Abstract
In Saccharomyces cerevisiae, silencing at the HM loci depends on Sir proteins, which are structural components of silenced chromatin. To explore the structure and assembly of silenced chromatin, the associations of Sir proteins with sequences across the HMR locus were examined by chromatin immunoprecipitation. In wild-type cells, Sir2p, Sir3p, and Sir4p were spread throughout and coincident with the silenced region at HMR. Sir1p, in contrast, associated only with the HMR-E silencer, consistent with its role in establishment but not maintenance of silencing. Sir4p was required for the association of other Sir proteins with silencers. In contrast, in the absence of Sir2p or Sir3p, partial assemblies of Sir proteins could form at silencers, where Sir protein assembly began. Spreading across HMR required Sir2p and Sir3p, as well as the deacetylase activity of Sir2p. These data support a model for the spreading of silenced chromatin involving cycles of nucleosome deacetylation by Sir2p followed by recruitment of additional Sir2p, Sir3p, and Sir4p to the newly deacetylated nucleosome. This model suggests mechanisms for boundary formation, and for maintenance and inheritance of silenced chromatin. The principles are generalizable to other types of heritable chromatin states.
INTRODUCTION
Regional repression, or silencing, involves the formation of a distinct, long-range chromatin structure that blocks transcription of genes within the silenced domain. The silenced state can be epigenetically inherited, implying that some aspect of the structure of silenced chromatin can template its own replication. Regional repression is an important means by which eukaryotic cells regulate gene expression. However, many aspects of the formation and function of repressive chromatin remain poorly understood.
The silencing of the mating type loci in Saccharomyces cerevisiae is mediated by silencers, known as E and I, that flank the two mating type loci, HMR and HML. These silencers consist of binding sites for at least two of three DNA binding proteins, ORC, Rap1p, and Abf1p. A working model for the assembly of silenced chromatin at the HM loci involves two steps (Hecht et al., 1995; Braunstein et al., 1996; Grunstein, 1998; Lustig, 1998; Moazed, 2001b). First, the four Sir proteins bind to the silencer binding proteins at the silencer. Then, Sir2p, Sir3p, and Sir4p spread in both directions from the silencers. This model is derived from several lines of evidence. For example, Sir1p binds Orc1p (Triolo and Sternglanz, 1996) and Sir3p and Sir4p bind Rap1p (Moretti et al., 1994; Cockell et al., 1995; Moretti and Shore, 2001). Also, physical interactions between Sir1p and Sir4p (Triolo and Sternglanz, 1996), Sir2p and Sir4p (Moazed et al., 1997; Ghidelli et al., 2001), and Sir3p and Sir4p (Hecht et al., 1996) have been observed. Sir3p and Sir4p also bind to the tails of histones H3 and H4 (Hecht et al., 1995), and this interaction may enable the spreading of Sir3p and Sir4p. Sir2p is assumed to spread along with Sir3p and Sir4p via its interaction with Sir4p. Sir2p, Sir3p, and Sir4p also associate with Rap1 proteins bound at telomeric repeats and extend several kilobase pairs beyond these telomeric sequences (Strahl-Bolsinger et al., 1997; Lieb et al., 2001), supporting the notion that Sir2p, Sir3p, and Sir4p do spread from sites of nucleation.
The N-terminal tails of histones H3 and H4 are hypoacetylated at the HM loci (Braunstein et al., 1993; Suka et al., 2001). Sir2p is an NAD+-dependent deacetylase (Imai et al., 2000; Landry et al., 2000; Smith et al., 2000), and is thought to deacetylate histones at silenced loci. The hypoacetylated nucleosomes, together with the Sir proteins, are thought to form an ordered, compact structure that is restrictive to transcription. In support of this model, silenced regions are less accessible to restriction nucleases (Loo and Rine, 1994) and display ordered, regularly spaced nucleosomes (Weiss and Simpson, 1998; Ravindra et al., 1999). This structure, and hence the silenced region, is limited in extent by the presence of “boundary” elements flanking the HM loci (Bi et al., 1999; Donze et al., 1999).
After silenced chromatin is assembled, it must be stably maintained during cell growth and inherited in both daughters upon cell division. Strains bearing some silencer mutations or sir1Δ are defective in the establishment of silencing and are composed of mixed populations of silenced and unsilenced cells (Pillus and Rine, 1989; Mahoney et al., 1991). Single-cell assays of these strains demonstrate that once silencing is established, the silenced chromatin is stably maintained during cell growth and inherited in both daughters upon cell division. Thus, the mechanisms of maintenance and inheritance of silenced chromatin differ from the mechanism of establishment. In contrast to Sir1p, the other Sir proteins are required for both the establishment and the maintenance of silenced chromatin.
Although this model for the assembly of silenced chromatin is widely described (Hecht et al., 1995; Braunstein et al., 1996; Grunstein, 1998; Lustig, 1998; Moazed, 2001b), it masks several unresolved issues. One issue is the source of specificity in silencing. Individually, ORC, Rap1p, and Abf1p bind hundreds of locations in the genome, yet silencing is restricted to HMR, HML, telomeres, and rDNA. Additionally, it is unclear how the Sir proteins can spread over several kilobase pairs of varied sequence, binding to the tails of histones H3 and H4 in nucleosomes, yet not associate indiscriminately with DNA throughout the genome. Furthermore, it is not known why four different Sir proteins are needed when one is sufficient to provide deacetylase activity. These issues motivated a comprehensive study of the associations of Sir proteins with silent loci.
MATERIALS AND METHODS
Yeast Strains and Plasmids
Strains used in this study were all derived from W303-1a (Table 1). JRY7131 was described previously (Kirchmaier and Rine, 2001). The sir1Δ::LEU2, sir1Δ::TRP1, sir2Δ::LEU2, sir2Δ::TRP1, sir3Δ::LEU2, sir3Δ::TRP1, sir4Δ::LEU2, and sir4Δ::TRP1 alleles were complete deletions of the open reading frames generated by one-step gene conversion. The SIR1–3xHA allele was derived from CFY416 (Gardner and Fox, 2001). The LEU2::sir2-N345A (Imai et al., 2000), hmr-e** (Axelrod and Rine, 1991), and hmr 331–324, 274–256 (hmr-eΔΔ) (Brand et al., 1987) alleles were described previously. Plasmid pCF448 expresses SIR1-3xHA in pRS316 (Gardner and Fox, 2001), and plasmid pJR1811 contains GAL4DBD-SIR1 fusion expressed from the MET3 promoter in pRS313 (Fox et al., 1997).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303 | ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| W303-AR | W303 MATa RDN::ADE2 sir2Δ::TRP1 LEU2::SIR2 | L. Guarente |
| W303-AR sir2-N345A | W303 MATa RDN::ADE2 sir2Δ::TRP1 LEU2::sir2-N345A | L. Guarente |
| W303-AR sir2Δ | W303 MATa RDN::ADE2 sir2Δ::TRP1 | L. Guarente |
| YGM1 | W303 MATα hmr-e (331-324 and 274-256; Rap1 and Abf1 binding sites deleted) | G. Micklem |
| JRY4013 | W303 MATα ADE2 lys2Δ | |
| JRY4563 | JRY4013 sir2Δ::TRP1 | |
| JRY4580 | JRY4013 sir4Δ::TRP1 | |
| JRY4605 | JRY4013 sir3Δ::TRP1 | |
| JRY4623 | JRY4013 sir1Δ::TRP1 | |
| JRY5472 | JRY4013 hmre∗∗ (Rap1 and Abf1 binding sites mutated) | |
| JRY7131 | W303 matΔ::ADE2 FRT-4xGal4-Rap1-Abf1 HMRa ΔI-FRT SIR1 LEU2::FLP1 [cir°] plus pJR1811 | |
| JRY7296 | JRY4013 sir1Δ::LEU2 sir2Δ::TRP1 | |
| JRY7297 | JRY4013 sir1Δ::LEU2 sir3Δ::TRP1 | |
| JRY7298 | JRY4013 sir2Δ::LEU2 sir3Δ::TRP1 | |
| JRY7299 | JRY4013 SIR1-3HA | |
| JRY7300 | JRY4013 SIR1-3HA sir2Δ::TRP1 | |
| JRY7301 | JRY4013 SIR1-3HA sir3Δ::TRP1 | |
| JRY7302 | JRY4013 SIR1-3HA sir4Δ::TRP1 | |
| JRY7303 | W303 MATα 4xGal4-Rap1-Abf1 HMRa ΔI sir2Δ::LEU2 SIR1 plus pJR1811 | |
| JRY7304 | W303 MATα 4xGal4-Rap1-Abf1 HMRa ΔI sir3Δ::LEU2 SIR1 plus pJR1811 | |
| JRY7305 | W303 MATα 4xGal4-Rap1-Abf1 HMRa ΔI sir4Δ::LEU2 SIR1 plus pJR1811 | |
| JRY7306 | W303 MATα 4xGal4-Rap1-Abf1 HMRa ΔI sir1Δ::LEU2 plus pJR1811 and pCF448 |
Chromatin Immunoprecipitation
Chromatin immunoprecipitations were performed as described previously (Rusche and Rine, 2001) using 10 OD equivalents of cells. DNA was sheared by sonication to an average size of 500–800 base pairs in all experiments. Each experiment was repeated at least once, isogenic duplicate strains were used to repeat some of the experiments, and results were uniformly reproducible. Antibodies were 4 μl of serum from rabbits inoculated with recombinant Sir proteins (rabbit 2931, LacZ-Sir2 fusion protein; rabbit 2934, LacZ-Sir3 fusion protein; rabbit 2913, C-terminal 46% of Sir4p; Axelrod 1991), 1 mg/ml rabbit polyclonal anti-di-acetyl-histone H3 (K9 and K14; 06-599; Upstate Biotechnology, Lake Placid, NY), or rabbit polyclonal anti-hemagglutinin (HA) tag (06-831; Upstate Biotechnology). The oligonucleotides used for polymerase chain reaction (PCR) are described in Table 2.
