Abstract
Site-specific recombination in genetically modified cells can be achieved by the activity of Cre recombinase from bacteriophage P1. Commonly an expression vector encoding Cre is introduced into cells; however, this can lead to undesired side-effects. Therefore, we tested whether cell-permeable Cre fusion proteins can be directly used for lox-specific recombination in a cell line tailored to shift from red to green fluorescence after loxP-specific recombination. Comparison of purified recombinant Cre proteins with and without a heterologous ‘protein transduction domain’ surprisingly showed that the unmodified Cre recombinase already possesses an intrinsic ability to cross the membrane border. Addition of purified recombinant Cre enyzme to primary bone marrow cells isolated from transgenic C/EBPαfl/fl mice also led to excision of the ‘floxed’ C/EBPα gene, thus demonstrating its potential for in vivo applications. We conclude that Cre enyzme itself or its intrinsic membrane-permeating moiety are attractive tools for direct manipulation of mammalian cells.
INTRODUCTION
The Cre/lox recombination system of the bacteriophage P1 is a widely used method to achieve conditional targeted deletions, inversion, insertions, gene activation, translocations and other modifications in chromosomal or episomal DNA (1–3). Cre recombinase specifically recognizes a 34 bp lox sequence and catalyzes DNA strand exchange between two of these sites. In cells harboring lox sites in their genome, site-specific recombination is usually induced by introducing an expression vector containing the Cre gene. This method has potential disadvantages: besides the risk of mutagenesis by random integration of the vector DNA into the host genome, the constitutive expression of Cre has been shown to be cytotoxic in many cell types (4,5).
We therefore sought to achieve site-specific recombination in eucaryotic cells by the simple addition of recombinant, purified Cre protein fused to peptides known to mediate the translocation of proteins across the membrane (6,7). Surprisingly, efficient recombination in mouse fibroblasts and primary bone marrow cells was already observed when Cre recombinase lacking such a ‘protein translocation domain’ was added. This indicates that Cre protein on its own has the capacity to cross the plasma membrane and translocate to the nucleus. Although the mechanism of delivery has to be elucidated, our results demonstrate that purified recombinant protein without an exogenous protein transduction domain can be efficiently used for specific manipulation of cells.
MATERIALS AND METHODS
Generation of the reporter cell line
A DNA fragment composed of 5′ loxP1 (5′-ATGATAACTTCGTATAATGTATGCTATACGAAGTTAT-NcoI-3′), the RFP gene (NcoI-RFP-BglII) isolated from pDsRed1-C1 (Clontech) and loxP2 (5′-BglII-TAGTCGACGCGTATAACTTCGTATAATGTATGCTATACGAAGTTATCAGGCCTGCACCCG-3′) was inserted into the retroviral vector SF91-eGFP (8,9), which had been pretreated with NcoI and subsequent mung- bean-nuclease (Promega) digestion. The resulting vector SF91-loxP1-RFP-loxP2-eGFP contained the loxP1-RFP-loxP2 sequence immediately downstream of a Kozak consensus sequence. To enhance protein expression, the woodchuck hepatitis B virus post-transcriptional regulatory element (wPRE) (10) was isolated from pKS-γwPRE (8) and inserted into the HindIII site of SF91-loxP1-RFP-loxP2-eGFP. The resulting vector was termed SFr (= SF91-loxP1-RFP-loxP2-eGFP+wPRE).
Packaging of SFr in retroviral particles pseudotyped with ecotropic envelope and infection of mouse SC-1 fibroblasts (ATCC no. CRL-1404) was performed as described (11). Clones with stably integrated SFr were obtained by sorting single cells.
