Abstract
Analysis of haplotypes is an important tool in population genetics, familial heredity and gene mapping. Determination of haplotypes of multiple single nucleotide polymorphisms (SNPs) or other simple mutations is time consuming and expensive when analyzing large populations, and often requires the help of computational and statistical procedures. Based on double PCR amplification of specific alleles, described previously, we have developed a simple, rapid and low-cost method for direct haplotyping of multiple SNPs and simple mutations found within relatively short specific regions or genes (micro-haplotypes). Using this method, it is possible to directly determine the physical linkage of multiple heterozygous alleles, by conducting a series of double allele-specific PCR amplification sets with simple analysis by gel electrophoresis. Application of the method requires prior information as to the sequence of the segment to be haplotyped, including the polymorphic sites. We applied the method to haplotyping of nine sites in the chicken HSP108 gene. One of the haplotypes in the population apparently arose by recombination between two existing haplotypes, and we were able to locate the point of recombination within a segment of 19 bp. We anticipate rapidly growing needs for SNP haplotyping in human (medical and pharmacogenetics), animal and plant genetics; in this context, the multiple double PCR amplifications of specific alleles (MD-PASA) method offers a useful haplotyping tool.
INTRODUCTION
Chromosomal segments within limited regions or genes (micro-haplotypes) maintain their integrity, unaffected by recombination, over very long periods. Consequently, intragenic or micro-haplotypes that are found to be identical can be assumed to be identical by descent (1–3). For this reason, haplotyping methods are used to compare DNA segments of non-familial individuals in cases where populations rather than segregating families are studied. Haplotyping greatly increases the genetic information content by converting essentially bi-allelic markers into multi-allellic systems (4,5). As such, micro-haplotypes have high polymorphic information content, allowing the inheritance of genomic elements of interest to be followed.
Determining single nucleotide polymorphism (SNP)-based haplotypes by direct sequencing or by genotyping of individual polymorphic sites requires either parental information or information from large numbers of individuals, at least some of whom must be homozygous. This procedure requires statistical analysis methods with computational aid, and may become burdensome (4,6–10). As an alternative, in this study a haplotyping method is presented by which haplotypes can directly be determined for specific individuals.
In recent years the importance of SNPs as genomic markers has risen (11–16). Many new methods have been developed to determine SNP alleles (see 1,12,17–26 for representative examples). PCR amplification of specific alleles (PASA) [also known as amplification refractory mutation system (ARMS) and allele specific amplification (ASA)] utilizes primers designed to match different SNP alleles and amplify them selectively (27–29). Lo et al. (30) described a double-ARMS method enabling haplotyping of two polymorphic sites. Independently, Sarkar and Sommer (31) described a double-PASA method for identifying haplotypes composed of two SNPs. Sarkar and Sommer (31) suggested that the method could be expanded to identify haplotypes of more than two SNPs. In the present study we follow up on this suggestion, and present a multiple-double PASA (MD-PASA) method for determining micro-haplotypes consisting of multiple polymorphic sites within regions of hundreds of bases. Higher allele specificity is supplemented to the MD-PASA method by using deliberately mismatched internal primers, following Okimoto and Dodgston (32).
While working on the chicken HSP108 gene, we found that a 653 bp segment including intron 8 is rich in SNPs and other simple polymorphisms. We demonstrate here the application of the MD-PASA method for directly determining nine-site micro-haplotypes in this segment.
MATERIALS AND METHODS
Theory
The double PASA (D-PASA) procedure. The D-PASA procedure (31), and the double ARMS procedure (30), for identifying haplotypes of two SNP sites, employed four separate PCR amplifications. Each of the four amplifications involved a combination of two pairs of allele-specific primers: a pair of forward (F) primers and a pair of reverse (R) primers. We will denote the primers using a letter for the site and a number for the allele. Thus, the primer A1F designates a forward primer specific to allele 1 of site A. The B site primers would then necessarily have to be reverse primers. The four primers are: A1F, A2F, B1R, B2R. Since the two sites being haplotyped are known, the four combinations are denoted by allele only. Thus, combination 1-1 represents the amplification with primers A1F, B1R; combination 1-2 represents the amplification with primers A1F, B2R, and so on. A product is obtained only when both primers match the existing alleles on the same DNA molecule (chromosome). Thus, by observing the reactions that yield products, the allelic combination on the two chromosomes is immediately determined. For detailed reference as to set up and validation of double allele specific amplification see Lo et al. (30) and Sarkar and Sommer (31).
The MD-PASA procedure. The MD-PASA begins with a simple D-PASA. This immediately provides two-site haplotypes, as above. This is followed by a second D-PASA, which involves one heterozygous site from the first D-PASA, and any other site. The combinations that yield products in the second amplification enable a three-site haplotype to be inferred. The process is now reiterated, adding a fourth and then fifth site, and so on until the entire haplotype is completed. In order to enable multiple site haplotyping to proceed to completion, the primer of the most 5′ heterozygous site must be a forward primer, and the primer of the most 3′ heterozygous site must be a reverse primer. The remaining ‘internal’ primers can be any combination of reverse and forward primers, which allows flexibility in primer design.