Table 2.
Oligonucleotides used in this study
| Region | Primer 1 | Primer 2 |
|---|---|---|
| HMR-E | ctaaatcgcatttcttttcgtccac | taacaaaaaccaggagtacctgcgc |
| HMR-GalSS | taataacaaacctctaatccggt | gcttggtaattttagatttgtacc |
| HMR-I | tgtcaccaacattttcgtatatggcg | ctaccacattatcaatccttgcatccag |
| HML-E | ggatggatctagggttttatgcc | tttggcccccgaaatcg |
| tagatttggcccccgaaatcg (Figures 3B and 6A) | ||
| HML-I | ccagctgagtaactaactctcatgg | gctgttacggagatgcaaagc |
| HMR-X/Ya | taccaacccatccgccg | tccgccatactacaaatatcatcc |
| HMR-Ya/Z1 | gtggcattactccacttcaagtaag | caagagcaagacgatgggg |
| al | ggatgatatttgtagtatggcgg | cccaaactcttacttgaagtgg |
| Region 1 | gcttgagcattgggcttctg | cgatgcaggcgacaccag |
| Region 2 | acaataacagacaagggcctacg | ggcgagaaaaacgccctg |
| Region 3 | agattcatatatcttcaaggggaacttcttgtac | tagtttcttaagtactaccggattagaggtttg |
| Region 4 | caaacctctaatccggtagtacttaaga | gtggacgaaaagaaatgcgatttagc |
| Region 5 | ctggatgcaaggattgataatgtggtag | catatacggtgttagaagatgacgc |
| Region 6 | aatccttgcgtttcagcttcc | tcgacgtcggatttgcg |
| Region 7 | gacacccaggttgccgc | tggtggcccatgccttg |
| Region 8 | caacatggtgttccaaagcac | gcagcttactcccaagagtgc |
| SSCI | gcttcggcccggttcca | cagcaagcatcttggtgcg |
| MAT | gttcaccctgtttccattggaa | gggtagagtcttattggcaaga |
Immunoblots
Logarithmically growing cells were suspended in SDS-PAGE sample buffer (25 mM Tris-HCl, pH 6.8, 2.5% SDS, 2.5% glycerol, 0.01% bromphenol blue, 1.25% β-mercaptoethanol) containing 0.1 mM N-tosyl-l-phenylalanine chloromethyl ketone, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml chymostatin, 2 μg/ml pepstatin A, 1 mM benzamidine, and 1× Complete protease inhibitors (Roche Applied Science, Indianapolis, IN) plus glass beads and vortexed for 3 min followed by incubation at 94°C for 1 min. Samples were separated on a 7.5% acrylamide gel and transferred to nitrocellulose. Protein blots were probed using antibodies against Sir2p (1:1000; described above), Sir3p (1:1000; described above), Sir4p (1:500; sc-6671; Santa Cruz Biotechnology, Santa Cruz, CA), tubulin (1:40,000, B206; Weinstein and Solomon, 1990), or phosphoglycerate kinase (1:250; A-6457; Molecular Probes, Eugene, OR). Immunoblotting was not successful with the rabbit polyclonal antibody against Sir4p (Axelrod, 1991).
RNA Blots
Total RNA was isolated from logarithmically growing cells as described previously (Schmitt et al., 1990). RNA was separated on 1.2% agarose-formaldehyde gels and transferred to Hybond XL membranes (Amersham Biosciences, Piscataway, NJ; Sambrook et al., 1989). To monitor silencing at HMR, the level of a1 mRNA relative to that of SCR1 mRNA was measured as described previously (Kirchmaier and Rine, 2001).
RESULTS
This study revealed new dimensions to the formation of silenced chromatin. We present evidence first that partial assemblies of Sir proteins could form at the HMR-E silencer and then that Sir2p, Sir3p, and Sir4p and deacetylated histones were distributed throughout the silenced domain. Building on these observations, we turn to the requirements for Sir protein spreading and the link between spreading and deacetylation. The partial assemblies of Sir proteins, which formed at the silencer, did not spread away from the silencer efficiently. Thus, the assembly and spreading steps of silenced chromatin could be separated. Moreover, the role of the deacetylase activity of Sir2p was restricted to the spreading step. Finally, we describe the assembly of Sir proteins at other silencers and the molecular explanation for why the HMR-I silencer is not sufficient to cause silencing. These discoveries suggest a mechanism for spreading of chromatin proteins, the principles of which may be applicable to other types of inherited chromatin states.
Partial Assemblies of Sir Proteins Could Form at HMR-E Silencer
The first step in the formation of silenced chromatin is predicted to be the association of Sir proteins with the silencer-binding proteins ORC, Rap1p, and Abf1p. However, it is not known whether the association of individual Sir proteins with the silencer can occur independently of other Sir proteins. Therefore, the association of each of the four Sir proteins with the HMR-E silencer was systematically examined in the presence or absence of other Sir proteins by chromatin immunoprecipitation. In these experiments, Sir1-HAp (Gardner and Fox, 2001) was immunoprecipitated with anti-HA tag antibodies, and the other Sir proteins were immunoprecipitated with antibodies raised against recombinant Sir proteins (Axelrod, 1991). Immunoprecipitated DNA was simultaneously amplified by PCR for the HMR-E silencer and the SSC1 promoter, a gene whose transcription is not controlled by Sir proteins. To interpret the data reliably, the input DNA was also amplified to determine the ratio of the two PCR products when the templates are present in a 1:1 ratio. Twofold serial dilutions of the starting material were performed to verify that the PCR yield was sensitive to the amount of starting DNA. Each immunoprecipitation was repeated at least once, and often three or four times.
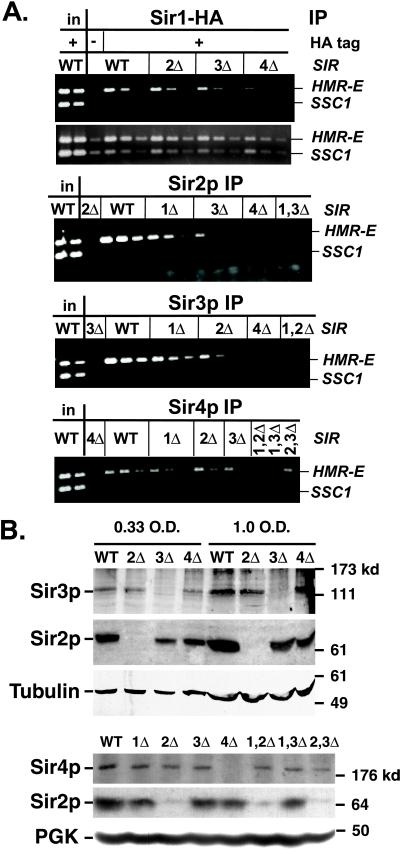
As expected, in wild-type cells, all four Sir proteins associated preferentially with HMR-E relative to the SSC1 negative control (Figure 1A, WT lanes; compare with input lanes). This silencer DNA coprecipitated specifically with the Sir proteins, because HMR-E DNA was not preferentially immunoprecipitated with anti-Sir antibodies from cells in which the corresponding Sir genes were deleted or were not tagged (Figure 1A, third lanes).
Figure 1.
Association of Sir proteins with the HMR-E silencer. (A) Chromatin immunoprecipitations from MATα SIR (JRY4013), sir2Δ (JRY4563), sir3Δ (JRY4605), sir4Δ (JRY4580), sir1Δ sir2Δ (JRY7296), sir1Δ sir3Δ (JRY7297), or sir2Δ sir3Δ (JRY7298) cells. Strains used in panels 1 and 2 also expressed SIR1–3xHA and were SIR (JRY7299), sir2Δ (JRY7300), sir3Δ (JRY7301), or sir4Δ (JRY7302). DNA immunoprecipitated with antibodies against HA tag (to monitor Sir1-HAp; panels 1 and 2), Sir2p (panel 3), Sir3p (panel 4), or Sir4p (panel 5) was analyzed by simultaneous amplification of HMR-E (top bands) and SSC1 (bottom bands). Panel 2 is a longer exposure of panel 1. For each sample, serial twofold dilutions of the template DNA are shown, starting with 1/19,000 of the input DNA or 2/75 (panels 1 and 2), 1/250 (panels 3 and 4), or 1/125 (panel 5) of the immunoprecipitated DNA. For the negative controls (lanes 3 in each panel), a single lane is shown, amplified from the largest fraction of the immunoprecipitated DNA used in that panel. These negative controls contained samples immunoprecipitated from strains lacking Sir2p (panel 3), Sir3p (panel 4), or Sir4p (panel 5), or bearing an untagged version of Sir1p (panels 1 and 2). (B) Expression of Sir proteins. Whole cell lysates were analyzed by immunoblots with antibodies against Sir2p (panels 2 and 5), Sir3p (panel 1), Sir4p, (panel 4) tubulin (panel 3), or phosphoglycerate kinase (panel 6). For panels 1–3, whole cell lysates were from SIR (JRY7131), sir2Δ (JRY7303), sir3Δ (JRY7304), or sir4Δ (JRY7305) cells. For panels 4–6, whole cell lysates were from strains used in Figure 1A and represent 0.75 OD equivalents. Sir2p was consistently reduced up to about twofold in sir3Δ and sir4Δ strains. Sir3p levels did not vary significantly in multiple cell lines tested.