Expression and purification of recombinant Cre proteins
For the expression of GST–Cre fusion proteins, a modified pGEX-2T plasmid, pGEX-2T(KT), containing the MCS from pEG(KT) (12), was used. An extended Cre (nlsCre) gene encoding the NLS of SV40 large T antigen and a c-myc epitope (3) was isolated from R518 (13) after XbaI digestion, fill-in of the protruding ends with the Klenow fragment of Escherichia coli DNA polymerase I and subsequent NcoI cleavage. This fragment was inserted into pGEX-2T(KT) which had been treated with XhoI, Klenow enzyme and NcoI, generating pGEX-nlsCre. The Tat-PTD oligonucleotides 5′-GATCCTATGGCCGTAAAAAGCGCCGTCAGCGCCGCCGTCC-3′ and 5′-CATGGGACGGCGGCGCTGACGGCGCTTTTTACGGCCATAG-3′ and the PreS2-TLM oligonucleotides 5′-GATCCCCGCTGTCTTCCATTTTCTCCCGTATCGGCGATCCGGGTGGCGGTTC-3′ and 5′-CATGGAACCGCCACCCGGATCGCCGATACGGGAGAAAATGGAAGACAGCGGG-3′ were annealed and ligated into the BamHI/NcoI-treated plasmid pGEX-nlsCre, leading to pGEX-PTDnlsCre and pGEX-TLMnlsCre, respectively.
Removal of sequences encoding the N- and C-terminal modifications of nlsCre was achieved by PCR, using the oligonucleotides 5′-GAAGAGGAAGGGATCCAATTTACTGACCG-3′ and 5′-CAGAAATAAGCTTTTGTTCCTAATCGCCATC-3′ on pGEX-nlsCre as template. The 1060 bp amplification product was cleaved with BamHI and HindIII ligated into pGEX-2T(KT) treated with BamHI and HindIII. All constructs were verified by DNA sequencing.
Protein expression was induced in E.coli BL21 [CodonPlus(DE3)-RP; Stratagene] with 0.5 mM IPTG for 8 h at 25°C. Cells were lysed in PBS, 0.5% Triton X-100, 0.5 mM Pefabloc (Roche Diagnostics) by sonification and GST fusion proteins were affinity purified using glutathione–Sepharose 4B (Amersham Pharmacia Biotech). Bound fusion proteins were incubated with 10 U/ml thrombin (Amersham) in PBS and eluted in PBS/100 mM Na2HPO4. Purity of Cre was >80%, as estimated by Coomassie staining after PAGE.
Cell culture and protein transduction
SC-1 reporter cells were cultivated as recommended by ATCC. For protein transduction, 1.5× concentrated MEM containing 15% fetal calf serum (FCS) (Sigma), 75 U/ml penicillin, 75 µg/ml streptomycin (Gibco BRL) was prepared and diluted with purified Cre proteins in PBS/100 mM Na2HPO4 to 1× medium, resulting in final concentrations of 0.8–5 µM Cre protein. After incubation for 3 or 6 h at 37°C, the Cre-containing medium was exchanged for MEM containing 10% FCS, 2 mM glutamate, 50 U/ml penicillin, 50 µg/ml streptomycin. Cells were analyzed 48 h later by fluorescence microscopy or by fluorescence-activated cell sorting (FACS) analysis using FACSCaliber and CellQuest software (Becton-Dickinson).
Mouse bone marrow cells were isolated and cultivated as described (11). Cre transduction was performed for 7 h in DMEM containing 2 mM glutamate, 5 ng/ml IL-6, 10 ng/ml IL-3, 50 ng/ml SCF, 50 U/ml penicillin, 50 µg/ml streptomycin.
PCR analysis
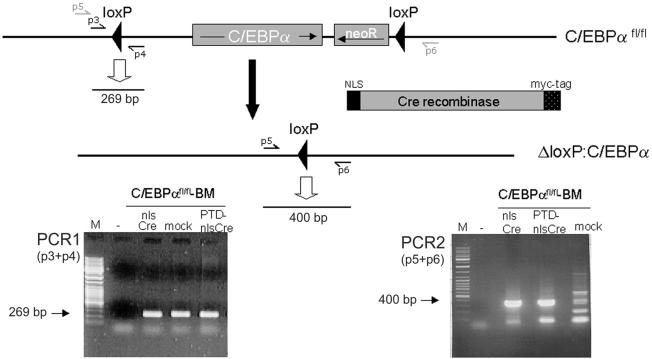
To detect recombination in SFr-transduced reporter cells, PCR reactions were performed using the oligonucleotides p1 (5′-ACGCCGAAACCGCGCCGCGCGTC-3′) and p2 (5′-GCAGATGAACTTCAGGGTCAGCTTGC-3′). Cre-mediated excision of RFP led to a 364 bp fragment (see Fig. 3B). Verification of the original C/EBPαfl/fl allele occured using p3 (5′-TGGCCTGGAGACGCAATGA-3′) and p4 (5′-CGCAGAGATTGTGCGTCTTT-3′) by amplification of a 269 bp product (see PCR1 in Fig. 5). Cre-mediated recombination was detected using p5 (5′-GCCTGGTAAGCCTAGCAATCCT-3′) and p6 (5′-TGGAAACTTGGGTTGGGTGT-3′), producing a 400 bp fragment (see PCR2 in Fig. 5). All PCRs were performed using Taq polymerase (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
Figure 3.
