Abstract
Two ferrocene-labelled analogues of dTTP, 5-(3-ferrocenecarboxamidopropenyl-1) 2′-deoxyuridine 5′-triphosphate (Fc1-dUTP) and 5-(3-ferroceneacet-amidopropenyl-1) 2′-deoxyuridine 5′-triphosphate (Fc2-dUTP) have been produced to demonstrate the incorporation of redox labels into DNA by polymerases. Cyclic voltammetry indicates that the ferrocenyl moieties display reversible redox behaviour in aqueous buffer with E1/2 values of 398 (Fc1-dUTP) and 260 mV (Fc2-dUTP) versus Ag/AgCl. Primer extension by the proofreading enzymes Klenow fragment and T4 DNA polymerase shows that Fc1-dUTP is efficiently incorporated into DNA during synthesis, including incorporation of two successive modified nucleotides. Production of a 998 bp amplicon by Tth DNA polymerase demonstrates that Fc1-dUTP is also a satisfactory substrate for PCR. Despite its structural similarity, Fc2-dUTP acts predominantly as a terminator with the polymerases employed here. UV melting analysis of a 37mer duplex containing five Fc1-dU residues reveals that the labelled nucleotide introduces only a modest helix destabilisation, with Tm = 71 versus 75°C for the corresponding natural construct. Modified DNA is detected at femtomole levels using a HPLC system with a coulometric detector. The availability of simple and effective enzymatic labelling strategies should promote the further development of electrochemical detection in nucleic acid analysis.
INTRODUCTION
Incorporation of reporter groups into nucleic acids plays a key role in most methods of genetic identification and analysis. Nucleic acid targets or hybridization probes may be tagged with reporters during chemical oligonucleotide synthesis, by enzymatic incorporation of labelled nucleotide substrates or by post-synthesis derivatization. Each type of reporter group and labelling approach has its relative merits for particular applications. For example, radioactive tags such as 32P have been successfully used for many years and continue to be widely employed, but radioactive methods suffer from potential health risks and attendant regulatory complications, relatively short isotope half-lives, radiolysis and generally cumbersome detection formats.
In response to such issues, application of non-isotopic labelling methods to nucleic acids has grown dramatically in the 20 years since Ward and co-workers introduced biotin-labelled UTP and dUTP. In these constructs, attachment of biotin to the pyrimidine C5 position via an allylamine linker allowed efficient incorporation into DNA and RNA by polymerases (1,2). The strong biotin–avidin interaction and an avidin–enzyme conjugate were subsequently employed to enable colorimetric detection. These and other biotinylated nucleotides are now in widespread use, along with alternative hapten-labelled nucleotides and antibody–enzyme conjugates such as the digoxigenin and dinitrophenol systems. Fluorescent tagging, with its advantages of high sensitivity and multicolour detection, has quickly come to dominate applications in nucleic acid sequencing and microarray expression analysis (3,4). A broad variety of fluorescent-tagged NTPs, dNTPs and ddNTPs are commercially available. Other competing technologies are under active development, for example, detection of nucleic acid mass tags by mass spectrometry has experienced a dramatic recent upsurge (5).
Electrochemical detection (ECD) offers a promising alternative to other approaches: it can be highly sensitive, rapid and amenable to inexpensive production in miniaturised (‘lab-on-a-chip’) formats. Several different ECD implementations are currently under development for nucleic acid analysis. In one approach, unlabelled nucleic acids are detected with attomole sensitivity by transition metal complex-mediated oxidation of G nucleobases at potentials around 1.1 V versus SHE (6). It is likely that this approach can be extended to the detection of A, C and T/U by labelling with nucleobase analogues possessing lower redox potentials than the natural species (7). In another method, the electrocatalytic signal from an intercalated dye is monitored to determine DNA duplex integrity (8). In a third, the ability of a naphthalene diimide derivative of ferrocene to preferentially bind double-stranded (ds)DNA via intercalation is used to detect hybridization (9).
Most other electrochemical applications are based upon introducing one or more copies of a conjugated redox label, typically metal complexes, metallocenes or quinones (10,11). Due to its stability, ready synthetic access and ease of redox tuning, labelling with ferrocene (Fc) has been the focus of significant attention. In early demonstrations of Fc redox tagging, 5′-aminohexyl oligonucleotides were chemically conjugated with carboxyl derivatives of Fc to enable ECD of hybridization (12,13) and PCR amplicons (14) at femtomole levels. Similarly, 5′-Fc nucleotides were immobilised at their 3′-ends for characterisation of surface monolayers (15). Internal post-synthetic labelling of DNA probes has been obtained by reaction with ferrocenecarboxaldehyde or aminoferrocene (16). For incorporation during chemical oligonucleotide synthesis, Fc phosphoramidites (17) and monomers with a ferrocenyl moiety linked to position 5 of 2′-dU (18) or the 2′-sugar position of dA and dC (19) have been described, as has on-column derivatization of iodo-dU with ferrocenyl propargylamide (20).
