Abstract
The gut microbiota, which includes prokaryotes, archaea, and eukaryotes such as yeasts, some protozoa, and fungi, significantly impacts fish by affecting digestion, metabolism, and the immune system. In this research, we combine various tasks carried out by various bacteria in the gut of fish. This study also examines the gut microbiome composition of marine and freshwater fish, identifying important bacterial species linked to different biological functions. The diversity within fish species highlights the importance of considering nutrition, habitat, and environmental factors in microbiological research on fish. The ever-changing gut microbiome of the fish indicates that microbial communities are specifically adapted to meet the needs of both the host and its environment. This indicates that the fish can adjust to a specific environment with the help of gut microbiota. This important research is crucial for comprehending the complex relationships between fish and their gut bacteria in different aquatic environments. These discoveries have implications for aquaculture practices, fisheries administration, and the broader ecological processes of both freshwater and marine environments. With further progress in this area of study, the knowledge acquired would offer a valuable standpoint to enhance our comprehension of aquatic microbiology and enhance the sustainability and nutrition of fish resources.
Keywords: gut microbiome, freshwater fishes, marine fishes, environmental factors, comparative analysis: aquaculture
The comparative analysis of marine and freshwater fish gut microbiomes reveals significant differences in microbial composition and diversity, highlighting the influence of distinct environmental factors on gut microbiota.
Introduction
The gut of an animal consists of trillions of diverse microorganisms that can have both positive and negative effects on the nutrition, immunity, and overall well-being of the host (Bairagi et al. 2002, Ray et al. 2012, Deb et al. 2020, Ghori et al. 2022, De Marco et al. 2023). Its structure is influenced by factors such as microbial diversity, spatial distribution, pH, and interactions with host enzymes (Jordaan and Bezuidenhout 2013, Kim et al. 2021). Microbes in the gut of fish engage in competition, generate antimicrobials, communicate, and consume each other, impacting the population dynamics and health of the host (Wang et al. 2018, Cui et al. 2022, Luan et al. 2023). Struggles between microorganisms, such as competition for resources and bacteriophage assaults, impact the equilibrium of microbes (Di Maiuta et al. 2013, Parris et al. 2019, Qi et al. 2023b). The fish has a unique assemblage of microorganisms residing within their gastrointestinal tract (Givens et al. 2015, Deb et al. 2020, Zou et al. 2020, Xi et al. 2023). Some of these microorganisms form a dynamic and symbiont relationship with the host and impact various aspects of fish biology such as digestion, absorption, synthesis of essential nutrients, antimicrobial peptides (AMPs), and bacteriocins’ cellular and humoral immunity (Roeselers et al. 2011, De Marco et al. 2023). In return, the host receives exogenous enzymes and nutrients, such as vitamins and fatty acids, which cannot be produced by the host body cells (Dhanasiri et al. 2011, Wu et al. 2024). A balanced microbiome composition reduces the colonization and proliferation of harmful pathogens and controls diseases (Fjellheim et al. 2007, Ou et al. 2024). Therefore, the microbiota of the gut is considered an “extra organ” owing to powerful microbial genes, and the role of microorganisms in digestion, immunity, and overall development (Bairagi et al. 2002, Dhanasiri et al. 2011, Feng et al. 2018, Butt and Volkoff 2019). Facultative anaerobes and aerobes are present in greater numbers in the fish gut in comparison to obligate anaerobes (Cahill 1990, Clements 1997, Izvekova et al. 2007, Trust et al. 2011). This is mainly because of the fish gut environment, which typically has higher oxygen levels, particularly in the front parts such as the stomach and nearby intestine (Nelson and Dehn 2010, Egerton et al. 2018). Facultative anaerobes can adjust to changing levels of oxygen, while obligate anaerobes prefer environments with no oxygen, such as the lower regions of mammalian intestines (André et al. 2021, Lu and Imlay 2021, Duncan et al. 2023). The gut microbiomes are divided into autochthonous (i.e. native bacteria or when they can attach and colonize the gut epithelial surface of the host) and allochthonous (i.e. foreign bacteria or when they accidentally enter the host gut and get removed after some time without colonizing) (Nayak 2010, Navarrete et al. 2013, Givens et al. 2015, Sharma et al. 2023). Therefore, a thorough understanding of the fish gut microbiome is very important in aquaculture because it can be helpful in the management of fisheries and conservation and has the potential to boost fish health and sustainable seafood production (Van Kessel et al. 2011). In aquaculture, it is crucial to preserve a balanced gut microbiome to prevent diseases that could have a severe impact on fish populations. Probiotic and prebiotic therapies are frequently employed to improve advantageous microbial populations, resulting in improved feed efficiency and decreased expenses in fish farming (Merrifield et al. 2010, Dutta 2015, Ghori et al. 2022, De Marco et al. 2023). Moreover, a well-balanced gut microbiome can aid in decreasing waste generation, thereby lessening the environmental consequences of aquaculture operations. According to Miranda et al. (2022), numerous fish species are at risk of extinction because of human activities and climate change, and yet little is known about their microbiota, making the study of intestinal microbiota crucial for the conservation of these species (Soh et al. 2024). The gut microbiome is crucial for the health and survival of fish, particularly in captive breeding programs aimed at species conservation (West et al. 2019, Ruiz et al. 2024). A properly cared for microbiome helps fish adjust to shifting environmental conditions, especially crucial with climate change and habitat damage. Having a strong gut microbiome can boost the chances of survival for fish being released back into their natural habitat by enhancing their overall health and ability to fight off diseases. Comparative analysis of the fish gut microbiome is an important field of research as it can unravel hidden realities that can help to understand the relations between microorganisms, and microbial interaction with their host besides functions and diversity of the complex microbiota.
The function and composition of the microbiome may vary from species to species like other aquatic and terrestrial animals (Sehnal et al. 2021, de Jonge et al. 2022). This intriguing scientific endeavour involves studying and comparing the composition, diversity, and functional roles of these microbial communities across a wide spectrum of fish, ranging from freshwater to marine species, and from herbivorous to carnivorous feeders (Givens et al. 2015). Thus, we aimed to gain a deeper understanding of how microbial communities have evolved in response to the specific dietary, environmental, and physiological adaptations of fish species. Through comparative analysis, researchers have uncovered the profound impact of the fish gut microbiome on various aspects of fish biology, including immunity, metabolism, growth, and even behaviour (Collazos et al. 1994, Aquac et al. 2023). The insights obtained from this study will not only contribute to our understanding of fish health and ecology, but also have immense promise for enhancing aquaculture practices, conserving endangered species, and advancing gut microbial biotechnological applications (Ghanbari et al. 2015).
Thorough research was conducted using appropriate keywords on online platforms such as Google Scholar, ResearchGate, Science Direct, Scopus, and regular Google searches to find accurate data. The Preffered Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) methodology used for systematic review has been depicted in Fig. 1. Certain pertinent articles that were connected to the keywords and subject have been incorporated in the research. Articles that are not pertinent, lack crucial information, are not in full text, and are off-topic were eliminated. Most of the literature examined was from the years 2015 to 2024, with some older literature included due to incomplete data.
Figure 1.
PRISMA methodology was followed during the literature survey.
Functional status of fish gut microbiome
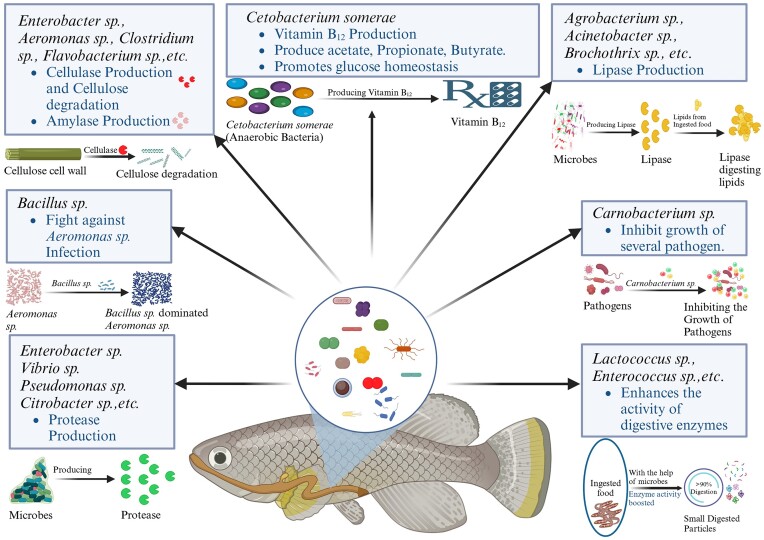
The study on fish is increasing progressively due to the demand for fish and fish-origin nutraceuticals The gut microbiota plays a crucial role in regulating the growth and production of fish, hence aiding in meeting the market demand for fish (Sullam et al. 2012, Wong et al. 2013, Butt and Volkoff 2019, Liu et al. 2021). Proper knowledge of bacterial function in a particular species of fish can help us develop efficient probiotic strains or synbiotics (as depicted in Fig. 2). For instance, Cetobacterium somerae, a Gram-negative micro aerotolerant bacterium present in the gastrointestinal tract (GI) tract of some freshwater fish such as tilapia and carp, produces large amounts of Vitamin B12 (Tsuchiya et al. 2008). The fish harbouring C. somerae in their gut, in general, did not require Vitamin B12 in their diet, whereas species such as catfish and Japanese eel that do not have C. somerae in their gut require Vitamin B12 (Tsuchiya et al. 2008, Jobling 2012). Cetobacterium somerae is crucial for fish, particularly those consuming plant-based diets or having low B12 levels, as it aids in protein fermentation and amino acid absorption for their growth and energy needs (Sugita et al. 1991, Li et al. 2015). It generates a large quantity of acetate, contributing to improved glucose regulation, enhanced gut barrier function, and increased resistance to diseases (Wang et al. 2021, Qi et al. 2023b). As a prevalent gut bacterium, it helps support a balanced ecosystem by beating harmful bacteria and creating substances such as short-chain fatty acids (SCFAs) (Sugita et al. 1991, Li et al. 2015, Bhardwaj et al. 2023). While it has positive effects on fish, the impact on human health in aquaculture environments is not clearly understood, and there may be risks of transmission (Finegold et al. 2003). The generation of gases such as hydrogen and methane in the process of fermentation is also a feature of C. somerae (Li et al. 2015). In general, this bacterium plays a role in supporting gut health, energy metabolism, and overall well-being in fish such as carp, tilapia, and catfish, underscoring its significance in keeping a balanced and healthy gut microbiome. More studies are required to comprehend how it could affect human health, and the dangers linked to its existence in aquaculture.
Figure 2.