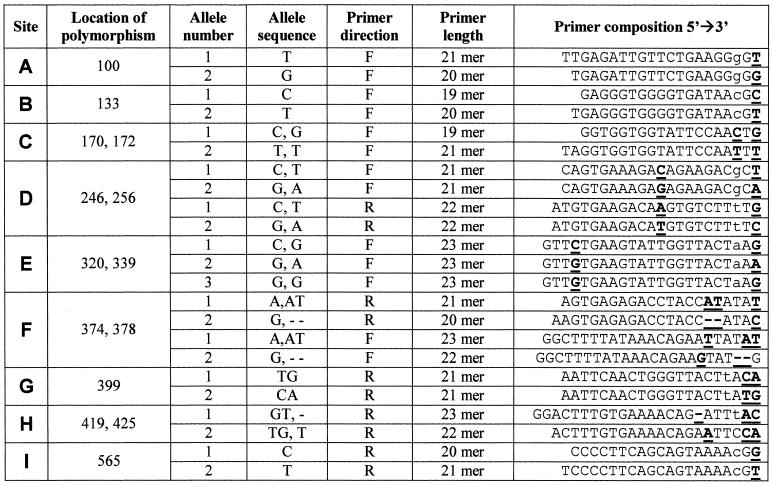
Allele-specific primers. Selections of polymorphic sites for haplotyping and direction of primers were made according to the DNA sequence (Fig. 1) and regular conditions for optimal primer design. We chose nine of the polymorphic sites for haplotyping, for which we designed five ‘forward’ and four ‘reverse’ primer-pairs, denoted ‘F’ and ‘R’, respectively. Primers were designated A to I, according to order of appearance in the sequence (Table 1). The primers were designed to specifically match SNP alleles (one or two nucleotides) at their 3′ end.
Figure 1.
Polymorphic sites haplotyped by MD-PASA. Nine polymorphic sites in intron 8 of the chicken HSP108 gene, denoted A to I, were chosen for micro-haplotyping. Alleles of each site are randomly placed with respect to a slash (/) with no inferred linkage to alleles of other sites. Sequences used for allele specific primers are marked by arrows which show the direction of the primer. When the primer sequence covered two polymorphic sites (sites C, D, E, F and H), linkage was assumed between the two alleles placed to the right of the slashes and between the two alleles placed to the left of the slashes.
Table 1. Primers used to amplify specific alleles of nine bi-allelic sites in intron eight of the chicken HSP108, by MD-PASA.
Polymorphic sites are denoted A to I. Locations of the polymorphisms, which the primers match, are in bases from the beginning of sequenced segments (Fig. 1). Allele-specific nucleotides are bold and underlined. Deliberate mismatches are in lower case.
In five of the sites, the primers overlap an additional SNP allele inner to the 3′ end, under the assumption that these two SNPs are proximate enough to be regarded as one bi-allelic site (primers for sites C, D, E, F and H) (Fig. 1 and Table 1). The particular allele combinations within the region relied on fragment sequence information for these sites. For one site (E), overlapping two SNPs, a third specific primer was designed, to help locate a site of recombination (see Results). Site-alleles and specific primers were denoted 1 or 2, arbitrarily, to simplify analysis.
In addition to the described D-PASA protocol (31) we introduced deliberate mismatches in the third position from the 3′ end of the primers. Okimoto and Dodgston (32) reported that this improves specificity, allows a wider range of allele-specific amplification conditions and improves reproducibility. Thus, as they recommended, an additional deliberate third position mismatch was designed in primers specific to one SNP allele at their 3′ end (primers for sites A, B, D, E, G and I) (Fig. 1 and Table 1).
For the nine chosen sites, we could have chosen, respectively, between one and eight forward and, accordingly, eight to one reverse primers; this allows flexibility in primer design. In addition to normal considerations in primer design, considerations were to allow reasonable size of product for amplification by PCR, and similar annealing temperatures for all primers.
