Abstract
Ssh1p of Saccharomyces cerevisiae is related in sequence to Sec61p, a general receptor for signal sequences and the major subunit of the channel that guides proteins across the membrane of the endoplasmic reticulum. The split-ubiquitin technique was used to determine whether Ssh1p serves as an additional receptor for signal sequences in vivo. We measured the interactions between the Nub-labeled Ssh1p and Cub-translocation substrates bearing four different signal sequences. The so-determined interaction profile of Ssh1p was compared with the signal sequence interaction profile of the correspondingly modified Nub-Sec61p. The assay reveals interactions of Ssh1p with the signal sequences of Kar2p and invertase, whereas Sec61p additionally interacts with the signal sequences of Mfα1 and carboxypeptidase Y. The measured physical proximity between Ssh1p and the β-subunit of the signal sequence recognition particle receptor confirms our hypothesis that Ssh1p is directly involved in the cotranslational translocation of proteins across the membrane of the endoplasmic reticulum.
INTRODUCTION
In Saccharomyces cerevisiae signal sequence-bearing proteins can be targeted to the membrane of the endoplasmic reticulum (ER) either co- or posttranslationally (Hann and Walter, 1991). The signal recognition particle (SRP) interacts with a subset of signal sequences shortly after their synthesis and thereby initiates their cotranslational translocation across the ER membrane (Keenan et al., 2001). Proteins bearing signal sequences that are not recognized by the SRP translocate at a significantly later state of their synthesis. The translocation of these proteins depends on the Sec62p/Sec63p complex, an assembly of four different proteins that is localized in the ER membrane (Deshaies et al., 1991; Brodsky and Schekman, 1993; Panzner et al., 1995). To distinguish whether a certain protein is targeted co- or posttranslationally the translocation of the protein is usually monitored in a strain harboring a mutation in one of the two targeting pathways. A hindrance of translocation in one of the two strains identifies the targeting pathway taken by this protein, provided that the protein cannot sidestep this obstruction by using the alternative way. According to this criterion the signal sequences of carboxypeptidase Y (CPY) and Mfα1 use the posttranslational mode, whereas the membrane proteins DPAP1 and Och1p strictly rely on the cotranslational targeting pathway (Ng et al., 1996). The preference for either of the two pathways correlates with the hydrophobicity of the respective signal sequence. Comparatively low hydrophobicity selects the postranslational pathway, whereas stronger hydrophobic signal sequences prefer to translocate via the SRP (Bird et al., 1987; Ng et al., 1996; Martoglio and Dobberstein, 1998). However, the spectrum of signal sequences covers hydrophobicities that lie between those of CPY and DPAP1. Mutations in one of the two targeting pathways influence the translocation of these proteins to only a certain degree. The translocation of Kar2p and invertase, for example, is severely affected but not completely abolished in a strain carrying a deletion of the SRP (Hann and Walter, 1991; Johnsson and Varshavsky, 1994b; Ng et al., 1996).
After being targeted to the ER membrane signal sequences are recognized by Sec61p (Jungnickel and Rapoport, 1995; Plath et al., 1998). Sec61p is the major constituent of the channel that guides the proteins across the membrane (Rothblatt et al., 1989; Görlich et al., 1992; Hanein et al., 1996; Beckmann et al., 1997; Menetret et al., 2000). Both targeting pathways converge at this point. The proteins targeted via the posttranslational pathway are translocated by the heptameric Sec-complex. This complex consists of the trimeric Sec61p and the tetrameric Sec62p/Sec63p complex (Deshaies et al., 1991; Brodsky and Schekman, 1993; Panzner et al., 1995). The cotranslational substrates are probably directly delivered to the trimeric Sec61-complex via SRP and the SRP receptor (SR) (Bacher et al., 1999; Johnson and van Waes, 1999; Song et al., 2000).
In yeast a second complex with high similarity to the trimeric Sec61p complex has been described previously (Finke et al., 1996). Ssh1p is related to Sec61p and forms a trimeric complex with Sbh2p and Sss1p. Sbh2p shares sequence similarity with Sbh1p, the β-subunit of the Sec61p complex, and Sss1p is present in both trimeric complexes as the γ-subunit (Esnault et al., 1993; Finke et al., 1996). The functions of Ssh1p are not immediately evident. A strain carrying a deletion of SSH1 shows no obvious translocation defects (Finke et al., 1996; Ng et al., 1996). However, a strain that combines a deletion of SSH1 with the sec61-2 temperature-sensitive allele is not viable at the permissive temperature for the sec61-2 allele (Finke et al., 1996). Furthermore, both Sec61p and Ssh1p bind to ribosomes and therefore seem to share a certain subset of functions (Prinz et al., 2000). Although the nature of these functions remains unknown the presence of an alternative Sec-complex gives reason to the assumption that the entire range of signal sequences is not distributed between two but between three different channels: the trimeric Sec61p complex, the heptameric Sec-complex, and the trimeric Ssh1p complex.
In this study we tested this hypothesis by using the split-ubiquitin (split-Ub) technique to monitor the in vivo flux of signal sequences across the different channels in the membrane of the ER. Using this assay we show that Ssh1p in contrast to Sec61p exclusively recognizes proteins bearing signal sequences of stronger hydrophobic character.
MATERIALS AND METHODS
Construction of Fusion Proteins
The construction of the signal sequence bearing Cub constructs of invertase and Mfα1 is described in Dünnwald et al. (1999). CPY30-CUB-Dha/URA3 was derived from construct XX of Johnsson and Varshavsky (1994a) and the Mfα137-CUB construct by replacing the ClaI-SalI fragment containing the Mfα137 sequence by the corresponding sequence of the CPY gene (PRC1). To obtain KAR220- and KAR240-CUB-URA3, the Mfα137 sequence of the Mfα137-CUB-URA3 was replaced by a ClaI SalI cut polymerase chain reaction (PCR) fragment obtained from genomic DNA of the yeast strain JD53 and the appropriate PCR primers. The PCR primers for KAR240 were as follows: 5′CCTCCATCGATATGTTTTTCAACAGACTAAG and 5′CCTCCGTCGACCCAATTTCAGTCTTACCATTTTT. The PCR primers for KAR220 were as follows: 5′CCTCCATCGATATGTTTTTCAACAGACTAAG and 5′CCTCCCGTCGACCCTCTAACTAAAACATTGG. The underlined sequences are the ClaI and SalI sites, respectively. Indicated in bold are the first or the last triplets from the sequence of KAR2. The expression of the Ura3p-based Cub fusions was mediated by the PCUP1 promoter. The expression of the Cub-dihydrofolate reductase fusions (DHFR-Dha) was mediated by the PADH1 promoter. The construction of the Nub fusions of SEC61, SEC62, BOS1, STE14, SED5, SSH1, SEC22, TPI1 are described in Wittke et al. (1999). The Nub-constructs of UBC6 were assembled from the PCUP1-Nub-cassette and a PCR fragment containing the open reading frame (ORF) of UBC6 and 188 nucleotides downstream of the STOP codon. A BamHI site was used to bring the Nub in frame with the PCR product. The linker between the last codon of Nub (bold letters) and the second codon of UBC6 (bold letters) reads GG ATCCCTGGGTCTGGGGCT. The BamHI site is underlined. To insert the ha-epitope between Nub and SEC61 or SSH1 we replaced the Nua moiety in the corresponding Nua-constructs by a newly created Nua-ha module. Nua-HA was constructed via PCR with PCUP1-Nua as a template and a PCR primer annealing to the sequence of the PCUP1 promoter and a primer annealing to the C-terminal coding sequence of Nub and additionally harboring the sequence encoding the HA epitope. This primer reads 5′CCCCGGATCCCAGCGTAATCTGGAACATCGTATGGGTACCCGATCCCTTCCTTGTCTTGAAT. The BamHI restriction site is underlined, the HA coding sequence is shown in bold letters, and the Nub sequence is shown in italic letters. The PCR product was ligated in front of the coding sequences of SEC61 and SSH1 by using the BamHI restriction site. The obtained fusion products were integrated into the genome of the yeast JD53 as detailed in Wittke et al. (1999). The correct integration was verified by a diagnostic PCR. All Nub-fusion proteins were expressed from the PCUP1 promoter in the pRS314 vector (Sikorski and Hieter, 1989).