Function of microorganisms in fish gut. Some bacteria showing probiotic and pro-health benefits have been highlighted. Microbial feed additives (probiotics and synbiotics) are available commercially to improve fish nutrition and health.
Many microbes are involved in the digestion process (Ray et al. 2012, Karasov and Douglas 2013, Ringø et al. 2016, Sehnal et al. 2021). The function is more clear in herbivorous and omnivorous fish that eat diets with cellulose and plant secondary compounds such as tannins, alkaloids, and flavonoids (Nelson et al. 1999, Francis et al. 2001, Li et al. 2016). Specific microbial communities are needed to break down complex carbohydrates and detoxify secondary metabolites. Bacteria such as Aeromonas sobria, A. veronii, A. hydrophila, A. jandaei, Enterobacter aerogenes, E. ludwigii, Clostridium sp., Citrobacter braakii, Raoultella ornithinolytica, Klebsiella variicola, Pseudomonas veronii, Erwinia billingiae, Enterococcus faecium, Brevibacillus laterosporus, Anoxybacillus sp., Bacillus megaterium, and Sediminibacterium salmoneum provide the necessary enzymes for plant-based diets (Bairagi et al. 2002, Saha et al. 2006, Ray et al. 2012, Ye et al. 2014, Li et al. 2016). The microbial consortia vary according to the host species, diet, habitat and environmental factors (Kumar et al. 2023). Some of the bacteria producing fibrolytic enzymes, reported from different fishes, have been summarized in Table 1. Many carnivorous fish feed on crustaceans that are digested by chitinase-producing gut bacteria such as Marinobacter lutaoensis, Pseudoalteromonas piscicida, Pseudomonas spp., Ferrimonas balearica, Enterovibrio norvegicus, Grimontia hollisae, Photobacterium damselae spp., Acinetobacter spp., Vibrio spp., Enterobacter spp., Aeromonas spp., Flavobacterium spp., and Photobacterium spp. (Ray et al. 2012).
Table 1.
List of microbes along with its functions reported by different researchers who conducted studies in different fishes.
| Sl. No. | Microbes | Functions/role | References |
|---|---|---|---|
| 1 | Aeromonas sobria, A. veronii, A. hydrophila, A. jandaei, Enterobacter sp., E. aerogenes, E. ludwigii, Clostridium sp., Citrobacter braakii, Raoultella ornithinolytica, Klebsiella variicola, Pseudomonas veronii, Erwinia billingiae, Enterococcus faecium, Brevibacillus laterosporus, Anoxybacillus sp., Bacillus megaterium, Sediminibacterium salmoneum | Cellulose degradation | (Bairagi et al. 2002, Saha et al. 2006, Ray et al. 2012, Wu et al. 2012, Ye et al. 2014, Li et al. 2016) |
| 2 | Cetobacterium somerae | Synthesizes Vitamin B12; produces acetate, propionate, and butyrate; and promotes glucose homeostasis | (Kim et al. 2021, Wang et al. 2021, Qi et al. 2023a) |
| 3 | Lactococcus lactis and Enterococcus faecalis | Enhance the activity of digestive enzyme | (Luan et al. 2023) |
| 4 | Lactococcus lactis | Promotes an increase in beneficial microbes and decrease pathogenic bacteria | (Luan et al. 2023) |
| 5 | Bacillus cereus and B. thuringiensis | Function against Aeromonas hydrophila infection | (Kong et al. 2021, Luan et al. 2023) |
| 6 | Carnobacterium sp. | Inhibits several pathogens | (Nayak 2010) |
| 7 | Aeromonas hydrophila, Aeromonas spp., Bacteroidaceae, Clostridium spp., Bacillus circulans, B. pumilus, B. cereus, Aeromonas spp., Enterobacteriaceae, Pseudomonas spp., Flavobacterium spp., Citrobacter freundii, B. subtilis, Brochothrix sp., Brochothrix thermosphacta | Amylase production | (Bairagi et al. 2002, Ray et al. 2012) |
| 8 | Enterobacter spp., Vibrio spp., Pseudomonas spp., Acinetobacter spp., Aeromonas spp., Flavobacterium balustinum, Bacillus cereus, B. circulans, B. pumilus, Citrobacter sp., Citrobacter freundii, B. licheniformis, B. subtilis | Protease production | (Bairagi et al. 2002, Ray, Ghosh and Ringø 2012) |
| 9 | Agrobacterium sp., Brevibacterium sp., Microbacterium sp., Staphylococcu sp., Vibrio spp., Acinetobacter spp., Enterobacteriaceae, Pseodomonas spp., Bacillus thuringiensis, B. cereus, Bacillus sp., Brochothrix sp., Brochothrix thermosphacta | Lipase production | (Bairagi et al. 2002, Ringø et al. 2010, Ray et al. 2012) |
| 10 | Marinobacter lutaoensis, Ferrimonas balearica, Pseudoalteromonas piscicida, Enterovibrio norvegicus, Grimontia hollisae, Photobacterium damselae spp. damselae, P. leiognathi, P. lipolyticum, P. phosphoreum, P. rosenbergii, Vibrio campbelli, V. chagasii, V. fischeri, V. fortis, V. gallicus, V. harveyi, V. natrigenes, V. nigripulchritudo, V. ordalii, V. parahaemolyticus, V. pomeroyi, V. ponticus, V. proteolyticus, V. rumoiensis, V. shilonii, V. tasmaniensis and V. tubiashii, Enterobacter spp., Vibrio spp., Pseudomonas spp., Aeromonas spp., Vibrio spp., Acinetobacter sp., Enterobacteriaceae, Flavobacterium sp., Photobacterium spp. | Chitinase production | (Ray, Ghosh and Ringø 2012) |
| 11 | Streptococcus sp., Leuconostoc sp., Pediococcus sp., Aerococcus sp., Enterococcus sp., Vagococcus sp., Carnobacterium sp., Carnobacterium divergens, C. piscicola, Lactobacillus spp., L. plantarum, L. rhamnosus, L. bulgaricus | Lactic acid fermentation and produce organic acids, hydrogen peroxide, and some other substances suppressing the growth of pathogenic microorganisms | (Ringø and Gatesoupe 1998, Gatesoupe 2007, Izvekova et al. 2007, Ringø et al. 2010) |
Various studies also reported about microbes fighting against harmful bacteria and diseases such as Streptococcus sp., Pediococcus spp., Aerococcus spp., Enterococcus spp., Vagococcus spp., Carnobacterium spp., Lactobacillus spp., and Bacillus spp., Leuconostoc spp., and Lactococcus lactis (Ringø and Gatesoupe 1998, Gatesoupe 2007, Izvekova et al. 2007, Nayak 2010, Ringø et al. 2010, Kong et al. 2021, Luan et al. 2023). Various types of bacteria found in the intestines create diverse bioactive substances and specific genes that play a crucial role in the production of secondary metabolites. In Table 2, some of the important genes are presented that are known to play a role in producing secondary metabolites within the microbiome of fish guts. Genes, including cobA, cobG, and cobT, are essential for synthesizing Vitamin B12 in fish and can be found in Cetobacterium somerae, Clostridium spp., and Propionibacterium spp. (Fang et al. 2017, Guo and Chen 2018, Balabanova et al. 2021, Qi et al. 2023b). The production of SCFAs, such as butyrate and propionate, depends on genes such as but and buk present in Clostridium and Bacteroides species (Vital et al. 2014, Tarnecki et al. 2017, Meng and Shu 2024). Fish use genes such as iucA and pvd from Pseudomonas and Vibrio species for siderophore biosynthesis to acquire iron (Ravel and Cornelis 2003, Hassan and Troxell 2013, Mydy et al. 2020). The production of antimicrobial peptides such as lantibiotics and nisin is crucial for preserving a balanced microbial population in fish, and it involves genes such as lantA and nisA that are present in Lactobacillus and Bacillus species (Siegers and Entian 1995, McAuliffe et al. 2001, Kuipers et al. 2011, Egerton et al. 2018). PKS and NRPS pathways in marine Streptomyces and Pseudomonas species synthesize secondary metabolites such as antibiotics and pigments, with regulation by PKS gene clusters. NRPS gene clusters are in charge of creating non-ribosomal peptides, which have the ability to serve as antibiotics or signalling molecules, impacting both microbial competition and fish health (Ray et al. 2012, Wang et al. 2014, Borsetto et al. 2019, Komaki et al. 2020, Yin et al. 2023b). Molecules involved in quorum sensing, such as acyl-homoserine lactones (AHLs) produced by genes luxI and luxR, enable bacteria such asVibrio, Aeromonas, and Pseudomonas to regulate activities such as biofilm formation and virulence factors (Miyashiro and Ruby 2012, Rajput and Kumar 2017). The tnaA gene encodes tryptophanase, which aids bacteria such as Escherichia coli and Lactobacillus in generating indole and its derivatives that impact gut barrier integrity and inflammation (Li and Young 2013, Boya et al. 2021). The genes cysJIH found in organisms such as Desulfovibrio play a role in generating hydrogen sulphide, which can exhibit anti-inflammatory properties when present in small amounts (Ostrowski et al. 1989, Álvarez et al. 2015). The srfA operon found in Bacillus and Pseudomonas helps in the production of biosurfactants, which support bacterial colonization and prevent biofilm formation (Kisil et al. 2023, Xu et al. 2023, Qi et al. 2023a). Genes involved in terpenoid biosynthesis, such as dxs and ispG, are responsible for signalling and possible antimicrobial roles in the GI tract of Streptomyces and Cyanobacteria (Xue et al. 2015, Marshall et al. 2023). These procedures demonstrate the various crucial functions that bacterial genes and molecules have in maintaining gut health and communication. In general, these processes are crucial for the well-being of fish, their energy metabolism, the health of their digestive system, and their defence against infections, demonstrating the complex relationships between microbes and genetic processes in aquatic settings.
Table 2.