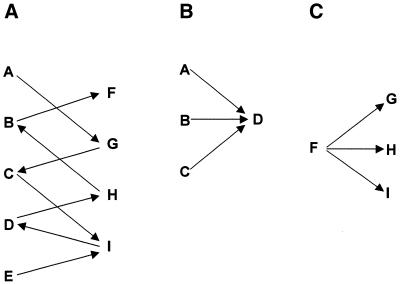
Haplotyping scheme design. In the present study, we proceeded by designing a standard haplotyping scheme, which we applied to all of the individuals to be haplotyped. Having five forward and four reverse primer pairs, 20 site pairings are theoretically possible, although only eight are required to determine the haplotype of nine polymorphic sites. The main goal in constructing a scheme is that all sites be covered through an appropriate arrangement of connecting sites, so that linkage between all alleles can be determined. Many different combinations of site couples can fulfill this goal, which allows flexibility in choosing a scheme. In the present study, the scheme shown in Figure 2A and other schemes (not shown) were implemented, but only results for the scheme shown in Figure 2A are given. In practice, direct identification of an individual’s two haplotypes requires a minimum of 4×(N – 1) PCR amplifications, for (N – 1) site couples, if all polymorphic sites are haplotyped.
Figure 2.
MD-PASA schemes. (A) An MD-PASA scheme of site couples was made to allow analysis of physical linkage between all nine sites. Order of amplifications would be, in this case, A-G, C-G, C-I, E-I, D-I, D-H, B-H and B-F. (B) A scheme for haplotype analysis of individuals who are heterozygous for sites A to D (like individual 3). (C) A scheme for haplotype analysis of individuals who are heterozygous for sites F to I (like individual 4).
Haplotyping can be done after genotyping all sites. In that case, only the individual’s heterozygous sites need to be included in its haplotyping scheme, thus reducing the total haplotyping load.
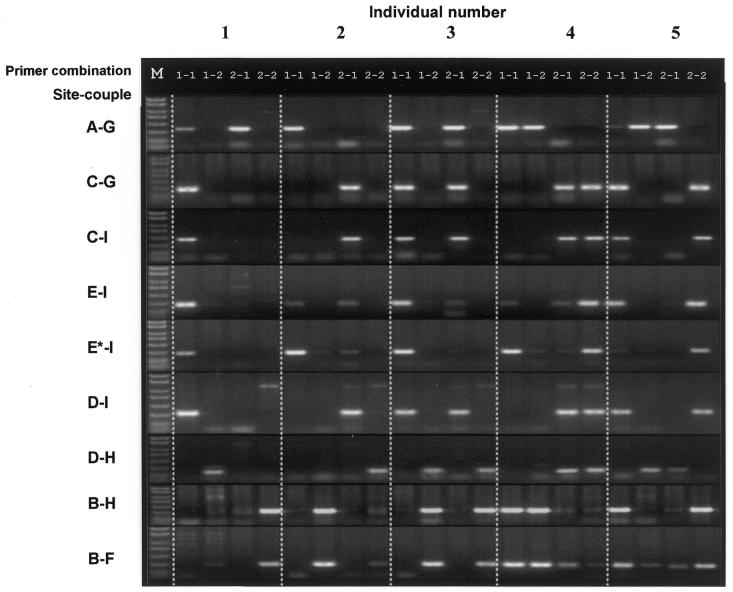
Analysis. Figure 3 presents the eight double-PASA products, after electrophoresis in an agarose gel. Site couples (D-PASAs) are according to the scheme in Figure 2A. The figure is laid out so that lane-quartets for each individual tested create a ‘column’. For each individual, alleles are determined from each quartet, going from the top of the column to the bottom.
Figure 3.
MD-PASA amplification results. Genomic DNA from five representative chickens was used for comparative micro-haplotyping. Presented here are examples of eight double amplifications for these individuals. Each individual (1–5) went through eight groups of separate two-site haplotypings (by D-PASA), each two-site haplotyping with four different allele specific primer couples, following the scheme in Figure 2A. In each group, four lanes (1-1, 1-2, 2-1 and 2-2 = ‘quartet’) represent a two-site haplotype for one individual. The position of product(s) within a quartet points to the allele specific primer pair(s) used to obtain the product(s), thus showing the linkage of alleles between the two polymorphic sites. For site E, two sub-sets of amplifications were made, after recognizing inconsistencies for individuals 2, 3 and 4, in comparison with other sites; in E-I, forward primers E1 and E2 (Table 1) were used; whereas in E*-I, primers E3 substituted primers E1 (Table 1). (DNA marker sizes in bp: 50, 150, 300, 500, 750, 1000, 1500 and 2000.)
Results of two-site haplotypings can be of three types. (i) An individual homozygous at both bi-allelic sites yields only one significant product on the electrophoresis gel. The PCR product points directly at the two alleles composing the homozygous haplotype (e.g. individual 1, for all D-PASAs, Fig. 3). (ii) Heterozygosity at one of the two sites results in two products. The two products are given by one primer for the homozygous site and two primers for the heterozygous site, thus identifying the two haplotypes (e.g. individuals 3 and 4 for A-G D-PASA, Fig. 3). (iii) When both bi-allelic sites are heterozygous, a PCR product is possible only for two alleles that are physically on the same DNA molecule. The two products point to the only possible two pairs of alleles, and so identify the two haplotypes (e.g. individual 5, Fig. 3).