SEC63-CUB-RURA3 was constructed using two primers to amplify the complete ORF of SEC63 with genomic DNA as a template. The PCR product was cut with BamHI and SalI and inserted between the CUB-RURA3 module and the PMET25 promoter in the vector pRS313 containing a CEN ARS element. The linker between the last codon (bold letters) of SEC63 and the first codon of CUB (bold letters) reads GAAGGC GGG TCG ACC GGT. The SalI site is underlined. The same PCR product was used to insert the ORF of SEC63 between the PGAL1 promoter and the coding sequence of the ha-epitope to create SEC63-ha in the vector pRS424. The plasmids expressing Sec62p or Ste14-Dha from the PGAL1 promoter on a pRS313 vector are described in Wittke et al. (1999). SRβ-CUB-RURA3 was constructed by PCR amplification of the last 479 base pairs of the coding sequence of SRP102 not including the stop codon by using genomic DNA of S. cerevisiae as a template. The ends of the PCR product contained restriction sites to allow the in-frame fusion with the CUB-RURA3 module located in the vector pRS303. The short linker sequence between the last codon of SRP102 and the first codon of CUB reads CTG TCC GGG TCG ACC GGT. The last codon of SRP102 and the first codon of CUB are in bold letters, and the SalI site is underlined. The vector was cut at its unique SphI site in the SRP102-containing sequence and transformed into the S. cerevisiae strain JD53 to yield, through homologous recombination, the integrated cassette that expressed SRβ-Cub-RUra3p from the native promoter. Integration was confirmed by diagnostic PCR. The unmodified SSH1 expressed from the PCUP1 promoter was obtained by PCR with genomic DNA as a template and two oligonucleotides priming at the start codon and 160 nucleotides downstream of the stop codon, respectively. The obtained fragment was cut with BamHI and SalI and inserted behind the PCUP1 promoter on a pRS315 vector.
Deletion of SSH1
The open reading frame of SSH1 was replaced by the dominant kan MX marker essentially as described by Güldener et al. (1996). The PCR primers used for the construction of the kan MX disruption cassette were as follows: 5′ TTTAGCACATTTGCCCCCGCCACTCTCCATTGTTTTAGTACCAGCTGAAGCTTCGTACGC and 5′ TACGTATATAAATGCGCGTAGCAGAGAGAATTTGATCTTC-TAGGCCACTAGTGGATCTG. Transformed yeast cells were selected for kan MX integration by Geneticin (Invitrogen, Paisley, Scotland), deletion was verified by diagnostic PCR, and the complementation of the small growth defect by the plasmid-borne SSH1.
Immunoblotting
Cell extraction for immunoblotting was performed essentially as described previously (Johnsson and Varshavsky, 1994b). All experiments were performed without adding additional amounts of copper to the medium. Proteins were fractionated by SDS-12.5% PAGE and electroblotted onto nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany), by using a semidry transfer system (Hoefer Pharmacia Biotech, San Francisco, CA). Blots were incubated with a monoclonal anti-ha antibody (Babco, Richmond, CA), or with the anti-Sec61p antibody. Bound antibody was visualized with horseradish peroxidase-coupled rabbit anti-mouse or goat anti-rabbit antibody, respectively (Bio-Rad, Hercules, CA), by using the chemiluminescence detection system (Pierce Chemical, Rockford, IL). The chemiluminescence was quantified with the aid of the lumi-imager system (Roche Applied Science, Mannheim, Germany). For comparing the amounts of expressed Nub-ha-Sec61p and Nub-ha-Ssh1p protein extracts were diluted with twofold PAGE sample buffer and heated for 20 min at 40°C before electrophoresis. The chemiluminescence was captured by a Hypofilm ECL (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom). The exposed film was scanned and quantified with the aid of the Aida/two-dimensional densitometry program (Raytest Isotopenmeβ, Straubenhardt, Germany).
Media and Interaction Assays
Yeast rich (YPD) and synthetic minimal media with 2% dextrose (SD) or 2% galactose (SG) followed standard recipes (Dohmen et al., 1995).
For interaction assays, S. cerevisiae cells were first grown at 30°C in liquid selective media containing uracil to an OD600 of 1. 4 μl of these cultures, and serial 1:10 dilutions in water were spotted on agar plates selecting for the presence of the fusion constructs and lacking uracil. All experiments were performed without adding additional amounts of copper to the medium. The same dilutions were also spotted onto plates containing uracil to check for cell numbers. The plates were incubated at 30°C for 2–5 d.