Sir1p has been proposed to bind Orc1p at the silencer independently of other Sir proteins and then, through its association with Sir4p, to increase the likelihood of the other Sir proteins assembling at the silencer (Chien et al., 1993; Triolo and Sternglanz, 1996; Gardner et al., 1999). Indeed, the association of Sir1-HAp with HMR-E was not noticeably affected in sir2Δ or sir3Δ cells and slightly, but reproducibly, reduced in sir4Δ cells (Figure 1A, panel 1). A longer exposure of this gel (Figure 1A, panel 2) demonstrates that the SSC1 internal control was recovered at similar levels from each immunoprecipitation. For brevity, longer exposures are not shown for the subsequent gels.
Deletion of Sir2p, Sir3p, or Sir4p results in loss of silencing at HM loci and, because the cells express both a and α genes, a 105-fold reduction in mating efficiency (Rine and Herskowitz, 1987). Therefore, stable silenced chromatin would not be expected in these cells, implying that in the absence of Sir2p, Sir3p, or Sir4p, the remaining two proteins might not associate with silent loci (see also Strahl-Bolsinger et al., 1997; Sekinger and Gross, 2001). Indeed, in a sir4Δ strain, Sir2p and Sir3p did not detectably associate with HMR-E (Figure 1A, panels 3 and 4, sir4Δ lanes). Immunoblot analysis revealed that both Sir2p and Sir3p were expressed efficiently in sir4Δ strains (Figure 1B; our unpublished data), establishing that their absence from the silencer did not reflect a dramatic reduction in abundance. Thus, Sir4p was crucial for the association of Sir2p and Sir3p.
In contrast to the sir4Δ strain, in sir2Δ and sir3Δ strains, the remaining two Sir proteins did associate with HMR-E, albeit at a reproducibly lower level than in wild-type strains (Figure 1A, panel 3, sir3Δ lanes; panel 4, sir2Δ lanes; and panel 5, sir2Δ and sir3Δ lanes). Therefore, partial assemblies of Sir proteins could form at the HMR-E silencer. The spreading of these partial assemblies is explored below.
Deletion of Sir1p results in two metastable populations of cells, silenced and not silenced (Pillus and Rine, 1989). Therefore, in a sir1Δ strain, the remaining Sir proteins would be expected to associate with the silencer in the fraction of the cells that are silenced. In the MATα sir1Δ strain used in this study, 33–42% of the HMR loci were silenced (determined by mating efficiency; our unpublished data). Therefore, roughly one-third as much HMR-E DNA was expected to precipitate compared with the wild-type strain. Indeed, Sir2p, Sir3p, and Sir4p associated with HMR-E with approximately one-half to one-quarter the efficiency observed in wild-type cells (Figure 1A, panels 3–5; the intensity of the first lane of the sir1Δ sample [1X] falls between the intensities of the second [1/2X] and third [1/4X] lanes of the wild-type sample). Similar results were obtained all four times that wild-type and sir1Δ cells were compared, and longer exposures indicate that similar levels of the internal control, SSC1, were recovered (our unpublished data). Thus, the fraction of sir1Δ cells in which the other Sir proteins were associated with the silencer was similar to the fraction of sir1Δ cells in which HMR was silenced, consistent with Sir1p promoting the loading of the other Sir proteins onto silencers.
To explore whether Sir1p, Sir2p, and Sir3p had overlapping or distinct roles in stabilizing the Sir protein complex at HMR-E, double mutant strains were constructed. In a sir2Δ sir3Δ strain, the level of Sir4p associated with the silencer was no less than the level in sir2Δ or sir3Δ strains (Figure 1A, panel 5; our unpublished data), consistent with Sir2p and Sir3p stabilizing the association of Sir4p with the silencer in a similar way. In contrast, sir1Δ sir2Δ or sir1Δ sir3Δ mutant strains lost all residual association of the remaining Sir proteins with HMR-E (Figure 1A, panels 3–5), implying that Sir1p acts in a different way than Sir2p or Sir3p in the assembly of Sir proteins at the silencer.
Sir2p, Sir3p, and Sir4p Were Associated with DNA throughout the Silenced Domain
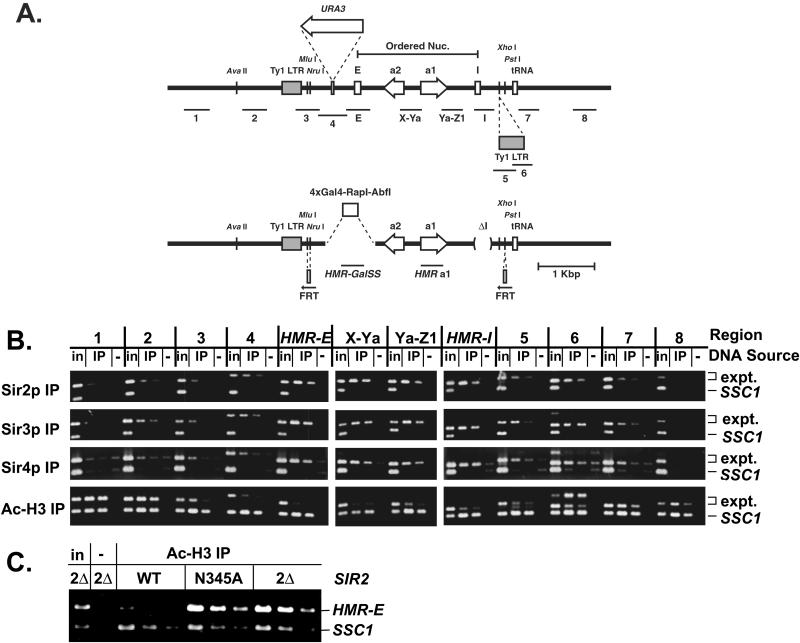
The limits of the silenced domain created by Sir proteins at HMR have been determined by nuclease protection and insertion of reporter genes at various positions (Loo and Rine, 1994; Donze et al., 1999). The silenced domain is continuous between the HMR-E and HMR-I silencers and extends in both directions beyond the silencers, ending at boundary elements on either side (Donze et al., 1999; Donze and Kamakaka, 2001). To determine whether Sir proteins were present throughout HMR and coincident with the silenced domain, DNA coimmunoprecipitated with Sir2p, Sir3p, or Sir4p from wild-type cells was analyzed by PCR for the presence of 12 different regions spanning and extending beyond the HMR locus (Figure 2A, top). The ratios of each region relative to SSC1 were compared in the input and immunoprecipitated samples to determine whether that region exhibited enhanced association with Sir proteins. In addition, because the same amount of input or immunoprecipitated DNA was used to amplify each region, the relative amounts of these regions in the immunoprecipitated sample could be determined by comparing the ratios of the immunoprecipitated sample to the input sample for each PCR product.
Figure 2.
Distribution of Sir proteins across HMR in a SIR strain. (A) Scale diagram of the HMR locus. Wild-type (top) and synthetic silencer-bearing (bottom) alleles are shown. The regions amplified are shown below each allele. The positions of the silencers, open reading frames, tRNA gene (boundary), and Ty1 LTRs (5′ LTR is a boundary) are shown on the line. The 3′ LTR is particular to these strains and is inserted at the known XhoI site. The Mlu I site just 3′ of the 5′ Ty1 LTR is less accessible to cleavage in SIR vs. sir strains, whereas the AvaII site 5′ of the Ty1 LTR is cut equally well in either strain (Loo and Rine, 1994). The region of ordered nucleosomes, the position at which URA3 was inserted (Donze et al., 1999), and the positions at which FRT sites were inserted are indicated. (B) Chromatin immunoprecipitations from MATα SIR cells (JRY4013). DNA immunoprecipitated with antibodies against Sir2p (panel 1), Sir3p (panel 2), Sir4p (panel 3), or di-acetylated histone H3 (panel 4) was analyzed by simultaneous amplification of the indicated region of HMR (top bands) and the SSC1 promoter (bottom bands). Negative controls (lanes −) had samples immunoprecipitated from strains lacking Sir2p (panel 1), Sir3p (panel 2), Sir4p (panel 3), or represent a mock precipitation with no antibody from SIR cells (panel 4). 1/19,000 of the input DNA, 1/250 or 1/500 (panels 1 and 2), 1/125 or 1/250 (panel 3), and 1/500 or 1/1000 (panel 4) of immunoprecipitated DNA was analyzed. The primers used to amplify region 6 generated a product of the predicted size (middle band) as well as a larger product of unknown identity, presumably involving another Ty1 LTR. (C) Chromatin immunoprecipitations from MATa SIR2, sir2-N345A, or sir2Δ cells (all W303-AR background). DNA was immunoprecipitated with antibodies against di-acetylated histone H3. Negative control (lane −) represents a mock precipitation with no antibody. The template DNA was serially diluted twofold, starting with 1/19,000 of the input DNA or 1/375 of the immunoprecipitated DNA.
Sir2p, Sir3p, and Sir4p preferentially associated with sequences at the HMR-E and HMR-I silencers and at two regions between the silencers compared with SSC1 (Figure 2B, panels 1–3, HMR-E, X-Ya, Ya-Z1, and HMR-I). Furthermore, these Sir proteins were equally associated with all four regions, supporting the notion that Sir2p, Sir3p, and Sir4p were evenly distributed between the silencers. This distribution pattern of Sir proteins in wild-type cells is compared with the distribution of Sir proteins in mutant cells below.