Some of the important genes known to play a role in the creation of secondary metabolites in the fish gut microbiome.
| Metabolites | Key genes | Bacteria | Function | Reference |
|---|---|---|---|---|
| Vitamin B12 (cobalamin) | cobA, cobG, cobL, cobM, cobT | Cetobacterium, Clostridium | Cobalamin biosynthesis (DNA synthesis, metabolism) | (Fang et al. 2017, Guo and Chen 2018, Balabanova et al. 2021, Qi et al. 2023b) |
| Short-chain fatty acids | but, buk, propionate CoA-transferase | Clostridium, Bacteroides | Butyrate, propionate, and acetate production (gut health) | (Vital et al. 2014, Tarnecki et al. 2017, Meng and Shu 2024) |
| Siderophores | iucA, iucB, pvd | Pseudomonas, Aeromonas | Iron acquisition via siderophore production | (Ravel and Cornelis 2003, Hassan and Troxell 2013, Mydy et al. 2020) |
| Antimicrobial peptides | lantA, lantB, lantC, nisA | Lactobacillus, Bacillus | Bacteriocin production (inhibition of pathogens) | (Siegers and Entian 1995 1995, McAuliffe et al. 2001, Kuipers et al. 2011, Egerton et al. 2018) |
| Polyketides/NRPs | PKS, NRPS | Streptomyces, Bacillus | Production of antibiotics and immunomodulatory compounds | (Ray et al. 2012, Wang et al. 2014, Borsetto et al. 2019, Komaki et al. 2020, Yin et al. 2023a) |
| Quorum sensing | luxI, luxR | Vibrio, Pseudomonas | Bacterial communication (biofilm formation, colonization) | (Miyashiro and Ruby 2012, Rajput and Kumar 2017) |
| Indole (tryptophan) | tnaA | E. coli, Lactobacillus | Gut health regulation and anti-inflammatory signalling | (Li and Young 2013, Boya et al. 2021) |
| Hydrogen sulphide | cysJIH | Desulfovibrio, Clostridium | Sulphate reduction (gut signalling, motility) | (Ostrowski et al. 1989, Álvarez et al. 2015) |
| Biosurfactants | srfAA, srfAB, srfAC | Bacillus, Pseudomonas | Surfactin production (colonization, biofilm inhibition) | (Kisil et al. 2023, Qi et al. 2023a, Xu et al. 2023) |
| Terpenoids | dxs, ispD, ispG | Streptomyces, Cyanobacteria | Signalling molecules and antimicrobial functions | (Xue et al. 2015, Marshall et al. 2023) |
The fish intestinal microbiome is an intricate network of symbiotic connections between the host and its microbial residents, encompassing mutualistic, commensal, and antagonistic relationships (Ray et al. 2012). Bacteria play a crucial role by breaking down complex nutrients and producing essential nutrients for the fish, showing the importance of nutrient availability and metabolism (Nayak 2010). The immune system of the host helps tolerate helpful bacteria and inhibits the growth of harmful bacteria (Bledsoe et al. 2022). Quorum sensing enables bacterial populations to communicate and synchronize behaviours such as forming biofilms (Miyashiro and Ruby 2012, Rajput and Kumar 2017, Moreno et al. 2024). Microorganisms compete and spread out in the gut, leading to niche separation, where various bacteria inhabit specific regions and carry out unique functions (Melo-Bolívar et al. 2019). Biofilm development on the intestinal lining provides protection for the host and bacteria against stressors and pathogens (Harika et al. 2020). In general, the microbiome of fish intestines is a constantly changing setting where different types of microbes engage in a fragile equilibrium of collaboration and rivalry.
Antagonistic relationships within the gut microbiome are crucial for upholding microbial equilibrium and hindering the excessive growth of harmful bacteria. These interactions involve the creation of antimicrobial substances such as bacteriocins and organic acids, which hinder the growth of other bacteria (Egerton et al. 2018). Competition for nutrients and space is another factor, as helpful bacteria outcompete harmful ones for limited resources and sites on the gut lining (Tarnecki et al. 2017). Furthermore, bacterial siderophore competition entails the production of molecules to procure iron, which restricts the proliferation of rival organisms (Nayak 2010). Quorum quenching is a different process in which specific bacteria break down signalling molecules produced by pathogens, interrupting their communication and decreasing their ability to cause harm (Rajput and Kumar 2017). In general, these hostile interactions contribute to supporting gut health by preserving a varied and well-balanced microbiome.
Comparative study of fish gut microbiome: freshwater versus marine water
Marine and freshwater fish have distinct gut microbiomes, influenced by the different environments (Li et al. 2017). Studies reveal that the gut microbiomes of freshwater fish and marine fish are dominated by the phyla Fusobacteria and Proteobacteria (Givens et al. 2015, Li et al. 2017, Deb et al. 2020). Common microbial species found in freshwater fish include Proteobacteria such as Aeromonas, Pseudomonas, and Enterobacter, Firmicutes such as Lactobacillus and Streptococcus, Actinobacteria, including Micrococcus, and Bacteroidetes such as Flavobacterium and Chryseobacterium (Sullam et al. 2012, Wu et al. 2012, Llewellyn et al. 2014, Givens et al. 2015, Deb et al. 2020). Marine fish often contain Proteobacteria species such as Vibrio, Photobacterium, and Shewanella, as well as Firmicutes, including Bacillus and Clostridium, and Bacteroidetes such as Cytophaga (Llewellyn et al. 2014, Givens et al. 2015, Egerton et al. 2018, Ou et al. 2021, Uniacke-Lowe et al. 2024). Planctomyces species, specifically Planctomycetes, are marine microorganisms with unique metabolic abilities such as anaerobic ammonium oxidation (Fuerst and Sagulenko 2011). The improvements in high-throughput sequencing techniques have led to the discovery of previously uncultured or poorly understood species in the digestive systems of freshwater and marine fish (Ghanbari et al. 2015, Rasmussen et al. 2022, Brar et al. 2023). A few instances include Cetobacterium somerae, which synthesizes Vitamin B12 in freshwater fish (Sugita et al. 1991); ZOR0006, discovered in carp and tilapia aiding in nutrient uptake (Zhou et al. 2023); and Endozoicomonas spp. in marine fish promoting gut health and immunity (Neave et al. 2016). Aliivibrio and Pseudoalteromonas species have important functions in the gut of marine fish, being involved in bioluminescence and interactions with the host, respectively (Klemetsen et al. 2021, Drønen et al. 2022). Researchers are still studying Tenericutes found in the intestines of marine fish to understand their ecological role as bacteria with smaller genomes, potentially adapted to live in hosts (Givens et al. 2015, Egerton et al. 2018). These results highlight the diverse and important bacteria present in fish guts across different environments.
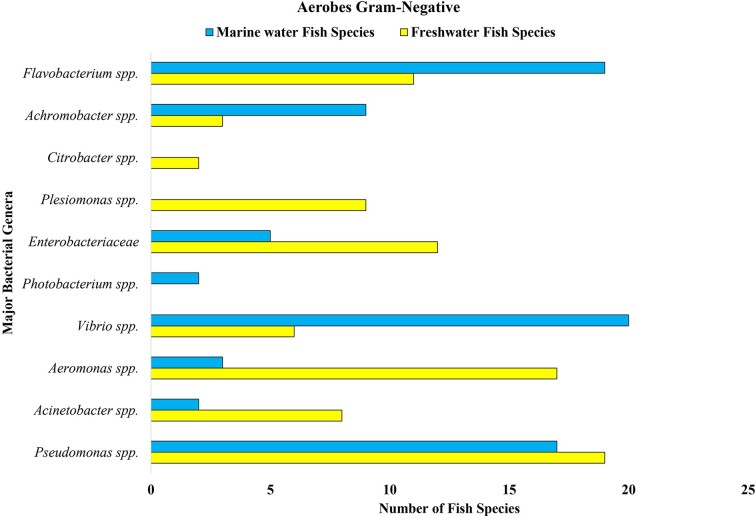
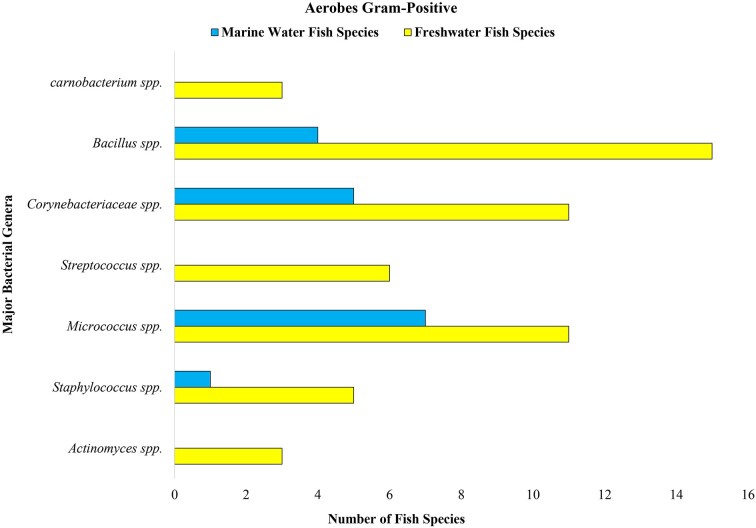
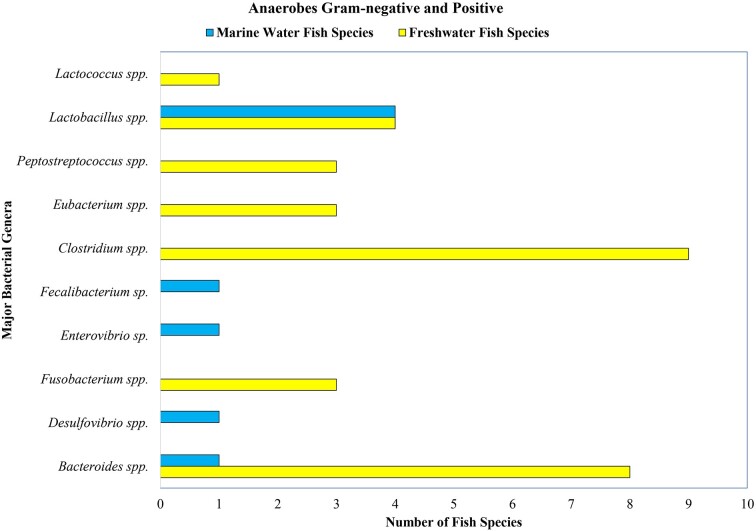
Some of the reported bacterial groups in marine and freshwater fish guts are presented in Figs 3, 4, and 5. According to Izvekova et al. (2007), these data are obtained as a result of isolation and identification by traditional techniques. The figures include only the popular groups whose composition varies according to habitat. In Fig. 3, it is found that the dominant aerobic Gram-negative bacteria of marine fish are Flavobacterium spp., Achromobacter spp., Photobacterium spp., Vibrio spp., and Pseudomonas spp., which proves the variation in the gut of marine and freshwater fish. Similarly, the aerobic Gram-positive bacterial data presented in Fig. 4 show that Bacillus spp., Cornebacteriaceae spp., Streptococcus spp., Lactococcus spp., Micrococcus spp., Staphylococcus spp., Actinomyces spp., and Carnobacterium spp. are present more in freshwater fish than in marine fish (Izvekova et al. 2007). Surprisingly, more anaerobes have been reported in freshwater fish, i.e. Eubacterium spp., Peptostreptococcus spp., Fusobacterium spp., Clostridium spp., and Bacteroides spp., than in marine fish (depicted in Fig. 5). This might be due to insufficient studies conducted on marine fish and the difficulty involved in isolating anaerobic bacteria. The reason for sharing these data is to demonstrate the difference in gut microbiome among fish living in various environments. Researchers compared the gut microbiomes of 51 fish species and found that 47 species had Gram-negative aerobes, 34 species had Gram-positive aerobes, 10 species had Gram-negative anaerobes, and 8 species had Gram-positive anaerobes (Table S1) (Izvekova et al. 2007). Nevertheless, with the recent development of techniques such as next-generation sequencing, pyrosequencing, etc. (Van Kessel et al. 2011, Terova et al. 2018), we now have a more reliable option for obtaining authentic data. Traditional methods such as isolation and identification, although time-consuming and laborious, still provide a basic understanding of microorganism composition and diversity. Gram-negative bacteria were found in more species and at similar rates in both freshwater and marine fish. Greater quantities of Gram-positive aerobic bacteria were present in freshwater fish and were also observed to host anaerobic bacteria, as depicted in Figs 3, 4, and 5. In order to gain more insight, we also contrasted certain data from freshwater and marine fish presented in Table 3.