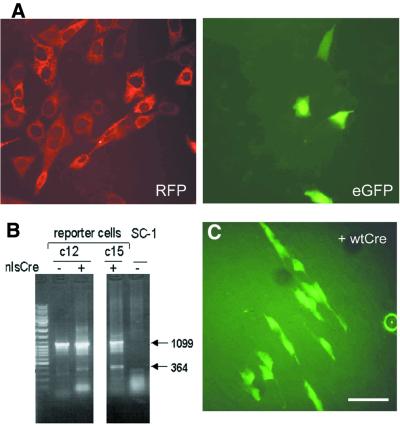
Recombination in mouse fibroblasts mediated by Cre protein without a heterologous protein translocation sequence. (A) Reporter cells with the stably integrated reporter cassette were incubated for 4 h in medium containing 0.8 µM recombinant nlsCre protein (see Fig. 2A). Forty-eight hours later, cells expressing eGFP were detected by fluorescence microscopy. (B) Recombination was confirmed using genomic DNA from untreated (–) and nlsCre-treated (+) cell populations as template for a PCR. Two PCR products are observed: a 1099 bp fragment resulting from the non-recombined, integrated reporter construct and an additional 364 bp fragment as a result of successful excision of the RFP gene. No amplification product was detected using the DNA of the parental SC-1 cell line. Direct sequencing of the 364 bp PCR product confirmed exact recombination at the loxP sites. The analysis was performed with two different reporter cell lines (c12 and c15). (C) Reporter cell clones were incubated for 3 h in medium containing 1 µM unmodified wtCre protein. Forty-eight hours later, eGFP expression was detected in 1.5% of the reporter cells. Thus, cellular uptake of nlsCre is independent of the SV40 NLS and the myc epitope. Bar, 20 µm.
Figure 5.
NlsCre-mediated recombination in mouse bone marrow (BM) cells. Aliqouts of 2 × 106 BM cells prepared from homozygous C/EBPαfl/fl mice, with both C/EBPα genes floxed (14), were incubated for 7 h in medium containing 1.5 µM nlsCre or PTD-nlsCre. As a negative control, an equivalent volume of PBS was added (mock). Genomic DNA prepared from these cells was used as a template for PCR reactions to confirm the presence of the C/EBPαfl/fl allele (PCR1) and to detect the recombined allele, produced by Cre/lox-specific excision of the C/EBPα gene (PCR2). The 269 bp product produced in PCR1 indicated the C/EBPαfl/fl knock-in allele. A 400 bp fragment, which was generated in the second PCR only when excision of C/EBPα occured, could only be detected when the BM cells were incubated with nlsCre or PTD-nlsCre. Sequencing of the 400 bp product confirmed that exact lox-specific recombination occurred. Therefore, primary bone marrow cells were genetically modified by external addition of nlsCre protein.
RESULTS
Generation of a reporter cell line for Cre activity
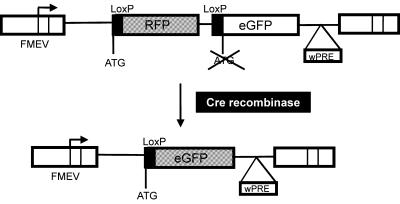
A retroviral reporter construct, SFr (SF91-loxP-RFP-loxP-eGFP+wPRE), was generated to allow fast and easy detection of Cre activity in the nucleus of target cells. SFr is based on the high level expressing SF91 retroviral vector backbone (8,9) and contains an expression cassette with two open reading frames (ORFs) encoding different reporter proteins. The 5′ ORF contains the RFP gene (DsRed1, encoding a red fluorescent protein) and is flanked by loxP sites. Cre-mediated excision of RFP leads to expression of the second reporter gene, which encodes eGFP (enhanced green fluorescent protein), by placing it in-frame to the start codon of the 5′ ORF (Fig. 1).