To determine whether the distribution of Sir proteins matched the previously defined silenced domain, sequences near the boundaries of the silenced domain were examined. The telomere-proximal boundary of the silenced domain at HMR is a tRNA gene (Donze and Kamakaka, 2001). In the strain used here, an additional 440 base pairs sequence corresponding to a Ty1 long terminal repeat (LTR) was discovered between HMR-I and the tRNA gene (Figure 2A, top). This Ty1 LTR did not block the spread of Sir proteins, because Sir2p, Sir3p, and Sir4p associated with regions overlapping the Ty1 LTR (Figure 2B, panels 1–3, regions 5 and 6). The boundary element at the tRNA gene did limit the spread of Sir2p, Sir3p, and Sir4p, because the associations of these Sir proteins with regions 20 base pairs and 1000 base pairs beyond the tRNA gene (regions 7 and 8) were greatly reduced relative to HMR-I.
The centromere-proximal boundary maps to a Ty1 LTR ∼900 base pairs outside of HMR-E, but it may not be a discrete element (Kamakaka and Donze, personal communication). Furthermore, although the silenced domain extends beyond the silencers in either direction, the composition of the chromatin may not be identical on the two sides of a silencer. For example, a URA3 gene inserted with its promoter only 200 base pairs 5′ of HMR-E and well within the defined silenced domain (Figure 2A, top) is silenced one-tenth as well as a URA3 gene inserted 3′ of HMR-E within the a2 open reading frame (Donze et al., 1999). Additionally, ordered nucleosomes, whose positioning depends on Sir protein function, are present between the silencers but not beyond HMR-E (Ravindra et al., 1999). In agreement with these observations, the associations of Sir2p, Sir3p, and Sir4p were diminished immediately outside of HMR-E and then tapered off gradually, terminating in the vicinity of the Ty1 LTR (Figure 2B, panels 1–3, regions 1–4, compare intensities of products from input and immunoprecipitation samples). In summary, Sir2p, Sir3p, and Sir4p were present at high levels between the silencers and were also present up to, but not beyond, the boundaries of the silenced domain.
Association of Sir Proteins Correlated with a Reduction in Acetylated Histones
The presence at HMR of histones H3 and H4 with hypoacetylated tails (Braunstein et al., 1993; Suka et al., 2001), together with the histone deacetylase activity of Sir2p, implied that Sir2p was the enzyme responsible for deacetylating the tails of histones H3 and H4. If so, the extent of the hypoacetylated domain at HMR should coincide with the distribution of Sir2p. Indeed, the regions with high association of Sir2p were less abundant in samples immunoprecipitated using antibodies against diacetylated histone H3 (Figure 2B, panel 4). A similar analysis examining a subset of these regions revealed a correlation between hypoacetylation of histone H4 and the association of Sir2p (our unpublished data). Furthermore, in cells expressing a catalytically inactive Sir2p, Sir2-N345Ap (Imai et al. 2000), histone H3 in the vicinity of HMR-E was acetylated (Figure 2C) despite the presence of the Sir2-N345Ap, Sir3p, and Sir4p at HMR-E (Figure 4C). Therefore, Sir2p was likely the deacetylase that acted at HMR.
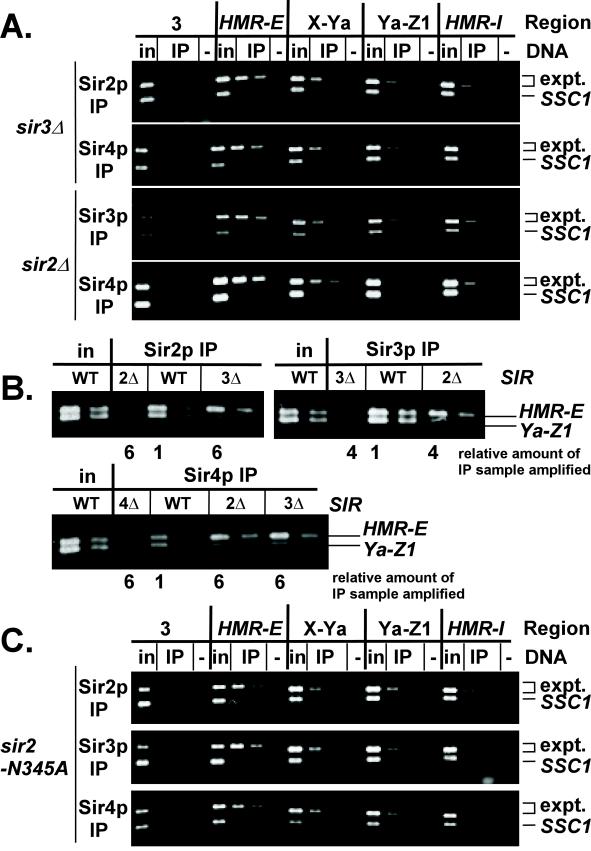
Figure 4.
Distribution of Sir proteins across HMR in sir strains. (A) Chromatin immunoprecipitations from MATα sir3Δ (JRY4605; panels 1 and 2) or sir2Δ (JRY4563; panels 3 and 4) cells. DNA immunoprecipitated with antibodies against Sir2p (panel 1), Sir3p (panel 3), or Sir4p (panels 2 and 4) was analyzed by simultaneous amplification of the indicated region of HMR (top bands) and the SSC1 promoter (bottom bands). Negative controls (lanes −) had samples immunoprecipitated from strains lacking Sir2p (panel 1), Sir3p (panel 3), or Sir4p (panels 2,4). 1/19,000 of the input DNA, 1/125 or 1/250 (panels 1 and 3), 1/50 or 1/100 (panel 2), or 2/125 or 1/125 (panel 4) of immunoprecipitated DNA was analyzed. (B) Chromatin immunoprecipitations from MATα SIR (JRY4013), sir2Δ (JRY4563), sir3Δ (JRY4605), or sir4Δ (JRY4580) cells. Immunoprecipitated DNA was analyzed by simultaneous amplification of HMR-E (top bands) and Ya-Z1 (bottom bands). The template DNA was serially diluted twofold, starting with 1/28,000 of the input DNA or 1/750 of the DNA immunoprecipitated from SIR cells. The relative amount of each immunoprecipitated sample amplified is indicated below the gel. (C) Chromatin immunoprecipitations from MATa sir2-N345A cells. Immunoprecipitated DNA was analyzed as in Figure 4A. Negative controls (lanes −) had samples immunoprecipitated from strains lacking Sir2p (panel 1), or represent a mock precipitation with no antibody (panels 2 and 3). 1/23,000 of the input DNA, 1/250 or 1/500 (panels 1 and 2), 2/125 or 1/125 (panel 3) of immunoprecipitated DNA was analyzed.
Sir1p Was Restricted to the Silencer
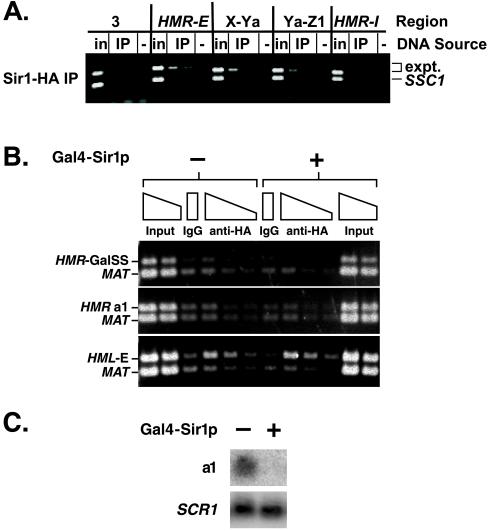
Because Sir1p is required for the establishment but not the maintenance of silencing, Sir1p has been thought to act only at the silencer. However, recent results suggest that Sir1p spreads throughout the silenced domain (Gardner and Fox, 2001). Therefore, the distribution of Sir1-HAp was examined across HMR. Unlike the other Sir proteins (Figure 2B), Sir1-HAp was not associated equally with both silencers and the two regions between but instead was associated primarily with the HMR-E silencer (Figure 3A).
Figure 3.
Distribution of Sir1p across HMR. (A) Chromatin immunoprecipitation from MATα SIR cells bearing an HA-tagged Sir1p (JRY7299). DNA immunoprecipitated with antibodies against HA tag was analyzed by simultaneous amplification of the indicated region of HMR (top bands) and the SSC1 promoter (bottom bands). Negative controls (lanes −) had samples immunoprecipitated from a strain (JRY4013) bearing an untagged Sir1p. 1/23,000 of the input DNA or 1/50 or 1/100 of immunoprecipitated DNA was analyzed. (B) Chromatin immunoprecipitation from cells bearing a synthetic silencer in place of HMR-E. Cells constitutively expressed Sir1-HAp in the absence (lanes 1–6) or presence (lanes 7–12) of Gal4-Sir1p. DNA immunoprecipitated with antibodies against HA tag was analyzed by simultaneous amplification of the synthetic silencer (panel 1), a1 open reading frame (panel 2), or HML-E (panel 3) and the MAT locus (lower bands). Negative controls (lanes IgG) had samples immunoprecipitated with rabbit sera from an unimmunized rabbit. 1/7500 and 1/15,000 of the input DNA or 1/100, 1/200, or 1/400 of the immunoprecipitated DNA was analyzed. In this experiment, the ratio of immunoprecipitated HMR to MAT DNA was smaller than the ratio of immunoprecipitated HMR to SSC1 DNA in previous experiments. Consequently, these images reflect longer exposures than those in other figures, and the negative control is more visible. (C) RNA was isolated from the same samples used in B in the absence (lane 1) or presence (lane 2) of Gal4-Sir1p and analyzed for a1 or SCR1 mRNA.