Current progress in the application of electrochemical methods to redox-tagged nucleic acids is encouraging. Capillary electrophoresis chips with integrated ECD (21), a ‘four colour’ strategy for electrochemical DNA sequencing (22) and microarray formats for mutant detection (23–25) have been demonstrated recently. While further development is required before these methods are ready for widespread use, the current need to prepare redox-labelled nucleic acids by phosphoramidite or post-synthesis chemistry significantly restricts the range of practical applications. By analogy to the development of dye terminator sequencing and two-colour cDNA labelling in the fluorescence arena, access to enzymatic redox tagging of nucleic acids should help to facilitate the broader diffusion of electrochemical approaches. The recently reported incorporation of Fc-ddUTP into the 3′-terminus of an oligonucleotide by terminal transferase (26) is the first step in this direction.
In this publication we report the preparation and application of redox-tagged dNTPs for random or site-specific redox labelling of nucleic acids. These analogues can be incorporated into polynucleotides by a number of polymerases in the course of conventional enzymatic synthesis. A high level of probe labelling can be achieved, allowing high sensitivity detection.
MATERIALS AND METHODS
Unless otherwise stated, reagents were obtained from Sigma-Aldrich or Bio-Rad and were used without further purification.
Oligonucleotides were purchased from Sigma Genosys and purified by denaturing PAGE (20% acrylamide/8 M urea) as described by Sambrook et al. (27).
The Klenow fragment of Escherichia coli DNA polymerase I (NEB), 3′→5′ exonuclease-deficient Klenow fragment (Progen Industries), T4 DNA polymerase (MBI Fermentas) and Tth DNA polymerase (Perkin Elmer) were purchased commercially.
1H and 31P NMR spectra were recorded on a Bruker DPX-300 spectrometer. Chemical shifts are reported in parts per million (δ) relative to an external standard.
UV spectra and melting behaviour were measured on a Cary 100 Bio spectrophotometer (Varian) equipped with a Thermal accessory.
HPLC separation and analyses were performed with an Akta Purifier system (Pharmacia Biotech). A reverse phase C18 column (Zorbax ODS, 250 × 9.4 mm) was utilised for preparative separations.
Analytical PAGE was performed with a Protean II xi Cell (Bio-Rad) with 20 cm glass plates. Gels were run at 600 V in 0.09 M Tris–borate, 2 mM EDTA buffer and stained with SYBR Green II (Molecular Probes) before scanning with a Fluor-S MultiImager (Bio-Rad). Analytical agarose gels were run at 5 V/cm in a Gello-tank cell (HyBaid) in 0.045 M Tris–borate, 1 mM EDTA buffer.
Synthesis of Fc1-dUTP and Fc2-dUTP
5-(3-Aminopropenyl-1) 2′-deoxyuridine 5′-triphosphate was prepared according to the reported procedure (1). A 45 µmol sample of 5-(trans-3-aminopropenyl-1) 2′-deoxyuridine 5′-triphosphate was evaporated twice from absolute ethanol to remove traces of water before dissolution in 1 ml of anhydrous DMF. A solution of 23 mg (0.1 mmol) ferrocenecarboxylic acid in DMSO and 37.9 mg (0.1 mmol) solid O-benzotriazol-1-yl-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU) was added to the nucleotide solution with stirring until dissolution of HBTU and the mixture incubated at room temperature overnight. The reaction mixture was diluted with 20 ml of 5 mM 2-mercaptoethanol in water and the yellow ferrocenecarboxylic acid precipitate removed with a 0.45 µm polypropylene membrane filter (Gelman Sciences). The filtrate was applied to a DEAE–cellulose column (1 × 25 cm) equilibrated with 5 mM aqueous 2-mercaptoethanol and separated with a linear gradient of TEAB (0–0.35 M, 500 ml) in 5 mM 2-mercaptoethanol at a 2 ml/min flow rate. The column eluate was detected at 260 and 440 nm, the large product peak eluting at 220 min. Product fractions were pooled, evaporated and purified by RP-HPLC with a linear gradient of acetonitrile (0–30% in 60 min) in 0.05 M LiClO4 at a flow rate of 4 ml/min. The major product peak eluted at 23 min. Solvent was removed by rotary evaporation, the residue dissolved in 0.5 ml water and the product precipitated by addition of 5 ml of 2% LiClO4 in acetone. The precipitate was washed with acetone and dried in air. For 5-(3-ferrocenecarboxamidopropenyl-1) 2′-deoxyuridine 5′-triphosphate (Fc1-dUTP) the yield was 14 µmol (30%). UV (H2O) λmax = 439 nm (ɛ = 300 M–1 cm–1). 1H NMR (D2O) δ 2.36 (m, H2′, 2H), 3.98 (d, J = 4.5 Hz, H9, 2H), 4.19 (m, H4′, H5′, 3H), 4.27 (s, C5H5 of Fc, 5H), 4.51 (s, H2″, 2H), 4.77 (m, H3′, 1H), 4.81 (s, H1″, 2H), 6.27 (t, J = 6 Hz, H1′, 1H), 6.39 (s, H7, 1H), 6.48 (t, J = 4.5 Hz, H8, 1H), 7.88 (s, H6, 1H). 31P NMR (D2O) δ –22.45 (t, J = 22 Hz, Pβ, 1P), –11.48 (d, J = 22 Hz, Pα, 1P), –9.15 (d, J = 22 Hz, Pg, 1P). ESI-MS calculated for C23H28N3O15P3Fe, 735.24; found m/z 734 [M – H]–.