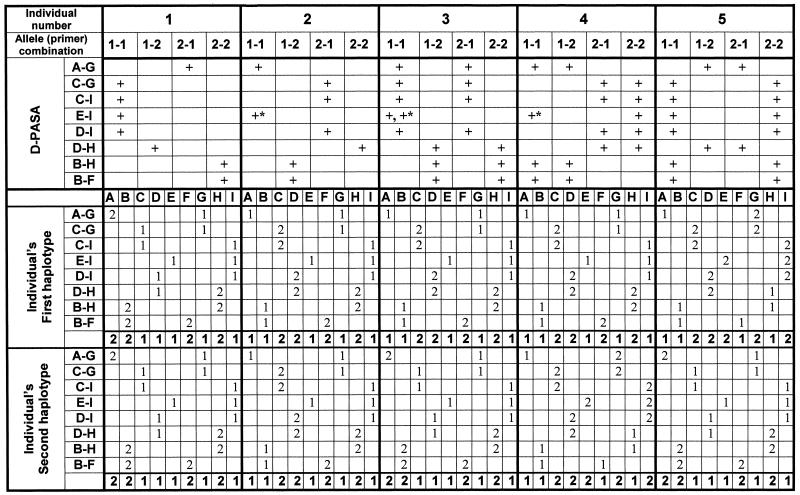
For purposes of analysis, we found it convenient to arrange results of all D-PASA amplifications for each individual as a column of eight lane-quartets and to show this in tabular form (compare Fig. 3 with the upper part of Table 2). For each individual, alleles are then determined from each D-PASA, following the connecting alleles. In this way, for each site pair, alleles are attributed to each site. Alleles for all sites are summed into two concluding haplotype codes for each individual. For homozygous individuals, the two concluding haplotypes would be the same. With the help of Table 1, haplotype codes are ‘deciphered’ into the actual haplotypes of SNP alleles, as shown in Figure 4.
Table 2. Analysis of MD-PASA products and identification of haplotypes.
For five individuals, results of the D-PASAs, as shown in Figure 3, were gathered and analyzed in this table. Each site pair was marked with a plus sign according to the allele-specific primers that yielded a significant amplification product. Physical linkages between the alleles were then identified stepwise, according to the order of site pairs, as set by the scheme in Figure 2A. Physical linkages are then concluded as nine-site haplotypes (in bold). The process is first applied to the homozygous individuals (here, numbers 1 and 2) and individuals that are double heterozygous in all site pairs (here, number 5). In other individuals, homozygous parts of the haplotypes are identified directly (E to I for individual 3 and A to D for individual 4), and the heterozygous parts are compared with already identified haplotypes in other individuals (E to I for individual 4 and A to D for individual 3). Results designated with a plus sign and asterisk (E to I for individuals 2, 3 and 4) were obtained by using primer E3 in D-PASA for site pair E-I (E*-I in Figure 3).
Figure 4.
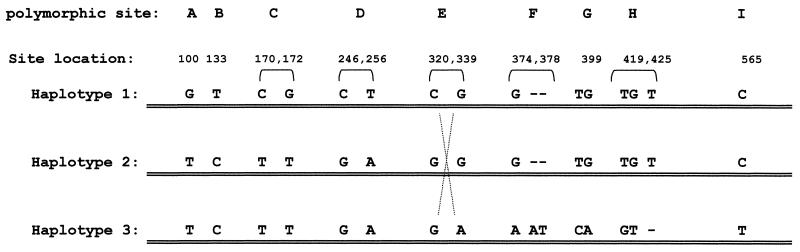
The three micro-haplotypes identified by MD-PASA. The three haplotypes, which were identified by MD-PASA analysis according to Table 2. Comparing the three haplotypes, we assume that a recombination event occurred between haplotypes 1 and 3, within the two SNPs of site E, to create haplotype 2 (dashed cross).
Experimental
Genomic DNA. Whole blood samples were obtained from five White-Rock chickens (Gallus gallus) (ANAK Breeders, Ltd, Netanya, Israel) and five local domestic chickens. Genomic DNA was isolated from 30 µl of whole fresh chicken blood by a mini-prep phenol-chloroform DNA extraction protocol, followed by ethanol precipitation (modification of 33).
PCR. Oligonucleotide primers were designed using the Generunner 3.05 software (Hastings Software Inc.), and purchased from GENSET, SA (Paris, France). Taq DNA polymerase and dNTPs were purchased from MBI FERMENTAS (Germany). To reduce reagent and genomic DNA requirements, PCR amplifications were carried out in a low total volume of 11 µl, which contained 10% reaction buffer (100 mM Tris–HCl pH 8.8, 500 mM KCl, 0.8% Nonidet P-40, 15 mM MgCl2), 0.4 µM of each primer, 0.175 mM dNTP mix, 0.25 U Taq DNA polymerase and ∼30 ng chicken genomic DNA.