RESULTS
Ssh1p Transiently Interacts with a Subclass of Signal Sequences
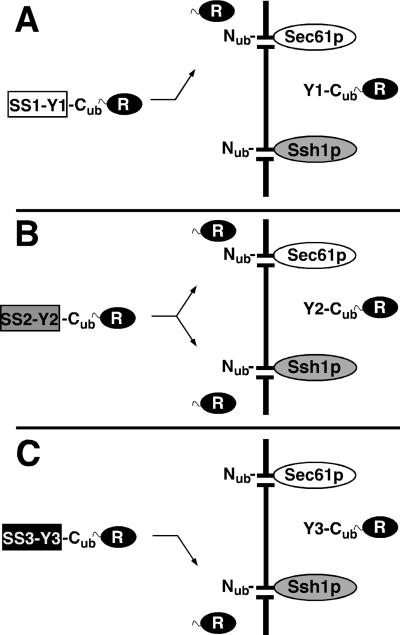
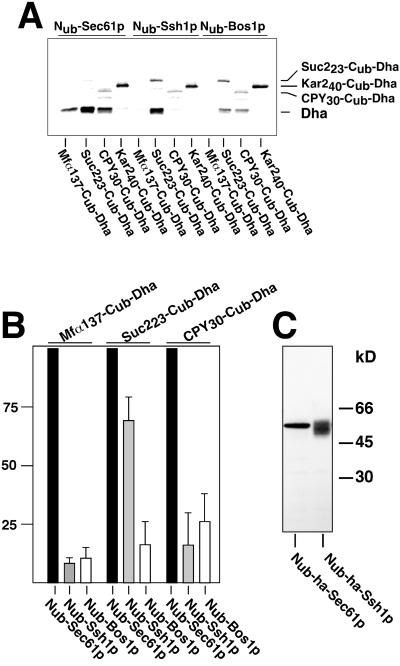
The split-Ub assay can detect the transient interactions that occur between a signal sequence-bearing protein and a component of the translocation machinery. For this application of the technique the N-terminal half of Ub (Nub) is fused to the N terminus of a membrane protein of the translocation machinery and the C-terminal half of Ub (Cub) is sandwiched between a signal sequence and a reporter protein. As soon as the signal sequence brings the attached Cub into proximity of the Nub, the two Ub halves will reconstitute the native-like Ub and the reporter will be cleaved off from the C terminus of Cub by the ubiquitin-specific proteases (Ubps) (Figure 1) (Johnsson and Varshavsky, 1994a; Dünnwald et al., 1999). Because the assay requires the cytosolic location of the reconstituted Ub only those interactions can be monitored that occur shortly before and during the translocation of the Cub on the cytosolic face of the membrane. Sec61p and Ssh1p share a sequence identity of 34% across the entire lengths of the proteins. The N terminus of Sec61p points into the cytosol of the cell (Wilkinson et al., 1996). The same topology was indirectly confirmed for Ssh1p (Wittke et al., 1999). The N termini of both proteins were labeled with Nub to directly compare their activities toward different signal sequence-bearing Cub substrates. We knew from our previous studies that both Nub-fusions displayed a comparable activity toward a Cub-fusion of Ste14p. Ste14p does not interact with Sec61p nor Ssh1p (Wittke at al., 1999). Consequently, a measured difference in cleavage of a tested Cub-fusion should indicate its specific interaction with either Nub-Sec61p or Nub-Ssh1p. We envision three scenarios if Ssh1p constitutes an independent translocation pore (Figure 1): 1) The Cub-Reporter is attached to a signal sequence that translocates exclusively via Sec61p. Here cleavage of the reporter is observed in cells cotransformed with Nub-Sec61p but not in cells carrying Nub-Ssh1p (Figure 1A). 2) The Cub-Reporter is attached to a signal sequence that translocates via Sec61p and Ssh1p. Here cleavage is observed in cells synthesizing either Nub-Sec61p or Nub-Ssh1p (Figure 1B). 3) The Cub-Reporter is attached to a signal sequence that translocates exclusively via Ssh1p. Here cleavage is only observed in cells carrying Nub-Ssh1p (Figure 1C). We chose the N-terminal sequences of the α-factor (Mfα1-), invertase (Suc2-), CPY-, and Kar2p (Kar2-) as the signal sequences of the Cub constructs. The lengths of the peptides (spacer sequence) that separate the hydrophobic core of the signal sequences from the Cub moiety are denoted as indices. We first used the ha-epitope–tagged mouse DHER (Dha) as a reporter protein. The Ubp-induced cleavage at the Cub-Dha junction was detected by immunoblotting with the ha-antibody after cell extraction and denaturing gel electrophoresis. Cleavage is indicated by the presence of the free Dha of ∼28 kDa. The translocated and uncleaved fraction of the Cub-fusion protein gives rise to a second band with a higher molecular mass. However, due to differences in stability, glycosylation, and secretion, the fraction of the translocated proteins cannot be quantitatively compared among the different signal sequence bearing Cub-fusions by Western blotting (Dünnwald et al., 1999). Figure 2A shows that the coexpression of Nub-Sec61p with the four different signal sequence-bearing substrates resulted in a significant accumulation of the cleaved Dha from Mfα137-Cub-Dha, Suc223-Cub-Dha, and CPY30-Cub-Dha. No cleavage was observed for Kar240-Cub-Dha and only the uncleaved fraction of Kar240-Cub-Dha could be detected on the Western blot. Nub-Ssh1p induced significant cleavage of Suc223-Cub-Dha only (Figure 2, A and B). No cleaved Dha was detectable in the extracts of the cells that contained Mfα137-Cub-Dha or Kar240-Cub-Dha. The slight accumulation of Dha observed upon coexpression of Nub-Ssh1p with CPY30-Cub-Dha was below the background that was determined by the coexpression of the Cub-translocation substrates with Nub-Bos1p, a membrane protein of the ER that is not involved in translocation (Figure 2B) (Dünnwald et al., 1999). To exclude that the different signal sequence interaction profiles of Nub-Sec61p and Nub-Ssh1p arise from a higher expression level of Nub-Sec61p, we inserted an ha-epitope between the Nub and the Sec61p- and Ssh1p coding sequence. After reducing the heterogeneity in the running behavior of Nub-Ssh1p during SDS-PAGE the quantification of the Western blots of the cell extracts revealed a nearly identical amount of the two fusion proteins in the cells (Figure 2C).
Figure 1.
Split-Ub assay was used to measure the flow of a signal sequence-bearing Cub fusion across Sec61p- or Ssh1p-translocation channels. Cells containing either Nub-Sec61p or Nub-Ssh1p were cotransformed with a signal sequence bearing SS-Y-Cub-reporter fusion. SS indicates the signal sequence of a protein that is translocated in yeast. Y indicates the peptide that is derived from this protein to separate the hydrophobic core of the signal sequence from the Cub. The numbers 1–3 indicate three different proteins, each translocating across the membrane by using a different combination of channels. (A) If the signal sequence SS1 is translocated exclusively via Sec61p only the Nub-moiety of the labeled Sec61p will get close to the Cub of the translocation substrate. Consequently, the reporter activity (R) will be cleaved off by the ubiquitin-specific proteases and released into the cytosol of only those cells that contain Nub-Sec61p. (B) Signal sequence SS2 that translocates via Sec61p or Ssh1p will induce the cleavage of the reporter in cells containing either Nub-Sec61p or Nub-Ssh1p. (C) Signal sequence SS3 that translocates exclusively via Ssh1p will induce cleavage of the attached Cub-Reporter only in cells containing Nub-Ssh1p but not in cells containing Nub-Sec61p.
Figure 2.