Figure 3.
Aerobic Gram-negative bacteria reported in the gut of wild freshwater and marine water fish.
Figure 4.
Aerobic Gram-positive bacteria reported in the gut of wild freshwater and marine water fish.
Figure 5.
Anaerobic bacteria reported in the gut of wild freshwater and marine water fish.
Table 3.
Presence of top bacteria in freshwater and marine water fish species.
| Fish species | Environment | Method of study | Diet | Dominant microbiota | Most abundant bacteria | Key functions | References | |
|---|---|---|---|---|---|---|---|---|
| Freshwater | Onchorhyncus mykiss (rainbow trout) | Farm | Whole shotgun metagenomic | Omnivorous (insect, plants) | Proteobacteria, Firmicutes, Actinobacteria | Mycoplasma, Cetobacterium, Lactococcus, Lactobacillus, Leuconostoc, Ureaplasma, Propionibacterium | Protein digestion, fermentation of carbohydrates | (Nayak 2010, Llewellyn et al. 2014a, Tarnecki et al. 2017, Betiku et al. 2023) |
| Ctenopharyngodon idellus (grass carp) | Farm | Pyrosequencing | Omnivorous (plants, detritus) | Firmicutes, Bacteroidetes, Cetobacterium | Streptococcus, Lactobacillus, Flavobacterium, Veillonella, Streptococcus, Pseudomonas, Anoxybacillus, Citrobacter, Clostridium, Leuconostoc | Fermentation of polysaccharides, Vitamin B12 synthesis | (Nayak 2010, Wu et al. 2012, Llewellyn et al. 2014b, Li et al. 2015, Tarnecki et al. 2017) | |
| Marine Water | Salmo salar (Atlantic salmon) | Farm | High-throughput sequencing | Carnivorous (fish, invertebrates) | Proteobacteria (Vibrio), Firmicutes | Janthinobacterium, Propionibacterium, Stenotrophomonas, Pseudomonas, Phyllobacterium, Delftia, Herbaspirillum, Burkholderia, Sphingomonas, Ochrobactrium, Variovorax, Microbacterium, Rhodococcus, Acinetobacter | Protein digestion, nitrogen metabolism | (Nayak 2010, Llewellyn et al. 2014b, Gajardo et al. 2016, Tarnecki et al. 2017, Rudi et al. 2018) |
| Acanthurus triostegus (surgeonfish) | Wild | 16S rRNA gene amplicon sequencing | Herbivorous (algae) | Firmicutes, Bacteroidetes, Cyanobacteria | Epulopiscium, Acinetobacter, Arcobacter, Arthrospira, Brevinema, Cetobacterium, Fusobacterium, Methylobacterium, Photobacterium, Pelomonas, Vibrio, Pseudoalteromonas | Algal polysaccharide breakdown, SCFA production | (Miyake et al. 2015, Tarnecki et al. 2017, Parata et al. 2020) | |
Freshwater fish gut microbiome
Based on the next-generation sequencing (NGS) technique, diverse groups of microbes have been detected in freshwater fishes. The core microbiomes are resistant to variation in diet and rearing density as claimed by researchers who experimented on GI microorganisms of Onchorhynchus mykiss (Wong et al. 2013). However, an alteration in diet can cause a change in the health status of fish (Wong et al. 2013). In herbivorous and omnivorous fish, the breakdown of cellulose can be enhanced by microbes, such as Bacillus circulans and B. megaterium (Saha et al. 2006). A study conducted on Carassius auraus gibrlio concluded that the first phylum of the microbe to develop in the gut is Proteobacteria (Li et al. 2017). However, the actual reason behind this fact is still unknown. We can assume that proteobacteria in environmental water enable the ability to interact with the host as bacteria are ubiquitous in water and are found to be the most abundant and diverse. They play an important role in nutrient cycling, decomposition, and organic matter breakdown. Common bacterial phyla found in water include Proteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, and Firmicutes (Cottrell et al. 2005, Jordaan and Bezuidenhout 2013, Xia et al. 2013, Savio et al. 2015, Brar et al. 2023). Proteobacteria are the most dominant gut species in freshwater fish followed by Firmicutes, Actinobacteria, and Bacteroidetes (Wu et al. 2012). Actinobacteria spp. are well-known producers of secondary metabolites such as hydrolytic enzymes, e.g. amylase, protease, and lipase. Studies have revealed that Actinobacteria spp. play an important role in the fermentation of a large variety of oligosaccharides in the gut (Ventura et al. 2007). Fusobacteria spp. are most frequent in freshwater fishes (Kim et al. 2021). The freshwater fish gut is dominated by the species of Enterobacter, Aeromonas, and Acinetobacter (Cahill 1990, Mondal et al. 2008, Tsuchiya et al. 2008, Deb et al. 2020, Suescun-Sepulveda et al. 2023). Intestinal microflora also includes species of Escherichia, Klebsiella, Proteus, Serrata, Aermonas, Alcaligenes, Eikenella, Bacillus, Listeria, Propionibacterium, Bacteroides, Citrobacter freundii, Hafnia alvei, Cytophaga/Flexibacter, Staphylococcus, Mycoplasma, Streptococcus, Lactococcus, Peptostreptococcus, Deefgea, Cetobacterium, Moraxella, and Pseudomonas (Austin 2002, Brown et al. 2018, Hernández et al. 2021, Singh et al. 2021 ).
A comparison of the gut microbiome of rainbow trout (Onchorhyncus mykiss) and grass carp (Ctenopharyngodon idella) was done to better understand the variation in fish gut microbiome (Table 3). Based on their habitat, feeding habits, and access to freshwater, the species were chosen.
Rainbow trout is a freshwater carnivorous fish. It feeds on a wide variety of aquatic insects and crustaceans as well as small fish and even land insects that wash up on the surface of the water. Their diet can vary depending on where they live and what food sources are available (Huyben et al. 2018). Rainbow trout prefer chilled water having temperatures from 10 to 15°C. They may seek out certain parts of their habitat that have optimal temperature ranges. Several bacterial phyla, including Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, often dominate the gut microbiome of rainbow trout (Betiku et al. 2023). However, these phylas’ relative abundance can change. Aeromonas, Pseudomonas, Acinetobacter, Shewanella, Clostridium, and Bacteroidetes are the common genera discovered in rainbow trout gut. Mycoplasma, Cetobacterium, Lactococcus, Lactobacillus, Leuconostoc, Ureaplasma, and Propionibacterium were reported as an abundant genus (Llewellyn et al. 2014, Betiku et al. 2023). The gut microbiota in rainbow trout helps with digestion, modulating the immune system, and potentially protecting against pathogens. Some intestinal bacteria help in the digestion and utilization of complex polysaccharides and the synthesis of vitamins (Mondal et al. 2008, Li et al. 2016, Podell et al. 2023, Qi et al. 2023b).
Grass carp like freshwater habitats such as rivers, lakes, ponds, and reservoirs. Because their diet comprises primarily of plant material, they are usually found in places with abundant aquatic vegetation. Grass carp are noted for their herbivorous feeding habits, in which they consume a variety of aquatic plants (Ray et al. 2012). It can tolerate a wide range of temperatures but prefers warmer water. Temperatures ranging from 20 to 30°C are ideal. Different bacteria reported from grass carp gut microbiome include Aeromonas, Bacillus, Clostridium, Bacteroides, and lactobacilli. Streptococcus, Lactobacillus, Flavobacterium, Veillonella, Pseudomonas, Anoxybacillus, Citrobacter, Clostridium, and Leuconostoc were reported as abundant microbial genera in the Ctenopharyngodon idella intestine (Wu et al. 2012, Llewellyn et al. 2014). These occurrences and family abundance can be modified by factors such as food and environmental conditions.
The contrast underscores how diet and environmental factors affect the gut microbiomes of both species. The carnivorous tendencies and preference for cool water of rainbow trout have led to the development of a gut microbiome that is well adapted for digesting protein and fat efficiently, as well as for maintaining robust immune defences. On the other hand, the grass carp’s plant-based diet and preference for higher water temperatures help to create a digestive system full of beneficial bacteria that specialize in breaking down tough plant fibers and producing important nutrients from plants.
Marine water fish gut microbiome
The higher concentration of salt in water creates a challenging environment for fish; similarly, there is a possibility of variation in the environmental microbes. Although at the phylum level Firmicutes, Proteobacteria, and Actinobacteria are the most abundant species in the fish gut, at a lower taxonomic level, variations are observed. The marine fish intestinal flora consists of dominant species of Vibrio, Pseudomonas, Achromobacter, Corynebacterium, Flavobacterium, and Micrococcus (Cahill 1990, Izvekova et al. 2007, Ou et al. 2021) as well as Aeromonas spp., Alcaligenes sp., Alteromonas sp., Micrococcus sp., Carnobacterium sp., Flavobacterium sp., Photobacerium sp., Pseudomonas spp., Staphylococcus sp., and Vibrio sp. (Austin 2002, Izvekova et al. 2007, Huang et al. 2020, Ou et al. 2021), whereas in freshwater fish the composition varies as shown in Figs 3, 4, and 5.
A comparison of the gut microbiomes of Atlantic salmon (Salmo salar) and surgeonfish (Acanthurus triostegus) was done to better comprehend the variations in fish gut microbiota (Table 3). The species were chosen based on their habitat, feeding habits, and access to saltwater (Egerton et al. 2018, Huang et al. 2020, Ou et al. 2021, De Marco et al. 2023).