The same procedure was used for the synthesis of 5-(3-ferroceneacetamidopropenyl-1) 2′-deoxyuridine 5′-triphosphate (Fc2-dUTP), except that ferroceneacetic acid was employed and the reaction mixture was incubated for 2 days (yield 8%). UV (H2O) λmax = 290 nm (ɛ = 8100 M–1 cm–1). 1H NMR (D2O) δ 2.35 (m, H2′, 2H), 3.31 (s, H12, 2H), 3.88 (d, J = 4.5 Hz, H9, 2H), 4.12 (m, H3′, H4′, H5′, 4H), 4.22 (s, C5H5 of Fc, 5H), 4.27 (s, H2″, 2H), 4.76 (s, H1″, 2H), 6.25 (t, J = 6 Hz, H1′, 1H), 6.29 (s, H7, 1H), 6.38 (t, J = 4.5 Hz, H8, 1H), 7.81 (s, H6, 1H). ESI-MS calculated for C24H30N3O15P3Fe, 749.27; found m/z 748 [M – H]–.
Cyclic voltammetry
Cyclic voltammograms were recorded with a BAS electrochemical analyser. The three-electrode system consisted of a glassy carbon working electrode, an Ag/AgCl (saturated KCl) reference electrode (Eref = 206 mV versus SHE) and a platinum counter electrode. Experiments were performed in a 5 ml electrochemical cell containing 0.8 mM Fc-NTP in 20 mM Tris–acetate, 100 mM KCl, 1 mM MgCl2, pH 7.4, at a scan rate of 20 mV/s. The scan range was from –0.1 to +0.8 V versus Ag/AgCl.
DNA primer extension
A DNA duplex consisting of the 18mer primer P18.1 (5′-CAACGTCCGAGCAGTACA) and the 40mer template T40.1 (5′-AAGCTCCTTAGTCTGTCAATGTACTGCTCGGACGTTGCGA) was prepared by annealing PAGE-purified oligonucleotides. The DNA duplex (2 µM) was incubated in 20 µl of polymerase reaction mixture [6.7 mM Tris–HCl pH 8.8, 6.6 mM MgCl2, 1 mM DTT, 16.8 mM (NH4)2SO4, 200 µM dNTPs and 0.25 U/µl Klenow fragment, exo– Klenow fragment or T4 DNA polymerase] at room temperature for 20 min. Reactions were stopped with an equal volume of gel loading buffer (98% formamide, 10 mM EDTA, pH 8.0, 0.025% bromophenol blue, 0.025% xylene cyanol FF), heated at 95°C for 2 min and subjected to denaturing PAGE.
PCR
A segment of the T4 DNA ligase gene cloned into plasmid pKL01 was used as a template for PCR amplification in the presence of Fc1-dUTP. The 25mer 5′-GCTGATGGAGCTCGGTGTTTTGCTT served as forward primer and the 31mer 5′- TATATAAGCTTCATAGACCAGTTACCTCATG as reverse primer, producing a 998 bp amplicon. Reaction mixtures (20 µl) contained 6.7 mM Tris–HCl pH 8.8, 1.66 mM (NH4)2SO4, 0.045% Triton X-100, 0.02 mg/ml gelatin, 2.5 mM MgCl2, 0.2 µM each primer, 20 µg/ml pKL01 plasmid, 0.2 mM dNTPs and 0.1 U/µl Tth DNA polymerase (exo–). In some reaction mixtures, dTTP was partially or fully substituted by Fc1-dUTP while maintaining the total [dTTP + Fc1-dTTP] concentration at 200 µM. PCR conditions were: 2 min at 94°C, then 22 cycles at 94°C for 30 s and 50°C for 1 min, and a final cycle at 70°C for 10 min. After amplification, 4 µl of gel loading buffer (30% glycerol, 0.25% bromophenol blue and 0.25% xylene cyanol FF) was added and samples were analysed on a 1% agarose gel.