Figure 1.
The reporter system for Cre activity. The expression cassette has the RFP gene in-frame to the ATG initiation codon with its adjacent loxP sequence. Translation of RFP is terminated at a UAG stop codon. The ORF of a humanized eGFP, lacking an initiation codon, is downstream of the distal loxP site. Following Cre-mediated excision of RFP, eGFP codons are placed in-frame of the proximal ATG-LoxP sequence and are consequently translated. The reporter cassette was inserted into the FMEV-based retroviral vector SF91, which allows stable integration in the host genome and high expression of the reporter genes (8). FMEV, Friend-MCF/MESV hybrid vectors; wPRE, woodchuck hepatitis B virus post-transcriptional regulatory element (10).
A mouse fibroblast cell line (SC-1) was transduced with SFr and single clones containing stably integrated reporter cassettes were isolated. No spontaneous recombination resulting in eGFP expression was detected. This reporter system permits the quantification of lox-specific recombination in live cells by fluorescence microscopy and FACS analysis.
Expression and purification of recombinant Cre proteins
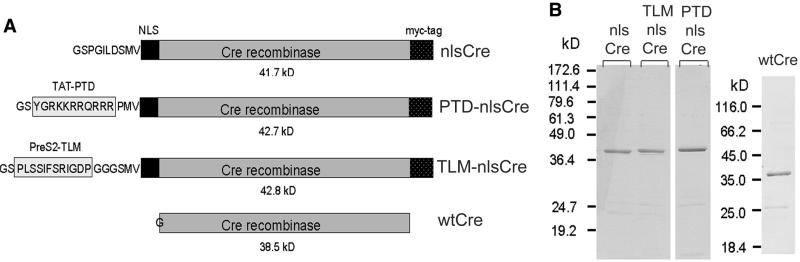
Our original intention to directly deliver Cre into cells led us to fuse DNA sequences encoding the protein transfer domains of HIV-1 Tat (Tat-PTD) and hepatitis B virus PreS2 (PreS2-TLM) to the Cre gene, generating PTD-nlsCre and TLM-nlsCre, respectively. The Cre polypeptide used for this purpose (further referred to as nlsCre) contained the nuclear localization sequence (NLS) of the simian virus 40 (SV40) large T antigen at its N-terminus and a c-myc (9E10) epitope at its C-terminus (3). Additionally, unmodified Cre protein (wtCre) was tested (Fig. 2A).
Figure 2.
Recombinant Cre proteins. The nlsCre protein contains the NLS of the SV40 large T antigen and the 9E10 c-myc epitope (myc-tag). TLM-nlsCre and PTD-nlsCre were generated by fusing the protein translocation sequences of the HBV PreS2 (6) or the HIV-1 Tat protein (7) to the N-terminus of Cre. Wild-type Cre protein (wtCre) was generated by removal of all C- and N-terminal heterologous sequences of nlsCre. (A) The composition of Cre variants following thrombin digestion. The N-terminal GS residues represent the P1′ and P2′ positions of the thrombin recognition site in the GST fusion proteins and are always present in the cleavage products. The only exchange in wtCre, which contains a serine residue at position 2, is therefore a glycine instead of a methionine residue at the first position. This is not expected to alter any property of the wild-type Cre recombinase. (B) Cre proteins were produced as GST fusion proteins in E.coli and affinity purified using glutathione–Sepharose. Elution of Cre proteins by thrombin cleavage resulted in highly enriched protein preparations of >80% purity. The eluates were separated by SDS–PAGE and proteins stained with Coomassie blue.
All Cre variants were produced as GST fusion proteins in E.coli, affinity purified and separated from GST by thrombin cleavage. The preparations used for the transduction experiments were >80% pure, as estimated by Coomassie staining of the separated proteins (Fig. 2B).