Atlantic salmon spend most of their lives in the Atlantic Ocean. They are recognized for their anadromous habit, which means that they travel from freshwater to the ocean and return at various phases of their lives. Atlantic salmon prefer cold, well-oxygenated waters. Their adaptation to varied environmental circumstances is demonstrated by their capacity to live in both freshwater and saltwater (Morales et al. 2022). Atlantic salmon are opportunistic eaters in the ocean, devouring a wide range of marine creatures. Pseudomonas, Janthinobacterium, Stenotrophomonas, Delfia, Herbaspirillum, Burkholderia, Sphingomonas, Propionibacterium, Ochrobacterium, Variovorax, Microbacterium, Phyllobacterium, Rhodococcus, and Acinetobacter are the abundant genera in the Salmo salar GI tract (Llewellyn et al. 2014, Gajardo et al. 2016).
Acanthurus triostegus, sometimes known as the Convict Tang, is commonly found in tropical marine settings with warm water temperatures. It thrives at the temperatures found in coral reef ecosystems. They are herbivorous and mostly eat algae. Epulopiscium, Acinetobacter, Arcobacter, Arthrospira, Brevinema, Cetobacterium, Fusobacterium, Methylobacterium, Photobacterium, Pelomonas, Vibrio, and Pseudoalteromonas are the most prevalent microbial genera found in the digestive tract of surgeonfish (Miyake et al. 2016, Ngugi et al. 2017a, Parata et al. 2020).
The gut microbiomes of both Atlantic salmon and Convict Tang species are varied and can be affected by what they eat and the surroundings they live in. Atlantic salmon, as anadromous fish, do well in cold waters and eat a range of marine animals because they are opportunistic feeders. The bacteria found in their gut microbiome, such as Pseudomonas and Burkholderia, help with absorbing nutrients (Moore et al. 2006, Wang et al. 2018). On the other hand, Convict Tang species live in tropical marine habitats and mainly eat algae. The bacteria found in their gut microbiome, such as Epulopiscium and Vibrio, are specifically designed to break down algal material (Thompson and Polz 2006, Miyake et al. 2016, Ngugi et al. 2017b, Sampaio et al. 2022). This points out how diet and environmental conditions affect the composition of the gut microbiome, with Atlantic salmon containing bacteria that digest protein and Convict Tang having bacteria that degrade algae, which helps them thrive in their specific diets and habitats.
Impact of environment on fish gut microbiome
The fish gut microbiome is critical to their health, development, and overall well-being. Quality of water, habitat and diet can all have a substantial impact on—composition and function of their gut microbiome (Sullam et al. 2012, Wong and Rawls 2012, Dehler et al. 2017, Huyben et al. 2018, Huang et al. 2020, Kim et al. 2021, Leeper et al. 2021, Karlsen et al. 2022, Brar et al. 2023, Herrera et al. 2023, Yin et al. 2023a, Kanika et al. 2024). Recent studies on the gut microbiota of tilapia concluded that the optimal composition and functions of the gut microbiota are not always accurately represented by the highest growth outcomes of the host (Ou et al. 2024). The negligent inclusion of macronutrients negatively affects the gut microbiota. Hence, it is important to take into account both growth performance and gut microbiota when assessing specific macronutrients (Ou et al. 2024). A study conducted on rainbow trout by changing the water temperature and diet found a decrease in the number of important microbes (order Lactobacillales) in the gut (Huyben et al. 2018). The studies also assumed that a high proportion of gut bacteria represented by Mycoplasma sp. (phylum Tenericutes) is nutrient-dependent, which means that these bacteria develop only in the presence of specific nutrients, because many studies on the same species did not report this bacteria (Huyben et al. 2018). A study was conducted in which the intestinal microbiota of Atlantic salmon was evaluated in two different habitats, namely a recirculated aquarium facility and an open freshwater loch cage. The researchers found variations in the composition of the microbiome such as the greater presence of phylum Tenericutes in aquarium fish samples, whereas Proteobacteria were more abundant in loch samples; similarly, Mycoplasmataceae (phylum Tenericutes) was the second most common family in aquarium fish samples but less common in loch fish samples (Dehler et al. 2017). A study conducted on yellowtail kingfish found that an increase in water temperature (26°C) caused changes in the microbial communities of young yellowtail kingfish, influencing their growth trajectory and immunological condition (Horlick et al. 2020). Temperature is essential in determining the composition of the gut microbiome in humans and fish (Wang et al. 2018, Sepulveda and Moeller 2020, Larios-Soriano et al. 2021). Elevated temperatures may boost metabolic rates, aiding heat-resistant microbes and harmful bacteria, whereas lower temperatures can slow microbial metabolism and benefit cold-adapted species (Abram et al. 2017, Huyben et al. 2018, Ghosh et al. 2022). Fish are significantly affected by temperature changes because they are cold-blooded, which can impact their health and size (Wu et al. 2022). Temperature, diet, and habitat all affect the gut microbiome of fish, leading to changes in metabolism, immune response, and overall health (Collazos et al. 1994, Horlick et al. 2020, Sepulveda and Moeller 2020, Li et al. 2023). Keeping the ideal water temperature is crucial in aquaculture to improve gut bacteria health, boost fish growth, and strengthen disease defences.
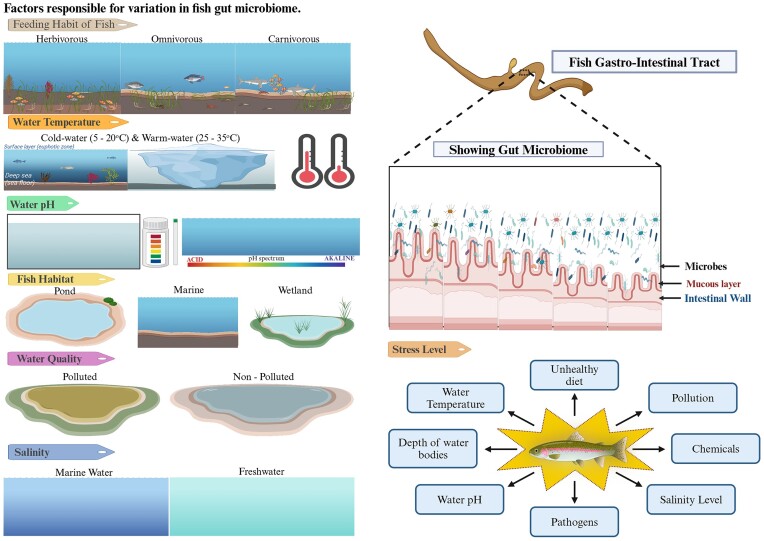
Our comparative study also indicates that environmental factors cause changes in gut microbial composition in the host. In Table 3, the most abundant bacterial genera are found to be different due to their different habits, habitats, and species they belong to. It is assumed that the gut microbiome helps in the adaptation of the host to different environments and requirements. Multiple factors such as environment, diet, host immunity, microbes, habit, habitat, water quality, etc. can make a host capable of sustainable survival. A clear visualization is presented in Fig. 6, which shows some factors responsible for variation in fish gut microbiome (Al-Harbi and Uddin 2004, Escalas et al. 2021, Kim et al. 2021, Podell et al. 2023, Bharti et al. 2023, Herrera et al. 2023, Sadeghi et al. 2023, Small et al. 2023, Viver et al. 2023).
Figure 6.
Some factors responsible for variation in fish gut microbiome.
Conclusion and future directions
There are notable shifts in the microbial communities of the gut microbiomes of marine and freshwater fish. These variations are influenced by both the diet and the surrounding water sources. In spite of these differences, there are certain resemblances in the gut microbiomes of marine and freshwater fish. Aeromonas, Vibrio, Pseudomonas, and other species are present in the GI tract of marine and freshwater fish, contributing to nutrition metabolism, fermentation, and overall gut health. Different microbes present in the intestines of fish create bioactive compounds by using specific genes. Examples include cobA, cobG, and cobT for Vitamin B12 creation; but and buk for short-chain fatty acid output; iucA and pvd for obtaining iron; lantA and nisA for generating antimicrobial peptides; PKS and NRPS routes for producing antibiotics and pigments; luxI and luxR for regulating population density; and tnaA for creating indole. These genetic mechanisms are essential for preserving the health of the fish gut and protecting against infections. The gut microbiome of fish is made up of symbiotic connections between the host and microbes, with bacteria breaking down nutrients and generating Vitamin B12. The immune system accepts beneficial bacteria and suppresses harmful ones, as quorum sensing enables bacteria to communicate. Microbes vie for space and nutrients, leading to the development of specific ecological niches. Antagonistic interactions in a balanced microbiome consist of creating antimicrobial substances, resource competition, and interrupting pathogen communication. Comprehending the fish microbiome is essential for grasping the intricate connections between microbes and their hosts. Continuing research is providing an understanding of the functional roles of these microorganisms and their effects on the health of fish in different aquatic environments. As advancements are made in the field, new findings can influence aquaculture, the management of fisheries, and our comprehension of aquatic ecology. More research is required to comprehend how host–microbiome interactions coevolve and adapt, as well as the specific roles of certain microorganisms in processing nutrients and regulating the immune system. It is important to study the gut microbiomes of wild fish populations in order to understand their natural microbial communities and ecological functions besides focusing on aquaculture or laboratory fish. This comparative study will help increase the understanding of aquatic microbiology and develop techniques to enhance the health and sustainability of fish populations in various aquatic habitats.
Supplementary Material
Acknowledgements
The authors duly acknowledge the Central University of Himachal Pradesh for providing lab facilities. K.T. acknowledges UGC-CSIR India for providing a Senior Research Fellowship. D.S. acknowledges the Department of Health Research, Government of India, for the Young Scientist Fellowship.
Contributor Information
Binoy Kumar Singh, Department of Animal Sciences, School of Life Sciences, Central University of Himachal Pradesh, Dharamshala 176206, India.
Kushal Thakur, Department of Animal Sciences, School of Life Sciences, Central University of Himachal Pradesh, Dharamshala 176206, India.
Hishani Kumari, Department of Animal Sciences, School of Life Sciences, Central University of Himachal Pradesh, Dharamshala 176206, India.
Danish Mahajan, Department of Animal Sciences, School of Life Sciences, Central University of Himachal Pradesh, Dharamshala 176206, India.
Dixit Sharma, Department of Animal Sciences, School of Life Sciences, Central University of Himachal Pradesh, Dharamshala 176206, India.
Amit Kumar Sharma, Department of Animal Sciences, School of Life Sciences, Central University of Himachal Pradesh, Dharamshala 176206, India.