HPLC–ECD
To generate the duplex for HPLC–ECD, the P18.1 and T40.1 oligonucleotides (4 µM each) were annealed before incubation in 240 µl of reaction mixture containing 6.7 mM Tris–HCl pH 8.8, 6.6 mM MgCl2, 1 mM DTT, 16.8 mM (NH4)2SO4, 200 µM dNTPs (except dTTP), 200 µM Fc1-dUTP and 0.25 U/µl Klenow fragment at room temperature for 20 min. Low molecular weight components were removed with a Bio-Spin 30 chromatography column (Bio-Rad). The eluate was successively extracted with equal volumes of phenol/chloroform (1:1) and chloroform. DNA was precipitated by addition of 10 vol 2% LiClO4 in acetone and centrifugation (12 000 g, 15 min). The resulting precipitate was dried in vacuo, redissolved in 200 µl HPLC buffer (50 mM LiClO4, 2.5% acetonitrile in water) and the DNA concentration determined spectrophotometrically by absorbance at 260 nm. Analyses were performed with a Shimadzu HPLC system equipped with a Coulochem II electrochemical detector (ESA Inc.). Samples were separated by isocratic elution of a reverse phase column (Vydac C18, 250 × 4 mm) at a flow rate of 1 ml/min with both optical (260 nm) and electrochemical (E = 0.7 V versus Ag/AgCl) detection.
UV melting
Fc1-labelled P18.1F/T40.1 duplex DNA for hybrid stability analysis was prepared from the P18.1/T40.1 primer–template as described above for HPLC–ECD. This sample contained five residues of Fc1-dU in place of dT, while a second sample containing only natural nucleotides was prepared as a control. DNA duplexes were dissolved at 0.6 µM in 1 ml of 0.3 M KH2PO4, pH 7.0, and transferred to standard quartz cuvettes. Absorbance at 260 nm was monitored while cell temperature was increased from 25 to 95°C at 1°C/min. The melting temperature Tm was taken as the maximum point in the derivative curve.
RESULTS
Synthesis and characterization
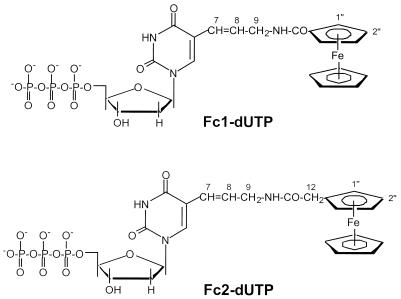
The ferrocene-labelled Fc1-dUTP and Fc2-dUTP derivatives (Fig. 1) were successfully synthesized by reaction of the 5-(3-aminopropenyl) nucleoside triphosphates with carboxyl-bearing ferrocenes in the presence of HBTU. This simple procedure generates a moderately rigid linkage between the nucleobase and redox label.
Figure 1.
Structures of Fc1-dUTP and Fc2-dUTP. Atoms corresponding to 1H NMR assignments are numbered by extension of the nucleobase nomenclature.
The products were purified to homogeneity by ion exchange chromatography followed by reverse phase (RP)-HPLC, producing reasonable but not high final yields of both products (30% for Fc1-dUTP and 8% for Fc2-dUTP). We also used the HBTU-mediated procedure to synthesize a ferrocenedicarboxylic acid derivative of dUTP, but this species was obtained in low yield and decomposed during purification.
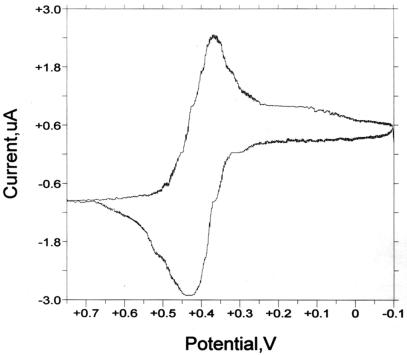
Fc1-dUTP and Fc2-dUTP have characteristic absorption spectra which correspond to a superposition of spectra for the modified nucleotide and ferrocene carboxamide constituents, displaying strong absorption in the UV region and a weak, broad peak characteristic of ferrocene near 440 nm. Cyclic voltammetry of Fc1-dUTP (Fig. 2) yielded a symmetric peak with E1/2 = 398 mV versus Ag/AgCl. The spacing between the half-wave potentials was 60 mV, consistent with reversible redox reaction of the Fc moiety in this aqueous buffer system. The redox potential of Fc1-dUTP was 90 mV greater than the potential of ferrocenecarboxylate (310 mV versus Ag/AgCl) measured in the same buffer (data not shown), reflecting the change of the pentadienyl ring substituent to one which is more electron-withdrawing (-COO– to -CONHR). The observed potential is close to that reported for a ferrocene carboxamide moiety attached to the 5′-end of DNA oligonucleotides in aqueous buffer at 406–425 mV versus Ag/AgCl (13). For Fc2-dUTP, an apparent E1/2 of 260 mV versus Ag/AgCl was obtained.
Figure 2.
Cyclic voltammogram of Fc1-dUTP in Tris–acetate/KCl/MgCl2 buffer, pH 7.4. Half-wave potential maxima occur at 366 and 426 mV versus Ag/AgCl, respectively.
