samples to the correct genotype and eliminates all discrepancies between duplicate genotypes in our study.
Previous studies have assessed the overall error rate by using duplicate genotypes (blind samples) and determine the number of discrepancies. In our study, we did not detect any discrepancies between the blind duplicates (55 samples) and no discrepancies were detected between the duplicates for each sample in the assay. Therefore, we decided to use an established genotyping method as standard for our comparisons. Our group has used the TaqMan method extensively in the past, and the error rate in genotype assignment has been estimated to be <0.05% (8). We therefore used genotype calls obtained by TaqMan and compared them with the genotype calls obtained by either monoplex or biplex invader assay for the same samples and the same SNPs. The results, summarized in Table 2, clearly illustrate that all three approaches are equally accurate. The results differ for <0.3% of all samples genotyped. Furthermore, several of the discrepancies can be explained by the failure of both the invader and the TaqMan assay to accurately detect heterozygotes. For the invader assay, this failure is not limited to the monoplex assay, but can be found for both the monoplex and the biplex invader assays. Therefore, it is conceivable that the remaining discrepancies can be explained by a failure of the TaqMan assay to detect both alleles, since in all cases the invader assays assign the samples to the heterozygous cluster, while the TaqMan results suggest that these samples are homozygous for one allele. The lack of an increased number of these types of discrepancies with the monoplex invader assay also supports earlier claims that there is no increased genotyping error due to the monoplex format (3).
Given the large number of samples typed in this study, we believe that our estimate of error inherent in SNP genotyping using the invader platform is more accurate than previously published estimates. Mein et al. (10) reported an average failure rate of 2.3% and an error rate of 0.8%. In our analysis, we determined an overall average failure rate for biplex assays of 1.64% using a small set of DNA samples and a failure rate of 3.5% in genotyping of large study cohorts (4.37% for monoplex invader assays). Over 60% of the observed failures can be explained by failed PCRs when the PCR products are quantified prior to the invader reaction (data not shown). However, in our comparisons, we found a lower error rate of 0.3% for biplex assays (0.13% for monoplex assays). This reduced error rate when compared with previous studies may be explained by the use of a drydown reagent plate for our studies, while previous assays were run using traditional reagents.
From our study, it is obvious that the biplex invader assay offers a high-throughput SNP genotyping platform that can be easily automated (for both set-up and analysis). The assay is highly accurate and a very simple and user-friendly alternative to existing SNP genotyping methods since standard reaction conditions are used. However, even the biplex invader assay in its current form still does not permit a genome-wide screen with an estimated 300 000 SNPs on a large number of samples. This can only be achieved when the requirement for PCR amplification can be eliminated and the overall reaction volume can be decreased, thus lowering the cost of the assay significantly. While the invader assay also works reliably using genomic DNA as template (11), the required DNA amount needs to be reduced to <1 ng/genotype before a genome-wide analysis using SNPs is feasible for most clinical studies (8). Despite these limitations, the biplex invader assay clearly offers an alternative to other currently available SNP genotyping methods and can be used efficiently with standard laboratory equipment, thus eliminating the need for costly investments prior to genotyping.
Recently, other novel genotyping approaches such as methods based on allele-specific PCR (15) have been proposed as another alternative to existing SNP genotyping methods discussed in this paper. However, these approaches, while facilitating assay set-up, also require an expensive PCR step. Thorough testing and comparison of these novel approaches under high-throughput operation to other existing methods will allow identification of the approaches most suited and cost-efficient for genome-wide SNP assocation analyses.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank V. Bustos, T. Lee, I. Moreno, K. Sheppard and D. Zierten for help with sequencing, and X. Liu, A. Indap, N. Vo and D. Flowers for computer assistance at the Stanford Human Genome Center. Thanks are due to B. Gau (Human and Molecular Genetics Center, Medical College of Wisconsin) for generating additional genotyping data to assess the biplex assay failure rate. We thank all participants in the SAPPHIRe study for their support. This paper is written on behalf of members of the Stanford, Asia, Pacific Program for Hypertension and Insulin Resistance (SAPPHIRe). This work was funded by a grant from the Family Blood Pressure Program of the National Heart, Lung and Blood Institute, National Institutes of Health.