Direct transfer of Cre protein into cells
We first tested the specificity of protein transduction by Cre. Therefore, low amounts of purified nlsCre, PTD-nlsCre or TLM-nlsCre proteins (with a final concentration of 1 µM in the culture medium) were initially incubated with the reporter cell line for 3 h. Cells were analyzed by fluorescence microscopy 48 h later. In all individual experiments with the different nlsCre variants, eGFP-expressing cells were detected, indicating successful transfer of active recombinase. In contrast, eGFP expression was never detected in any single cell of the mock-treated reporter cell line. Quantification of eGFP-positive cells by fluorescence-activated cell cytometry (FACS) revealed a low but reproducible ratio of eGFP-positive reporter cells incubated with nlsCre, PTD-nlsCre and TLM-nlsCre, respectively (Table 1, rows 1–3). This strongly suggested that nlsCre without a defined protein transduction domain readily crossed the plasma membrane and induced recombination in the reporter cells. The efficiency was at least as high as that of the nlsCre variants fused to the Tat or PreS2 protein transduction domains.
Table 1. Recombination mediated by low concentrations of different Cre proteins.
| Cre protein | Recombination frequency |
|---|---|
| nlsCre | 3.2% (1.9–5.4%; n = 6) |
| PTD-nlsCre | 2.5% (1.9–3.5%; n = 6) |
| TLM-nlsCre | 2.1% (0.5–3.9%; n = 4) |
| wtCre | 1.5% (0.6–2.5%; n = 7) |
Reporter cells were incubated for 3–4 h with 0.8–1 µM nlsCre, PTD-nlsCre, TLM-nlsCre or wtCre. Forty-eight hours later, the percentage of eGFP-expressing cells representing the recombination frequency was determined by FACS analysis. At least two independent protein preparations were tested for each Cre variant.
To confirm the specificity of nlsCre-mediated recombination, different reporter cell clones were incubated with 0.8 µM nlsCre for 4 h. After 48 h, 3–4% eGFP-expressing cells were detected (Fig. 3A). DNA derived from these cell populations was used in a PCR, shown in Figure 3B. A 364 bp product indicative of the lox-specific excision of the RFP gene was obtained from nlsCre-treated cells, but not from the parental reporter cells (Fig. 3B). The identity of the PCR products was confirmed by direct sequencing. Thus, the addition of bacterially expressed, purified nlsCre recombinase was necessary and sufficient to induce recombination at the single nucleotide level in cultured cells.
We next asked whether the SV40-NLS or the c-myc epitope, present in nlsCre, mediated protein transduction. Using the same conditions as described above, wtCre protein was added to the reporter cells. Recombination was reproducibly observed in ∼1.5% of reporter cells (Fig. 3C and Table 1, row 4). Application of wtCre protein at higher concentrations led to an increased recombination frequency (10%, Table 2). This indicates that the wild-type Cre protein itself already possesses an intrinsic ability to pass the membrane barrier, to enter the nucleus and catalyze lox-specific recombination.
Table 2. Recombination mediated by wtCre protein.
| Concentration of wtCre protein | Recombination frequency |
|---|---|
| 0.8 µM | 1.5 |
| 1.4 µM | 4.5 |
| 3.5 µM | 8.0 |
| 5.8 µM | 10.1 |
Reporter cells were incubated for 3 h with different concentrations of wtCre. Forty-eight hours later, the percentage of eGFP-expressing cells representing the recombination frequency was determined by FACS analysis.
High efficiency of recombination by purified Cre protein
Although unmodified Cre recombinase can pass the membrane barrier and mediate lox-specific recombination, we concentrated our efforts on nlsCre, since presence of the additional NLS possibly accelerates transfer into the nucleus.
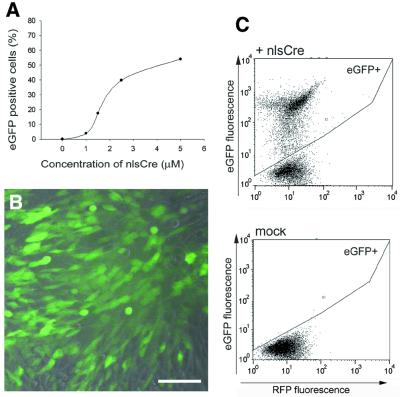
As expected, increasing nlsCre concentrations mediated increased recombination efficiencies. Using 5 µM nlsCre, a recombination rate of >50% was achieved (Fig. 4). Thus, Cre protein lacking any heterologous protein transduction domains can induce recombination in eucaryotic cells with high efficiency.