Enzymatic nucleotide incorporation
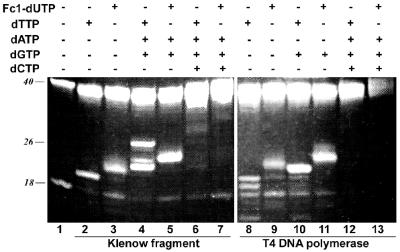
The substrate properties of Fc1-dUTP were tested in DNA polymerase-catalysed primer extension assays using the P18.1/T40.1 primer–template pair shown in Figure 3. The sequence of the template allows the progress of primer extension to be controlled by omitting one or more dNTPs from the reaction mixture. Results of incubating primer–template with E.coli DNA polymerase I Klenow fragment or T4 DNA polymerase are shown in Figure 4. Addition of unlabelled dTTP to the reaction mix results in extension of the P18.1 primer (lane 1) by 2 nt (lanes 2 and 8). The product heterogeneity displayed by T4 DNA polymerase in lane 8 is caused by its stronger 3′→5′ exonuclease activity, which is also evident in lanes 9–11. When Fc1-dUTP replaces dTTP, both DNA polymerases incorporate two consecutive Fc1-dU residues into the 3′-end of the primer (lanes 3 and 9). Because the incorporated Fc1-dU residue has a molecular weight almost twice that of the dT residue (574 versus 321 Da) and the bulky adduct also alters the hydrodynamic properties of the chain, the mobility of the Fc1-dU-containing primer is significantly lower than that of its natural counterpart. There is an indication that a small fraction of the primer is not extended by Klenow fragment (lane 3), but this behaviour is not consistent across the gel.
Figure 3.
The P18.1/T40.1 primer–template pair, with expected extension products. Residues incorporated by primer extension are capitalized, while T residues that can be substituted by Fc1-dU are underlined. UV melting and ECD experiments were performed on the P18.1F/T40.1 duplex.
Figure 4.
Incorporation of Fc1-dUTP into DNA by Klenow fragment and T4 DNA polymerase. The P18.1/T40.1 primer–template pair of Figure 3 was incubated with DNA polymerase and different sets of dNTPs as indicated. DNA fragment lengths in nucleotides are shown on the left.
Some modified nucleoside triphosphates have the properties of terminators, their incorporation into DNA preventing or dramatically slowing further extension. To examine this issue, we incubated the primer–template with Fc1-dUTP and one or two other dNTPs required for limited primer extension. In the presence of Fc1-dUTP, dGTP and dATP, the chain is successfully extended by 4 nt following Fc1-dU incorporation by Klenow fragment (lane 5). Similarly, T4 DNA polymerase extends the primer by 3 nt in the presence of Fc1-dUTP and dGTP (lane 11). This allows us to conclude that Fc1-dUTP is both efficiently incorporated and does not significantly inhibit further extension. Of some interest, Klenow fragment displays cleaner extension behaviour with the Fc1-dUTP/dATP/dGTP mixture (lane 5) than with dTTP/dATP/dGTP (lane 4), where misincorporation of C23 allowed the formation of a minor 26mer product which terminated at the next ‘stop’ position, C27. Incubation of the primer–template with all four natural dNTPs (lanes 6 and 12) or Fc1-dUTP plus three dNTPs (lanes 7 and 13) allowed run-off extension of the primer, yielding bands that overlapped with the P40.1 primer band at 40 nt. Again, no visible termination sites are registered when Fc1-dUTP replaces dTTP.
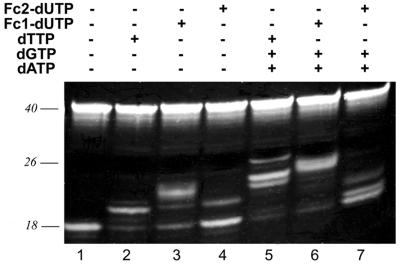
Experiments with Klenow fragment and T4 DNA polymerase reveal that Fc2-dUTP is a much less satisfactory substrate than Fc1-dUTP for these polymerases. This is also true for the non-proofreading exo– Klenow fragment. As shown in Figure 5, under conditions where extension of primer P18.1 in the presence of dTTP or Fc1-dUTP yielded predominantly the expected 18 + 2 nt products (Fig. 5, lanes 2 and 3, respectively), Fc2-dUTP caused the low level incorporation of only 1 nt (Fig. 5, lane 4). Similarly, in the presence of dTTP (Fc1-dUTP)/dGTP/dATP, where 18 + 4 nt products are expected, both dTTP (Fig. 5, lane 5) and Fc1-dUTP (Fig. 5, lane 6) produced the expected products predominantly, while Fc2-dUTP yielded the single base extended product and misincorporation products (Fig. 5, lane 7).
Figure 5.
Comparion of the substrate behaviour of Fc2-dUTP with exo– Klenow fragment. The P18.1/T40.1 primer–template pair was incubated with substrates and polymerase as indicated.