REFERENCES
- 1.Sachidanandam R., Weissman,D., Schmidt,S.C., Kakol,J.M., Stein,L.D., Marth,G., Sherry,S., Mullikin,J.C., Mortimore,B.J., Willey,D.L. et al. (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 2.Risch N. and Merikangas,K. (1996) The future of genetic studies of complex human diseases. Science, 273, 1516–1517. [DOI] [PubMed] [Google Scholar]
- 3.Kwok P.-Y. (2001) Genetic association by whole-genome analysis? Science, 294, 1669–1670. [DOI] [PubMed] [Google Scholar]
- 4.Kwok P.-Y. (2000) High-throughput genotyping assay approaches. Pharmacogenomics, 1, 95–100. [DOI] [PubMed] [Google Scholar]
- 5.Livak K.J., Marmaro,J. and Todd,J.A. (1995) Towards fully automated genome-wide polymorphism screening. Nature Genet., 9, 341–342. [DOI] [PubMed] [Google Scholar]
- 6.Ranade K., Hsiung,A.C., Wu,K.D., Chang,M.S., Chen,Y.T., Hebert,J., Chen,Y.I., Olshen,R., Curb,D., Dzau,V. et al. (2000) Lack of evidence for an association between alpha-adducin and blood pressure regulation in Asian populations. Am. J. Hypertens., 13, 704–709. [DOI] [PubMed] [Google Scholar]
- 7.Province M.A., Boerwinkle,E., Chakravarti,A., Cooper,R., Fornage,M., Leppert,M., Risch,N and Ranade,K. (2000) Lack of association of the angiotensinogen-6 polymorphism with blood pressure levels in the comprehensive NHLBI Family Blood Pressure Program, National Heart, Lung and Blood Institute. J. Hypertens., 18, 867–876. [DOI] [PubMed] [Google Scholar]
- 8.Ranade K., Chang,M.S., Ting,C.T., Pei,D., Hsiao,C.F., Olivier,M., Pesich,R., Hebert,J., ChenY.I., Dzau,V.J. et al. (2001) High-throughput genotyping with single nucleotide polymorphisms. Genome Res., 11, 1262–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyamichev V., Mast,A.L., Hall,J.G., Prudent,J.R., Kaiser,M.W., Takova,T., Kwiatkowski,R.W., Sander,T.J., de Arruda,M., Arco,D.A. et al. (1999) Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol., 17, 292–296. [DOI] [PubMed] [Google Scholar]
- 10.Mein C.A., Barratt,B.J., Dunn,M., Siegmund,T., Smith,A.N., Esposito,L., Nuland,S., Stevens,H.E., Wilson,A.J., Phillips,M.S. et al. (2000) Evaluation of single nucleotide polymorphism typing with invader on PCR amplicons and its automation. Genome Res., 10, 330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan D., Nuce,B. and Arvan,D. (1999) Non-PCR-dependent detection of the factor V leiden mutation from genomic DNA using a homogeneous invader microtiter plate assay. Mol. Diagn., 4, 135–144. [DOI] [PubMed] [Google Scholar]
- 12.Beasley E.M., Myers,R.M., Cox,D.R. and Lazzeroni,L.C. (1999) Statistical refinement of primer design parameters. In Innis,M., Gelfand,D. and Sninsky,J. (eds), PCR Applications: Protocols for Functional Genomics. Academic Press, San Diego, CA, pp. 55–71.
- 13.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 14.Nickerson D.A., Tobe,V.O. and Taylor,S.L. (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res., 25, 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myakishev M.V., Khripin,Y., Hu,S. and Hamer,D.H. (2001) High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res., 11, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.