Specific amplification conditions. As noted above, all allele specific primers were designed to have roughly the same annealing temperature (according to the classical A/T = 2°C, C/G = 4°C calculation), so the same PCR protocol was used for each site pair, enabling the same thermocycler to amplify more than one site pair simultaneously. A 5 min denaturing step at 95°C was followed by 30 cycles of: 1 min at 95°C, 1 min at 59°C and 1 min at 72°C. A final 8 min step at 72°C allowed final elongation. Amplification reactions were performed on an MJ Research (Watertown, MA, USA) 96-well thermocycler.
Gel electrophoresis. PCR amplification products were mixed with 1 µl loading buffer (60% glycerol, trace bromphenol blue) and run through a 1.5–1.8% agarose gel, stained with ethidium bromide. Electrophoresis was at 70–80 V for 35–40 min, after which gels were photographed and analyzed.
RESULTS
Eight D-PASA analyses, each consisting of four two-site amplifications, were made for each of the 10 chickens. Site pairs were A-G, C-G, C-I, E-I, D-I, D-H, B-H and B-F, according to the scheme in Figure 2A. We also implemented other schemes with the same level of success (data not shown).
Figure 3 presents the eight D-PASA products of five representative individuals after electrophoresis in agarose gel. These specific individuals were chosen since they exhibit five different possible results (all of which are repeated in the remaining five individuals). Primer combinations are represented by 1-1, 1-2, 2-1 or 2-2 for each D-PASA. Individuals 2, 3 and 4 did not show clear conclusive results with regard to the amplification reaction involving sites E and I (E-I, Fig. 3). Since site E was a double SNP, it was suspected that one of the primers might not exactly match the intra-primer allele combination found in these individuals, and therefore a third primer for allele 3 was designed (Table 1), which resolved inconclusive results in these individuals (Fig. 3). At site E, individuals 1 and 5 were genotyped for alleles 1 and 2 (D-PASA designated E-I, Fig. 3), consistent with the results pattern of other D-PASAs; individual 2 showed significant product only for allele 3 (E*-I) and was genotyped as homozygous for it. Individual 3 showed significant products for alleles 1 (E-I) and 3 (E*-I), and was genotyped as heterozygous for them; individual 4 showed significant products for alleles 2 (E-I) and 2 and 3 (E*-I), and was genotyped as heterozygous for them.
The upper part of Table 2 shows the results of the eight separate two-site haplotypings, for the five representative individuals, for which electrophoresis gels are presented in Figure 3. A plus sign represents a significant amplification product for allele combination(s) in a site couple. The upper part of Table 2 is, in fact, a tabular representation of the results shown in Figure 3. The lower part shows the inferred haplotypes of each of the D-PASA amplifications.
Individuals 1 and 2 were homozygous, each for a different haplotype, designated haplotypes 1 and 2, respectively. Individual 5 was heterozygous at all sites, and on MD-PASA analysis was found to carry haplotype 1 and a third haplotype, designated haplotype 3. Individual 3 was heterozygous at sites A to D and homozygous at sites E to I. In effect, this meant that it was heterozygous at the sites of all forward primers and homozygous at those of all reverse primers (with no heterozygous connecting site for sites A to D). As a result, the MD-PASA scheme procedure was not informative with respect to haplotypic structure for this individual. However, the MD-PASA did provide genotypes for all sites in individual 3. By consideration of the genotypes for the homozygous and heterozygous sites, it was easy to infer that individual 3 was heterozygous for haplotypes 1 and 2 (Table 2). To verify this, a reverse primer was prepared for site D, for direct analysis of sites A to D by MD-PASA (Fig. 2B). The MD-PASA results confirmed the inferred haplotypes (data not shown). Individual 4 was homozygous at sites A to D and heterozygous at sites E to I. In this case, therefore, the individual was homozygous for all forward primers and heterozygous for all reverse primers so that, once again, the MD-PASA scheme was not informative with respect to haplotypic structure (with no heterozygous connecting site for sites F to I). Here too, by consideration of genotypes for the homozygous and heterozygous sites, it was easy to infer that individual 4 was heterozygous for haplotypes 2 and 3. In this case, a forward primer was prepared for site F, for direct analysis of sites F to I by MD-PASA (Fig. 2C). Here too, the MD-PASA results confirmed the inferred haplotypes (data not shown).
DISCUSSION
We chose to demonstrate the application of the MD-PASA method we developed for haplotype analysis of 10 non-related individuals, of which results from five specific ones are presented. The haplotype analysis by MD-PASA as initially schemed was essential to determine the haplotypes of individual 5, who was heterozygous for all sites included in haplotype analysis. Individuals 1 and 2 were homozygous for all sites, and therefore MD-PASA was not needed for haplotype analysis. Their haplotypes could easily have been inferred directly from sequence analysis or by individual genotyping of each site. In our case, genotyping analysis was obtained by MD-PASA, and these two individuals served as controls. Individuals 3 and 4 were analyzed first through the genotype information alone provided by the MD-PASA. Having this information, we designed two more primer pairs, so that MD-PASA could be applied for direct analysis of haplotypes from heterozygous parts.