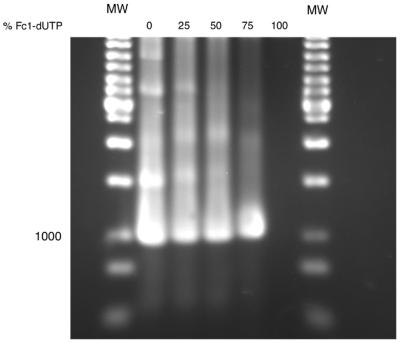
With the primer extension results in hand, a preliminary examination of Fc1-dUTP as a substrate for PCR was undertaken. Full substitution of dTTP by Fc1-dUTP supported very minor formation of a 998 bp amplicon by Tth DNA polymerase under the conditions employed here. However, when dTTP was substituted by Fc1-dUTP at the 25, 50 or 75% level, efficient synthesis of the correct amplicon was observed (Fig. 6). The agarose gel mobility of the amplicon showed a progressive decrease with increasing Fc1-dUTP:dTTP ratios, indicating extensive Fc1-dU incorporation.
Figure 6.
PCR ampification of a 998 bp target in the presence of Fc1-dUTP.
Hybrid stability
Modification of natural nucleic acid components may severely affect the stability of the DNA duplex; this issue is very important for all applications involving DNA hybridization. To examine the effect of Fc1-dU residues on DNA duplex stability, we measured the melting temperature of a labelled P18.1F/T40.1 duplex, the P18.1F strand containing five residues of Fc1-dU out of seven total ‘T’ residues (Fig. 3). Melting of a natural all-dT counterpart was performed for comparison. The experimental melting temperature Tm of the Fc1-dU-modified DNA hybrid (71°C) was found to be 4°C lower than that of the normal duplex (75°C), a relatively small change given the high degree of substitution. This allows us to conclude that modification by ferrocene at the C5 position of dU does not significantly disrupt the native structure of DNA.
Electrochemical detection of Fc1-dU-labelled DNA
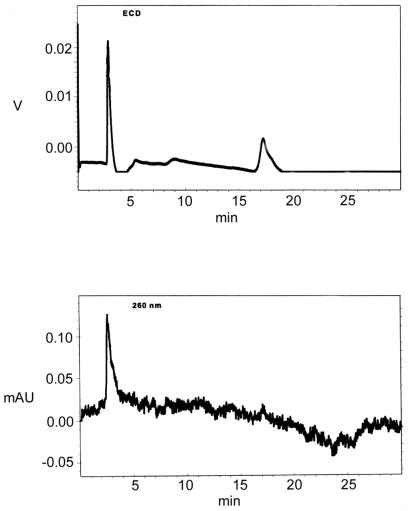
The ferrocene-labelled P18.1F/T40.1 duplex was employed for a comparison of electrochemical and UV detection at 260 nm in the course of RP-HPLC. Various quantities of P18.1F/T40.1 duplex were injected into the system and isocratically eluted, with typical elution times of 17.5 min. Only picomole quantities of DNA were reliably detectable at 260 nm. As shown in Figure 7 (lower), the sensitivity of UV detection was well exceeded at 60 fmol duplex. In contrast, ECD allowed femtomole levels to be observed with a good signal-to-noise ratio, consistent with previous reports on ferrocene-tagged DNA (12). Unlabelled DNA produced no ECD signal at the voltage employed (0.7 V versus Ag/AgCl).
Figure 7.
RP-HPLC elution profile of 60 fmol Fc1-dU-labelled P18.1F/T40.1 duplex with UV (lower) and ECD (upper) detection.
DISCUSSION
We have presented redox-tagged dNTPs for labelling nucleic acids by common DNA polymerases with a view to facilitating the preparation of electrochemically detectable nucleic acid probes. Ferrocene has been employed as the redox-active moiety due to its electrochemical properties and ready availability; there is a substantial history of ferrocene application to nucleic acid detection in the literature (9,12–20,22–24,26). In these previous reports, ferrocene was attached either to the terminus of a strand post-synthesis or incorporated during chemical oligonucleotide synthesis. Internal labelling of any sequence by primer extension should permit a wide range of applications.
Synthesis
We have chosen to first synthesise derivatives of dUTP modified at position C5 of the pyrimidine ring (Fig. 1) using nucleoside triphosphate precursors. These precursors are inexpensive and straightforward to prepare (1,2), avoiding the triphosphorylation of derivatives (28) in the process. The synthetic yield of Fc1-dUTP is satisfactory and almost certainly superior to the yield of a multistep synthesis. Our initial yield of Fc2-dUTP was modest, but sufficient quantities were obtained to establish its properties, which are not suitable for processive incorporation by the polymerases examined here. The linker moiety of Fc1-dUTP is six bonds in contour length and is relatively but not completely rigid. Shorter linkers can be conceived, but a three bond acetylene linker attached to the C5 position of dU is known to display an unfortunate isomerization into a cyclised form which hybridises preferably with dG instead of dA (18).
Enzymatic incorporation
As expected, Fc1-dUTP proved to be a good substrate for common DNA polymerases, thus allowing a high degree of labelling. The C5 modification does not often interfere with incorporation of modified nucleotides; even dUTP derivatives with bulky C5 substituents have been successfully employed as substrates for these enzymes (29). High quality substrate behaviour appears to hold also for the analogous Fc1-ribose derivative; we have observed high efficiency RNA labelling with T7 RNA polymerase (W.A.Wlassoff and G.C.King, unpublished results).