Signal sequence specificity of Ssh1p. (A) Immunoblot analysis of yeast cells containing either Nub-Sec61p, Nub-Ssh1p, or Nub-Bos1p and expressing the four different signal sequence-bearing Cub constructs. Uncleaved and cleaved Dha reporter was detected by an anti-ha antibody after SDS-PAGE and transfer onto nitrocellulose of whole cell extracts. (B) Quantification of three independent experiments is shown. In each experiment the amount of cleaved Dha that was induced by coexpressing Nub-Sec61p was set to 100. The amount of cleaved Dha that was induced by Nub-Ssh1p and Nub-Bos1p was calculated in reference to the Nub-Sec61p–induced cleavage for each of the signal sequence-bearing Cub-Dha separately, except for Kar240-Cub-Dha that yielded no cleavage with any of the Nub-fusions. (C) Comparison of the expression levels of Nub-ha-Sec61p and Nub-ha-Ssh1p. Proteins extracts of yeast cells expressing Nub-ha-Sec61p or Nub-ha-Ssh1p were separated by SDS-PAGE and transferred onto nitrocellulose. The amounts of protein were estimated after anti-ha antibody treatment via the chemiluminescence generated by the conjugated second antibody. To better focus the Nub-ha-Ssh1p during SDS-PAGE, the extracts were heated in sample buffer at 40°C for 20 min before loading on the gel.
The lack of cleaved Dha in cells coexpressing Kar240-Cub-Dha and Nub-Sec61p or Nub-Ssh1p might indicate that Kar2p does not translocate via Sec61p or Ssh1p (Figure 2, A and B). We considered this very unlikely. As suggested by our experience with the invertase signal sequence, longer spacer sequences allow the still attached ribosomes to dock to the translocation channel before the Cub-moiety of the fusion protein is translated. As a consequence the Cub is not accessible for interactions with the Nub in the cytosol of the cell (Johnsson and Varshavsky, 1994b; Dünnwald et al., 1999). We therefore tested a second Kar2-Cub construct (Kar220-Cub) that retained only 20 residues of the sequence of Kar2p between the hydrophobic core of the signal sequence and the Cub moiety.
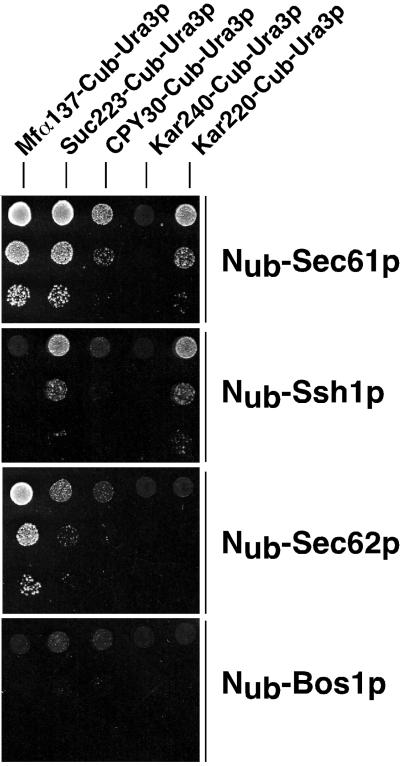
To avoid any artificial Nub-Cub reassociation and subsequent reporter cleavage after membrane rupture during protein extraction, we switched to the enzyme Ura3p as a reporter for the interaction assay. The localization of the enzyme Ura3p allows monitoring the interaction between translocation substrates and the components of the translocation machinery by a simple growth assay (Dünnwald et al., 1999). On Nub-induced cleavage, the Ura3p is released into the cytosol and enables the otherwise URA3-deficient cells to grow on medium lacking uracil (SD-Ura). We therefore cotransformed the corresponding signal sequence bearing Cub-URA3 plus the newly constructed Kar220-Cub-URA3 into cells expressing Nub-Sec61p or Nub-Ssh1p. The coexpression of the different Cub-Ura3p constructs confirmed and extended the results obtained with the Dha reporter (Figure 3). Good growth of the Nub-Sec61p–containing cells expressing Mfα137-Cub-Ura3p, Suc223-Cub-Ura3p, and the weaker but still significant growth of the cells coexpressing CPY30-Cub-Ura3p indicated the proximity between these signal sequences and Sec61p during the process of translocation (Figure 3). As expected from the results with Dha as the reporter, Kar240-Cub-Ura3p yielded no growth in the presence of Nub-Sec61p. However, the coexpression of Nub-Sec61p and Kar220-Cub-Ura3p resulted in solid growth of the cells on SD-Ura. The specificity of each interaction signal was tested by comparing it with the growth of the cells coexpressing the signal sequence bearing Cub-Ura3p and Nub-Bos1p. None of the five different cotransformants yielded growth of the cells on SD-Ura (Figure 3).
Figure 3.
Ssh1p interacts with the signal sequences of invertase and Kar2p. Yeast cells that contained different signal sequence bearing Cub-Ura3p and Nub-Sec61p, Nub-Ssh1p, Nub-Sec62p, or Nub-Bos1p were grown to an OD600 of 1. Then 4 μl of two serial 1:10 dilutions were spotted onto media lacking uracil and tryptophan and histidine to select for the presence of the plasmids expressing the fusion proteins. Growth was recorded after 3 d at 30°C. The growth of the cells is a measure of the interaction between the signal sequence of the Cub-Ura3p and the cotransformed Nub-fusion protein.
Nub-Ssh1p–containing cells grew on SD-Ura when coexpressing Suc223-Cub-Ura3p and Kar220-Cub-Ura3p (Figure 3). Interestingly, the interaction signal derived from Suc223-Cub-Ura3p is less pronounced in the Nub-Ssh1p than in Nub-Sec61p–containing cells, whereas the signals derived from Kar220-Cub-Ura3p are very similar in cells expressing Nub-Ssh1p or Nub-Sec61p (Figure 3). No interaction signals were detected upon coexpression of Nub-Ssh1p and Mfα137-Cub-Ura3p, CPY30-Cub-Ura3p, or Kar240-Cub-Ura3p (Figure 3). Because the well-established role of Sec61p in post- and cotranslational translocation of proteins across the ER membrane is reflected by its recognition of all four different signal sequences, we conclude that Ssh1p interacts only with the signal sequences of invertase and Kar2p. These two sequences display a higher hydrophobicity than the corresponding sequences of Mfα1 and CPY (Ng et al., 1996; see DISCUSSION). A significant fraction of Kar2p and invertase is known to be targeted to the ER membrane via the SRP (Hann and Walter, 1991; Johnsson and Varshavsky, 1994b; Ng et al., 1996). In contrast, Mfα1 and CPY are targeted via the tetrameric Sec62p/Sec63p complex. In this complex only Sec62p is known to be exclusively involved in the posttranslational translocation, whereas Sec63p fulfills a further role in the cotranslational translocation of proteins (Deshaies and Schekman, 1989; Ng et al., 1996; Brodsky et al., 1995; Young et al., 2001). We coexpressed Nub-Sec62p together with the different signal sequence bearing Cub-Ura3p to compare the signal sequence interaction profiles of Sec62p and Ssh1p. Nub-Sec62p revealed a strong interaction with Mfα137-Cub-Ura3p (Figure 3). No interactions were observed between Sec62p and the signal sequences of Kar2p and CPY. However, the weak interaction that is measured between Nub-Sec62p and Suc223-Cub-Ura3p is above the background that was determined by the growth of cells coexpressing Suc223-Cub-Ura3p together with Nub-Bos1p. This weak interaction was not detected by the growth assay in a previous study (Dünnwald et al., 1999). Whereas Sec61p and Ssh1p showed an overlapping specificity toward the more hydrophobic signal sequences of invertase and Kar2p, none of the signal sequence bearing Cub constructs that are recognized by Ssh1p induced a strong interaction signal with Nub-Sec62p and vice versa.