In all, therefore, a total of three haplotypes were found, in 10 individual chickens. We demonstrated here that it is possible to identify every existing haplotype in a population, by applying the MD-PASA method to heterozygous SNP sites. Having no initial information as to the existing haplotypes, an efficient approach would then be to genotype all sites first, for representative individuals. Having information as to homozygous and heterozygous sites, a site-pairing scheme should be plotted accordingly (only for heterozygous sites), to be applied for all individuals tested. In cases of double heterozygosity, D-PASA will also convey haplotype information. A second step of the MD-PASA would be needed to connect the information of partial haplotypes that could not be inferred by the initial step of D-PASAs to identify the complete haplotypes.
Examining the three haplotypes, it is evident that haplotypes 1 and 3 differ at all polymorphic sites. One half of haplotype 2 is identical to haplotype 3 (sites A to D) and the other half is identical to haplotype 1 (sites F to I). At site E, all three haplotypes carry different alleles. This can be interpreted to indicate that haplotype 2 was formed by a recombination event between haplotypes 1 and 3, with the point of recombination within site E. Thus, the point of recombination has been mapped between the two sites in positions 320 and 339 (Fig. 1).
For each D-PASA, four primers and four amplifications are required. In comparison, allele specific amplification (ASA, PASA, ARMS) for genotyping of two separate sites requires four amplifications, four allele specific primers and one or two common opposing primers (27–29). Therefore, haplotyping information can be collected by D-PASAs, at no ‘extra cost’. In addition, in D-PASA two specific primers act in each amplification to yield a product, thus creating a synergistic effect of improved specificity (30; Y.Eitan and Y.Kashi, unpublished data). Calibration of reaction conditions were rather simply achieved, as all primers were designed to work roughly in the same annealing temperature; primers ‘covered’ more than one SNP, or contained a deliberate mismatch. The deliberate mismatches in the third primer position proved to be important for increasing specificity and reliability of the MD-PASA method.
The information obtained by the MD-PASA method for 10 individual chickens, as described, was confirmed in each case by direct sequencing of the analyzed DNA segment. In addition, the MD-PASA method was validated by the fact that allele specific primers for sites which ‘participated’ in more than one amplification set (i.e. connecting sites) showed consistent results (Fig. 3). Also, homozygous individuals served both as negative and positive controls (Fig. 3).
In order to examine the reproducibility of the MD-PASA method, we applied it successfully to determine micro-haplotypes in two additional chicken genes, in a large number of individuals from three different breeds (Y.Eitan and Y.Kashi, manuscript in preparation).
The density of SNPs in the human genome is estimated to be one SNP per kb on average (34). MD-PASA can be readily applied to identify haplotypes of SNPs that are in this distance range. The distance between the farthest SNPs haplotyped by D-PASA in the present study was ∼460 bp (sites A and I, data not shown). The MD-PASA method was developed using the chicken genome as a model; however, there does not appear to be any impediment to its implementation in mammalian or human genomes. Application of MD-PASA for haplotyping distant SNPs requires the use of allele-specific primers in both directions for each of the SNP sites haplotyped. In this way the use of MD-PASA can be extended to haplotype segments up to a few kilobases long.
CONCLUSION
Our method demonstrates the possibility and simplicity of directly determining a micro-haplotype consisting of numerous polymorphic sites, requiring genomic DNA samples only from the individuals of interest. It does not require information from population studies or pedigree analysis (1,35). It also requires only the basic thermocycler and gel apparatus available in most laboratories, and is relatively low cost. The method can be applied to any series of SNPs or simple mutations from any diploid organism. The only requirements are (i) that the sequence of the segment to be haplotyped should be known, including the polymorphic sites, and (ii) that the largest distance between adjoining heterozygous sites in the haplotyped individuals should be within the range that can be amplified by PCR.
In this study, we found three haplotypes in a relatively inbred population of chickens. Human populations are known to be much more heterogeneous, and therefore more difficult for inferring haplotypes (11,34,36–40). This emphasizes the importance of the MD-PASA as a method for haplotyping.
SNPs have recently become the most widely used polymorphic marker in genomic research. We anticipate that there will be rapidly growing needs for SNP haplotyping, in human, animal and plant genetics (e.g. medicine, pharmacogenetics, evolution, ecology and breeding). In this context, the MD-PASA method offers a useful haplotyping tool.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by the Technion Otto Meyerhof Center for Biotechnology, established by the Minerva Foundation, Germany.