Sunil Kumar, Department of Animal Sciences, School of Life Sciences, Central University of Himachal Pradesh, Dharamshala 176206, India.
Birbal Singh, ICAR—Indian Veterinary Research Institute (IVRI), Regional Station, Palampur 176061, India.
Pranay Punj Pankaj, Department of Zoology, Nagaland University (A Central University), Lumami 798627, India.
Rakesh Kumar, Department of Animal Sciences, School of Life Sciences, Central University of Himachal Pradesh, Dharamshala 176206, India.
Author contributions
Binoy Kumar Singh (Writing – original draft), Kushal Thakur (Writing – original draft), Hishani Kumari (Formal analysis), Danish Mahajan (Formal analysis), Dixit Sharma (Writing – review & editing), Amit Kumar Sharma (Writing – review & editing), Sunil Kumar (Conceptualization), Birbal Singh (Writing – review & editing), Pranay Punj Pankaj (Formal analysis), and Rakesh Kumar (Conceptualization)
Conflict of interest
None declared.
Funding
None declared.
References
- Abram QH, Dixon B, Katzenback BA. Impacts of low temperature on the teleost immune system. Biology. 2017;6:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Harbi A, Uddin M. Seasonal variation in the intestinal bacterial flora of hybrid tilapia (Oreochromis niloticus × Oreochromis aureus) cultured in earthen ponds in Saudi Arabia. Aquaculture. 2004;229:37–44. [Google Scholar]
- Álvarez R, Neumann G, Frávega J et al. CysB-dependent upregulation of the Salmonella Typhimurium cysJIH operon in response to antimicrobial compounds that induce oxidative stress. Biochem Biophys Res Commun. 2015;458:46–51. [DOI] [PubMed] [Google Scholar]
- André A, Debande L, Marteyn B. The selective advantage of facultative anaerobes relies on their unique ability to cope with changing oxygen levels during infection. Cell Microbiol. 2021;23:1–8. 10.1111/cmi.13338. [DOI] [PubMed] [Google Scholar]
- Austin B. The bacterial microflora of fish. ScientificWorldJournal. 2002;2:558–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairagi A, Ghosh KS, Sen SK et al. Enzyme producing bacterial flora isolated from fish digestive tracts. Aquac Int. 2002;10:109–21. [Google Scholar]
- Balabanova L, Averianova L, Marchenok M et al. Microbial and genetic resources for cobalamin (Vitamin B12) biosynthesis: from ecosystems to industrial biotechnology. Int J Mol Sci. 2021;22:4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betiku O, Yeoman C, Gaylord T et al. Evidence of a divided nutritive function in rainbow trout (Oncorhynchus mykiss) midgut and hindgut microbiomes by whole shotgun metagenomic approach. Aquac Rep. 2023;30:101601. [Google Scholar]
- Bhardwaj S, Thakur K, Sharma AK et al. Regulation of omega-3 fatty acids production by different genes in freshwater fish species: a review. Fish Physiol Biochem. 2023;49:1005–16. 10.1007/s10695-023-01236-y. [DOI] [PubMed] [Google Scholar]
- Bharti M, Nagar S, Negi R. Riverine pollution influences the intraspecific variation in the gut microbiome of an invasive fish, Cyprinus carpio (Linn., 1758). 3 Biotech. 2023;13:10. 10.1007/s13205-023-03747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bledsoe JW, Pietrak MR, Burr GS et al. Functional feeds marginally alter immune expression and microbiota of Atlantic salmon (Salmo salar) gut, gill, and skin mucosa though evidence of tissue-specific signatures and host–microbe coadaptation remain. Anim Microbiome. 2022;4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsetto C, Amos GCA, da Rocha UN et al. Microbial community drivers of PK/NRP gene diversity in selected global soils. Microbiome. 2019;7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya BR, Kumar P, Lee J-H et al. Diversity of the tryptophanase gene and its evolutionary implications in living organisms. Microorganisms. 2021;9:2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar B, Kumar R, Sharma D et al. Metagenomic analysis reveals diverse microbial community and potential functional roles in Baner rivulet, India. J Genet Eng Biotechnol. 2023;21:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar B., Thakur K., Mahajan D., Mapping water quality parameters in Baner and Gaj rivulets: insights into the potential impact on river Beas in Himachal Pradesh using ArcGIS. World Water Policy. 2023;9:550–90. 10.1002/wwp2.12122. [DOI] [Google Scholar]
- Brown R, Wiens G, Salinas I. Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2019;86;497–506. 10.1016/j.fsi.2018.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt RL, Volkoff H. Gut microbiota and energy homeostasis in fish. Front Endocrinol. 2019;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill MM. Bacterial flora of fishes: a review. Microb Ecol. 1990;19:21–41. [DOI] [PubMed] [Google Scholar]
- Clements KD. Fermentation and gastrointestinal microorganisms in fishes. In: Mackie RI, White BA (eds.), Gastrointestinal Microbiology: Volume 1 Gastrointestinal Ecosystems and Fermentations. Boston, MA: Springer US, 1997,156–98. [Google Scholar]
- Collazos ME, Ortega E, Barriga C. Effect of temperature on the immune system of a cyprinid fish (Tinca tinca, L). Blood phagocyte function at low temperature. Fish Shellfish Immunol. 1994;4:231–8. [Google Scholar]
- Cottrell MT, Waidner LA, Yu L et al. Bacterial diversity of metagenomic and PCR libraries from the Delaware River. Environ Microbiol. 2005;7:1883–95. [DOI] [PubMed] [Google Scholar]
- Cui X, Zhang Q, Zhang Q et al. Research progress of the gut microbiome in hybrid fish. Microorganisms. 2022;10:891. 10.3390/microorganisms10050891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge N, Carlsen B, Christensen MH et al. The gut microbiome of 54 mammalian species. Front Microbiol. 2022;13:886252. 10.3389/fmicb.2022.886252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco G, Cappello T, Maisano M. Histomorphological changes in fish gut in response to prebiotics and probiotics treatment to improve their health status: a review. Animals. 2023;13:2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb S, Das L, Das SK. Composition and functional characterization of the gut microbiome of freshwater pufferfish (Tetraodon cutcutia). Arch Microbiol. 2020;202:2761–70. [DOI] [PubMed] [Google Scholar]
- Dehler CE, Secombes CJ, Martin SAM. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture. 2017;467:149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasiri AKS, Brunvold L, Brinchmann MF et al. Changes in the intestinal microbiota of wild Atlantic cod Gadus morhua L. upon captive rearing. Microb Ecol. 2011;61:20–30. [DOI] [PubMed] [Google Scholar]
- Di Maiuta N, Schwarzentruber P, Schenker M et al. Microbial population dynamics in the faeces of wood-eating loricariid catfishes. Lett Appl Microbiol. 2013;56:401–7. [DOI] [PubMed] [Google Scholar]
- Drønen K, Roalkvam I, Nilsen H et al. Presence and habitats of bacterial fish pathogen relatives in a marine salmon post-smolt RAS. Aquac Rep. 2022;26:101312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Conti E, Ricci L et al. Links between diet, intestinal anaerobes, microbial metabolites and health. Biomedicines. 2023;11:1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D. Selection and probiotic characterization of exoenzyme-producing bacteria isolated from the gut of Catla catla (Actinopterygii: Cypriniformes: Cyprinidae). Acta Ichthyol Piscat. 2015;45:373–84. [Google Scholar]
- Egerton S, Culloty S, Whooley J et al. The gut microbiota of marine fish. Front Microbiol. 2018;9:873. 10.3389/fmicb.2018.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalas A, Auguet J-C, Avouac A et al. Ecological specialization within a carnivorous fish family is supported by a herbivorous microbiome shaped by a combination of gut traits and specific diet. Front Mar Sci. 2021;8:622883. 10.3389/fmars.2021.622883. [DOI] [Google Scholar]
- Fang H, Kang J, Zhang D. Microbial production of vitamin B12: a review and future perspectives. Microb Cell Fact. 2017;16:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Chen W-D, Wang Y-D. Gut microbiota: an integral moderator in health and disease. Front Microbiol. 2018;9:151. 10.3389/fmicb.2018.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold SM, Vaisanen M-L, Molitoris DR et al. Cetobacterium somerae sp. nov. from human feces and emended description of the genus Cetobacterium. Syst Appl Microbiol. 2003;26:177–81. [DOI] [PubMed] [Google Scholar]
- Fjellheim AJ, Playfoot KJ, Skjermo J et al. Vibrionaceae dominates the microflora antagonistic towards Listonella anguillarum in the intestine of cultured Atlantic cod (Gadus morhua L.) larvae. Aquaculture. 2007;269:98–106. [Google Scholar]
- Francis G, Makkar HPS, Becker K. Antinutritional factors present in plant-derived alternate fish feed ingredients and their effects in fish. Aquaculture. 2001;199:197–227. [Google Scholar]
- Fuerst JA, Sagulenko E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Micro. 2011;9:403–13. [DOI] [PubMed] [Google Scholar]
- Gajardo K, Rodiles A, Kortner TM et al. A high-resolution map of the gut microbiota in Atlantic salmon (Salmo salar): a basis for comparative gut microbial research. Sci Rep. 2016;6:30893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatesoupe F-J. Updating the importance of lactic acid bacteria in fish farming: natural occurrence and probiotic treatments. J Mol Microbiol Biotechnol. 2007;14:107–14. [DOI] [PubMed] [Google Scholar]
- Ghanbari M, Kneifel W, Domig KJ. A new view of the fish gut microbiome: advances from next-generation sequencing. Aquaculture. 2015;448:464–75. [Google Scholar]
- Ghori I, Tubassam M, Ahmad T et al. Gut microbiome modulation mediated by probiotics: positive impact on growth and health status of Labeo rohita. Front Physiol. 2022;13:949559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SK, Wong MK-S, Hyodo S et al. Temperature modulation alters the gut and skin microbial profiles of chum salmon (Oncorhynchus keta). Front Mar Sci. 2022;9:1027621. 10.3389/fmars.2022.1027621. [DOI] [Google Scholar]
- Givens CE, Ransom B, Bano N et al. Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar Ecol Prog Ser. 2015;518:209–23. [Google Scholar]
- Guo M, Chen Y. Coenzyme cobalamin: biosynthesis, overproduction and its application in dehalogenation—a review. Rev Environ Sci Biotechnol. 2018;17:259–84. [Google Scholar]
- Harika K, Shenoy VP, Narasimhaswamy N et al. Detection of biofilm production and its impact on antibiotic resistance profile of bacterial isolates from chronic wound infections. J Glob Infect Dis. 2020;12:129–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan HM, Troxell B. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol. 2013;3:59. 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández A, Rojas D, Wuertz S et al. Dietary filamentous fungi and duration of feeding modulates gut microbial composition in rainbow trout (Oncorhynchus mykiss). Front Mar Sci. 2021;8:1–11.35685121 [Google Scholar]