Ssh1p Is in Vicinity of SRP-Receptor
The signal sequence interaction profile of Ssh1p suggests that Ssh1p might be involved in the cotranslational translocation of proteins across the ER membrane (Figure 3). To further substantiate this hypothesis, we measured the proximity between Ssh1p and the β-subunit of the SRP receptor (SRβ). Biochemical data showed that SRβ plays a role in coordinating the transfer of the signal sequence from the SRP to the translocation channel (Fulga et al., 2001). One consequence of this activity should be a physical proximity between SRβ and components of the channel. The N-terminal part of SRβ anchors the protein in the ER membrane and directs its C-terminal domain into the cytosol (Ogg et al., 1998). If Ssh1p is indeed involved in cotranslational protein translocation it should ideally be as close to members of the SRP pathway as Sec61p. Furthermore, Ssh1p should be closer to SRβ than the proteins that are not involved in cotranslational protein translocation. We estimated the proximity between SRβ and Ssh1p by comparing the growth of cells containing SRβ-Cub-RUra3p and Nub-Ssh1p with the growth of cells containing SRβ-Cub-RUra3p and a panel of other Nub-labeled proteins. In this variation of the split-Ub assay the reporter RUra3p is immediately degraded by the enzymes of the N-end rule pathway after being cleaved from Cub (Wittke et al., 1999). Proximity between a pair of Nub- and Cub-labeled fusion proteins is therefore indicated by the nongrowth of the corresponding yeast transformants on SD-Ura. The Nub mutants Nua and Nug have a lower affinity to Cub than the wild-type Nub (Nui). An Nub-fusion very close to a certain Cub-fusion will therefore induce cleavage of the Cub-linked reporter not only as its Nui- but also as its Nua- and potentially even as its Nug-derivative (Wittke et al., 1999). Figure 4 shows that Nub-Ssh1p interacts with SRβ-Cub-RUra3p as strongly as Nub-Sec61p. According to this interaction assay both Nua-fusion proteins display a physical proximity to SRβ-Cub-RUra3p. Membrane proteins of the ER that are not involved in translocation, such as Nua-Bos1p, Nua-Ubc6p, Nua-Ste14, and Nua-Sec22p, are not close to SRβ-Cub-RUra3p (Shim et al., 1991; Sommer and Jentsch, 1993; Ballensiefen et al., 1998; Romano and Michaelis, 2001). Importantly, Nua-Sec62p as a component of the postranslational translocation pathway is more distant to SRβ than Nua-Sec61p or Nua-Ssh1p in our assay.
Figure 4.

Ssh1p is close to the β-subunit of the SRP receptor. Cells expressing SRβ-Cub-RUra3p from its own promoter were cotransformed with the Nui-, Nua-, and Nug-fusions of the translocation components Sec61p, Sec62p, and Ssh1p. Cells were grown in media containing uracil to an OD600 of ∼1. Then 4 μl of the cultures was spotted on plates lacking uracil. The plates were also lacking histidine and tryptophan to select for the presence of the plasmids expressing the fusion proteins. Growth was recorded after 5 d at 30°C. Nongrowth indicates interaction between the corresponding Nub-fusion and SRβ-Cub-RUra3p. Also included in this analysis were the Nub-fusions of the ER membrane proteins Bos1p, Ste14p, Sec22p, and Ubc6p; the Nub-fusion of the Golgi-protein Sed5p; and the cytosolic enzyme Tpi1p.
Flow of Translocated Proteins Is Not Measurably Changed in an ssh1Δ Strain
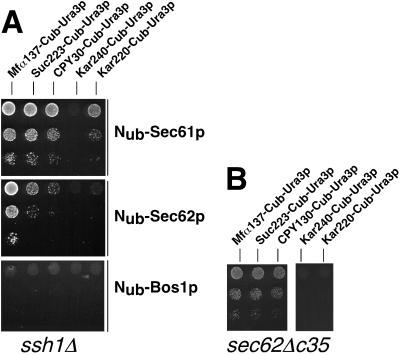
We constructed an ssh1Δ strain to ask whether the interactions between Nub-Sec61p or Nub-Sec62p and the signal sequence bearing Cub-Ura3p are changed upon deleting SSH1. The ssh1Δ strain showed a slightly reduced growth at 17, 25, and 37°C but grew nearly indistinguishable from the wild-type JD53 at 30°C. The expression of a plasmid-borne Nub-Ssh1p or the unmodified Ssh1p could compensate for the growth defect of this strain, thus proving the functionality of Nub-Ssh1p (our unpublished observation). The signal sequence bearing Cub-Ura3p constructs were coexpressed with Nub-Sec61p, Nub-Se62p, and Nub-Bos1p in the ssh1Δ strain. The interaction assay between the Nub-fusion proteins and the signal sequence bearing Cub-Ura3p revealed no major differences between the wild-type and the ssh1Δ strain (Figure 5A). All four signal sequences still interact with Nub-Sec61p, whereas Nub-Sec62p strongly interacts with Mfα137-Cub-Ura3p only. A slight increase in interaction between Nub-Sec62p and Suc223-Cub-Ura3p might be inferred from Figure 5A. However, the effect is very small. Thus, the experiment does not detect a major rerouting from the co- to the posttranslational pathway of protein translocation upon deletion of SSH1. Interestingly, Nub-Bos1p showed again no significant interaction with any of the signal sequence bearing Cub-Ura3p. This observation implies that there is also no major translocation defect and consequently no cytosolic accumulation of any of the tested translocation substrates in the ssh1Δ strain. To prove that these artificial constructs can principally detect a defect in translocation, we expressed all five Cub-Ura3p in a strain that harbors only a partially functional allele of Sec62p (sec62ΔC35-Dha) (Wittke et al., 2000). A translocation defect will result in the cytosolic accumulation of Ura3p activity even in the absence of any coexpressed Nub fusion. The cytosolic Ura3p fusion protein will enable the cells to grow on SD-Ura (Johnsson and Varshavsky, 1994b; Ng et al., 1996). This effect is detected by the growth of the transformed strain on SD-Ura for the Mfα-, invertase-, and the CPY-signal sequence–bearing constructs (Figure 5B). The sec62ΔC35-Dha allele displayed no measurable defect in the translocation of the two Kar2-Cub-Ura3p constructs (Figure 5B). We conclude that the deletion of SSH1 did not cause a severe accumulation of any of the signal sequence-bearing Cub-Ura3p in the cytosol of the cell, thus confirming the results derived from more direct translocation assays (Ng et al., 1996).