Figure 4.
Efficient recombination induced by recombinant nlsCre protein. (A) RFP-expressing reporter cells were incubated for 6 h in medium containing 1.0, 1.5, 2.5 or 5 µM purified nlsCre protein. The proportion of eGFP-expressing cells (in which recombination has occurred) was determined 48 h later by FACS analysis. The plotted values are representative of two independent experiments. Fluorescence microscopy (B) and dot blot analysis of RFP/eGFP-expressing cells detected by FACS (C) of reporter cells treated with 2.5 µM nlsCre protein. Bar, 40 µm.
Specific recombination in transgenic mouse bone marrow cells
To examine the ability of externally added nlsCre protein to induce site-specific recombination in primary cells, bone marrow cells from C/EBPαfl/fl mice (14) were tested. This mouse strain carries a C/EBPα locus engineered to contain flanking loxP sites (C/EBPαfl/fl). Bone marrow cells isolated from these mice were incubated with 1.5 µM nlsCre for 7 h. To verify lox-specific recombination on the C/EBPαfl/fl locus, PCRs were performed on isolated genomic DNA with subsequent sequencing of the amplification products (Fig. 5). Site-specific recombination leading to a 400 bp PCR product (Fig. 5, PCR2) was only observed after treatment of bone marrow cells with nlsCre. Sequence analysis of the PCR product confirmed the specificity of recombination. Thus, purified nlsCre was also capable of entering primary bone marrow cells and mediated site-specific recombination.
DISCUSSION
We present here an alternative strategy to induce site-specific recombination by simple addition of recombinant, purified Cre recombinase. Direct protein transduction of nlsCre circumvents the introduction of Cre-encoding nucleic acids into cells, thereby eliminating the risk of insertional mutagenesis and the proposed genomic instability induced by continuous Cre expression (4,5). Our results demonstrated that the translocation of Cre lacking a heterologous protein translocation motif was at least as efficient as the transfer of Cre fused to the protein transduction domain of Tat (PTD) or that of PreS2 (TLM). We also showed that even unmodified Cre recombinase, differing only at its N-terminal amino acid, was able to cross cellular membranes, enter the nucleus and catalyze recombination at lox sites.
In a recent article published during revision of this work, Peitz et al. compared the transduction efficiency of different Cre proteins (15). Whereas the transduction efficiency of recombinant Cre protein containing the SV40 NLS was very similar to the results presented here, Peitz et al. did not achieve recombination in >5% of fibroblasts, even after 16 h incubation with >9 µM Cre protein lacking the SV40 NLS, arguing against an intrinsic capability of Cre to transduce cells. In our study, however, the recombination frequency correlates with the concentration of wtCre and reaches ∼10% after 3 h incubation with wtCre, suggesting that the NLS is not essential for the passage of Cre through the plasma membrane but enhances recombination at another level (e.g. by localizing most of the Cre protein into the nucleus).