Due to the heterogeneity in size of sheared DNA in chromatin immunoprecipitation assays, this experiment could not distinguish between low-efficiency spreading of Sir1-HAp and coprecipitation of large DNA fragments with Sir1-HAp cross-linked to the silencer. Therefore, the ability of Sir1p to spread was reinvestigated using a strain in which the HMR-E silencer was replaced with a synthetic silencer, consisting of Rap1p and Abf1p binding sites as well as four Gal4p binding sites in place of the ORC binding site (Figure 2A, bottom). This strain expressed both a Gal4-Sir1 fusion protein, which binds to the synthetic silencer and is required for silencing, and Sir1-HAp, which cannot bind to this synthetic silencer lacking ORC binding sites. If additional Sir1p molecules were recruited to the HMR locus beyond those that function at the silencer, then Sir1-HAp would be associated with the HMR locus. However, Sir1-HAp was not detectably associated either with the synthetic silencer or internal to HMR (Figure 3B). In contrast, Sir1-HAp did associate with the HML-E silencer, presumably through ORC bound to HML-E, indicating that the immunoprecipitation was successful. Gal4-Sir1p mediated silencing at HMR, because transcription of a1 mRNA was repressed (Figure 3C) and Sir2p, Sir3p, and Sir4p were associated with and spread across HMR (our unpublished data). Thus, Sir1p acted primarily at the silencer and little, if any, Sir1p spread from the HMR-E silencer.
Spreading of Sir Proteins Required Sir3p and Deacetylase Activity of Sir2p
The ability of some Sir proteins to associate with the HMR-E silencer in the absence of others (Figure 1A) raised the question of whether Sir proteins require one another to spread. For example, because Sir3p and Sir4p bind to both the HMR-E silencer and the tails of histones H3 and H4 in the absence of Sir2p (Figure 1A; Hecht et al., 1995), Sir3p and Sir4p might spread in the absence of Sir2p. Therefore, the distributions of Sir2p, Sir3p, and Sir4p across HMR were examined in sir2Δ and sir3Δ strains. In contrast to the wild-type strain (Figure 2B), these Sir proteins associated primarily with the HMR-E silencer in these mutants (Figure 4A). To facilitate the comparison of Sir protein distributions in wild-type and sir− cells, DNA immunoprecipitated from these strains was also analyzed by simultaneous amplification of the HMR-E silencer and an internal region, Ya-Z1 (Figure 4B). The amount of template DNA was adjusted to maintain the HMR-E product at a constant level. The internal region was relatively less abundant in samples from sir2Δ or sir3Δ cells compared with wild-type samples, confirming the inefficient spreading. Thus, Sir2p, Sir3p, and Sir4p were mutually dependent on one another for stable association with nonsilencer DNA. The chromatin immunoprecipitation assay cannot reveal whether transient, low-affinity interactions occur between Sir proteins and chromatin at HMR. However, even if such interactions do occur, they are not stable and thus no spreading was detected. The precipitation of some small amounts of the internal regions X-Ya and Ya-Z1 was likely due to limitations in shearing the DNA before immunoprecipitation, as seen above with Sir1-HAp. The greatly reduced association of Sir2p, Sir3p, and Sir4p with HMR-I was surprising because both the HMR-E and HMR-I silencers had previously been thought to be sites at which Sir proteins assemble (see below).
There are at least two ways in which Sir2p may facilitate the spreading of Sir3p and Sir4p. First, Sir2p could be an essential structural component of the silenced chromatin, without which the chromatin is not stable. Alternatively, its deacetylase activity may be required for Sir3p and Sir4p to spread. To distinguish between these alternatives, the spreading of Sir2p, Sir3p, and Sir4p was examined in strains expressing a catalytically inactive point mutant of Sir2p, Sir2-N345Ap. This mutant protein is expressed efficiently and is likely to be structurally intact (Imai et al., 2000; Min et al., 2001). The catalytically inactive Sir2p associated with HMR-E (Figure 4C, panel 1), therefore, deacetylation at the silencer was not required for loading. However, mutant Sir2p did not spread efficiently. Likewise, in this strain, Sir3p and Sir4p also failed to spread efficiently (Figure 4C, panels 2 and 3). Therefore, the histone deacetylase activity of Sir2p was required for the stable spreading of Sir2p, Sir3p, and Sir4p.
The separation of Sir protein loading and spreading also enabled further investigation of the role of Sir1p in establishing silencing. The experiments described above demonstrated that Sir1p facilitated the association of Sir proteins with the HMR-E silencer (Figure 1A). In principle, Sir1p might also facilitate establishment by activating the spreading of Sir proteins from the silencer, perhaps by inducing a conformational change or catalyzing a posttranslational modification on one of the other silencing proteins. However, no reduction in the spreading of Sir2p, Sir3p, or Sir4p was detected in sir1Δ compared with wild-type cells either by examining each region of HMR individually or by examining the relative proportions of sequences at the HMR-E silencer and an internal region, Ya-Z1 (our unpublished data). Therefore, Sir1p acted at the nucleation step but not at the spreading step of silenced chromatin formation.
In summary, efficient spreading of Sir2p, Sir3p, and Sir4p required the presence of all three proteins at the silencer and a catalytically active Sir2p but did not require Sir1p. Furthermore, when spreading was blocked by the absence of one protein, the maximal association of Sir proteins was at the HMR-E silencer. Therefore, the HMR-E silencer was a nucleation site for the assembly of Sir proteins and was the site from which spreading occurred.
HMR-I Silencer Did Not Efficiently Nucleate Assembly of Sir Proteins
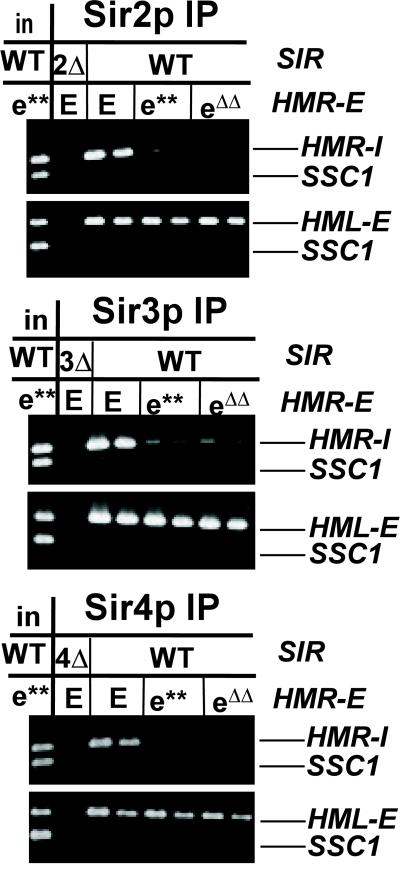
The HMR-I silencer cannot silence independently of the HMR-E silencer (Brand et al., 1985). Sir1p did not efficiently associate with HMR-I (Figure 3A) and in sir2Δ, sir2-N345A, and sir3Δ strains, the remaining Sir proteins did not associate with HMR-I, whereas they did associate with HMR-E (Figure 4, A and C). These observations implied that the HMR-I silencer was not sufficient for silencing because it could not efficiently nucleate the assembly of Sir proteins. In such a scenario, Sir proteins would only arrive at the HMR-I silencer after their loading and spreading from HMR-E. To test this idea, the association of Sir2p, Sir3p, and Sir4p with HMR-I was examined in strains bearing mutations (e**) (Axelrod and Rine, 1991), or deletions (eΔΔ; Brand et al., 1987), in the Rap1p and Abf1p binding sites of the HMR-E silencer. These mutations prevented the efficient binding of Sir2p, Sir3p, and Sir4p to HMR-E and reduced the mating efficiency to 3 × 10−3 (e**) or 3 × 10−4 (eΔΔ) relative to wild-type (our unpublished data). In contrast to a strain bearing a wild-type HMR-E silencer, in strains bearing mutations in the HMR-E silencer, the association of Sir2p, Sir3p, and Sir4p with HMR-I was significantly reduced (Figure 5, panels 1, 3, and 5, compare lanes 3 and 4 with 5–8). These Sir proteins associated with the HML-E silencer at wild-type levels in these strains (Figure 5, panels 2, 4, and 6), indicating that the immunoprecipitation was successful. Therefore, Sir2p, Sir3p, and Sir4p did not associate with HMR-I unless HMR-E was functional, implying that these Sir proteins spread from HMR-E to HMR-I rather than initially assembling at HMR-I.
Figure 5.
Association of Sir proteins with HMR-I silencer. Chromatin immunoprecipitations from MATα SIR cells bearing wild-type (JRY4013) or mutated HMR-E silencers (e** = JRY5472, eΔΔ = YGM1). Immunoprecipitated DNA was analyzed by simultaneous amplification of HMR-I (panels 1, 3, and 5) or HML-E (panels 2, 4, and 6) and the SSC1 promoter (bottom bands). 1/19,000 of the input DNA or 1/250 or 1/500 (panels 1–4) or 1/125 or 1/250 (panels 5 and 6) of the immunoprecipitated DNA was analyzed.