Figure 5.
Flow of signal sequences is not redirected in ssh1Δ cells. (A) Analysis was as in Figure 3 but in cells containing a deletion of SSH1. (B) Translocation defect is detected by the signal sequence bearing Cub-Ura3p fusions in cells carrying a sec62 allele. The indicated Cub-Ura3p fusions were expressed in cells carrying the sec62ΔC35-DHA allele expressed from the PCUP1 promotor. A partial translocation arrest is indicated by the growth of the cells on SD-Ura after 3 d at 30°C. The media were also lacking histidine to select for the presence of the plasmids.
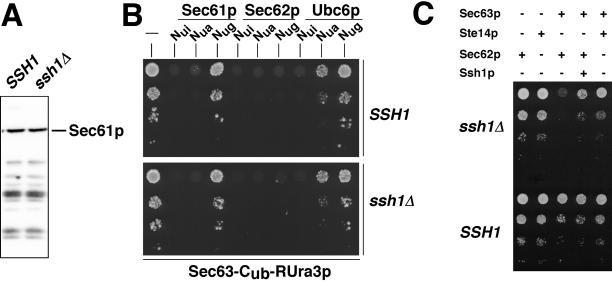
Concentration of Sec61p Is Not Measurably Changed in an ssh1Δ Strain
The lack of significant defects in protein translocation in our ssh1Δ strain is the more surprising because we have shown that Ssh1p recognizes a certain subset of signal sequences (Figures 2 and 3). It is possible that the cell can compensate for the lack of Ssh1p in more than one way. One mechanism might include the up-regulation of Sec61p to provide more channels in the ER membrane. We tested this hypothesis by comparing the amount of Sec61p in a wild-type and an ssh1Δ strain. We found no significant difference between the two yeast strains (Figure 6A). An alternative mechanism of compensating for the loss of SSH1 might use a readjustment in the composition of the Sec-complexes. Specifically, a weaker binding between Sec61p and the tetrameric Sec62p/Sec63p complex might free more Sec61p for the cotranslational way of protein translocation. Very similar to the measurements between SRβ-Cub-RUra3p and the different Nub-fusion proteins we compared the interaction of Sec63-Cub-RUra3p with Nub-Sec61p and also with Nub-Sec62p in a wild-type and the ssh1Δ-strain (Wittke et al., 1999). We monitored the interaction between the Nub-modified Ubc6p and Sec63-Cub-RUra3p as a pair of proteins whose proximity should be unaffected by a deletion of SSH1. Taking the growth on SD-Ura as a measure of interaction strength we found that our assay did not detect a difference between the ssh1Δ strain and the isogenic JD53 strain in the binding of Sec63-Cub-RUra3p to Nub-Sec61p or Nub-Sec62p (Figure 6B). Specifically, the nongrowth of the wild-type and the ssh1Δ cells containing Sec63-Cub-RUra3p and Nug-Sec62p on SD-Ura indicates a very strong interaction between the two molecules. The nongrowth of wild-type and the ssh1Δ cells containing Sec63-Cub-RUra3p and Nua-Sec61p indicates an interaction that is weaker but still specific compared with the growth of the cells containing Sec63-Cub-RUra3p and Nua-Ubc6p (Figure 6C). Similar to Nub-Ubc6p, Nub-Ssh1p behaves as a typical ER membrane protein in this assay (Wittke et al., 1999). However, because the measurements are purely qualitative, we cannot exclude that a slight but still significant readjustment of the Sec-complex upon deleting SSH1 might have gone unnoticed in our assay.
Figure 6.
Concentration of Sec61p and its interaction with Sec63p are not measurably changed in ssh1Δ cells. (A) Whole cell extracts from a wild-type and an ssh1Δ strain were probed with an anti-Sec61p antibody after SDS-PAGE and transfer onto nitrocellulose. A representative blot from one of three experiments is shown. (B) Interactions between Sec63p and Sec61p, Sec62p and Ubc6p were analyzed with the split-Ub assay in wild-type and ssh1Δ cells. Cells containing Sec63-Cub-RUra3p and coexpressing Nui-, Nua-, or Nug-fusions of the indicated ER membrane proteins were grown in uracil-containing medium to an OD600 of ∼1. Then 4 μl of these cultures and 4 μl of three serial 1:10 dilutions were spotted on plates lacking uracil and histidine and tryptophan to select for the plasmids. Growth was recorded after 3 d at 30°C. Nongrowth indicates proximity between the Nub- and Cub-labeled fusion proteins. (C) Overexpression of Sec62p and Sec63p is toxic in ssh1Δ cells. Cells containing the indicated combination of plasmid-borne proteins were grown to an OD600 of ∼1 in media lacking leucine, histidine, and tryptophan to select for the presence of the plasmids and glucose to repress the expression of Sec62p, Sec63-ha, or Ste14-Dha. Then 4 μl of these cultures and 4 μl of three serial 1:10 dilutions were spotted on plates lacking leucine, histidine, and tryptophan to select for the plasmids and galactose to induce the PGAL1 driven expression of Sec62p, Sec63-ha, or Ste14-Dha. Growth was recorded after 3 days at 30°C. Cells were also spotted on glucose-containing medium to check for equal cell numbers (our unpublished observation).
ssh1Δ Strain Is Less Resistant to Changes in Concentration of Components of Protein Translocation Machinery
If the cell does not react to the loss of Ssh1p by either increasing the amount of Sec61p or decreasing its association with the tetrameric Sec-complex, the correct balance between free and Sec62p/Sec63p-bound Sec61p might become more fragile in a cell lacking SSH1. To test this hypothesis we overexpressed Sec62p and an ha-tagged version of the Sec63p (Sec63-ha) in the wild-type and the ssh1Δ strain to limit the amount of free Sec61p in both cell types. Compared with the wild-type strain, the strain lacking SSH1 did barely grow upon overexpression of SEC62p and Sec63-ha (Figure 6C). On introducing a plasmid-borne copy of SSH1, the ssh1Δ cells regained growth that was only slightly slower than the growth of the corresponding wild-type strain. The overexpression of Sec63-ha together with the membrane protein Ste14-Dha did affect the growth of the wild-type and the ssh1Δ cells less severely and to a similar extent (Figure 6C). Expression of Sec62p or Ste14-Dha alone had no effect on the growth of the cells (Figure 6C).