Efficient recombination was achieved both in a cultured cell line as well as in primary bone marrow cells without the need for serum deprivation as reported by others (6,16). The successful transfer of Cre protein into primary bone marrow allows the targeting of hematopoietic stem cells. These quiescent cells are not suited for manipulation by conventional gene transfer technologies. The cell cycle independence of Cre has been previously shown by the rapid catalysis of genomic DNA recombination in post-mitotic cells (17,18). Therefore, this method has the potential of reversing genetic alterations introduced into hematopoietic stem cells during gene therapy. The direct transfer of Cre protein will certainly extend the spectrum of its already wide applications.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Heinz Lother for critical advice and tefan Albert for providing the modified pGEX-2T(KT) expression vector. We are also very grateful to Julie Lekstrom-Himes and Dan Tenen for providing us with the C/EBPαfl/fl mouse strain. This work was supported by the Bundesministerium für Bildung und Forschung/Vision7 and the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Testa G. and Stewart,A.F. (2000) Creating a transloxation. Engineering interchromosomal translocations in the mouse. EMBO Rep., 1, 120–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauer B. (1998) Inducible gene targeting in mice using the Cre/lox system. Methods, 14, 381–392. [DOI] [PubMed] [Google Scholar]
- 3.Bergemann J., Kuhlcke,K., Fehse,B., Ratz,I., Ostertag,W. and Lother,H. (1995) Excision of specific DNA-sequences from integrated retroviral vectors via site-specific recombination. Nucleic Acids Res., 23, 4451–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silver D.P. and Livingston,D.M. (2001) Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell, 8, 233–243. [DOI] [PubMed] [Google Scholar]
- 5.Loonstra A., Vooijs,M., Beverloo,H.B., Allak,B.A., van Drunen,E., Kanaar,R., Berns,A. and Jonkers,J. (2001) Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl Acad. Sci. USA, 98, 9209–9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oess S. and Hildt,E. (2000) Novel cell permeable motif derived from the PreS2-domain of hepatitis-B virus surface antigens. Gene Ther., 7, 750–758. [DOI] [PubMed] [Google Scholar]
- 7.Nagahara H., Vocero-Akbani,A.M., Snyder,E.L., Ho,A., Latham,D.G., Lissy,N.A., Becker-Hapak,M., Ezhevsky,S.A. and Dowdy,S.F. (1998) Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nature Med., 4, 1449–1452. [DOI] [PubMed] [Google Scholar]
- 8.Schambach A., Wodrich,H., Hildinger,M., Bohne,J., Krausslich,H.G. and Baum,C. (2000) Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol. Ther., 2, 435–445. [DOI] [PubMed] [Google Scholar]
- 9.Hildinger M., Abel,K.L., Ostertag,W. and Baum,C. (1999) Design of 5′ untranslated sequences in retroviral vectors developed for medical use. J. Virol., 73, 4083–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zufferey R., Donello,J.E., Trono,D. and Hope,T.J. (1999) Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol., 73, 2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klump H., Schiedlmeier,B., Vogt,B., Ryan,M., Ostertag,W. and Baum,C. (2001) Retroviral vector-mediated expression of HoxB4 in hematopoietic cells using a novel coexpression strategy. Gene Ther., 8, 811–817. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell D.A., Marshall,T.K. and Deschenes,R.J. (1993) Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast, 9, 715–722. [DOI] [PubMed] [Google Scholar]
- 13.Prassolov V., Meyer,J., Brandenburg,G., Hannemann,J., Bergemann,J., Ostertag,W. and Stocking,C. (2001) Functional identification of secondary mutations inducing autonomous growth in synergy with a truncated interleukin-3 receptor: implications for multi-step oncogenesis. Exp. Hematol., 29, 756–765. [DOI] [PubMed] [Google Scholar]
- 14.Zhang P., Iwasaki-Arai,J., Lodie,T., Fenyus,M.L., Lekstrom-Himes,J., Foerster,I., Akashi,K. and Tenen,D.G. (2001) C/ebp alpha deficiency blocks granulocytic differentiation at the common myeloid progenitor stage in both adult and fetal liver myelopoiesis. Blood, 98, 792a–793a. [Google Scholar]
- 15.Peitz M., Pfannkuche,K., Rajewsky,K. and Edenhofer,F. (2002) Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl Acad. Sci. USA, 99, 4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jo D., Nashabi,A., Doxsee,C., Lin,Q., Unutmaz,D., Chen,J. and Ruley,H.E. (2001) Epigenetic regulation of gene structure and function with a cell-permeable Cre recombinase. Nat. Biotechnol., 19, 929–933. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y.H., Sauer,B., Johnson,P.F. and Gonzalez,F.J. (1997) Disruption of the c/ebp alpha gene in adult mouse liver. Mol. Cell. Biol., 17, 6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Krushel,L.A. and Edelman,G.M. (1996) Targeted DNA recombination in vivo using an adenovirus carrying the cre recombinase gene. Proc. Natl Acad. Sci. USA, 93, 3932–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]