- Herrera M, Heras J, Catabay C. et al. Dietary-induced changes in the hindgut microbiome and metabolism of a marine herbivorous fish. Physiology. 2023;38:5733889. 10.1152/physiol.2023.38.S1.5733889. [DOI] [Google Scholar]
- Horlick J, Booth MA, Tetu SG. Alternative dietary protein and water temperature influence the skin and gut microbial communities of yellowtail kingfish (Seriola lalandi). PeerJ. 2020;8:e8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Sham RC, Deng Y et al. Diversity of gut microbiomes in marine fishes is shaped by host-related factors. Mol Ecol. 2020;29:5019–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyben D, Sun L, Moccia R et al. Dietary live yeast and increased water temperature influence the gut microbiota of rainbow trout. J Appl Microbiol. 2018;124:1377–92. [DOI] [PubMed] [Google Scholar]
- Izvekova GI, Izvekov EI, Plotnikov AO. Symbiotic microflora in fishes of different ecological groups. Izv Akad Nauk Ser Biol. 2007;34:610–8. [PubMed] [Google Scholar]
- Jobling M. National Research Council (NRC): nutrient requirements of fish and shrimp. Aquac Int. 2012;20:601–2. [Google Scholar]
- Jordaan K, Bezuidenhout CC. The impact of physico-chemical water quality parameters on bacterial diversity in the Vaal River, South Africa. Water SA. 2013;39:385–96. [Google Scholar]
- Kanika N, Hou X, Liu H et al. Specific gut microbiome's role in skin pigmentation: insights from SCARB1 mutants in Oujiang colour common carp. J Appl Microbiol. 2024;135:lxae226. [DOI] [PubMed] [Google Scholar]
- Karasov WH, Douglas AE. Comparative digestive physiology. Compr Physiol. 2013;3:741–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsen C, Tzimorotas D, Robertsen E et al. Feed microbiome: confounding factor affecting fish gut microbiome studies. ISME Commun. 2022;2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PS, Shin N-R, Lee J-B et al. Host habitat is the major determinant of the gut microbiome of fish. Microbiome. 2021;9:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisil OV, Trefilov VS, Sadykova VS et al. Surfactin: its biological activity and possibility of application in agriculture. Appl Biochem Microbiol. 2023;59:1–13. [Google Scholar]
- Klemetsen T, Karlsen C, Willassen N. Phylogenetic revision of the genus Aliivibrio: intra- and inter-species variance among clusters suggest a wider diversity of species. Front Microbiol. 2021;12:626759. 10.3389/fmicb.2021.626759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki H, Sakurai K, Hosoyama A et al. Diversity of PKS and NRPS gene clusters between Streptomyces abyssomicinicus sp. nov. and its taxonomic neighbor. J Antibiot. 2020;73:141–51. [DOI] [PubMed] [Google Scholar]
- Kong Y, Li M, Chu G et al. The positive effects of single or conjoint administration of lactic acid bacteria on Channa argus: digestive enzyme activity, antioxidant capacity, intestinal microbiota and morphology. Aquaculture. 2021;531:735852. [Google Scholar]
- Kuipers A, Rink R, Moll GN. Genetics, biosynthesis, structure, and mode of action of lantibiotics. In: Drider D, Rebuffat S (eds.), Prokaryotic Antimicrobial Peptides: From Genes to Applications. New York, NY: Springer, 2011,147–69. [Google Scholar]
- Kumar J, Sharma N, Singh SP. Genome-resolved metagenomics inferred novel insights into the microbial community, metabolic pathways, and biomining potential of Malanjkhand acidic copper mine tailings. Environ Sci Pollut Res. 2023;30:50864–82. [DOI] [PubMed] [Google Scholar]
- Larios-Soriano E, Re-Araujo AD, Gómez-Gil B et al. Reciprocal effect of temperature and dietary lipids on metabolic performance and gut microbiota of yellowtail kingfish (Seriola lalandi) juveniles. Aquac Res. 2021;52:6189–204. [Google Scholar]
- Leeper A, Ekmay R, Knobloch S et al. Torula yeast in the diet of Atlantic salmon Salmo salar and the impact on growth performance and gut microbiome. Sci Rep. 2021;12:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Young KD. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiol Read Engl. 2013;159:402–10. [DOI] [PubMed] [Google Scholar]
- Li H, Wu S, Wirth S et al. Diversity and activity of cellulolytic bacteria, isolated from the gut contents of grass carp (Ctenopharyngodon idellus) (Valenciennes) fed on Sudan grass (Sorghum sudanense) or artificial feedstuffs. Aquac Res. 2016;47:153–64. [Google Scholar]
- Li J, Bates KA, Hoang KL et al. Experimental temperatures shape host microbiome diversity and composition. Global Change Biol. 2023;29:41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Long M, Gatesoupe F-J et al. Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing. Microb Ecol. 2015;69:25–36. [DOI] [PubMed] [Google Scholar]
- Li X, Zhou L, Yu Y et al. Composition of gut microbiota in the gibel carp (Carassius auratus gibelio) varies with host development. Microb Ecol. 2017;74:239–49. [DOI] [PubMed] [Google Scholar]
- Liu Q, Lai Z, Gao Y et al. Connection between the gut microbiota of largemouth bass (Micropterus salmoides) and microbiota of the pond culture environment. Microorganisms. 2021;9:1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn MS, Boutin S, Hoseinifar SH et al. Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front Microbiol. 2014a;5:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Imlay JA. When anaerobes encounter oxygen: mechanisms of oxygen toxicity, tolerance and defence. Nat Rev Micro. 2021;19:774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y, Li M, Zhou W et al. The fish microbiota: research progress and potential applications. Engineering. 2023;29:137–46. 10.1016/j.eng.2022.12.011. [DOI] [Google Scholar]
- Marshall B, Amritkar K, Wolfe M et al. Evolutionary flexibility and rigidity in the bacterial methylerythritol phosphate (MEP) pathway. Front Microbiol. 2023;14:1286626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe O, Ross RP, Hill C. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol Rev. 2001;25:285–308. [DOI] [PubMed] [Google Scholar]
- Melo-Bolívar JF, Pardo RYR, Hume ME et al. Establishment and characterization of a competitive exclusion bacterial culture derived from Nile tilapia (Oreochromis niloticus) gut microbiomes showing antibacterial activity against pathogenic Streptococcus agalactiae. PLoS One. 2019;14:e0215375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Shu Q. Novel primers to identify a wider diversity of butyrate-producing bacteria. World J Microbiol Biotechnol. 2024;40:76. [DOI] [PubMed] [Google Scholar]
- Merrifield DL, Dimitroglou A, Foey A et al. The current status and future focus of probiotic and prebiotic applications for salmonids. Aquaculture. 2010;302:1–18. [Google Scholar]
- Miranda R, Miqueleiz I, Darwall W et al. Monitoring extinction risk and threats of the world's fishes based on the Sampled Red List Index. Rev Fish Biol Fisheries. 2022;32:975–91. [Google Scholar]
- Miyake S, Ngugi DK, Stingl U. Diet strongly influences the gut microbiota of surgeonfishes. Mol Ecol. 2015;24:656–72. [DOI] [PubMed] [Google Scholar]
- Miyake S, Ngugi DK, Stingl U. Phylogenetic diversity, distribution, and cophylogeny of giant bacteria (Epulopiscium) with their surgeonfish hosts in the Red Sea. Front Microbiol. 2016;7:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro T, Ruby EG. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol Microbiol. 2012;84:795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S, Roy T, Sen S et al. Distribution of enzyme-producing bacteria in the digestive tracts of some freshwater fish. Acta Icth Et Piscat. 2008;38:1–8. [Google Scholar]
- Moore ERB, Tindall BJ, Martins D et al. Nonmedical: pseudomonas. In: Dworkin M, Falkow S, Rosenberg E et al. (eds.), The Prokaryotes: A Handbook on the Biology of Bacteria Volume 6: Proteobacteria: Gamma Subclass. New York, NY: Springer, 2006,646–703. [Google Scholar]
- Morales M, Valenzuela-Miranda D, Núñez-Acuña G et al. Atlantic salmon (Salmo salar) transfer to seawater by gradual salinity changes exhibited an increase in the intestinal microbial abundance and richness. Microorganisms. 2022;11:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Y, Moreno-Mesonero L, Soler P et al. Influence of drinking water biofilm microbiome on water quality: insights from a real-scale distribution system. Sci Total Environ. 2024;921:171086. [DOI] [PubMed] [Google Scholar]
- Mydy LS, Bailey DC, Patel KD et al. The siderophore synthetase IucA of the aerobactin biosynthetic pathway uses an ordered mechanism. Biochemistry. 2020;59:2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete P, Fuentes P, De la Fuente L et al. Short-term effects of dietary soybean meal and lactic acid bacteria on the intestinal morphology and microbiota of Atlantic salmon (Salmo salar). Aquac Nutr. 2013;19:827–36. [Google Scholar]
- Nayak SK. Role of gastrointestinal microbiota in fish. Aquac Res. 2010;41:1553–73. [Google Scholar]
- Neave MJ, Apprill A, Ferrier-Pagès C et al. Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl Microbiol Biotechnol. 2016;100:8315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JA, Dehn AM. 10–the GI tract in air breathing. In: Grosell M, Farrell AP, Brauner CJ (eds), Fish Physiology. Vol 30. Academic Press, Netherlands: Elsevier Science, 2010:395–433. [Google Scholar]
- Nelson JA, Wubah DA, Whitmer ME et al. Wood-eating catfishes of the genus Panaque: gut microflora and cellulolytic enzyme activities. J Fish Biol. 1999;54:1069–82. [Google Scholar]
- Ngugi DK, Miyake S, Cahill M et al. Genomic diversification of giant enteric symbionts reflects host dietary lifestyles. Proc Natl Acad Sci USA. 2017;114:E7592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski J, Wu JY, Rueger DC et al. Characterization of the cysJIH regions of Salmonella typhimurium and Escherichia coli B: DNA sequences of cysI and cysH and a model for the siroheme-Fe4S4 active center of sulfite reductase hemoprotein based on amino acid homology with spinach nitrite reductase. J Biol Chem. 1989;264:15726–37. [PubMed] [Google Scholar]
- Ou W, Guo Z, Pan Y et al. Advances in the effects of dietary macronutrients on the gut microbiota of tilapia (Oreochromis spp.). Microorganisms. 2024;12:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou W, Yu G, Zhang Y et al. Recent progress in the understanding of the gut microbiota of marine fishes. Mar Life Sci Technol. 2021;3:434–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parata L, Nielsen S, Xing X et al. Age, gut location and diet impact the gut microbiome of a tropical herbivorous surgeonfish. FEMS Microbiol Ecol. 2020;96:fiz179. [DOI] [PubMed] [Google Scholar]