DISCUSSION
Sequence analysis of complete genomes revealed homologs of many known proteins whose functions are not immediately apparent in spite of their similarity. Ssh1p is related in sequence to Sec61p and is organized in a very similar trimeric complex. This, and its confirmed location in the ER membrane, led to the assumption that Ssh1p is involved in certain aspects of protein translocation (Finke et al., 1996). Surprisingly, a deletion of the gene did not reveal any severe translocation defects, and the hypothesis that Ssh1p interacts with signal sequences and is actively involved in translocation remained unproven (Finke et al., 1996; Ng et al., 1996). In this work we show in vivo that Ssh1p recognizes a subset of the signal sequences that are also recognized by Sec61p but not by Sec62p. Our data therefore provide direct evidence that Ssh1p is involved in the cotranslational mode of protein translocation across the membrane of the ER.
Ssh1p Recognizes Signal Sequences
We made use of the split-Ub technique to demonstrate proximity between signal sequence bearing Cub-fusions and the Nub-modified Ssh1p. The assay showed that Ssh1p is close to the signal sequence of invertase and Kar2p, whereas no interaction could be detected for the signal sequences of CPY and Mfα1 (Figures 2 and 3). The relevance, especially of the failure to detect interactions between Ssh1p and the signal sequences of CPY and Mfα1, is strengthened by the observation that all four signal sequences interact with Sec61p (Figures 2 and 3). Sec61p and Ssh1p, being closely related in sequence are organized in very similar trimeric complexes and have similar activities as Nub-fusions toward unrelated Cub-substrates (Wittke et al., 1999). We therefore interpret the finding that Sec61p interacts with the signal sequences of Mfα1 and CPY, whereas Ssh1p does not as a true reflection of the in vivo specificity of the two proteins toward these sequences. The signal sequences that are recognized by Ssh1p seem more hydrophobic than those that do not bind to Ssh1p. Counting the hydrophobic residues in a window of 11 we find that the signal sequence of Kar2p has an uninterrupted stretch of 11 hydrophobic amino acids. Invertase has a stretch of 10 hydrophobic residues that is interrupted by a glycine in position 8 of this stretch. Mfα1 has a stretch of nine hydrophobic residues that is interrupted by two serines in positions 7 and 8, and CPY has a stretch of six hydrophobic residues that is interrupted by two glycines and three hydroxylated residues in positions 3, 5, 8, 9, and 10. We conclude that the signal sequences of Mfα1 and CPY are less hydrophobic than the signal sequences of invertase and Kar2p. Ng et al. (1996) have convincingly shown that more hydrophobic signal sequences are targeted via SRP, whereas less hydrophobic signal sequences are targeted via the tetrameric Sec62p/Sec63p complex. Although the translocation of Kar2p and invertase are affected by a deletion of the SRP, the translocation of Mfα1 and CPY is not (Ng et al., 1996). The profile of signal sequences interacting with Ssh1p strongly suggests that the SRP and its receptors contact Ssh1p during the targeting process. The physical proximity that we measured between Ssh1p and SRβ confirms this prediction (Figure 4).
The following simple model integrates the data of our study: Sec61p as the pore-forming subunit of the trimeric and heptameric Sec-complexes is involved in co- and posttranslational translocation and therefore recognizes all four different signal sequences (Deshaies and Schekman, 1987; Matlack et al., 1998; Pilon et al., 1998). Ssh1p has a more restricted specificity and is only involved in the SRP-dependent protein translocation. Consequently, Ssh1p only interacts with the signal sequences of invertase and Kar2p. Sec62p as part of the Sec62p/Sec63p complex that is exclusively involved in posttranslational translocation should therefore recognize the signal sequences that do not interact with Ssh1p. This prediction is fulfilled by our data concerning the Mfα1 signal sequence but not concerning the signal sequence of CPY (Figure 3). We can rationalize our failure to measure the postulated interaction between Nub-Sec62p and CPY30-Cub-RUra3p in two ways. 1) The identity of the binding site(s) for signal sequences on the heptameric Sec-complex is still not completely defined. Sec62p might be responsible for the recognition of Mfα1, whereas a different component of the Sec-complex, for example, Sec72p, might be the primary acceptor site for CPY (Feldheim and Schekman, 1994; Matlack et al., 1997). 2) We note that Nub-Sec61p gives a weaker interaction signal with CPY30-Cub-RUra3p than with the corresponding Mfα1 construct (Figure 3). The relatively hydrophilic character of the signal sequence of CPY might cause a weaker interaction and a shorter residence time at the Sec-complex. As a consequence, the interaction between Sec62p and CPY might fall below the sensitivity of our assay.
On the Function of Ssh1p
The interaction of Ssh1p with a certain subset of signal sequences strongly suggests but does not unequivocally prove that Ssh1p is also directly involved in the translocation of those proteins across the membrane. Instead, Ssh1p might be an additional receptor that keeps the signal sequences bound to the membrane as long as no Sec61p is available for their translocation. These and related objections to Ssh1p being a channel withstanding, our data show that Ssh1p acts as an additional receptor in the cotranslational mode of protein translocation (Plath et al., 1998). The question whether Ssh1p is a true channel can only be answered with the help of an in vitro system for cotranslational protein translocation in yeast.
A further unresolved issue is the lack of severe translocation defects in our ssh1Δ strain. Although initially surprising, the capacity of the SRP targeting pathway might suffice to overcome a shortage of Ssh1p-channels by pausing and thereby slowing down the translation of those proteins (Mason et al., 2000). In support of this notion a very recent report by Wilkinson et al. (2001) demonstrated a genetic link between a mutation in one of the components of the yeast SRP and a deletion of SSH1 leading to a synthetic lethality in the respective strain. The authors also demonstrate a remarkable capacity of ssh1Δ cells to adapt to and to suppress the initially observed translocation defects. These features might explain the lack of severe defects in our ssh1Δ strain. However, Wilkinson et al. (2001) noticed that adaptation correlates with the frequent occurrence of the petite phenotype in their W303 strain, whereas the ssh1Δ strain used in this study repeatedly grew well on glycerol or galactose-containing media (Figure 6C; our unpublished observation). The obvious difference between the two strains in responding to a deletion of SSH1 might reflect subtle differences in the genotypes of the two different strains.
In discussing the consequences of deleting SSH1 one has to be aware that the spectrum of proteins that were tested for translocation in an ssh1Δ strain still represents only a small fraction of all translocated proteins. Because it is now well established that different signal sequences show different requirements and kinetics for being targeted to the channel, it is possible that a still undiscovered fraction of proteins travels preferentially via Ssh1p across the membrane (Figure 1C). The slight growth defect of an ssh1Δ strain that is not completely cured by the ectopic overexpression of Sec61p hints at the existence of such a subset of Ssh1p-dependent translocation substrates (our unpublished data).
As more Sec61p and Ssh1p related proteins are identified in other organisms the flow of proteins across these different potential channels needs to be addressed. By allowing estimation of the contribution of the different components of the system, including its redundant members, the split-Ub technique can be used to analyze this flow in living cells.
Table 1.