A Synthetic Silencer Had Similar Requirements for Loading and Spreading
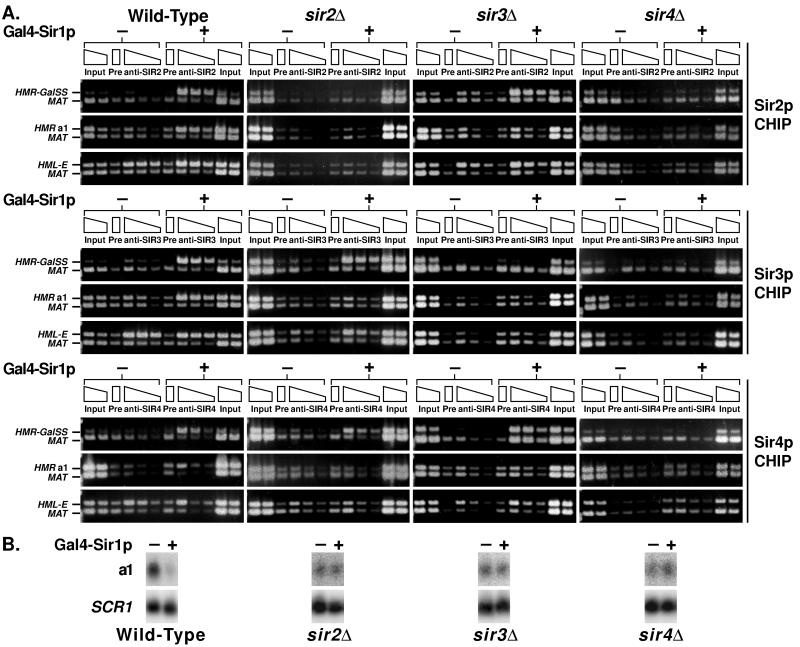
Many of the key experiments exploring the mechanism of silencing have used simplified synthetic silencers in place of HMR-E. For example, to explore the spreading of Sir1p (Figure 3B), a synthetic silencer to which Gal4-Sir1p can be tethered was used (Figure 2A, bottom). To determine whether such a silencer behaves similarly to the natural HMR-E silencer, chromatin immunoprecipitation experiments were conducted in cells expressing or not expressing Gal4-Sir1p (Figure 6A). In this case, MAT served as the negative control locus. The association of Sir2p, Sir3p, and Sir4p with the synthetic silencer required the expression of Gal4-Sir1p (Figure 6A, wild-type column, top gel of each trio), consistent with Sir1p playing a critical role in the establishment of silencing by facilitating the assembly of a Sir protein complex at the silencer.
Figure 6.
Loading and spreading of Sir proteins at a synthetic silencer. (A) Chromatin immunoprecipitations from SIR, sir2Δ, sir3Δ, or sir4Δ strains (left-to-right columns; JRY7131, JRY7303, JRY7304, and JRY7305, respectively). Cells contained 4xGal4-RAP1-ABF1 HMRa ΔI at HMR, and Gal4-Sir1p was repressed (−, first six lanes) or expressed (+, second six lanes). Wild-type Sir1p was constitutively expressed and acted at HML but not the synthetic silencer at HMR. DNA immunoprecipitated with antibodies against Sir2p (top), Sir3p (middle), or Sir4p (bottom) was analyzed by simultaneous amplification of the synthetic silencer (top band, top gels), the a1 open reading frame (top band, middle gels), or HML-E (top band, bottom gels) and sequences adjacent to MAT (bottom band, all gels). Negative controls (lanes Pre) had samples immunoprecipitated with rabbit sera from unimmunized rabbits. 1/7500, 1/15,000, or 1/30,000 of the input DNA or 1/100, 1/200, or 1/400 of the immunoprecipitated DNA was analyzed. In this experiment, the ratio of immunoprecipitated HMR to MAT DNA was smaller than the ratio of immunoprecipitated HMR to SSC1 DNA in previous experiments. Consequently, these images reflect longer exposures than those in other figures, and the negative control is more visible. (B) RNA was isolated from the same samples used in part A in the absence (lane 1) or presence (lane 2) of Gal4-Sir1p and analyzed for a1 or SCR1 mRNA.
As at HMR-E, the other Sir proteins associated with the synthetic silencer in the absence of Sir2p or Sir3p (Figure 6A, sir2Δ and sir3Δ column, top gels; compare the ratios of the HMR-Ga1SS to MAT PCR products from mutants with wild-type), but required Sir4p (Figure 6A, sir4Δ column, top gels). Spreading, as measured by association with the a1 gene at HMR, did not occur unless all Sir proteins were present (Figure 6A, sir2Δ and sir3Δ columns, middle gels). The lack of Sir2p, Sir3p, or Sir4p resulted in loss of silencing (Figure 6B), as expected. Therefore, the interdependencies for loading and spreading were similar at the wild-type and synthetic silencers.
Loading of Sir Proteins at HML-E and HML-I Silencers Had Similar Requirements to HMR-E
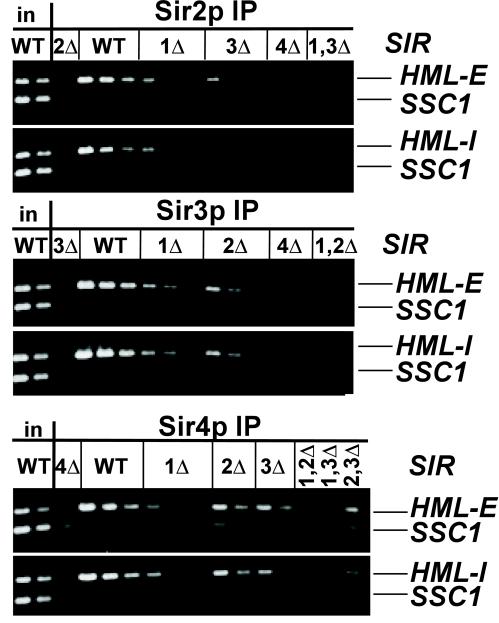
To determine whether the architecture and assembly of silenced chromatin at HML silencers is similar to HMR-E, the association of Sir2p, Sir3p, and Sir4p with both HML silencers was examined. As at the HMR-E silencer, association of these Sir proteins with both HML silencers required Sir4p but not Sir2p or Sir3p (Figures 6A, bottom gels, and 7). Also, as at HMR, the association of Sir proteins at the HML silencers was reduced in a sir1Δ strain (Figure 7). The same dilutions of the same samples were used in both Figures 1A and 7, revealing that in a sir1Δ strain there was relatively more Sir2p, Sir3p, and Sir4p at HMR-E than at either HML silencer. Therefore, the requirements for assembly of the Sir protein complex were similar at all of the silencers examined, with the exception of HMR-I, which did not efficiently nucleate the assembly of Sir proteins. The only detectable difference among the other silencers was the degree of dependence on Sir1p to recruit the remaining Sir proteins.
Figure 7.
Association of Sir proteins with HML silencers. Chromatin immunoprecipitations from MATα strains described in Figure 1. Immunoprecipitated DNA was analyzed by simultaneous amplification of HML-E (panels 1, 3, and 5) or HML-I (panels 2, 4, and 6) and the SSC1 promoter (bottom bands). The template DNA was serially diluted twofold, starting with 1/19,000 of the input DNA or 1/250 (panels 1 and 2) or 1/125 (panel 3) of immunoprecipitated DNA.
DISCUSSION
This study established that partial assemblies of Sir proteins could form at the silencers but could not spread efficiently from the silencers. Thus, the assembly and spreading steps of silenced chromatin formation were separable, thereby enabling a more precise determination of the step at which a protein or activity acts. For example, Sir1p facilitated assembly of Sir proteins at a silencer, whereas the deacetylase activity of Sir2p was required for spreading. The separation of the assembly and spreading steps also revealed that Sir proteins assembled at the HMR-E but not the HMR-I silencer. Previous studies concluded that deletion of one Sir protein resulted in the loss of all Sir proteins at the silent loci. However, those studies used primers that either amplified regions that did not include the silencers (Strahl-Bolsinger et al., 1997) or only examined sir4Δ cells (Sekinger and Gross, 2001).
Model for Spreading of Sir Proteins
The efficient spreading of Sir2p, Sir3p, and Sir4p at HMR required Sir2p and Sir3p (Figures 4A and 6A) and the deacetylase activity of Sir2p (Figure 4C). Furthermore, this study revealed that deacetylation was carried out by Sir2p (Figure 2C) and was an integral part of the spreading process and not a later step that occurred after Sir2p had spread. These findings suggest that the decrease in the association of catalytically defective Sir2p with telomeres and rDNA (Tanny et al., 1999) probably resulted from the disruption of spreading. Although Sir2 protein levels were slightly reduced in the absence of Sir3p (Figure 1B; our unpublished data), it is most likely that the disruption of Sir2p's ability to spread resulted from the absence of Sir3p rather than the reduction in Sir2p because silencing, and hence spreading, occurs in diploid cells heterozygous for null alleles of SIR2 (Rine and Herskowitz, 1987) as well as on HMR-bearing multicopy plasmids in strains expressing wild-type levels of Sir proteins (Abraham et al., 1984).
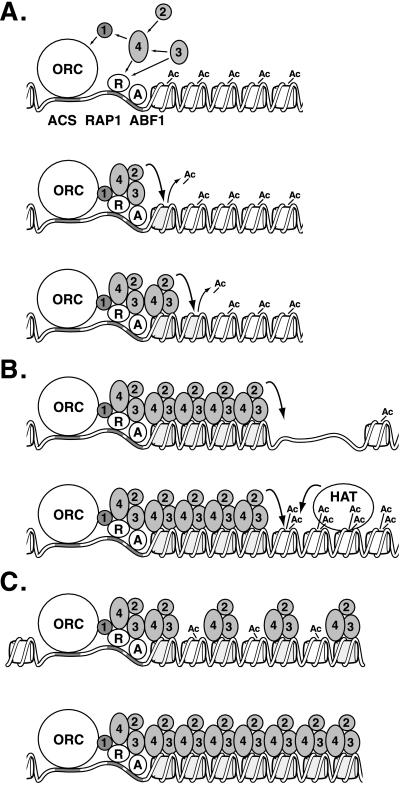
The requirement for a deacetylase in the spreading process inspires a “sequential deacetylation” model for the propagation of silenced chromatin along the DNA (Figure 8A). In this model, the assembly of a Sir protein complex at the silencer brings Sir2p in juxtaposition to the neighboring nucleosome, which, like most nucleosomes in S. cerevisiae (Waterborg, 2000), presumably has acetylated histone tails. Sir2p then deacetylates these tails, creating new high-affinity binding sites for Sir3p and Sir4p, which bind hypoacetylated tails of histones H3 and H4 more tightly than acetylated tails (Hecht et al., 1995). Sir3p and Sir4p then bind this newly deacetylated nucleosome adjacent to the silencer, bringing another Sir2p molecule. This new Sir2p deacetylates the next nucleosome, allowing further association of Sir3p and Sir4p. Finally, interactions among the Sir proteins result in the formation of an ordered, compact structure. For simplicity, this last step is not represented in the figure. This model borrows from and extends earlier models for spreading (Braunstein et al., 1996; Grunstein, 1998; Moazed, 2001a).