Nonetheless, the substrate quality of any particular nucleotide derivative is essentially unpredictable, varying from construct to construct and polymerase to polymerase, especially when labelled versus natural substrate selectivity is at issue (30). The behaviour of Fc2-dUTP is a case in point; it differs from Fc1-dUTP by a single methylene group (Fig. 1), but acts as a poorly incorporated terminator (Fig. 5). The cleaner incorporation of Fc1-dUTP into DNA compared to dTTP displayed by Klenow fragment in Figure 4 (lane 5 versus lane 4) is partly a function of the relatively long incubation time and high dNTP concentrations used in this particular experiment. Coupled with the slowed incorporation and hence increased active site residency of the modified substrate and its products, there is less opportunity for misincorporation. This behaviour is indicative of a context-dependence of substrate quality, the Fc derivative in this case being the better substrate in terms of ‘one-colour’ primer extension fidelity.
Our initial experiments indicated that Fc1-dUTP is a satisfactory substrate for PCR by Tth DNA polymerase (Fig. 6). The polymerase-dependence of incorporation noted above means that a range of polymerases and conditions must be explored to determine optimal PCR performance. These issues are under further investigation.
Electrochemical detection
ECD can be employed as the end stage of liquid chromatography, capillary and microchannel electrophoresis and in microarrays. It has been demonstrated that ECD is very sensitive, being able to measure attomole to zeptomole quantities of sample in nanolitre to picolitre volumes. In nucleic acid monitoring, electrochemical methods have been used to detect redox-labelled DNA in HPLC (12), electrophoresis (21,22) and microarray formats (23–25). We have employed HPLC–ECD in this paper. The separation power of this method is low in comparison to electrophoresis, but is adequate for demonstration purposes and certain practical applications (W.A.Wlassoff, D.DiGiusto and G.C.King, unpublished results). While access to appropriate instrumentation is a significant problem in the biological and to a certain extent even in the chemical sciences, we believe that biologically oriented detection systems will be available in the near future.
Conclusion
Conjugation of a redox label such as ferrocene with nucleoside triphosphates opens the possibility of ready access to electrochemical methods for molecular biologists, analogous to the diffusion of fluorescence methodologies over the past decade. Further development of polymerase substrates and electrochemical detection systems should facilitate this diffusion.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Rachel Codd (University of Sydney) and Justin Gooding for assistance with cyclic voltammetry, Martin Zarka for assistance with HPLC–ECD and David Andrews for encouragement.
REFERENCES
- 1.Langer P.R., Waldrop,A.A. and Ward,D.C. (1981) Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc. Natl Acad. Sci. USA, 78, 6633–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward D.C., Waldrop,A.A. and Langer,P.R. (1982) European Patent EP0063879.
- 3.Fuller C.W. (2000) The biochemistry of DNA sequencing with DNA polymerases. Ligand Assay, 23, 249–255. [Google Scholar]
- 4.Schulze A. and Downward,J. (2001) Navigating gene expression using microarrays—a technology review. Nature Cell Biol., 3, E190–E195. [DOI] [PubMed] [Google Scholar]
- 5.Jackson P.E., School,P.F. and Groopman,J.D. (2000) Mass spectrometry for genotyping: an emerging tool for molecular medicine. Trends Biotechnol., 16, 117–121. [DOI] [PubMed] [Google Scholar]
- 6.Thorp H.H. (1998) Cutting out the middleman—DNA biosensors based on electrochemical oxidation. Trends Biotechnol., 16, 117–121. [Google Scholar]
- 7.Baik M.H., Silverman,J.S., Yang,I.V., Ropp,P.A., Szalai,V.A., Yang,W.T. and Thorp,H.H. (2001) Using density functional theory to design DNA base analogues with low oxidation potentials. J. Phys. Chem. B, 105, 6437–6444. [Google Scholar]
- 8.Boon E.M., Ceres,D.M., Drummond,T.G., Hill,M.G. and Barton,J.K. (2000) Mutation detection by electrocatalysis at DNA-modified electrodes. Nat. Biotechnol., 18, 1096–1100. [DOI] [PubMed] [Google Scholar]
- 9.Takenaka S., Yamashita,K., Takagi,M., Uto,Y. and Kondo,H. (2000) DNA sensing on a DNA probe-modified electrode using ferrocenylnaphthalene diimide as the electrochemically active ligand. Anal. Chem., 72, 1334–1341. [DOI] [PubMed] [Google Scholar]