Yeast strains
| Strain | Relevant genotype | Source/comment |
|---|---|---|
| JD53 | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 | Dohmen et al., 1995 |
| NJY73-1 | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 NUI-BOS1::pRS304 | Derivative of JD53; this work |
| NJY73-A | MAT his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 NUA-BOS1::pRS304 | This work |
| NJY73-G | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 NUG-BOS1::pRS304 | This work |
| NJY61-1 | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 NUI-SEC61::pRS304 | Wittke et al., 1999 |
| NJY61-A | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 NUA-SEC61::pRS304 | Wittke et al., 1999 |
| NJY61-G | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 NUG-SEC61::pRS304 | Wittke et al., 1999 |
| NJY78-1 | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 NUI-SSH1::pRS304 | Wittke et al., 1999 |
| NJY78-A | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 NUA-SSH1::pRS304 | Wittke et al., 1999 |
| NJY78-G | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 NUG-SSH1::pRS304 | Wittke et al., 1999 |
| NJY144 | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 SRP102-CUB-RURA3:: pRS303 | Derivative of JD53; this work |
| NJY126Δ35 | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 SEC62::KAN+ PCUP1SEC62ΔC35:pRS314 | Wittke et al., 2000 |
| NJY145 | MATαhis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 SSH1::KAN+ | This work |
ACKNOWLEDGMENTS
We thank Drs. Walter Mothes and Tom Rapoport for the Se61p antiserum. We thank Silke Müller for excellent technical assistance and Drs. K. Johnsson and J. Müller for critically reading the manuscript. We thank Oliver Kötting for constructing the SEC63- and SSH1-containing plasmids. This work was supported by a grant to N.J. from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10–0518. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0518.
REFERENCES
- Bacher G, Pool M, Dobberstein B. The ribosome regulates the GTPase of the beta-subunit of the signal recognition particle receptor. J Cell Biol. 1999;146:723–730. doi: 10.1083/jcb.146.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballensiefen W, Ossipov D, Schmitt HD. Recycling of the yeast v-SNARE Sec22p involves COPI-proteins and the ER transmembrane proteins Ufe1p and Sec20p. J Cell Sci. 1998;111:1507–1520. doi: 10.1242/jcs.111.11.1507. [DOI] [PubMed] [Google Scholar]
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Bird P, Gething MJ, Sambrook J. Translocation in yeast and mammalian cells: not all signal sequences are functionally equivalent. J Cell Biol. 1987;105:2905–2914. doi: 10.1083/jcb.105.6.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci USA. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol. 1987;105:633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. SEC62 encodes a putative membrane protein required for protein translocation into the yeast endoplasmic reticulum. J Cell Biol. 1989;109:2653–2664. doi: 10.1083/jcb.109.6.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- Dünnwald M, Varshavsky A, Johnsson N. Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol Biol Cell. 1999;10:329–344. doi: 10.1091/mbc.10.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault Y, Blondel MO, Deshaies RJ, Scheckman R, Kepes F. The yeast SSS1 gene is essential for secretory protein translocation and encodes a conserved protein of the endoplasmic reticulum. EMBO J. 1993;12:4083–4093. doi: 10.1002/j.1460-2075.1993.tb06092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D, Schekman R. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J Cell Biol. 1994;126:935–943. doi: 10.1083/jcb.126.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke K, Plath K, Panzner S, Prehn S, Rapoport TA, Hartmann E, Sommer T. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 1996;15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Sinning I, Dobberstein B, Pool MR. SRβ coordinates signal sequence release from SRP with ribosome binding to the translocon. EMBO J. 2001;20:2338–2347. doi: 10.1093/emboj/20.9.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kalies KU, Rapoport TA. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D, Matlack KE, Jungnickel B, Plath K, Kalies KU, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Hann B, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Johnson AE, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA. 1994a;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. Ubiquitin-assisted dissection of protein transport across membranes. EMBO J. 1994b;13:2686–2698. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- Mason N, Ciufo LF, Brown JD. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack KE, Mothes W, Rapoport TA. Protein translocation: tunnel vision. Cell. 1998;92:381–390. doi: 10.1016/s0092-8674(00)80930-7. [DOI] [PubMed] [Google Scholar]
- Matlack KE, Plath K, Misselwitz B, Rapoport TA. Protein transport by purified yeast Sec complex and Kar2p without membranes. Science. 1997;277:938–941. doi: 10.1126/science.277.5328.938. [DOI] [PubMed] [Google Scholar]
- Menetret J, Neuhof A, Morgan DG, Plath K, Radermacher M, Rapoport TA, Akey CW. The structure of ribosome-channel complexes engaged in protein translocation. Mol Cell. 2000;6:1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg SC, Barz WP, Walter P. A functional GTPase domain, but not its transmembrane domain, is required for function of the SRP receptor beta-subunit. J Cell Biol. 1998;142:341–354. doi: 10.1083/jcb.142.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Pilon M, Römisch K, Quach D, Schekman R. Sec61p serves multiple roles in secretory precursor binding and translocation into the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:3455–3473. doi: 10.1091/mbc.9.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Prinz A, Hartmann E, Kalies KU. Sec61p is the main ribosome receptor in the endoplasmic reticulum of Saccharomyces cerevisiae. Biol Chem. 2000;381:1025–1029. doi: 10.1515/BC.2000.126. [DOI] [PubMed] [Google Scholar]
- Romano JD, Michaelis S. Topological, and mutational analysis of Saccharomyces cerevisiae Ste14p, founding member of the isoprenylcysteine carboxyl methyltransferase family. Mol Biol Cell. 2001;12:1957–1971. doi: 10.1091/mbc.12.7.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J, Newman AP, Ferro-Novick S. The BOS1 gene encodes an essential 27-kD putative membrane protein that is required for vesicular transport from the ER to the Golgi complex in yeast. J Cell Biol. 1991;113:55–64. doi: 10.1083/jcb.113.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Song W, Raden D, Mandon EC, Gilmore R. Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 2000;100:333–343. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson BM, Critchley AJ, Stirling CJ. Determination of the transmembrane topology of yeast Sec61p, an essential component of the endoplasmic reticulum translocation complex. J Biol Chem. 1996;271:25590–25597. doi: 10.1074/jbc.271.41.25590. [DOI] [PubMed] [Google Scholar]
- Wilkinson BM, Tyson JR, Stirling CJ. Ssh1p determines the translocation capacities of the yeast endoplasmic reticulum. Dev Cell. 2001;1:401–409. doi: 10.1016/s1534-5807(01)00043-0. [DOI] [PubMed] [Google Scholar]
- Wittke S, Dünnwald M, Johnsson N. Sec62p, a component of the endoplasmic reticulum protein translocation machinery, contains multiple binding sites for the Sec-complex. Mol Biol Cell. 2000;11:3859–3871. doi: 10.1091/mbc.11.11.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittke S, Lewke N, Müller S, Johnsson N. Probing the molecular environment of membrane proteins in vivo. Mol Biol Cell. 1999;10:2519–2530. doi: 10.1091/mbc.10.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BP, Craven RA, Reid PJ, Willer M, Stirling CJ. Sec63p, and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001;20:262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]