Figure 8.
Models for Sir protein action. (A) Formation of silenced chromatin. Sir proteins (1–4) associate with ORC, Rap1p (R), and Abf1p (A) DNA binding proteins through protein–protein interactions (straight arrows). Sir2p then deacetylates the H3 and H4 tails of the neighboring nucleosome (line 2), creating binding sites for Sir3p and Sir4p (line 3). Sir2p deacetylates the next nucleosome, and the process repeats itself. (B) Boundaries of silenced chromatin. Sir protein spreading is interrupted by a gap in the nucleosome array (line 1) or a highly acetylated nucleosome (line 2). (C) Inheritance of silenced chromatin. The newly replicated silenced locus contains a mixture of hypoacetylated nucleosomes associated with Sir proteins and newly synthesized acetylated nucleosomes (line 1). Sir2p deacetylates neighboring nucleosomes, creating binding sites for Sir3p and Sir4p and reforming silenced chromatin.
This sequential deacetylation model has two key features. The first is the use of Sir3p and Sir4p as an adaptor to link Sir2p to chromatin. This feature solves the fundamental problem of restricting silencing to specific loci. Sir2p relies on the Sir3p-Sir4p adaptor to bring it into proximity to its substrate. However, Sir3p and Sir4p themselves cannot stably associate with nucleosomes in the absence of the Sir2p deacetylase, which generates a binding site for Sir4p and Sir3p on histone tails. Thus, the Sir proteins are mutually dependent on one another for their stable association with nucleosomes, preventing silenced chromatin formation from initiating at sites other than those with the means to recruit Sir proteins.
The second key feature of the sequential deacetylation model is that Sir2p must act stoichiometrically rather than processively. Moreover, Sir2p has low deacetylase activity in vitro compared with its paralog Hst2p (Landry et al., 2000; Smith et al., 2000). Sir2p may be maximally active only when assembled with other Sir proteins into silenced chromatin and brought into proximity to its substrate. These features probably prevent Sir2p from indiscriminately deacetylating nucleosomes throughout the genome. This sequential deacetylation model does not exclude the possibility that the Sir2p deacetylase might have additional functions, such as using the energy released by NAD+ hydrolysis to promote spreading (Moazed, 2001b).
The inability of Sir3p to spread from the HMR-E silencer in the absence of Sir2p (Figures 4A and 6A) seems to conflict with the reported ability of Sir3p, when overexpressed, to form an “extended” silenced chromatin at telomeres that is reported to contain Sir3p but not Sir2p or Sir4p (Strahl-Bolsinger et al., 1997). This extension of telomeric silencing upon overexpression of Sir3p does require both Sir2p and Sir4p (Renauld et al., 1993), adding to the puzzle of how Sir3p would spread farther than Sir2p or Sir4p. Perhaps Sir2p and Sir4p are present in extended chromatin but are not accessible to the antibodies due to the excess Sir3p.
The ability to promote the spreading of chromatin-associated proteins may be a general property of the Sir2p family of deacetylases. For example, Sir2p may facilitate a similar spreading process at the rDNA, but use adaptor proteins other than Sir3p and Sir4p. Similarly, Hst1p, a paralog of Sir2p, may promote the spreading of Sum1-1p. SUM1-1 is a change-of-function mutation that leads to silencing of the HM loci in the absence of Sir proteins (Klar et al., 1985; Laurenson and Rine, 1991). Sum1-1p associates with Hst1p (Rusche and Rine, 2001; Sutton et al., 2001), much as Sir4p associates with Sir2p, and Sum1-1p spreads across HMR much as the Sir proteins do (Rusche and Rine, 2001). Interestingly, Sum1-1p does not spread when Hst1p is absent (Rusche and Rine, unpublished data), implying that, like Sir2p, Hst1p regulates the spreading of chromatin-associated proteins.
Protein–Protein Interactions at the Silencer
Past models of the architecture of Sir proteins at the silencer have been inferred primarily from pairwise physical and genetic interactions between individual components of the complex. This study, using chromatin immunoprecipitation, identified some of the protein–protein interactions required for the structure and implied an order of assembly, as outlined below. The interdependencies of Sir proteins to associate with a silencer were examined at HMR-E, HML-E, HML-I, and a synthetic silencer (Figures 1A, 6A, and 7). Importantly, although these silencers have different combinations of ORC, Rap1p, and Abf1p binding sites, all displayed the same requirements for the loading of Sir proteins. Sir4p was essential for loading Sir2p and Sir3p, Sir1p either improved the efficiency of or was essential for loading the other Sir proteins, and Sir2p and Sir3p were less important. Sir1p could associate with the HMR-E silencer independently of any individual other Sir protein.
In the simplest model for the architecture of the Sir protein complex at HMR-E (Figure 8A, top line), Sir1p binds independently of other Sir proteins to Orc1p (Triolo and Sternglanz, 1996; Gardner et al., 1999), and perhaps Rap1p as well (Chien et al., 1993). The association of Sir1p with the silencer was reproducibly reduced upon deletion of Sir4p (Figure 1A), indicating that this association may be stabilized by, although not completely dependent on, contact with Sir4p, consistent with two-hybrid observations (Triolo and Sternglanz, 1996).
In this model, Sir4p is central, making contacts with all three other Sir proteins as well as Rap1p. This placement was consistent with the deletion of Sir4p causing the most severe reductions in association of the other Sir proteins with silencers (Figures 1A, 6A, and 7). Sir4p associates with the silencer through Sir1p and Rap1p, and Sir3p associates through Rap1p. Although pairwise interactions of Sir4p or Sir3p with Rap1p are observed in vitro, we speculate that in the cell, pairwise interactions between Sir4p or Sir3p and Rap1p or Sir1p are individually inadequate for a stable association. Interactions among at least three of these proteins may be required to form a stable complex. This requirement, together with the low probability of a three-way collision, can explain why individual Rap1p or ORC binding sites are not silencers, because both DNA binding proteins would be needed to stabilize interactions among the Sir proteins. In support of this speculation, Sir3p does not associate with Rap1p at the silencer in the absence of Sir4p (Figures 1A, 6A, and 7). In contrast, Sir4p does associate with the silencer-bound Rap1p in the absence of Sir3p, but not if Sir1p is also absent (Figures 1A, 6A, and 7). In this scenario, Sir1p facilitates the assembly of the Sir protein complex by anchoring Sir4p at the silencer, allowing it to bind without Sir3p. Sir3p may also associate with Abf1p (Dhillon and Kamakaka, 2000). However, this association is unstable in the absence of Sir4p (Figure 1A and 6A). Finally, the interaction between Sir2p and Sir4p (Moazed et al., 1997; Ghidelli et al., 2001) suggests that Sir4p brings Sir2p to the silencer.
The reduced recovery of HMR-E silencer DNA in immunoprecipitations from sir2Δ or sir3Δ cells compared with wild-type cells probably reflects the absence of spreading to neighboring nucleosomes. With fewer proteins bound per locus, fewer epitopes would be available for antibody binding. Consistent with this conclusion, the associations of Sir3p and Sir4p with the HMR-E silencer were reduced nearly as much in sir2-N345A as in sir2Δ cells (our unpublished data).
The four endogenous silencers at the HM loci differ in strength and composition of binding sites. Of the four, the HMR-I silencer is the only one that cannot establish silencing independently (Brand et al., 1985). Interestingly, HMR-I was also the only silencer that did not associate with Sir1p (Figures 1A and 3; our unpublished data) and lacked the ability to recruit other Sir proteins efficiently (Figure 5). Consistent with these observations, Sir-dependent nuclease protection at HMR is abolished when the HMR-E silencer is deleted, even although the HMR-I silencer remains (Loo and Rine, 1994). Thus, the ability of a silencer to recruit Sir proteins correlated with the ability to establish silencing.
Boundaries of the Silent Domain at HMR
This study established that Sir2p, Sir3p, and Sir4p were found throughout the silenced domain at HMR but not outside its previously defined borders (see also Lieb et al., 2001). The association of Sir2p, Sir3p, and Sir4p with sequences throughout the silenced domain is the most compelling evidence for these Sir proteins being key structural components of silenced chromatin. Additionally, Sir2p, Sir3p, and Sir4p did not seem to spread from the HMR-E silencer equally in both directions. DNA sequences between the silencers were much more abundant in the immunoprecipitated samples than were sequences outside of HMR-E yet still within the silenced domain. The reduced immunoprecipitation of regions outside the silencers but within the silenced domain was interesting and could result from Sir proteins either being associated in only a fraction of the cells or being present in all cells but at a reduced density. This result and others (Shei and Broach, 1995; Donze et al., 1999; Ravindra et al., 1999) imply that silenced chromatin is not equivalent on the two sides of the HMR-E silencer. The origin and significance of this pattern awaits further study.