- 10.Sloop F.V., Brown,G.M., Sachleben,R.A., Garrity,M.L., Elbert,J.E. and Jacobson,K.B. (1994) Metalloorganic labels for DNA sequencing and mapping. New J. Chem., 18, 317–326. [Google Scholar]
- 11.Beilstein A.E., Tierney,M.T. and Grinstaff,M.W. (2000) Site-specifically labeled metallo-oligodeoxynucleotides. Comments Inorg. Chem., 22, 105–127. [Google Scholar]
- 12.Takenaka S., Uto,Y., Kondo,H., Ihara,T. and Takagi,M. (1994) Electrochemically active DNA probes: detection of target DNA sequences at femtomole level by high-performance liquid chromatography with electrochemical detection. Anal. Biochem., 218, 436–443. [DOI] [PubMed] [Google Scholar]
- 13.Ihara T., Maruo,Y., Takenaka,S. and Takagi,M. (1996) Ferrocene-oligonucleotide conjugates for electrochemical probing of DNA. Nucleic Acids Res., 24, 4273–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uto Y., Kondo,H., Abe,M., Suzuki,T. and Takenaka,S. (1997) Electrochemical analysis of DNA amplified by the polymerase chain reaction with a ferrocenylated oligonucleotide. Anal. Biochem., 250, 122–124. [DOI] [PubMed] [Google Scholar]
- 15.Mucic R.C., Herrlein,M., Mirkin,C.A. and Letsinger,R.L. (1996) Synthesis and characterization of DNA with ferrocenyl groups attached to their 5′-termini—electrochemical characterization of a redox-active nucleotide monolayer. Chem. Commun., 555–557. [Google Scholar]
- 16.Xu C., Cai,H., He,P. and Fang,Y. (2001) Electrochemical detection of sequence-specific DNA using a DNA probe labeled with aminoferrocene and chitosan modified electrode immobilized with ssDNA. Analyst, 126, 62–65. [DOI] [PubMed] [Google Scholar]
- 17.Wiessler M. and Schutte,D. (1997) European Patent WO9709337.
- 18.Yu C.J., Yowanto,H., Wan,Y., Meade,T.J., Chong,Y., Strong,M., Donilon,L.H., Kayyem,J.F., Gozin,M. and Blackburn,G.F. (2000) Uridine-conjugated ferrocene DNA oligonucleotides: unexpected cyclization reaction of the uridine base. J. Am. Chem. Soc., 122, 6767–6768. [Google Scholar]
- 19.Yu C.J., Wang,H., Wan,Y., Yowanto,H., Kim,J.C., Donilon,L.H., Tao,C., Strong,M. and Chong,Y. (2001) 2′-Ribose-ferrocene oligonucleotides for electronic detection of nucleic acids. J. Org. Chem., 66, 2937–2942. [DOI] [PubMed] [Google Scholar]
- 20.Beilstein A.E. and Grinstaff,M.W. (2000) On-column derivatization of oligodeoxynucleotides with ferrocene. Chem. Commun., 6, 509–510. [Google Scholar]
- 21.Woolley A.T., Lao,K.Q., Glazer,A.N. and Mathies,R.A. (1998) Capillary electrophoresis chips with integrated electrochemical detection. Anal. Chem., 70, 684–688. [DOI] [PubMed] [Google Scholar]
- 22.Brazill S.A., Kim,P.H. and Kuhr,W.G. (2001) Capillary gel electrophoresis with sinusoidal voltammetric detection: a strategy to allow four-‘color’ DNA sequencing. Anal. Chem., 73, 4882–4890. [DOI] [PubMed] [Google Scholar]
- 23.Umek R.M., Lin,S.W., Vielmetter,J., Terbrueggen,R.H., Irvine,B., Yu,C.J., Kayyem,J.F., Yowanto,H., Blackburn,G.F., Farkas,D.H. and Chen,Y.-P. (2001) Electronic detection of nucleic acids. A versatile platform for molecular diagnostics. J. Mol. Diagn., 3, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C.J., Wan,Y., Yowanto,H., Li,J., Tao,C., James,M.D., Tan,C.L., Blackburn,G.F. and Meade,T.J. (2001) Electronic detection of single-base mismatches in DNA with ferrocene-modified probes. J. Am. Chem. Soc., 123, 11155–11161. [DOI] [PubMed] [Google Scholar]
- 25.Yamashita K., Takagi,M., Uchida,K., Kondo,H. and Takenaka,S. (2001) Visualization of DNA microarrays by scanning electrochemical microscopy. Analyst, 126, 1210–1211. [DOI] [PubMed] [Google Scholar]
- 26.Anne A., Blanc,B. and Moiroux,J. (2001) Synthesis of the first ferrocene-labeled dideoxynucleotide and its use for 3′-redox end-labelling of 5′-modified single-stranded oligonucleotides. Bioconjug. Chem., 12, 396–405. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Burgess K. and Cook,D. (2000) Synthesis of nucleoside triphosphates. Chem. Rev., 100, 2047–2059. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Z.R., Chao,J., Yu,H. and Waggoner,A.S. (1994) Directly labeled DNA probes using fluorescent nucleotides with different length linkers. Nucleic Acids Res., 22, 3418–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandis J.W. (1999) Dye structure affects Taq DNA polymerase terminator selectivity. Nucleic Acids Res., 27, 1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]