- Parris DJ, Morgan MM, Stewart FJ. Feeding rapidly alters microbiome composition and gene transcription in the clownfish gut. Appl Environ Microb. 2019;85:15. 10.1128/AEM.02479-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Oliver A, Kelly L et al. Herbivorous fish microbiome adaptations to sulfated dietary polysaccharides. Appl Environ Microbiol. 2023;89:e0215422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Liu W, He X et al. A review on surfactin: molecular regulation of biosynthesis. Arch Microbiol. 2023a;205:313. [DOI] [PubMed] [Google Scholar]
- Qi X, Zhang Y, Zhang Y et al. Vitamin B12 produced by Cetobacterium somerae improves host resistance against pathogen infection through strengthening the interactions within gut microbiota. Microbiome. 2023b;11:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput A, Kumar M. In silico analyses of conservational, functional and phylogenetic distribution of the LuxI and LuxR homologs in Gram-positive bacteria. Sci Rep. 2017;7:6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen JA, Villumsen KR, Ernst M et al. A multi-omics approach unravels metagenomic and metabolic alterations of a probiotic and synbiotic additive in rainbow trout (Oncorhynchus mykiss). Microbiome. 2022;10:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Cornelis P. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 2003;11:195–200. [DOI] [PubMed] [Google Scholar]
- Ray AK, Ghosh K, Ringø E. Enzyme-producing bacteria isolated from fish gut: a review. Aquac Nutr. 2012;18:465–92. [Google Scholar]
- Ringø E, Gatesoupe F-J. Lactic acid bacteria in fish: a review. Aquaculture. 1998;160:177–203. [Google Scholar]
- Ringø E, Løvmo L, Kristiansen M et al. Lactic acid bacteria vs. pathogens in the gastrointestinal tract of fish: a review. Aquac Res. 2010;41:451–67. [Google Scholar]
- Ringø E, Zhou Z, Vecino JLG et al. Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story?. Aquac Nutr. 2016;22:219–82. [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ et al. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011;5:1595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudi K, Angell IL, Pope PB et al. Stable core gut microbiota across the freshwater-to-saltwater transition for farmed Atlantic salmon. Appl Environ Microb. 2018;84:e01974–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Gisbert E, Andree KB. Impact of the diet in the gut microbiota after an inter-species microbial transplantation in fish. Sci Rep. 2024;14:4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi J, Chaganti S, Johnson T et al. Host species and habitat shape fish-associated bacterial communities: phylosymbiosis between fish and their microbiome. Microbiome. 2023;11:258. 10.1186/s40168-023-01697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Roy RN, Sen SK et al. Characterization of cellulase-producing bacteria from the digestive tract of tilapia, Oreochromis mossambica (Peters) and grass carp, Ctenopharyngodon idella (Valenciennes). Aquac Res. 2006;37:380–8. [Google Scholar]
- Sampaio A, Silva V, Poeta P et al. Vibrio spp.: life strategies, ecology, and risks in a changing environment. Diversity. 2022;14:97. [Google Scholar]
- Savio D, Sinclair L, Ijaz UZ et al. Bacterial diversity along a 2600 km river continuum. Environ Microbiol. 2015;17:4994–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal L, Brammer-Robbins E, Wormington AM et al. Microbiome composition and function in aquatic vertebrates: small organisms making big impacts on aquatic animal health. Front Microbiol. 2021;12:567408. 10.3389/fmicb.2021.567408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda J, Moeller AH. The effects of temperature on animal gut microbiomes. Front Microbiol. 2020;11:384. 10.3389/fmicb.2020.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Thakur K, Pankaj PP et al. International Journal of Oceanography & Aquaculture committed to create value for researchers gut microbiota of Salmo trutta fario and Oncorhynchus mykiss: implications for fish health and aquaculture management gut microbiota of Salmo trutta fario and Oncorhynchus mykiss: implications for fish health and aquaculture management. Int J Oceanogr Aquac. 2023;7:000242. 10.23880/ijoac-16000242. [DOI] [Google Scholar]
- Siegers K, Entian KD. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl Environ Microb. 1995;61:1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Karimi S, Vidakovic A et al. (October 21, 2021) Dietary filamentous fungi and duration of feeding modulates gut microbial composition in rainbow trout (Oncorhynchus mykiss). Front Mar Sci. 2021;8:728569. 10.3389/fmars.2021.728569. [DOI] [Google Scholar]
- Small C, Beck E, Currey M et al. Host genomic variation shapes gut microbiome diversity in threespine stickleback fish. mBio. 2023;14:e0021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh M, Tay YC, Lee CS et al. The intestinal digesta microbiota of tropical marine fish is largely uncultured and distinct from surrounding water microbiota. NPJ Biofilms Microbiomes. 2024;10:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suescun-Sepulveda J, Rondón F, Fuentes LJ. Diversity of culturable bacteria from freshwater bodies from a altitudinal gradient in eastern Cordillera of Colombia. FEMS Microbiol Lett. 2023;17:370. 10.1093/femsle/fnad037. [DOI] [PubMed] [Google Scholar]
- Sugita H, Miyajima C, Deguchi Y. The vitamin B12-producing ability of the intestinal microflora of freshwater fish. Aquaculture. 1991;92:267–76. [Google Scholar]
- Sullam KE, Essinger SD, Lozupone CA et al. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol. 2012;21:3363–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnecki AM, Burgos FA, Ray CL et al. Fish intestinal microbiome: diversity and symbiosis unravelled by metagenomics. J Appl Microbiol. 2017;123:2–17. [DOI] [PubMed] [Google Scholar]
- Terova G, Rimoldi S, Ascione C et al. Next generation sequencing for gut microbiome characterization in rainbow trout (Oncorhynchus mykiss) fed animal by-product meals as an alternative to fishmeal protein sources. PloS ONE. 2018;13:e0193652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Polz MF. Dynamics of Vibrio Populations and Their Role in Environmental Nutrient Cycling. Washington, DC, USA: ASM Press, 2006;190–203. [Google Scholar]
- Trust T, Bull L, Currie B et al. Obligate anaerobic bacteria in the gastrointestinal microflora of the grass carp (Ctenopharyngodon idella), goldfish (Carassius auratus), and rainbow trout (Salmo gairdneri). J Fish Res Board Can. 2011;36:1174–9. [Google Scholar]
- Tsuchiya C, Sakata T, Sugita H. Novel ecological niche of cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett Appl Microbiol. 2008;46:43–8. [DOI] [PubMed] [Google Scholar]
- Uniacke-Lowe S, Stanton C, Hill C et al. The marine fish gut microbiome as a source of novel bacteriocins. Microorganisms. 2024;12:1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel MA, Dutilh BE, Neveling K et al. Pyrosequencing of 16S rRNA gene amplicons to study the microbiota in the gastrointestinal tract of carp (Cyprinus carpio L.). AMB Exp. 2011;1:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch A et al. Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio. 2014;5:e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viver T, Ruiz A, Bertomeu E et al. Food determines ephemerous and non-stable gut microbiome communities in juvenile wild and farmed Mediterranean fish. Sci Total Environ. 2023;889:164080. [DOI] [PubMed] [Google Scholar]
- Wang A, Zhang Z, Ding Q et al. Intestinal cetobacterium and acetate modify glucose homeostasis via parasympathetic activation in zebrafish. Gut Microbes. 2021;13:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AR, Ran C, Ringø E et al. Progress in fish gastrointestinal microbiota research. Rev Aquac. 2018;10:626–40. [Google Scholar]
- Wang H, Fewer DP, Holm L et al. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci USA. 2014;111:9259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Waite DW, Deines P et al. The microbiome in threatened species conservation. Biol Conserv. 2019;229:85–98. [Google Scholar]
- Wong S, Rawls JF. Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol Ecol. 2012;21:3100–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Waldrop T, Summerfelt S et al. Aquacultured rainbow trout (Oncorhynchus mykiss) possess a large core intestinal microbiota that is resistant to variation in diet and rearing density. Appl Environ Microb. 2013;79:4974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Pan M, Zan Z et al. Regulation of lipid metabolism by gut microbiota in aquatic animals. Rev Aquac. 2024;16:34–46. [Google Scholar]
- Wu S, Wang G, Angert ER et al. Composition, diversity, and origin of the bacterial community in grass carp intestine. PLoS One. 2012;7:e30440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li X, Zhu Y et al. Effects of water temperature on the growth, antioxidant capacity, and gut microbiota of percocypris pingi juveniles. Fishes. 2022;7:374. [Google Scholar]
- Xi Y, Jiang X, Xie X et al. Viromics reveals the high diversity of viruses from fishes of the Tibet highland. Microbiol Spectr. 2023;11:e00946–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia N, Xia X, Zhu B et al. Bacterial diversity and community structure in the sediment of the middle and lower reaches of the Yellow River, the largest turbid river in the world. Aquat Microb Ecol. 2013;71:43–55. [Google Scholar]
- Xu Y, Wu J-Y, Liu Q-J et al. Genome-wide identification and evolutionary analyses of SrfA operon genes in Bacillus. Genes. 2023;14:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Abdallah II, Haan IEM et al. Enhanced C30 carotenoid production in Bacillus subtilis by systematic overexpression of MEP pathway genes. Appl Microbiol Biotechnol. 2015;99:5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Amberg J, Chapman D et al. Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J. 2014;8:541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin B, Tan S, Pan K et al. Effects of diet composition on gut microbiome and mercury biotransformation in the gobyfish. Sci Total Environ. 2023a;892:164776. [DOI] [PubMed] [Google Scholar]
- Yin Q-J, Ying T-T, Zhou Z-Y et al. Species-specificity of the secondary biosynthetic potential in Bacillus. Front Microbiol. 2023b;14:1271418. 10.3389/fmicb.2023.1271418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yan J, Zhou X et al. Rice-fish symbiosis improves the muscle nutrition and intestinal flora diversity of tilapia. Israeli Journal of Aquaculture-Bamidgeh, 2024;76:223–32. 10.46989/001c.118696. [DOI] [Google Scholar]
- Zou S, Gong L, Khan TA et al. Comparative analysis and gut bacterial community assemblages of grass carp and crucian carp in new lineages from the Dongting Lake area. MicrobiologyOpen. 2020;9:e996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.