The sequential deacetylation model outlined above can explain how heterochromatin boundaries function (Figure 8B). The model implies that silenced chromatin must spread sequentially from nucleosome to nucleosome due to the limited range of the Sir2p deacetylase. Therefore, disrupting the chain of deacetylatable nucleosomes would interrupt the spread of Sir proteins (see also Bi and Broach, 2001). For example, a gap could be created in the nucleosome array by a DNA-binding protein that displaces histones. Similarly, a localized histone acetyltransferase, as is observed at some promoters, could effectively limit the spread of silenced chromatin by acetylating histones more effectively than Sir2p can deacetylate them. In fact, DNA binding proteins and tethered acetyltransferases do act as barrier elements (Bi and Broach, 1999; Fourel et al., 1999; Donze and Kamakaka, 2001). Interestingly, the distribution of Sum1-1p at HMR is virtually identical to that of Sir2p, Sir3p, and Sir4p (Rusche and Rine, unpublished data), implying that boundary elements are not specific to a particular type of silencing complex.
Model for Maintenance and Epigenetic Inheritance of a Chromatin Structure
Once silenced chromatin has been established, this chromatin structure is maintained during cell growth and inherited upon cell division (Pillus and Rine, 1989; Mahoney et al., 1991). Both of these phenomena could occur through the sequential deacetylation mechanism of spreading. Maintenance would occur by the spreading of Sir2p, Sir3, and Sir4p back into gaps in which silencing has been disrupted.
The model presented above for the epigenetic inheritance of silenced chromatin (Figure 8C; see also Braunstein et al., 1996) is rooted in how nucleosomes are affected by replication. Passage of a replication fork causes the two H2A-H2B dimers to disassociate from both the H3-H4 tetramer and the DNA. The H3-H4 tetramers remain associated with DNA (Kimura and Cook, 2001) and are randomly distributed to the two sister molecules (Jackson and Chalkley, 1985). Thus, if an epigenetic mark were associated with the histones, it would logically be on H3 and/or H4. At the silent loci, DNA replication would distribute H3-H4 tetramers with hypoacetylated tails to both sister chromatids. Sir proteins may remain associated with these histone tails or may quickly reassociate with the tails after passage of the replication fork. After replication the resulting nucleosome array would then be completed by the incorporation of newly synthesized and acetylated histones. However, these nucleosomes with acetylated tails would be adjacent to the Sir2p deacetylase along with Sir3p and Sir4p bound to the old H3-H4 tetramers. Sir2p would then deacetylate these tails, creating new binding sites for Sir3p and Sir4p, restoring the silenced state to the underlying gene. Cooperativity between the nucleosome-bound Sir complexes, silencer binding proteins, and unbound Sir proteins may help direct Sir3p and Sir4p to those newly deacetylated histones in heterochromatin rather than deacetylated histones elsewhere in the genome. In fact, silencer binding proteins probably stabilize the chromatin structure (Cheng and Gartenberg, 2000), and it is likely that in the absence of a silencer, silenced chromatin may initially be inherited but is not stable long enough to be detected (Holmes and Broach, 1996).
Perspective
The mechanisms for spreading and inheritance outlined above could apply to multiple types of epigenetically heritable states, including other types of regional repression and centromere inheritance (see also Jenuwein, 2001). There are two fundamental features required to permit both the spreading of a chromatin state from the site of initiation and the inheritance of that chromatin state: a mark that is inherited on both duplexes following replication and the ability of that mark to recruit an enzyme or complex that makes an additional mark. In the Sir-based mechanism described above, the hypoacetylated H3-H4 tetramer is the mark, providing a high-affinity binding site for the Sir complex, which, in turn, creates new marks. In the case of mating-type silencing in Schizosaccharoyces pombe or position effect variegation in Drosophila, histone H3 methylated at lysine 9 is a mark, which is recognized and bound by a chromodomain-containing protein, in partnership with a histone methyltransferase capable of adding additional marks (Jenuwein, 2001). Finally, in many organisms centromeres seem to be epigenetically inherited and are marked by the presence of an H3-like protein, CENP-A (Sullivan et al., 2001). The inheritance of this mark from one generation to the next predicts that there is a CENP-A chaperone that assembles new CENP-A–containing nucleosomes near existing ones.
ACKNOWLEDGMENTS
We thank Catherine Fox and Leonard Guarente for strains and plasmids; Rohinton Kamakaka and David Donze for communicating unpublished results; members of the laboratory for helpful discussions; and Michael Botchan, Alexa Franco, Paul Kaufmann, Michael Kobor, Judith Sharp, and reviewers for helpful comments and suggestions. This work was supported by postdoctoral fellowships from the Damon Runyon-Walter Winchell Cancer Research Fund (to L.N.R.), the National Institutes of Health (F32GM19392, to A.L.K.), and American Cancer Society (PF-01-126-01-MBC, to A.L.K.), and a grant from the National Institutes of Health (GM-31105, to J.R.). Core support was provided by National Institute of Environmental Health Sciences Mutagenesis Center grant E50 1896.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–03–0175. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–03–0175.
REFERENCES
- Abraham J, Nasmyth KA, Strathern JN, Klar AJ, Hicks JB. Regulation of mating-type information in yeast. Negative control requiring sequences both 5′ and 3′ to the regulated region. J Mol Biol. 1984;176:307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- Axelrod AR. Role of a Cell-Cycle Gene in Transcriptional Silencing. Ph.D. Thesis. Berkeley, CA: University of California, Berkeley; 1991. [Google Scholar]
- Axelrod A, Rine J. A role for CDC7 in repression of transcription at the silent mating-type locus HMR in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1080–1091. doi: 10.1128/mcb.11.2.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Braunstein M, Shei G, Broach J. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc Natl Acad Sci USA. 1999;96:11934–11939. doi: 10.1073/pnas.96.21.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Broach JR. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 1999;13:1089–1101. doi: 10.1101/gad.13.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Broach JR. Chromosomal boundaries in S. cerevisiae. Curr Opin Genet Dev. 2001;11:199–204. doi: 10.1016/s0959-437x(00)00179-9. [DOI] [PubMed] [Google Scholar]
- Brand AH, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Brand AH, Micklem G, Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987;51:709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Rose A, Holmes S, Allis C, Broach J. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TH, Gartenberg MR. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 2000;14:452–463. [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig AJ, Gasser SM. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N, Kamakaka RT. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol Cell. 2000;6:769–780. doi: 10.1016/s1097-2765(00)00076-9. [DOI] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CA, Ehrenhofer-Murray AE, Loo S, Rine J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science. 1997;276:1547–1551. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- Gardner KA, Fox CA. The Sir1 protein's association with a silenced chromosome domain. Genes Dev. 2001;15:147–157. doi: 10.1101/gad.852801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K, Rine J, Fox C. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics. 1999;151:31–44. doi: 10.1093/genetics/151.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidelli S, Donze D, Dhillon N, Kamakaka RT. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 2001;20:4522–4535. doi: 10.1093/emboj/20.16.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Holmes SG, Broach JR. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 1996;10:1021–1032. doi: 10.1101/gad.10.8.1021. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong C, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jackson V, Chalkley R. Histone segregation on replicating chromatin. Biochemistry. 1985;24:6930–6938. doi: 10.1021/bi00345a027. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier AL, Rine J. DNA replication-independent silencing in S. cerevisiae. Science. 2001;291:646–650. doi: 10.1126/science.291.5504.646. [DOI] [PubMed] [Google Scholar]
- Klar A, Kakar S, Ivy J, Hicks J, Livi G, Miglio L. SUM1, an apparent positive regulator of the cryptic mating-type loci in Saccharomyces cerevisiae. Genetics. 1985;111:745–758. doi: 10.1093/genetics/111.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov S, Heller R, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenson P, Rine J. SUM1–1: a suppressor of silencing defects in Saccharomyces cerevisiae. Genetics. 1991;129:685–696. doi: 10.1093/genetics/129.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- Lustig AJ. Mechanisms of silencing in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1998;8:233–239. doi: 10.1016/s0959-437x(98)80146-9. [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Marquardt R, Shei GJ, Rose AB, Broach JR. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu R-M. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001a;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- Moazed D. Enzymatic activities of Sir2 and chromatin silencing. Curr Opin Cell Biol. 2001b;13:232–238. doi: 10.1016/s0955-0674(00)00202-7. [DOI] [PubMed] [Google Scholar]
- Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Moretti P, Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Ravindra A, Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol Cell Biol. 1999;19:7944–7950. doi: 10.1128/mcb.19.12.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Rine J. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 2001;15:955–967. doi: 10.1101/gad.873601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schmitt M, Brown T, Trumpower B. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Gross DS. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell. 2001;105:403–414. doi: 10.1016/s0092-8674(01)00329-4. [DOI] [PubMed] [Google Scholar]
- Shei GJ, Broach JR. Yeast silencers can act as orientation-dependent gene inactivation centers that respond to environmental signals. Mol Cell Biol. 1995;15:3496–3506. doi: 10.1128/mcb.15.7.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- Sutton A, Heller RC, Landry J, Choy JS, Sirko A, Sternglanz R. A novel form of transcriptional silencing by Sum1–1 requires Hst1 and the origin recognition complex. Mol Cell Biol. 2001;21:3514–3522. doi: 10.1128/MCB.21.10.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- Waterborg JH. Steady-state levels of histone acetylation in Saccharomyces cerevisiae. J Biol Chem. 2000;275:13007–13011. doi: 10.1074/jbc.275.17.13007. [DOI] [PubMed] [Google Scholar]
- Weinstein B, Solomon F. Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae: differences between α-tubulin and β-tubulin. Mol Cell Biol. 1990;10:5295–5304. doi: 10.1128/mcb.10.10.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol Cell Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]