REFERENCES
- 1.Liu J.S., Sabatti,C., Teng,J., Keats,B.J.B. and Risch,N. (2001) Bayesian analysis of haplotypes for linkage disequilibrium mapping. Genome Res., 11, 1716–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tadmouri G.O., Garguier,N., Demont,J., Perrin,P. and Basak,A.N. (2001) History and origin of β-thalassemia in Turkey: sequence haplotype diversity of β-globin genes. Hum. Biol., 73, 661–674. [DOI] [PubMed] [Google Scholar]
- 3.Lyttle T.W. (1993) Cheaters sometimes prosper: distortion of mendelian segregation by meiotic drive. Trends Genet., 9, 205–210. [DOI] [PubMed] [Google Scholar]
- 4.Martin E.R., Scott,W.K., Nance,M.A., Watts,R.L., Hubble,J.P., Koller,W.C., Lyons,K., Pahwa,R., Stern,M.B., Colcher,A. et al. (2001) Association of single-nucleotide polymorphisms of the tau gene with late-onset Parkinson disease. J. Am. Med. Assoc., 286, 2245–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Judson R., Stephens,J.C. and Windemuth,A. (2000) The predictive power of haplotypes in clinical response. Pharmacogenomics, 1, 15–26. [DOI] [PubMed] [Google Scholar]
- 6.Clark V.J., Metheny,N., Dean,M. and Peterson,R.J. (2001) Statistical estimation and pedigree analysis of CCR2-CCR5 haplotypes. Hum. Genet., 108, 484–493. [DOI] [PubMed] [Google Scholar]
- 7.Rohde K. and Fuerst,R. (2001) Haplotyping and estimation of haplotype frequencies for closely linked biallelic multilocus genetic phenotypes including nuclear family information. Hum. Mutat., 7, 289–295. [DOI] [PubMed] [Google Scholar]
- 8.Schork N.J., Fallin,D. and Lanchbury,J.S. (2000) Single nucleotide polymorphisms and the future of genetic epidemiology. Clin. Genet., 58, 250–264. [DOI] [PubMed] [Google Scholar]
- 9.Fallin D. and Schork,N.J. (2000) Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data. Am. J. Hum. Genet., 67, 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long A.D. and Langley,C.H. (1999) The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res., 9, 720–731. [PMC free article] [PubMed] [Google Scholar]
- 11.Olivier M., Bustos,V.I., Levy,M.R., Smick,G.A., Moreno,I., Bushard,J.M., Almendras,A.A., Sheppard,K., Zierten,D.L., Aggarwal,A. et al. (2001) Complex high-resolution linkage disequilibrium and haplotype patterns of single-nucleotide polymorphisms in 2.5 Mb of sequence on human chromosome 21. Genomics, 78, 64–72. [DOI] [PubMed] [Google Scholar]
- 12.Mir K.U. and Southern,E.M. (2000) Sequence variation in genes and genomic DNA: methods for large-scale analysis. Annu. Rev. Genomics Hum. Genet., 1, 329–360. [DOI] [PubMed] [Google Scholar]
- 13.Vilain E. (1998) CYPs, SNPs and molecular diagnosis in the postgenomic era. Clin. Chem., 44, 2403–2404. [PubMed] [Google Scholar]
- 14.Wang D.G., Fan,J.B., Siao,C.J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J. et al. (1998) Large-scale identification, mapping, and genotyping of single nucleotide polymorphisms in the human genome. Science, 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- 15.Gu Z., Hillier,L. and Kwok,P.-Y. (1998) Single nucleotide polymorphism hunting in cyberspace. Hum. Mutat., 12, 221–225. [DOI] [PubMed] [Google Scholar]
- 16.Landergren U., Nilsson,M. and Kwok,P.-Y. (1998) Reading bits of genetic information: methods for single-nucleotide polymorphism analysis. Genome Res., 8, 769–776. [DOI] [PubMed] [Google Scholar]
- 17.Ahmadian A., Gharizadeh,B., Gustafsson,A.C., Sterky,F., Nyrén,P., Uhlén,M. and Lundeberg,J. (2000) Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem., 280, 103–110. [DOI] [PubMed] [Google Scholar]
- 18.Chen J.W., Iannone,M.A., Li,M.S., Taylor,J.D., Rivers,P., Nelsen,A.J., Slentz-Kesler,K.A., Roses,A. and Weiner,M.P. (2000) A microsphere-based assay for multiplex single nucleotide polymorphism analysis using single base extention. Genome Res., 10, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Levine,L and Kwok,P.-Y. (1999) Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res., 9, 492–498. [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis M.C. (2000) “Spot-on” SNP genotyping. Genome Res., 10, 895–897. [DOI] [PubMed] [Google Scholar]
- 21.Germer S. and Higuchi,R. (1999) Single-tube genotyping without oligonucleotide probes. Genome Res., 9, 72–78. [PMC free article] [PubMed] [Google Scholar]
- 22.Germer S., Holland,M.J. and Higuchi,R. (2000) High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res., 10, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin T.J. and Smith,L.M. (2000) Single-nucleotide polymorphism analysis by MALDI¯TOF mass spectrometry. Trends Biotechnol., 18, 77–84. [DOI] [PubMed] [Google Scholar]
- 24.Holloway J.W., Beghe,B., Turner,S., Hinks,L.J., Day,I.N.M. and Howell,W.M. (1999) Comparison of three methods for single nucleotide polymorphism typing for DNA bank studies: sequence-specific oligonucleotide probes hybridization, TaqMan liquid phase hybridization, and microarray diagonal gel electrophoresis (MADE). Hum. Mutat., 14, 340–347. [DOI] [PubMed] [Google Scholar]
- 25.Hoogendoorn B., Owen,M.J., Oefner,P.J., Williams,N., Austin,J. and O’Donovan,M.C. (1999) Genotyping single nucleotide polymorphisms by primer extension and high performance liquid chromatography. Hum. Genet., 104, 89–93. [DOI] [PubMed] [Google Scholar]
- 26.Moran P., Dightman,D.A. and Park,L.K. (1998) Nonelectrophoretic genotyping using allele-specific PCR and a dsDNA-specific dye. Biotechniques, 24, 206–212. [DOI] [PubMed] [Google Scholar]
- 27.Sommer S.S., Cassady,J.D., Sobell,J.L. and Bottema,C.D. (1989) A novel method for detecting point mutations or polymorphisms and its application to population screening for carriers of phenylketonuria. Mayo Clin. Proc., 64, 1361–1372. [DOI] [PubMed] [Google Scholar]
- 28.Okayama H., Curiel,D.T., Brantly,M.L., Holmes,M.D. and Crystal,R.G. (1989) Rapid, nonradioactive detection of mutations in the human genome by allele-specific amplification. J. Lab. Clin. Med., 114, 105–113. [PubMed] [Google Scholar]
- 29.Newton C.R., Graham,A., Heptinstall,L.E., Powell,S.J., Summers,C., Kalsheker,N., Smith,J.C. and Markham,A.F. (1989) Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res., 17, 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo Y.M., Patel,P., Newton,C.R., Markham,A.F., Fleming,K.A. and Wainscoat,J.S. (1991) Direct haplotype determination by double ARMS: specificity, sensitivity and genetic applications. Nucleic Acids Res., 19, 3561–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkar G. and Sommer,S.S. (1991) Haplotyping by double PCR amplification of specific alleles. Biotechniques, 10, 436–440. [PubMed] [Google Scholar]
- 32.Okimoto R. and Dodgston,J.B. (1996) Improved PCR amplification of multiple specific alleles (PAMSA) using internally mismatched primers. Biotechniques, 21, 20–26. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Dawson E., Chen,Y., Hunt,S., Smink,L.J., Hunt,A., Rice,K., Livingston,S., Bumpstead,S., Bruskiewich,R., Sham,P. et al. (2001) A SNP resource for human chromosome 22: extracting dense clusters of SNPs from the genomic sequence. Genome Res., 11, 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lagziel,A., Lipkin,E. and Soller,M. (1996) Association between SSCP haplotypes at the bovine growth hormone gene and milk protein percentage. Genetics, 142, 945–951. [Corrigendum (1997) Genetics, 146, 442.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shifman,S. and Darvasi,A. (2001) The value of isolated populations. Nature Genet., 28, 309–310. [DOI] [PubMed] [Google Scholar]
- 37.Reich D.E., Cargill,M., Bolk,S., Ireland,J., Sabeti,P.C., Richter,D.J., Lavery,T., Kouyoumjian,R., Farhadian,S.F., Ward,R. and Lander,E.S. (2001) Linkage disequilibrium in the human genome. Nature, 411, 199–204. [DOI] [PubMed] [Google Scholar]
- 38.Boehnke M. (2000) A look at linkage disequilibrium. Nature Genet., 25, 246–247. [DOI] [PubMed] [Google Scholar]
- 39.Taillon-Miller P., Bauer-Sardina,I., Saccone,N.L., Putzel,J., Laitinen,T., Cao,A., Kere,J., Pilia,G., Rice,J.P. and Kwok,P.Y. (2000) Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nature Genet., 25, 324–328. [DOI] [PubMed] [Google Scholar]
- 40.Clark A.G., Weiss,K.M., Nickerson,D.A., Taylor,S.L., Buchanan,A., Stengard,J., Salomaa,V., Vartiainen,E., Perola,M., Boerwinkle,E. and Sing,C.F. (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am. J. Hum. Genet., 63, 595–612. [DOI] [PMC free article] [PubMed] [Google Scholar]